Lentiviral Vpr usurps Cul4–DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle (original) (raw)
Abstract
The replication of viruses depends on the cell cycle status of the infected cells. Viruses have evolved functions that alleviate restrictions imposed on their replication by the host. Vpr, an accessory factor of primate lentiviruses, arrests cells at the DNA damage checkpoint in G2 phase of the cell cycle, but the mechanism underlying this effect has remained elusive. Here we report that Vpr proteins of both the human (HIV-1) and the distantly related simian (SIVmac) immunodeficiency viruses specifically associate with a protein complex comprising subunits of E3 ubiquitin ligase assembled on Cullin-4 scaffold (Cul4–DDB1[VprBP]). We show that Vpr binding to Cul4–DDB1[VprBP] leads to increased neddylation and elevated intrinsic ubiquitin ligase activity of this E3. This effect is mediated through the VprBP subunit of the complex, which recently has been suggested to function as a substrate receptor for Cul4. We also demonstrate that VprBP regulates G1 phase and is essential for the completion of DNA replication in S phase. Furthermore, the ability of Vpr to arrest cells in G2 phase correlates with its ability to interact with Cul4–DDB1[VprBP] E3 complex. Our studies identify the Cul4–DDB1[VprBP] E3 ubiquitin ligase complex as the downstream effector of lentiviral Vpr for the induction of cell cycle arrest in G2 phase and suggest that Vpr may use this complex to perturb other aspects of the cell cycle and DNA metabolism in infected cells.
Keywords: Cullin 4
The replication of HIV and other lentiviruses is restricted to some degree by the cell cycle status of the target cells. Although lentiviruses can infect nondividing cells, and thus establish stable reservoirs in CD4 T cells and terminally differentiated macrophages, they fail to replicate in quiescent cells (1). The observation that cells in G2 phase support lentivirus replication more efficiently than those in G1 phase further illustrates how the cell cycle status influences primate lentiviruses (2). HIV replication is also constrained by host cell mechanisms that detect and repair damaged DNA. For instance, the reverse-transcribed retroviral genomes are targeted for degradation by DNA repair proteins XPB and XPD (3), and the final steps of the integration of the reverse-transcribed retroviral genome into chromosomal DNA are thought to be catalyzed by DNA repair enzymes of the host cell (4). Viruses have evolved functions that partially alleviate various restrictions imposed on their replication by the host. In particular, accessory proteins of primate lentiviruses, such as Vpr, Vif, Nef, and Vpu, execute several such functions (5).
Vpr accessory proteins are multifunctional regulators located in the nuclei of the infected cells (6). Although Vpr is not required for lentivirus replication in cultured cells, its conservation in HIV-1, HIV-2, and simian immunodeficiency viruses (SIV) indicates that a strong selective pressure to preserve these proteins must operate in vivo. Indeed, Vpr was shown to accelerate progression to AIDS in rhesus macaques infected with SIV in the absence of a closely related SIV Vpx protein (7). How Vpr contributes to lentiviral pathogenesis is still under intense investigation. One conserved function of lentiviral Vpr is its ability to arrest the infected cells in the G2 phase of the cell cycle (8, 9). Vpr was also reported to suppress HIV-1 mutation rate, and this effect was correlated with its interaction with host-derived uracil DNA glycosylase, UNG2 (10). This enzyme is involved in the base excision repair pathway that specifically removes uracil from DNA (11). A recent study suggested that Vpr directs degradation of UNG2 to stabilize HIV reverse-transcription products (12). Finally, Vpr has been implicated as one of the factors that facilitates infection of terminally differentiated macrophages possibly by promoting entry of the HIV preintegration complex into the nucleus (13).
The most extensively studied Vpr function is its ability to arrest cycling cells in the G2 phase of the cell cycle (8, 9). One candidate mechanism for this function is suggested by the observation that Vpr activates the ataxia telengiectasia and Rad3-related (ATR) protein kinase (14). ATR triggers checkpoint signaling on genotoxic stress to stop the progression of the cell cycle until the damaged DNA is repaired (15). Recent evidence suggests that Vpr leads to ATR activation by interfering with the DNA replication machinery of the infected cell (16) and implicates Cullin 4 ubiquitin ligase containing VprBP/DCAF1 as potentially important for this effect (17, 18). It should be pointed out, however, that Vpr was also reported to interact with signaling molecules downstream of ATR, such as 14–3-3 proteins and Cdc25C, raising the possibility that additional mechanisms may be usurped by the viral protein to perturb the progression of the cell cycle (19–21).
