Distinct Expression Patterns of Natural Antisense Transcripts in Arabidopsis (original) (raw)
Abstract
It has been shown that overlapping cis-natural antisense transcripts (cis-NATs) can form a regulatory circuit in which small RNAs derived from one transcript regulate stability of the other transcript, which manifests itself as anticorrelated expression. However, little is known about how widespread antagonistic expression of cis-NATs is. We have determined how frequently cis-NAT pairs, which make up 7.4% of annotated transcription units in the Arabidopsis (Arabidopsis thaliana) genome, show anticorrelated expression patterns. Indeed, global expression profiles of pairs of cis-NATs on average have significantly lower pairwise Pearson correlation coefficients than other pairs of neighboring genes whose transcripts do not overlap. However, anticorrelated expression that is greater than expected by chance is found in only a small number of cis-NAT pairs. The degree of anticorrelation does not depend on the length of the overlap or on the distance of the 5′ ends of the transcripts. Consistent with earlier findings, cis-NATs do not exhibit an increased likelihood to give rise to small RNAs, as determined from available small RNA sequences and massively parallel signature sequencing tags. However, the overlapping regions of cis-NATs appeared to be enriched for small RNA loci compared to nonoverlapping regions. Furthermore, expression of cis-NATs was not disproportionately affected in various RNA-silencing mutants. Our results demonstrate that there is a trend toward anticorrelated expression of cis-NAT pairs in Arabidopsis, but currently available data do not produce a strong signature of small RNA-mediated silencing for this process.
Much of gene expression is primarily regulated at the level of transcription. Over the last few years, however, it has become increasingly apparent that posttranscriptional regulation at the RNA level is more widespread and important than previously assumed (Behm-Ansmant and Izaurralde, 2006; Brodersen and Voinnet, 2006; Newbury, 2006). While various types of regulatory RNA molecules have been shown to exist, arguably the most prominent ones are micro-RNAs (miRNAs; Bartel, 2004; Jones-Rhoades et al., 2006; Vazquez, 2006). MiRNAs are derived from larger transcripts generated by RNA polymerase II and found in both animals and plants. The primary transcript is processed to give rise to a short 20- to 24-nucleotide-long RNA molecule, the miRNA, which, by annealing to partially complementary sites of mRNAs, can lead to either cleavage of the mRNA or translational inhibition. Another type of small RNA that regulates the stability of transcripts is represented by short interfering RNAs (siRNAs). In contrast to miRNAs, siRNAs are always perfectly complementary to their targets. One source of siRNAs are double-stranded RNAs generated by transcription of a locus in both the sense and antisense orientation (Kumar and Carmichael, 1998; Vanhee-Brossollet and Vaquero, 1998). Such antisense transcripts were first observed in transgenic experiments, but natural antisense transcripts (NATs) also occur. There are two classes of NATs: cis-NATs, which are formed by antisense transcription at the same genomic locus, and trans-NATs, where sense and antisense transcripts are derived from different loci.
Large-scale genome projects have revealed the common occurrence of overlapping gene pairs in most species analyzed (Lehner et al., 2002; Shendure and Church, 2002; Osato et al., 2003; Yelin et al., 2003; Wang et al., 2005). The reported frequencies for overlapping gene pairs found in different species vary, depending on sample size and other search parameters, but usually range between 5% and 10% of all neighboring gene pairs. In the human genome, 4% to 9% of all transcript pairs overlap, while in the murine genome 1.7% to 14% have been identified as overlapping. A particularly extreme case is Drosophila, where up to 22% of all neighboring genes have been reported to overlap. Across the various species, the majority of overlapping gene pairs is transcribed in convergent orientation, thus representing true cis-NAT pairs.
NATs have been implicated in such diverse processes as transcription occlusion, RNA interference, alternative splicing, RNA editing, DNA methylation, and genomic imprinting (Farrell and Lukens, 1995; Sureau et al., 1997; Billy et al., 2001; Tufarelli et al., 2003; Kim et al., 2004; Jen et al., 2005; Wang et al., 2005). In the plant kingdom, cis-NATs have been analyzed in rice (Oryza sativa) and in Arabidopsis (Arabidopsis thaliana; Osato et al., 2003; Yamada et al., 2003; Jen et al., 2005; Wang et al., 2005). Analysis of the Arabidopsis transcriptome by means of whole-genome tiling arrays has revealed antisense expression of 7,600 transcripts, corresponding to roughly 25% of all annotated genes (Yamada et al., 2003). A few additional studies have addressed the question of antisense gene pairs in Arabidopsis in detail. Wang et al. (2005) identified 1,340 potential cis-NAT pairs in Arabidopsis and confirmed expression of sense and antisense transcripts of 957 cis-NAT pairs using sequence information of Arabidopsis full-length cDNAs and massively parallel signature sequencing (MPSS) data (Meyers et al., 2004a, 2004b). Using qualitative criteria, these authors concluded that the majority of cis-NATs showed highly anticorrelated expression. In an independent study, Jen et al. (2005) reported the existence of 1,083 transcript pairs that overlapped in antisense orientation. They further uncovered a possible role of convergent overlapping gene pairs in alternative splicing and polyadenylation but did not find any evidence for anticorrelated expression greater than expected by chance, which is in disagreement with the findings of Wang et al. (2005). Finally, in an elegant set of experiments, SRO5 and P5CDH, a pair of cis-NATs, were shown to have antagonistic functions in the regulation of salt tolerance in Arabidopsis (Borsani et al., 2005). In response to salt stress, SRO5 mRNA is induced, and a 24-nucleotide-long siRNA is formed from the region of overlap with P5CDH, dependent on components that are also involved in the generation of siRNAs from transgene-derived double-stranded RNAs, such as DICER-LIKE2 (DCL2) and RNA-DEPENDENT RNA POLYMERASE6 (RDR6). Subsequently, 21 nucleotide siRNAs are formed by DCL1-dependent processing of P5CDH transcripts. Finally, 1,320 putative trans-NATs have been recently identified in the Arabidopsis genome (Wang et al., 2006). Interestingly, a large number of transcripts was predicted to have both trans- and cis-NATs, suggesting that antisense transcripts can form a complex regulatory network.
Making use of large collections of microarray data, we have analyzed the extent to which cis-NATs in Arabidopsis show anticorrelated expression, as reported under salt stress for the SRO5 and P5CDH paradigm. We find that cis-NATs on average are significantly more anticorrelated than nonoverlapping neighboring genes, but clear global anticorrelated expression is restricted to a small subset of cis-NAT pairs, solving conflicting results that had previously been published (Jen et al., 2005; Wang et al., 2005). Available data sets do not point to small RNAs being increased in cis-NATs, nor is expression of cis-NATs typically affected by mutations in genes necessary for the biogenesis of small RNAs, suggesting that cis-NATs do not always enter the RNA-silencing pathway.
RESULTS AND DISCUSSION
Antisense Transcript Pairs in Arabidopsis
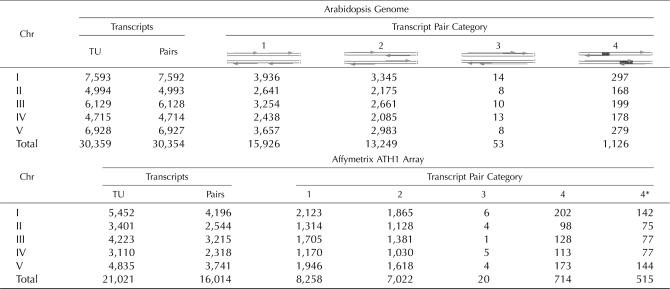
As a first step toward analyzing the transcriptional regulation of NATs derived from the same or adjacent loci, we categorized the transcription units of the Arabidopsis genome, as annotated by The Arabidopsis Information Resource (TAIR), release 6 (Haas et al., 2005). The Arabidopsis genome contains 30,359 transcription units that can be grouped into 30,354 transcript pairs (Table I). Transcript pairs were further broken down into four major categories, depending on which strand neighboring transcripts were located on and whether transcripts were overlapping or not. The majority of transcript pairs were found to be nonoverlapping, with 15,926 pairs transcribed from the same strand (category 1) and 13,249 from opposite strands (category 2). We found only 53 overlapping transcript pairs where both transcripts originated from the same strand (category 3). In contrast, we identified 2,243 overlapping transcripts originating from opposite strands forming 1,126 NAT pairs (cis-NAT; category 4), equaling 3.7% of all transcript pairs. The majority of the cis-NAT pairs were simple pairs, with only eight triplets and a single quadruplet identified.
Table I.