The 3D structure of HIV-1 Vpr implies that it functions as an adaptor protein (22). Hence, we used a proteomic approach to identify the key cellular proteins that Vpr binds to subvert DNA replication and cell cycle regulation. Here we show that Vpr binds and deregulates an unstudied Cul4 E3 ubiquitin ligase complex containing damaged DNA binding protein 1 (DDB1), WD40-repeat containing VprBP, and DDA1 subunits. The DDA1–DDB1–VprBP–Vpr complex we purified is probably the most abundant Vpr-containing protein complex in the cell. The previously characterized Cul4 ligase complexes control DNA replication and DNA repair through ubiquitination of key substrates in these pathways (23–28). The Cul4 complex targeted by Vpr appears to regulate the G1 phase of the cell cycle and is essential for completion of DNA replication in S phase. We further demonstrate that, through the interaction with this Cul4 complex, Vpr arrests cells in the G2 phase. Together our studies identify the immediate downstream effector of Vpr for G2 cell cycle arrest and suggest additional scenarios for how Vpr may use this complex to perturb other aspects of the cell cycle and DNA metabolism in infected cells.
Results
HIV-1 and SIV Vpr Proteins Bind a Common Set of Proteins.
To address the molecular mechanisms used by the lentiviral Vpr proteins, we purified protein complexes containing HIV-1 Vpr or its orthologue encoded by the pathogenic SIVmac 239 strain. Vpr proteins were tagged at their N-termini with FLAG and HA epitopes in tandem and expressed stably in U937 monocytes or transiently in human embryonic kidney 293 T (HEK293T) cells. Next, Vpr and their associated proteins were purified from detergent extracts by sequential immunoprecipitations with α-HA- and then α-FLAG-epitope antibodies, followed each time by elution with the respective peptide epitope.
The immunoprecipitates were analyzed by Multidimensional Protein Identification Technology (MudPIT). MudPIT is a combination of chromatographic and mass spectrometric procedures that allow unbiased and sensitive identification of proteins in complex mixtures. Three relatively abundant polypeptides, DDB1, DDA1, and VprBP, that were specifically associated with Vpr proteins from both HIV-1 and SIVmac, which are two distantly related primate lentiviruses (29), in both U937 and HEK293T cells and absent in purifications from negative control cells were thus identified (SI Table 1). The conservation of these interactions suggested that they are important mediators of a common Vpr function. Therefore, we focused our studies on these proteins.
DDA1, DDB1, and VprBP Form a Ternary Complex That Vpr Binds.
The finding that Vpr binds DDB1, VprBP, and DDA1 linked Vpr to Cullin-4 RING E3 (Cul4 E3) ubiquitin ligase complexes. DDB1 is an obligatory subunit of Cul4 E3 ligases (30). These enzymes regulate DNA repair and replication and cell cycle progression through ubiquitination of key substrates in these processes. VprBP, a known HIV-1 Vpr binding protein (31), and DDA1 have been found recently to bind DDB1 (26); however, their normal functions remain unknown. Therefore, to gain further insights into Vpr interactions with these polypeptides, we characterized Vpr association with VprBP, DDB1, and DDA1.
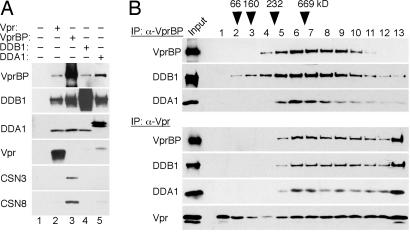
The observations that VprBP and DDA1, as well as DDB1, all copurified with Vpr, and that they were recovered at similar relative abundance based on relative spectral counts (see SI Table 1), suggested that they form a ternary complex, which is then targeted by Vpr. To address these possibilities, FLAG-tagged VprBP, DDB1, DDA1, and Vpr were individually expressed in HEK293T cells. Detergent extracts from the transfected and control cells were immunoprecipitated with α-FLAG beads and immune complexes analyzed by immunoblotting. As shown in Fig. 1A, each protein was capable of associating with each other (lanes 3–5). Next, the complexes purified via VprBP and HIV-1 Vpr (lanes 2 and 3) were separated on 10–40% glycerol gradients by ultracentrifugation. As shown in Fig. 1B, DDB1 (≈127 kDa molecular mass), DDA1 (11.8 kDa), and VprBP (169 kDa) all peaked in fractions 6 and 7, indicating that they form a ternary complex of ≈300–400 kDa (Fig. 1B Upper). Notably, the bulk of the Vpr-bound VprBP, DDB1, and DDA1 cosedimented with similar velocities (Fig. 1B Lower). Their distributions, however, were shifted toward the bottom of the gradient, compared with those in the absence of Vpr, suggesting that the viral protein may recruit additional components to these complexes. We conclude that VprBP, DDB1, and DDA1 form a ternary complex, which Vpr binds, and that the viral protein may modulate interactions between the DDA1–DDB1–VprBP complex and other factors.
Fig. 1.
Vpr binds DDA1–DDB1–VprBP complex. (A) Biochemical interactions among Vpr, VprBP, DDB1, and DDA1. Detergent extracts from HEK293T cells transiently expressing FLAG-tagged HIV-1 Vpr (lane 2), VprBP (lane 3), DDB1 (lane 4), or DDA1 (lane 5) subunits were immunoprecipitated (IP) with α-FLAG beads, and immunoprecipitates were analyzed by immunoblotting for VprBP, DDB1, DDA1, Vpr, and COP9 signalosome subunits CSN3 and CSN8. (B) Vpr binds the DDA1–DDB1–VprBP complex. FLAG-VprBP (Upper) and FLAG-Vpr (Lower) containing complexes were sedimented through 10–40% glycerol gradients. Aliquots of fractions collected from the tops of the gradients were immunoblotted for the indicated proteins.
Vpr Elevates Neddylation of Cul4 in Cul4–DDB1[VprBP] E3 Complex.
Surprisingly, although DDB1 functions through participation in the Cul4 E3 ligase platform (30), our MudPIT analyses detected only a few Cul4-derived peptides in α-Vpr immune complexes. This observation suggested that Cul4 is not a stoichiometric subunit of the Vpr complex we purified. To address a link to Cul4 ubiquitin ligase, we further characterized the interaction between Vpr and Cul4. Myc-tagged Cul4A was coexpressed together with VprBP and/or HIV-1 Vpr by transient transfection in HEK293T cells. In some experiments, Vpr, or VprBP, were FLAG-tagged to facilitate their immunoprecipitation. α-FLAG immune complexes prepared from transfected cells were analyzed by immunoblotting for Cul4. As shown in Fig. 2A, Cul4 was found in FLAG–VprBP immune complexes, thus confirming that VprBP participates in Cul4 E3 ligase complex (lanes 6 and 7) (26). Significantly, Cul4 was also found in FLAG–Vpr immune complexes, but only when VprBP was also ectopically expressed, thereby confirming that Vpr association with Cul4 is bridged by VprBP (compare lanes 3 and 2).
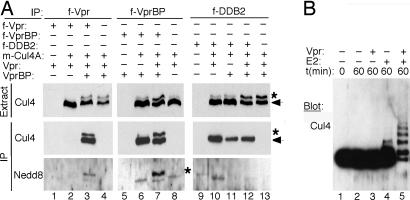
Fig. 2.
Vpr elevates neddylation and intrinsic ubiquitin-ligase activity of the Cul4–DDB1[VprBP] E3 complex. (A) Vpr binds to and specifically elevates neddylation of Cul4A associated with VprBP. Myc-tagged Cullin 4A (_m_-Cul4) was coexpressed with Vpr, VprBP, and DDB2, in various combinations, in HEK293T cells. FLAG-tagged (f) versions of these proteins were used in some experiments to facilitate their immunoprecipitation. _m_-Cul4A (arrow) and its neddylated form (∗) were detected in detergent extracts (Extr) and α-FLAG immune complexes (IP) by immunoblotting with α-myc and α-Nedd8 antibodies, respectively. (B) Vpr stimulates intrinsic ubiquitin ligase activity of the Cul4–DDB1[VprBP] E3 complex. _m_-Cul4–DDB1[VprBP] complexes were assembled with or without Vpr in HEK293T cells, purified via their FLAG-VprBP subunits, and incubated with E1, ubiquitin, and/or E2 as indicated. Cul4A and its ubiquitinated forms were detected by immunoblotting for Cul4.
The ubiquitin ligase activity of cullin complexes is regulated by cycles of Nedd8 moiety attachment to the cullin scaffold and its removal (32). The neddylated forms of cullins migrate slower than their unmodified forms during SDS/PAGE electrophoresis. Strikingly, ectopic expression of Vpr and VprBP together elicited a slower migrating Cul4A species readily detectable in detergent extracts prepared from the transfected cells (Fig. 2A top gels, lanes 3, 4, 7, 12, and 13). This Cul4 species was less pronounced in control experiments where Vpr, or VprBP, were expressed separately (lanes 2, 6, 8, 10, and 11). Immunoprecipitation experiments revealed that both Cul4A forms were associated with VprBP (Fig. 2A, middle gels, lane 7) and with Vpr, but only when VprBP was also ectopically expressed (compare lanes 2 and 3). Immunoblotting of the immune complexes for Nedd8 confirmed that the slower migrating Cul4A species was indeed the neddylated form (Fig. 2A, bottom gels, lanes 3 and 7).
The data described above demonstrated that Vpr can increase Cul4 neddylation through its interaction with VprBP, but they did not rule out that Vpr causes generalized neddylation of Cul4 present in other Cul4–DDB1 complexes. To address this latter possibility, we tested the effect of Vpr on the Cul4A component of the previously well characterized Cul4 E3 complex that contains DDB2 subunit instead of VprBP and mediates repair of damaged DNA (Cul4-DDN1[DDB2]) (23). As shown in Fig. 2A, Vpr did not induce detectable neddylation of Cul4A associated with DDB2, regardless of whether VprBP was also ectopically expressed (middle gels, lanes 10 and 12). We conclude that Vpr specifically binds the Cul4–DDB1[VprBP] complex and increases the levels of neddylated Cul4A in that complex only.
Vpr Elevates Intrinsic Ubiquitin Ligase Activity of Cul4–DDB1[VprBP] E3.