Categories of adjacent transcript pairs in Arabidopsis
TU, Transcribed unit; categories: 1, neighboring genes on the same strand, no overlap; 2, neighboring genes on opposite strands, no overlap; 3, neighboring genes on the same strand, overlap; 4, neighboring genes on opposite strands, overlap; TAIR6 genome annotation and TAIR ATH1 probe set mapping were used; 4*, gene pairs in category 4 whose overlap is supported by spliced or long ORF cDNA and/or EST clones. Chr, Chromosome. [See online article for color version of this table.]
To investigate the expression profiles of the cis-NATs, we mapped the TAIR6 transcription units onto the Affymetrix ATH1 microarray. We found that 21,021 (out of 30,359) transcripts were represented by the array. Of these, 16,014 were arranged in adjacent pairs, which correspond to about one-half of all transcript pairs encoded by the Arabidopsis genome. There was no substantial difference between adjacent nonoverlapping transcripts transcribed from the same strand (8,258; 51.8%) or from opposite strands (7,022; 53.0%). In contrast, overlapping transcripts derived from the same strand were slightly underrepresented (20; 37.7%), while cis-NATs were slightly overrepresented (714; 63.4%). The latter make up 4.4% of all transcript pairs mapped onto the ATH1 array. Because of the low number of transcript pairs in category 3, these were dropped from further analysis. Mapping information of the four different transcript pair categories onto the Arabidopsis genome and the ATH1 array can be found in Supplemental Tables S1 and S2, respectively.
One concern with cis-NAT predictions is that the transcript ends reported in the TAIR6 annotation might not necessarily be correct (Haas et al., 2005). We therefore manually inspected all 714 potential cis-NATs that are present on the Affymetrix ATH1 array for support by cDNA and/or EST clones that include either spliced introns or large (at least 100 codons long) open reading frames (ORFs). We found that of the 714 potential cis-NATs, only 515 (72.1%; 1,027 transcripts in total) are currently supported by cloned mRNAs with an overlap of at least one base (Supplemental Table S3). Subsequent analysis focused primarily on this set of cis-NATs.
The number of cis-NATs identified is slightly higher than what had previously been reported (Jen et al., 2005; Wang et al., 2005). The discrepancies are likely due to the different methods used to map cis-NATs onto the genome and changes in gene annotation introduced with the latest genome release. A limitation of this analysis, one has to keep in mind, is that the current annotation of the Arabidopsis genome may still lack the extreme 5′ and 3′ ends for many transcripts. As a consequence, our analysis might underestimate the number of Arabidopsis cis-NATs. Even so, the ATH1 array is a fair representation of the different transcript pair categories in Arabidopsis, allowing us to use expression data sets based on this array to examine the expression profiles of cis-NATs in detail.
An Excess of Negative Correlation Coefficients of cis-NAT Expression
To examine if there is a general difference between the expression profiles of cis-NATs and nonoverlapping transcript pairs, we calculated the pairwise Pearson correlation coefficient (PCC) for these transcript pairs from four publicly available data sets generated by the AtGenExpress initiative. The first set comprised data from 234 arrays that capture expression of 78 different tissue samples assayed in triplicate throughout development (Schmid et al., 2005). The original data set included also pollen samples, but because many genes show either very high or very low expression levels in this tissue type and pairs are therefore more likely to be perfectly correlated or anticorrelated by chance than in other samples, we omitted the pollen samples for this analysis. The second set of 236 arrays, from duplicate samples, reflects responses to hormones and related substances (mostly created by RIKEN; Kiba et al., 2005; Nakabayashi et al., 2005; Nemhauser et al., 2006). The two final sets, of 136 arrays each, had been used to measure the response to various abiotic stresses in shoots and roots, respectively, with duplicate samples (Kilian et al., 2007). We analyzed the shoot and or root data separately to minimize effects of tissue-specific expression.