The increase in Cul4 neddylation suggested that Vpr may modulate the catalytic activity of the Cul4–DDB1[VprBP] E3 ubiquitin ligase complex. To address this possibility we assembled the Cul4–DDB1[VprBP] complexes with or without HIV-1 Vpr, purified them via their FLAG-VprBP subunits, and compared their intrinsic ubiquitin ligase activities in an in vitro assay. Because the identity of the relevant substrates of these E3 complexes is not yet known, we measured their abilities to autoubiquitinate Cul4 (23). As shown in Fig. 2B, the Cul4–DDB1[VprBP] E3 complex assembled with HIV-1 Vpr possessed a robust autoubiquitinating activity, and this activity required exogeneous E2 (lane 5). In contrast, the catalytic activity of the E3 complex that did not contain Vpr was relatively low (lane 4). We conclude that Vpr stimulates intrinsic ubiquitin ligase activity of the Cul4–DDB1[VprBP] E3 complex.
VprBP Is Required for the Ability of Vpr to Accumulate Cells in G2.
As aforementioned, Cul4 E3 ubiquitin ligases are thought to control DNA replication and progression of the cell cycle (23–28). We hypothesized that Vpr usurps VprBP and the Cul4–DDB1[VprBP] E3 complex to perturb these processes. Therefore, experiments were performed to characterize the normal function of VprBP.
Initial fluorescence microscopy and laser scanning cytometry experiments revealed that VprBP is located primarily in the nucleus and is expressed throughout the cell cycle (SI Fig. 5). To gain insight into the normal function of VprBP, we knocked down its expression by using RNA interference (RNAi). Human osteosarcoma U2OS cells were transduced with TRIP lentiviral vectors expressing short hairpin RNAs (shRNA) specific for VprBP or a control shRNA specific for DOCK2 guanine nucleotide exchange factor, which is expressed only in hematopoietic cells (33). Three days later, the transduced cells were infected again, this time with a bicistronic TEIG lentiviral vector expressing HIV-1 Vpr and GFP or with a control empty TEIG vector. After 3 more days, the transduced populations were analyzed for VprBP expression to confirm that VprBP levels were depleted and for DNA content to evaluate the effect of Vpr expression on the cell cycle. Immunoblot analysis revealed that VprBP expression levels were greatly reduced by RNAi (Fig. 3D). Analysis of DNA profiles of the cell populations demonstrated that Vpr arrested the control DOCK2 knockdown cells in G2 (Fig. 3A). In contrast, the cell cycle profiles of VprBP-depleted cells were virtually identical regardless of whether Vpr was expressed in these cells. These data suggested that VprBP-depleted cells are resistant to the effect of Vpr.
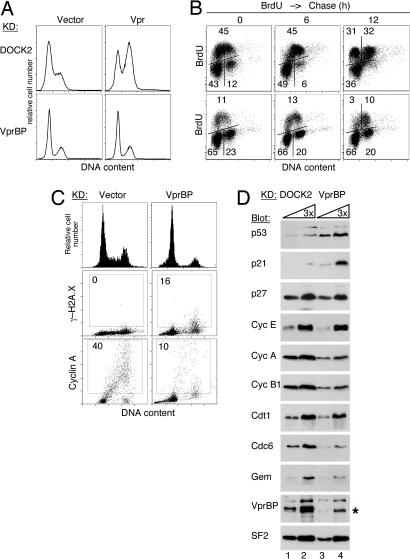
Fig. 3.
VprBP depletion leads to G1 and G2 phase arrests. (A) The effect of RNAi to VprBP on the ability of Vpr to arrest cells in G2. U2OS cells expressing shRNAs to VprBP (Lower) or DOCK2 (Upper) were transduced with a lentiviral TEIG vector expressing HIV-1 Vpr (Vpr) or a control empty vector (Vector). Three days later, the cells were stained with PI, and the DNA content was analyzed by flow cytometry. (B) VprBP-depleted cells arrest in G1 and G2. VprBP-depleted (Lower) and control cell populations (Upper) were labeled with BrdU for 1 h. BrdU incorporation and DNA content were analyzed by flow cytometry either immediately after BrdU labeling or after 6 and 12 h chase. Bivariant distributions of BrdU incorporation and DNA content are shown. The percent fractions of cells in the G1 (Lower Left), S (Upper), and G2 (Lower Right) phases are indicated. (C) Histone γ-H2A.X and cyclin A expression in VprBP-depleted (VprBP) and control cells (vector) were revealed by indirect fluorescence. Cells were counterstained with DAPI, and fluorescent signals were imaged with an iCys laser scanning cytometer. Histograms of DNA content and bivariate distributions of γ-H2A.X or cyclin A fluorescence versus DNA content are shown. (D) Levels of cyclins, checkpoint, and replication licensing proteins in lysates of VprBP-depleted and control cells were analyzed by immunoblotting with antibodies to the indicated proteins. Splicing factor 2 (SF2) was used as a loading control. The asterisk indicates a band reacting nonspecifically with the α-VprBP IgG. Lanes 1 and 3 contain 3-fold dilutions of the amounts loaded in lanes 2 and 4, respectively.
VprBP-Depleted Cells Arrest in G1 and G2 Phases.
We next studied the effect of VprBP knockdown on progression of the cell cycle in U2OS cells. VprBP-depleted and control DOCK2 knockdown cells were cultured for 1 h in the presence of BrdU to label S-phase cells. Next, the cells were cultured without BrdU for ≤12 h to allow time for progression of the BrdU-labeled cells through the S and G2 phases and mitosis. Subsequently, BrdU incorporation and DNA content in the cell populations were quantified by flow cytometry. Several aberrations in cell cycle profiles of the VprBP-depleted cells were revealed by these analyses. Strikingly, VprBP depletion led to a dramatic accumulation of cells in the G1 phase. As shown in Fig. 3B Lower, 65% of VprBP-depleted cells were found in G1 compared with 43% in the control population. Consistently, only a few VprBP-depleted cells appeared to replicate their DNA, as seen from a relatively low frequency of the BrdU-positive cells (11% vs. 45% in the control population). Moreover, an abnormally high proportion of VprBP-depleted cells was found in the G2 phase (23% vs. 12%). Notably, relatively few of the VprBP-depleted cells that were in late S or G2 at the time of BrdU labeling divided within the following 12-h period, compared with nearly all such cells in the control population. Overall, these data reveal that the majority of the VprBP-depleted cells were arrested in the G1 and G2 phases, thus suggesting the activation of DNA damage checkpoints. Additionally, the fact that VprBP-depleted U2OS cells were not cycling explains why Vpr was unable to cause their accumulation in G2 (Fig. 3A).
VprBP Depletion Leads to the Activation of DNA Damage Response.
To assess whether VprBP depletion led to the activation of DNA damage checkpoints, we characterized the expression of serine 139-phosphorylated histone H2A.X variant (γ-H2A.X). H2A.X is phosphorylated early on in response to DNA damage and is involved in the recruitment of repair proteins to the vicinity of DNA lesions (34). As shown in Fig. 3C, levels of γ-H2A.X expression were elevated in the G2-phase VprBP-depleted cells compared with control cells. This observation supports the notion that a subpopulation of VprBP-depleted U2OS cells is arrested in the G2 phase at the DNA damage checkpoint.
Next, we analyzed expression of key checkpoint proteins and cell cycle regulators by immunoblotting (Fig. 3D). Notably, the steady-state levels of p53 and p21 cyclin-dependent kinase inhibitor, which mediate p53-dependent DNA damage checkpoint (35), were both increased in VprBP-depleted cells. The levels of S-phase cyclin A and of cyclin B1 were relatively low, in agreement with a low frequency of S-phase cells (see Fig. 3 B–D). The expression levels of cdc6 and geminin, which control replication complex assembly and replication licensing (36), were also relatively low, consistent with low expression levels of these proteins in G1 cells. Together our observations provide evidence that depletion of VprBP leads to the activation of cellular response to DNA damage and arrest of the cells in the G1 and G2 phases at DNA damage checkpoints.
HIV-1 Vpr Arrests Cell Cycle by Interacting with the Cul4–DDB1[VprBP] E3 Complex.
Observations from our genetic and biochemical studies indicated that VprBP influences the transition from S to G2 phase probably through modulation of the DNA replication and/or damage repair processes. Significantly, RNA interference mediated depletion of the DDA1 and DDB1 subunits of the E3 complex targeted by Vpr also leads to cell cycle perturbations and activation of DNA damage response (SI Figs. 6 and 7) (28). This evidence strongly suggested that Vpr arrests cells in the G2 phase through its interaction with the DDA1–DDB1–VprBP complex and its associated Cul4 E3 ubiquitin ligase. To address this possibility, a panel of HIV-1 vpr alleles were tested for their abilities to arrest cells in G2, associate with VprBP and DDB1, and elevate Cul4A neddylation via VprBP.
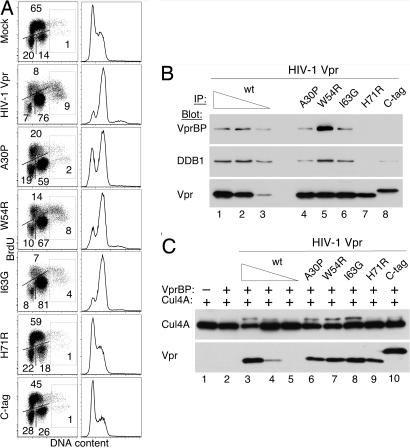
U2OS cells were transduced with retroviral MIG vectors expressing wild-type or mutant HIV-1 Vpr proteins, and their cell cycle profiles were analyzed 3 days later. As shown in Fig. 4A, ≈75% of cells transduced with wild-type vpr were arrested in G2. Notably, Vpr also caused accumulation of cells with >4n DNA content, suggesting that the viral protein can interfere with replication licensing (36). Next, we tested two mutations, one substituting arginine for histidine H71 (H71R) and the other attaching a tandem FLAG-HA epitope tag to the C terminus of the Vpr molecule (C-tag). Both mutations disrupted the ability of Vpr to arrest cells in the G2 phase, in agreement with a previous report (37). Notably, neither of the two proteins was able to associate with VprBP or DDB1, or to increase the levels of neddylated Cul4A (Fig. 4 B and C). Substitution of proline for alanine A30 (A30P) slightly diminished the ability of Vpr to arrest cells in G2 and also slightly decreased Vpr binding to VprBP and DDB1 and Cul4 neddylation. In contrast, no such effects were seen with two other mutations (W54R and I63G) that did not affect the ability of Vpr to arrest cells in G2. Of note, the W54R substitution was found to elevate the association of the viral protein with VprBP and DDB1, but did not elevate Cul4 neddylation above the level seen with wild-type Vpr. Thus, the binding of Vpr to VprBP probably is not sufficient to increase Cul4 neddylation, and some additional Vpr function is likely required for this effect. These data provide genetic and biochemical evidence in support of a model in which Vpr arrests cells in G2 by targeting the Cul4–DDB1[VprBP] E3 ubiquitin ligase.