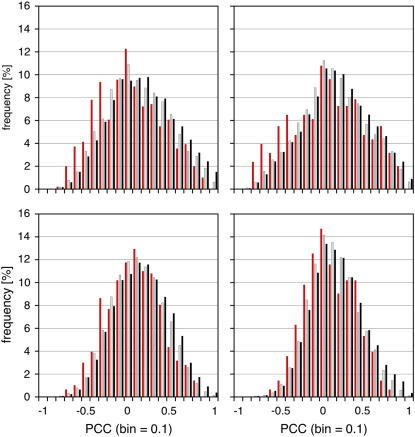
In all four data sets, the pairwise PCCs of cis-NATs are skewed toward negative values (Fig. 1) when compared to nonoverlapping transcript pairs located on either the same or opposite strands. This shift in distribution was statistically significant in all four data sets using a two-sided, two-sample Welch t test (Table II) regardless of whether all cis-NATs supported by the TAIR6 annotation (714; Table I, category 4) or only the manually curated set (515; Table I, category 4*) were used. Similar results were obtained using pairwise Spearman's rank correlation coefficients (SCCs), which are less sensitive to outliers (Supplemental Table S4). Comparisons of the PCC and SCC values by scatter plot analysis revealed a high degree of similarity, with _R_2 values ranging between 0.71 an 0.83, indicating the robustness of the anticorrelation we observed (Supplemental Fig. S1).
Figure 1.
Distribution of pairwise PCCs for expression of pairs of adjacent genes. PCCs of transcript pairs in categories 1 (same strand, no overlap; black), 2 (opposite strand, no overlap; gray), and 4* (opposite strand, overlap; red) were calculated for four Affymetrix ATH1 microarray data sets (development, hormones, abiotic stress/root, and abiotic stress/shoot) created by the AtGenExpress initiative.
Table II.
Statistical analysis of PCC distributions
P values for differences in the distribution of PCC were calculated using two-sided, two-sample Welch t test.
| Data Set | P Value | ||
|---|---|---|---|
| Category 1 versus 2 | Category 1 versus 4* | Category 2 versus 4*a | |
| Development | 0.5662 | 1.00−05 | 1.97−05 |
| Hormones | 0.5536 | 8.06−06 | 1.02−05 |
| Abiotic stress, root | 0.6453 | 1.43−04 | 2.45−04 |
| Abiotic stress, shoot | 0.6727 | 3.40−04 | 6.02−04 |
| Birnbaum | 0.3581 | 9.00−09 | 2.63−08 |
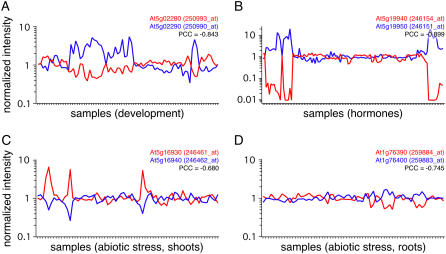
In contrast, distributions between nonoverlapping transcript pairs located on either the same or the opposite strand were not significantly different in any of the data sets. Figure 2 shows the expression profiles of the cis-NATs with the lowest PCCs for the individual microarray experiments.
Figure 2.
Expression profiles of selected cis-NATs. The NATs with the strongest anticorrelation (lowest PCC) in a particular data set are shown as examples.
One limitation of the AtGenExpress data sets is that they lack cellular resolution. We therefore analyzed microarray data Birnbaum et al. (2003) obtained from various cell types and regions of the root after cell sorting. We found that the distribution of PCC and SCC values of cis-NATs was skewed toward negative values when compared to nonoverlapping transcripts (Supplemental Fig. S2). As was the case for the AtGenExpress data sets, this shift toward negative correlation was found to be statistically significant (Table II; Supplemental Table S4), suggesting that the bias toward anticorrelation we observed in the AtGenExpress data set reflects true anticorrelation of cis-NATs within the same cells, as would be expected for direct regulatory effects.
The fact that we found on average statistically significant lower PCCs for cis-NATs suggests that expression of one of the transcripts in these pairs can influence expression of the other. However, the PCCs for the majority of cis-NATs fell in the same range as nonoverlapping transcript pairs, suggesting strong mutual regulation for only a subset of cis-NATs. Thus, anticorrelated expression is much less widespread than previously suggested based on MPSS expression data from 14 cDNA libraries in which, for the majority of cis-NATs, coexpression in the same tissue was rarely found (Wang et al., 2005).
It has experimentally been demonstrated that SRO5 and P5CDH, a pair of cis-NATs, have antagonistic functions in the regulation of salt tolerance in Arabidopsis (Borsani et al., 2005). We therefore examined the expression profiles of these two genes in greater detail and found that global expression of P5CDH and SRO5 is not highly anticorrelated (Supplemental Fig. S3). The strongest anticorrelation was found in the hormone data set with a PCC of −0.546, while in the development and the abiotic stress data sets derived from shoots, anticorrelation was weaker, with PCC values of −0.171 and −0.178, respectively. In roots, the expression of the two genes actually is positively correlated (PCC = 0.208) across the various stress treatments, suggesting that mutual regulation of these two genes is restricted to specific conditions.