Fig. 4.
Effects of mutations on Vpr abilities to arrest cells in the G2 phase, bind VprBP and DDB1, and elevate Cul4 neddylation. (A) Mutations disrupt the ability of Vpr to arrest cells in G2. U2OS cells were transduced with retroviral vectors expressing wild type (HIV-1 Vpr), the indicated mutant forms of HIV-1 Vpr, or a control empty vector (mock). Two days later, cells were labeled with BrdU, and their cell cycle profiles were analyzed by flow cytometry. (Left) Quantification of cells in the G1, S, and G2/M phases is shown. Cells with >4N DNA content are boxed. (Right) DNA content histograms also are shown. (B) Vpr mutations disrupt binding to DDB1 and VprBP. FLAG-tagged wild-type and mutant HIV-1 Vpr proteins were immunoprecipitated from transiently transfected HEK293T cells, and immune complexes were analyzed by immunoblotting for VprBP, DDB1, and Vpr. (C) Vpr mutants unable to arrest cells in G2 do not increase Cul4 neddylation. Wild-type and mutant HIV-1 Vpr proteins were transiently coexpressed with myc-Cul4A and VprBP in HEK293T cells, as described in the legend for Fig. 2A. Detergent extracts were immunoblotted for Cul4A and its neddylated form with α-myc antibody.
Discussion
In a search for downstream effectors of lentiviral Vpr, we purified an abundant Vpr-associated protein complex and identified its components as subunits of a specific ubiquitin ligase complex assembled on Cul4 scaffold (Cul4–DDB1[VprBP]). The VprBP subunit of this complex contains canonical WD40/WDXR motifs, similar to known Cul4 substrate receptors, such as DDB2, CSA, and Cdt2, which use these motifs to dock to DDB1 (23, 38). DDB1, in turn, connects them to Cul4. Thus, by analogy to these polypeptides, VprBP probably functions as a substrate receptor for Cul4 (17, 26, 38). Importantly, our data show that Vpr positively regulates the ubiquitin ligase activity of this specific E3 complex probably by elevating neddylation of Cul4. Covalent conjugation of a Nedd8 moiety to cullin is thought to mediate the recruitment of the E2-conjugating enzyme for the ubiquitin transfer reaction, thereby up-regulating ubiquitin ligase activity of the E3 complex (23, 39, 40). Regarding the mechanism that underlies the increased neddylation of Vpr-associated Cul4, Vpr could promote Nedd8 ligation to Cul4 or stabilize the neddylated Cul4 by inhibiting Nedd8 deconjugation by COP9 signalosome or another isopeptidase (40). Of note, we observed that COP9 subunits were undetectable in the E3 complexes associated with Vpr, but present in those assembled in the absence of Vpr expression (see Fig. 1, lanes 2 and 3; and data not shown). This evidence suggests that Vpr up-regulates the catalytic activity of Cul4–DDB1[VprBP] by interfering with Nedd8 deconjugation by COP9.
Two lines of evidence suggest that Vpr usurps the Cul4–DDB1[VprBP] E3 to arrest cells in the G2 phase. First, our data link VprBP and its associated Cul4 E3 ubiquitin ligase to the regulation of DNA replication and the cell cycle. Second, we found that mutant Vpr proteins deficient for binding the Cul4–DDB1[VprBP] complex and elevating neddylation of its associated Cul4 are unable to arrest cells in G2. These findings support a model in which Vpr perturbs the normal function of Cul4–DDB1[VprBP] E3, and thereby interferes with the completion of DNA replication, which leads to the activation of DNA damage checkpoint and cell cycle arrest in G2 phase.
Available evidence suggests three not exclusive scenarios for how Vpr may interfere with the normal function of the Cul4–DDB1[VprBP] E3 ubiquitin ligase. First, our finding that Vpr specifically stimulates catalytic activity of the Cul4–DDB1[VprBP] E3 implies that the viral protein may cause premature ubiquitination of proteins that are natural substrates of this ubiquitin ligase complex and mediate DNA replication/repair. However, the ability of Vpr to activate Cul4 is probably not sufficient to explain how it arrests cells in G2 because we observed that the Vpx protein, a Vpr orthologue encoded by HIV-2 and SIVsm viruses, also binds and stimulates neddylation of Cul4 in the Cul4–DDB1[VprBP] complex, but does not arrest cells in G2 (data not shown). Second, Vpr may recruit novel protein substrates that normally are not recruited by VprBP for ubiquitination by Cul4. This possibility is supported by recent evidence that Vpr directs ubiquitination of UNG2 and SMUG and that this involves Cullin 4 (12, 18). However, neither of these uracil glycosylases is essential for cell cycle progression, and their depletion does not explain how Vpr arrests cells in G2. Nevertheless, these data set a precedent to suggest that Vpr may direct inappropriate ubiquitination of a yet unknown replication factor(s) and thereby lead to G2 arrest. Third, Vpr could disrupt recruitment and ubiquitination of a physiological Cul4–DDB1[VprBP] E3 substrate that is involved in DNA replication. This possibility is consistent with our observation that RNAi-mediated loss-of-VprBP function leads to G2 arrest. It is conceivable that a combination of two or more of the above effects leads to the activation of DNA damage checkpoint and triggers G2 arrest.
How Vpr, by subverting the VprBP-linked Cul4 E3 ubiquitin ligase, benefits replication of primate lentiviruses is not known, but can be speculated on given the existing data. Our evidence firmly links VprBP to the control of G1-phase progression. Significantly, previous studies established that a certain threshold of cellular activation is needed to support productive lentiviral infection. Although quiescent cells in the G0 state are not permissive, just a partial stimulation that drives them into the G1 phase without triggering proliferation is frequently sufficient to establish competence for lentiviral replication (41, 42). Because cells in late G1 are permissive, whereas those in early G1 are not, the position of cells in the G1 phase probably determines how efficiently they support viral replication (41). Thus, Vpr could benefit HIV replication by manipulating progression through G0/early G1 phase in noncycling cells and/or minimally activated cells. Such an effect would benefit the virus early in natural infection, when the levels of immune activation are low and the majority of the infected cells are thought to be quiescent.
Another possibility is suggested by the observation that the depletion of VprBP expression levels by RNAi leads to the activation of the DNA damage checkpoint in G2. This evidence links VprBP to the control of DNA synthesis and/or repair processes that are essential for the completion of S phase. Notably, recent studies have begun to reveal the unexpected complexity of the interactions between the replicating retroviral genomes and cellular machineries that repair damaged DNA. For example, integration of cDNA copies of retrovirus genomes is thought to be aided by cellular machineries that mediate repair of damaged DNA (4). However, it appears that enzymes that normally mediate repair of damaged DNA target the incoming retroviral nucleic acids to inhibit, rather than promote, the infection (3, 43). Moreover, retroviral genomes are substrates for DNA editing enzymes and additional repair reactions (10, 12, 44). It will be important to determine whether the Cul4–DDB1[VprBP] complex controls any of these processes. Ultimately, the identification of substrate proteins recruited for ubiquitination by VprBP, both in the absence and presence of Vpr, should lead to a better understanding of the roles of Cul4–DDB1[VprBP] E3 in DNA metabolism, cell cycle control, and lentivirus infection.
Materials and Methods
Expression Vectors and Viruses.
HIV-1 NL4–3 and SIVmac 239 vpr tagged with N-terminal FLAG-HA-AU1 (hfa) epitopes in tandem (45) and other epitope-tagged cDNAs were cloned into pBABE-puro, pCG, MIG, and TEIG bicistronic vectors expressing GFP (46). shRNAs targeting sequences, listed in supporting information (SI) Text, were subcloned into TRIP lentiviral vectors (47). MIG and TEIG are modified MSCV and TRIP vectors containing a polylinker, followed by internal ribosome entry site element and GFP cassette derived from pCG. VSV-G pseudotyped viral particles were produced in transiently transfected HEK293T cells, and viral titers to U2OS cells were determined by flow-cytometry analysis of GFP/CFP expression.
Immunoaffinity Purification of Epitope-Tagged Proteins and MudPIT Analysis.
Protein complexes were purified by two sequential immunoprecipitations via the FLAG and HA epitope tags from 7 g to 12 g of U937 cells stably expressing hfa-tagged Vpr proteins (or control cells), or transiently transfected HEK293T cells as described previously (45). MudPIT analysis of protein complexes was performed as previously described (48, 49) and is described in detail in SI Text. Normalized spectral abundance factors were calculated for each detected protein as described (50).
Flow-Cytometry Analysis.
To visualize cells in S phase, cells were labeled with BrdU for 30 to 60 min. BrdU was detected with APC-conjugated α-BrdU antibody (Becton Dickinson, San Jose, CA) and DNA counterstained with 0.1 mg/ml propidium iodide (PI). For DNA content analysis only, cells were fixed with ethanol, and DNA was then stained with PI. The cells were analyzed on a Becton Dickinson LSRII Flow Cytometer, and data were processed with FlowJo software.
Transient Transfections, Immunoprecipitations, and Immunoblotting.