Anticorrelation across the Different Microarray Data Sets
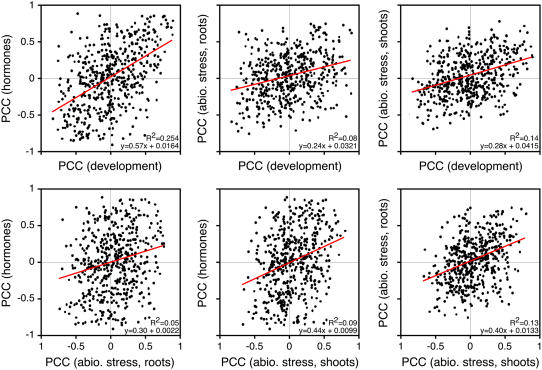
We next analyzed whether the same cis-NAT pairs always displayed strong negative anticorrelation in the various data sets. We found that across the different data sets, the most strongly anticorrelated cis-NATs varied (Fig. 3) and that there was only weak overall correlation between the individual experiments. The highest correlation was found between the development and hormone data sets with _R_2 = 0.25. For the remaining comparisons, the _R_2 value ranged from 0.05 to 0.14. Of the 515 manually curated cis-NAT pairs analyzed, only six showed an average PCC of less than −0.5 in all four microarray experiments. Of these, only two had PCC values lower than −0.5 in every individual experiment (Supplemental Table S3). These findings are consistent with the idea that gene expression is primarily regulated at the transcriptional level by factors such as tissue identity, hormone status, or stress, and that only under specific conditions clear anticorrelation is seen. This finding also implies that the simple presence of an antisense transcript is not sufficient for the negative cross regulation, suggesting that the effectiveness of posttranscriptional RNA regulation by RNA interference greatly varies.
Figure 3.
Correlation of the PCCs for 515 cis-NATs between different microarray data sets. [See online article for color version of this figure.]
Anticorrelation of Antisense Transcripts Is Not Predicted by Extent of Overlap or Promoter Distance
One obvious parameter that might influence the degree of mutual regulation could be the length of the overlapping region. We therefore analyzed whether the PCC for a given cis-NAT pair was correlated with the length of the overlap but found no evidence for such a relationship (Fig. 4) We next determined whether the distance between the 5′ ends of the transcripts of cis-NATs was indicative for the degree of negative correlation found, with the idea that proximity of promoters could cause positive correlation in expression. However, similar to the length of the overlap, the distance of 5′ ends of cis-NAT pairs had no effects on their PCCs (data not shown), indicating that varying promoter distance is unlikely to confound the conclusions about transcript overlap and anticorrelated expression.
Figure 4.
Scatter plot showing independence of PCCs for 515 cis-NATs and length of transcript overlap.
cis-NAT Transcripts and RNA Silencing
One possible mechanism that might cause negative correlation of cis-NAT RNA accumulation could be the formation of double-stranded RNAs from the overlapping mRNA regions and subsequent processing to siRNAs by DCL proteins. The resulting siRNAs could in turn lead to the destruction of one of the transcripts by an RNA interference-like mechanism, as demonstrated for P5CDH and SRO5 (Borsani et al., 2005).
To analyze the contribution of siRNAs to anticorrelated expression of cis-NATS, we examined the distribution of small RNA loci across the genome (Gustafson et al., 2005; Lu et al., 2005). Specifically, we asked whether MPSS tags or small RNA sequences from several source tissues are enriched in the overlapping regions of cis-NATs, as would be expected if double-stranded RNAs were the cause for down-regulation of one of the transcripts (Rajagopalan et al., 2006; Kasschau et al., 2007). Analysis was carried out for all cis-NATs based on the TAIR6 annotation (1,136 gene pairs), as well as for those cis-NATs that are present on the ATH1 array before (714 gene pairs) and after manual curation (515 gene pairs). Results are summarized in Table III. For detailed information on the mapping of unique small RNA loci to the Arabidopsis transcriptome, see Supplemental Data S1.
Table III.