Detergent extracts from transiently transfected HEK293T cells were immunoprecipitated with α-FLAG M2 affinity gel (Sigma–Aldrich, St. Louis, MO) (45). For preparation of lysates, U2OS cells were harvested by trypsinization, washed with PBS, and boiled in five volumes of Laemmli sample loading buffer. Antibodies used for immunoblot analyses are listed in SI Text.
Fluorescent Microscopy.
U2OS cells grown on coverslips were fixed in 2% paraformaldehyde in PBS for 15 min and permeabilized with 0.5% Triton X in PBS for 5 min on ice. Incubations with primary and secondary antibodies were performed as described (46). Cells were counterstained with 0.5 μg/ml DAPI (Sigma–Aldrich). Slides were imaged with a Zeiss Axioplan2 or iCys laser scanning cytometer (Carl Zeiss, Thornwood, NY).
In Vitro Ubiquitin Ligase Activity Assay.
To measure ubiquitin ligase activity, Cul4–DDB1[VprBP] complexes assembled in the absence or presence of HIV-1 NL43 Vpr expression in HEK293T cells were purified by immunoprecipitation via their FLAG-VprBP subunits and incubated at 30°C for 60 min with 0.2 μg of UbaI E1, 0.03 μg of UbcH5b, 5 μg of ubiquitin (BIOMOL Research Laboratories, Plymouth Meeting, PA) in the medium of 50 mM Tris·HCl (pH 8.0), 5 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT.
Supplementary Material
Supporting Information
Acknowledgments
We thank Linda Van Aelst for discussions and critical reading of the manuscript, Bruce Stillman for stimulating discussions and support, David Spector and Wolfgang Lukowitz for comments on the manuscript, Takahiro Nagase (DNA Research Institute, Chiba, Japan) for KIAA0080 cDNA, and Bruno Verhasselt (University of Gent, Gent, Belgium) for the TRIP vector. This work was supported by Public Health Service Grant AI-42561 (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Yamashita M, Emerman M. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 3.Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R. Proc Natl Acad Sci USA. 2006;103:4622–4627. doi: 10.1073/pnas.0509828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turlure F, Devroe E, Silver PA, Engelman A. Front Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 5.Frankel AD, Young JA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Le Rouzic E, Benichou S. Retrovirology. 2005;2:11. doi: 10.1186/1742-4690-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs JS, Lackner AA, Lang SM, Simon MA, Sehgal PK, Daniel MD, Desrosiers RC. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogel ME, Wu LI, Emerman M. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krokan HE, Drablos F, Slupphaug G. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 12.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. J Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. J Biol Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman ES, Chen J, Andersen JL, Ardon O, Dehart JL, Blackett J, Choudhary SK, Camerini D, Nghiem P, Planelles V. Mol Cell Biol. 2004;24:9286–9294. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 18.Schrofelbauer B, Hakata Y, Landau NR. Proc Natl Acad Sci USA. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh WC, Manel N, Emerman M. Virology. 2004;318:337–349. doi: 10.1016/j.virol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Kino T, Gragerov A, Valentin A, Tsopanomihalou M, Ilyina-Gragerova G, Erwin-Cohen R, Chrousos GP, Pavlakis GN. J Virol. 2005;79:2780–2787. doi: 10.1128/JVI.79.5.2780-2787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morellet N, Bouaziz S, Petitjean P, Roques BP. J Mol Biol. 2003;327:215–227. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- 23.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, McCall CM, Ohta T, Xiong Y. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 25.Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin J, Arias EE, Chen J, Harper JW, Walter JC. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. Proc Natl Acad Sci USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovejoy CA, Lock K, Yenamandra A, Cortez D. Mol Cell Biol. 2006;26:7977–7990. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apetrei C, Robertson DL, Marx PA. Front Biosci. 2004;9:225–254. doi: 10.2741/1154. [DOI] [PubMed] [Google Scholar]
- 30.Petroski MD, Deshaies RJ. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 31.Zhao LJ, Mukherjee S, Narayan O. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 32.Wei N, Deng XW. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 33.Fukui Y, Hashimoto O, Sanui T, Oono T, Koga H, Abe M, Inayoshi A, Noda M, Oike M, Shirai T, Sasazuki T. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 34.Ward IM, Chen J. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 35.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 36.Blow JJ, Dutta A. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau NR. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 39.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 41.Korin YD, Zack JA. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. J Exp Med. 1999;189:1735–1746. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lloyd AG, Tateishi S, Bieniasz PD, Muesing MA, Yamaizumi M, Mulder LC. PLoS Pathog. 2006;2:e40. doi: 10.1371/journal.ppat.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 45.Janardhan A, Swigut T, Hill B, Myers MP, Skowronski J. PLoS Biol. 2004;2:65–76. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lock M, Greenberg ME, Iafrate AJ, Swigut T, Muench J, Kirchhoff F, Shohdy N, Skowronski J. EMBO J. 1999;18:2722–2733. doi: 10.1093/emboj/18.10.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stove V, Van de Walle I, Naessens E, Coene E, Stove C, Plum C, Verhasselt B. J Virol. 2005;79:11422–11433. doi: 10.1128/JVI.79.17.11422-11433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Florens L, Washburn MP. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 50.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information