Density of small RNA loci in cis-NATs and nonoverlapping gene pairs
Density of small RNAs (loci per kilobase pair) according to the TAIR6 annotation and those cis-NATs present on the ATH1 array before (ATH1) and after manual curation (curated) of the overlapping region for MPSS (Meyers et al., 2004a, 2004b; Lu et al., 2005) and small RNA data sets (Gustafson et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007). Calculations were preformed based on all gene pairs (top) and those gene-pairs that actually contain small RNA loci (bottom). See supplemental data for detailed information.
| Annotation | Source | Nonoverlapping Gene Pairs | cis-NATs | ||
|---|---|---|---|---|---|
| Total | Nonoverlapping | Overlapping | |||
| TAIR6 | Small RNA | 1.467 | 0.388 | 0.315 | 1.126 |
| MPSS | 0.234 | 0.065 | 0.058 | 0.137 | |
| ATH1 | Small RNA | 0.808 | 0.295 | 0.290 | 0.448 |
| MPSS | 0.139 | 0.061 | 0.0059 | 0.096 | |
| Curated | Small RNA | 0.800 | 0.304 | 0.305 | 0.337 |
| MPSS | 0.138 | 0.061 | 0.062 | 0.064 | |
| TAIR6 | Small RNA | 2.523 | 0.711 | 0.630 | 4.949 |
| MPSS | 0.772 | 0.321 | 0.347 | 1.904 | |
| ATH1 | Small RNA | 1.455 | 0.552 | 0.575 | 2.777 |
| MPSS | 0.585 | 0.304 | 0.331 | 1.799 | |
| Curated | Small RNA | 1.442 | 0.571 | 0.579 | 3.970 |
| MPSS | 0.581 | 0.313 | 0.322 | 3.781 |
We found that over all 1,126 cis-NAT pairs predicted by the TAIR6 annotation, small RNAs were not enriched in cis-NATs when compared to nonoverlapping neighboring genes pairs (Table III, top half). For example, we observed 1.467 small RNA loci/kb genomic sequence in nonoverlapping gene pairs, but we found only 0.388 loci/kb in the cis-NATs. However, if small RNAs were present in cis-NATs at all, they appeared to be enriched in the overlapping region of cis-NAT pairs (1.126 loci/kb) when compared to the nonoverlapping region (0.315 loci/kb). Similar results were obtained when we restricted the analysis to those cis-NATs that are present on the ATH1 arrays (714) and were confirmed by manual curation (515). In all instances, no enrichment of small RNAs in cis-NATs was observed. If one takes into account that not all gene pairs in a given category contain small RNA loci, the outcome differs in that small RNAs were found to be enriched in the overlapping region of cis-NATs (4.949 loci/kb) compared to nonoverlapping gene pairs (2.523 loci/kb) by a factor of approximately 2 (Table III, lower half). Together, these findings point to the fact that siRNA-mediated silencing does not play a major role in the global regulation of cis-NAT expression, at least not under those conditions examined in published small RNA-sequencing projects (Gustafson et al., 2005; Lu et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007).
Further support for this notion came from analyzing microarray data of mutants affected in the biogenesis of small RNAs (Allen et al., 2005). We found that cis-NATs accounted for 2.1% to 5.7% of all transcripts that changed significantly between wild type and the different mutants (Table IV). Given that cis-NATs supported by mRNAs make up 4.5% of all probe sets present on the ATH1 array (1,027/22,810 probe sets), this is approximately what one would expect by chance, indicating that cis-NATs are not more likely to be regulated by small RNAs than nonoverlapping transcripts. Taken together, we could not find positive evidence for a pervasive role of small RNAs in the regulation of antisense transcripts.
Table IV.
Analysis of differential cis-NAT expression in RNA silencing mutants
Transcripts that changed significantly in a given genotype relative to the wild-type control are indicated.
| Genotype | All Transcripts | cis-NATs (4) | cis-NATs (4*) |
|---|---|---|---|
| dcl1-7 | 981 | 56 (5.7%) | 43 (4.4%) |
| dcl2-1 | 145 | 7 (4.8%) | 3 (2.1%) |
| dcl3-1 | 221 | 14 (6.3%) | 8 (3.6%) |
| hen1-1 | 893 | 44 (4.9%) | 34 (3.8%) |
| hst-15 | 895 | 55 (6.1%) | 45 (5.0%) |
| hyl1-2 | 291 | 22 (7.6%) | 16 (5.5%) |
| rdr1-1 | 105 | 8 (7.6%) | 6 (5.7%) |
| rdr2-1 | 166 | 9 (5.4%) | 5 (3.0%) |
| rdr6-15 | 397 | 22 (5.5%) | 17 (4.3%) |
CONCLUSION
Our results paint the most detailed picture of the global regulation of cis-NATS in plants so far. While we could show that cis-NAT pairs tend to have more anticorrelated expression patterns than nonoverlapping neighboring transcripts, we found that pronounced anticorrelation across many samples can only be found in a small subset of cis-NATs. Along these lines, we found that discrete cis-NAT pairs show anticorrelated expression in different experiments, suggesting that independent transcriptional regulation of both members of a pair has a strong influence on cis-NAT expression. The negative correlation of cis-NATs was also observed in a cell type-specific data set, indicating that cis-NATs affect each others' expression in individual cells. The observation that small RNA loci, representing mainly siRNAs, were underrepresented in cis-NATs along with the fact that mutations in the RNA silencing machinery did not have a significant effect on cis-NAT expression confirm this notion and complement previous suggestions that small RNAs and RNA interference are important for only a subset of cis-NATs (Lu et al., 2005).
However, there is at least one known example in which small RNAs derived from cis-NATs have been shown to be important in mutually antagonistic expression, namely, the SRO5 and P5CDH pair of cis-NATs involved in Arabidopsis salt tolerance (Borsani et al., 2005). When exposed to salt stress, SRO5 message is induced, leading to formation of small RNAs and activation of an RNA-silencing pathway that ultimately leads to down-regulation of the P5CDH transcript. As pointed out before, no small RNA MPSS tag from wild-type tissue maps to the overlapping region of the two transcripts, consistent with the inducible nature of this particular siRNA. Borsani et al. (2005) have also suggested that microarrays are imperfect for assessing mutually antagonistic effects, if 3′ products are largely stable. Indeed, SRO5 and P5CDH are only weakly anticorrelated in our data sets and are not significantly different from nonoverlapping transcripts. Nevertheless, the significant shift in correlation coefficients of cis-NATs toward negative values when compared with nonoverlapping transcripts indicates that coordinated expression of cis-NAT can be detected by microarrays, even if the mechanism by which this is achieved is still unclear. These strongly anticorrelated cis-NATs will be attractive targets for further mechanistic studies.
MATERIALS AND METHODS
Mapping of Transcript Pairs
The XML file containing the latest annotation (version 6) of Arabidopsis (Arabidopsis thaliana) pseudochromosomes was downloaded from the TAIR FTP server (ftp://ftp.arabidopsis.org/home/tair/). Start and stop position of the transcription units along with information on the strand that encodes a mRNA and the gene description were extracted. We used Perl scripts to categorize pairs of adjacent transcripts, depending on overlap and whether they were transcribed from the same strand. In a first step, we defined all antisense transcripts that overlapped for at least one base as predicted by the TAIR6 annotation as potential cis-NATs. In a second step, all predicted cis-NATs were manually inspected, and only those that were supported by spliced cDNA and/or EST clones were analyzed further. Single exon genes and gene models not supported by any mRNA were required to be clearly coding (≥100 codon ORF) to be included in the final cis-NATs list.
Determining Correlation Coefficients
Mapping information of transcripts onto the Affymetrix ATH1 array was obtained from TAIR as well. We only used those probe sets that mapped to a single transcription unit. In those few cases where a transcription unit was represented by more than one specific probe set, we retained for further analysis only one of the probe sets at random. Pairwise PCCs and pairwise SCCs were calculated using programs written in Java. Histograms (bin size 0.1), ranking, and comparisons of PCCs between individual microarray data sets were created in Microsoft Excel.
Microarray Analysis
All microarray data used are publicly available. Data for correlation analysis were from the AtGenExpress initiative (available from TAIR). Microarray data of small RNA biogenesis mutants (Allen et al., 2005) were obtained from National Center for Biotechnology Information (NCBI)-GEO (GSE2473). Microarray data were normalized using gcRMA (Wu et al., 2004) implemented in GeneSpring 7.1 (Agilent Technologies). Genes that were differentially expressed between controls and mutants affected in the biogenesis of small RNAs were identified using the Rank Product (Breitling et al., 2004) package implemented in R (http://www.R-project.org). Percentage false positives (pfp) were calculated based on 100 permutations. Only probe sets with a pfp <0.05 for a given comparison were carried forward. In addition to a pfp <0.05, we required a minimum of 2-fold change in expression estimate for a probe set to be considered to be robustly differentially expressed.
Mapping MPSS Tags and Small RNA Sequences to cis-NATs
All MPSS tags and small RNA sequences used are publicly available. MPSS tags were downloaded from the Arabidopsis MPSS database (http://mpss.udel.edu/at/; Meyers et al., 2004a, 2004b; Lu et al., 2005). Small RNAs sequences (ecotype Columbia) from several source tissues were described previously and are accessible at NCBI-GEO [GSE5228 and GSE6682] or the Arabidopsis Small RNA Project Database (http://asrp.cgrb.oregonstate.edu/db/; Gustafson et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007). All tags and sequences were blasted against the Arabidopsis genome to identify positions of perfect matches. MPSS tags or small RNA sequences mapping to a single locus were analyzed for position information in relation to cis-NATs using PERL scripts. MPSS tags or small RNA sequences were counted if any portion of the locus overlapped the region of interest.
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. Correlation of PCC and SCC values for 515 manually confirmed cis-NATs.
- Supplemental Figure S2. Distribution of PCC values in microarray data from sorted root cells.
- Supplemental Figure S3. Expression profiles of SRO5 and P5CDH.
- Supplemental Table S1. List of adjacent transcript pairs in Arabidopsis.
- Supplemental Table S2. List of adjacent transcript pairs in Arabidopsis, mapped onto the Affymetrix ATH1 array.
- Supplemental Table S3. List of cis-NAT pairs.
- Supplemental Table S4. Statistical analysis of pairwise SCC distributions.
- Supplemental Data S1. RNA mapping.
Supplementary Material
[Supplemental Data]
Acknowledgments
We are indebted to Blake Meyers for making the MPSS data of small RNAs available as a database dump. The initial generation of AtGenExpress microarray data was supported by the Deutsche Forschungsgemeinschaft through a grant to L. Nover, T. Altmann, and D.W., and by the Max Planck Society. J.U.L. is an EMBO Young Investigator, and D.W. is a director of the Max Planck Institute.
1
This work was supported by the Max Planck Society, by the National Science Foundation (grant no. MCB–0618433 to J.C.C.), and by the U.S. Department of Agriculture (grant no. 2005–35319–15280 to J.C.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Markus Schmid (markus.schmid@tuebingen.mpg.de).
[C]
Some figures in this article are displayed in color online but in black and white in the print edition.
[W]
Online version contains Web-only data.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Izaurralde E (2006) Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev 20 391–398 [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573 83–92 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Farrell CM, Lukens LN (1995) Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the alpha 1 (I) collagen gene. J Biol Chem 270 3400–3408 [DOI] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD (2005) ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res 33 D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith RK Jr, Maiti R, Chan AP, Yu C, Farzad M, Wu D, et al (2005) Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6 R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57 19–53 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His->Asp phosphorelay circuitry. Plant Cell Physiol 46 339–355 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A (2004) Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Carmichael GG (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev 62 1415–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Williams G, Campbell RD, Sanderson CM (2002) Antisense transcripts in the human genome. Trends Genet 18 63–65 [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S (2004. a) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004. b) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22 1006–1011 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Newbury SF (2006) Control of mRNA stability in eukaryotes. Biochem Soc Trans 34 30–34 [DOI] [PubMed] [Google Scholar]
- Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, et al (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5 R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Shendure J, Church GM (2002) Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol 3 research0044.1–research0044.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Soret J, Guyon C, Gaillard C, Dumon S, Keller M, Crisanti P, Perbal B (1997) Characterization of multiple alternative RNAs resulting from antisense transcription of the PR264/SC35 splicing factor gene. Nucleic Acids Res 25 4513–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34 157–165 [DOI] [PubMed] [Google Scholar]
- Vanhee-Brossollet C, Vaquero C (1998) Do natural antisense transcripts make sense in eukaryotes? Gene 211 1–9 [DOI] [PubMed] [Google Scholar]
- Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11 460–468 [DOI] [PubMed] [Google Scholar]
- Wang H, Chua NH, Wang XJ (2006) Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol 7 R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6 R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer FA (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99 909–917 [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846 [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21 379–386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]