The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination (original) (raw)
Abstract
Termination of RNA polymerase II transcription frequently requires a poly(A) signal and cleavage/polyadenylation factors. Recent work has shown that degradation of the downstream cleaved RNA by the exonuclease XRN2 promotes termination, but how XRN2 functions with 3′-processing factors to elicit termination remains unclear. Here we show that XRN2 physically associates with 3′-processing factors and accumulates at the 3′ end of a transcribed gene. In vitro 3′-processing assays show that XRN2 is necessary to degrade the downstream RNA, but is not required for 3′ cleavage. Significantly, degradation of the 3′-cleaved RNA was stimulated when coupled to cleavage. Unexpectedly, while investigating how XRN2 is recruited to the 3′-processing machinery, we found that XRN2 associates with p54nrb/NonO(p54)–protein-associated splicing factor (PSF), multifunctional proteins involved in several nuclear processes. Strikingly, p54 is also required for degradation of the 3′-cleaved RNA in vitro. p54 is present along the length of genes, and small interfering RNA (siRNA)-mediated knockdown leads to defects in XRN2 recruitment and termination. Together, our data indicate that p54nrb/PSF functions in recruitment of XRN2 to facilitate pre-mRNA 3′ processing and transcription termination.
Keywords: RNA polymerase II, pre-mRNA 3′ processing, XRN2, p54nrb/NonO, PSF
The RNA polymerase II (Pol II) transcription cycle involves a series of steps that include initiation, elongation, and termination. Pol II terminates transcription by releasing the DNA template and the nascent transcript. Despite its apparent simplicity, the mechanism of termination has proven elusive. It has been known for some time that an intact mRNA 3′-processing signal is necessary for transcription termination of protein coding genes (Whitelaw and Proudfoot 1986; Logan et al. 1987; Connelly and Manley 1988). Consistent with this, a number of the factors required for 3′ processing are necessary for subsequent termination (e.g.,see Birse et al. 1998; Dichtl et al. 2002). Additionally, considerable evidence suggests that cleavage/polyadenylation occurs cotranscriptionally (e.g., see Yonaha and Proudfoot 1999; Rigo et al. 2005). The C-terminal domain (CTD) of the Pol II largest subunit, which itself is necessary for efficient 3′ processing (Hirose and Manley 1998), is thought to mediate these interactions by association with several polyadenylation factors (McCracken et al. 1997; Barilla et al. 2001; Fong and Bentley 2001). Thus, it appears that a functional coupling exists between 3′-end processing and termination, but how these events are linked has been a matter of some debate (for reviews, see Luo and Bentley 2004; Buratowski 2005; Rosonina et al. 2006).
Two basic models have been suggested to explain the role of 3′ processing in transcription termination. In one, it has been proposed that termination is triggered by a conformational change of the Pol II elongation complex (EC) that occurs after transcribing the poly(A) site (referred to as the “allosteric model”). Consistent with this model, visualization of Drosophila chromosomes by electron microscopy revealed that a majority of full-length transcripts was released from template DNA in the apparent absence of 3′ cleavage, suggesting that prior endonucleotic cleavage at the poly(A) site is not a prerequisite for termination (e.g., see Osheim et al. 1999). Studies using a purified Pol II EC showed that the CTD-interacting domain (CID) of polyadenylation factor Pcf11 bridges the CTD to the nascent transcript, and this ternary complex can dismantle a paused Pol II EC (Zhang et al. 2005). Subsequent studies showed that the CTD-binding surface of the Pcf11 CID overlaps the RNA-binding surface, and CTD phosphopeptides compete with RNA for the interaction with Pcf11 (Hollingworth et al. 2006). Therefore, it is not clear whether this ternary complex stably exists and would be capable of inducing the putative conformational change required for termination.
A second model proposed that exonucleotic degradation of the uncapped 3′ transcript created by endonucleotic cleavage at the poly(A) site in some fashion signals to the downstream Pol II to terminate transcription (Connelly and Manley 1988). This model requires prior endonucleotic cleavage of the nascent RNA, and degradation of the downstream RNA then triggers template release by Pol II. This model has been supported by the recent demonstration that the 5′–3′ exonuclease XRN2 in human and Rat1 in yeast are required for efficient termination (Kim et al. 2004; West et al. 2004). Previous studies in yeast showed that Rat1 is a nuclear 5′–3′ exoribonuclease that possesses multiple functions in mRNA nuclear export as well as maturation of rRNA and small nucleolar RNA (snoRNA) (Petfalski et al. 1998). XRN1 is an evolutionally conserved 5′-to-3′ exoribonuclease that mainly functions in the cytoplasm (Bashkirov et al. 1997), and loss of XRN1 results in defects in mRNA turnover (e.g., see Bousquet-Antonelli et al. 2000). XRN1 can also function in nuclear rRNA and snoRNA processing (e.g., see Petfalski et al. 1998), but cannot normally substitute for XRN2/Rat1 in termination (Luo et al. 2006).
While RNA interference (RNAi) knockdown of XRN2 and genetic inactivation of Rat1 results in termination defects (Kim et al. 2004; West et al. 2004; Luo et al. 2006), the mechanism of XRN2/Rat1-dependent termination remains unclear. Given that Rat1 containing a point mutation that disrupts catalytic activity was termination defective, degradation of the cleaved downstream transcript appears crucial for termination (Kim et al. 2004). However, recent studies performed by Luo et al. (2006) reported that the 5′–3′ exoribonuclease activity provided by XRN1, fused with a nuclear localization signal to enhance nuclear accumulation, did not rescue the termination defects in rat1 mutant cells, even though both Rat1 and XRN1 were able to induce cotranscriptional degradation of 3′ downstream products. It is thus unclear whether degradation of nascent transcripts directly triggers termination or whether a function of Rat1 in addition to, or instead of, its exoribonuclease activity is critical for termination. Indeed, Luo et al. (2006) suggested that XRN2/Rat1 associates with 3′-processing factors and plays a functional role in 3′ processing.
Rat1 copurifies with Rtt103, a Ser2 P–CTD-binding protein, and both were isolated in tandem affinity purification (TAP) of cleavage factor Pcf11 (Kim et al. 2004). Rat1 and Rtt103 both localize throughout promoter and coding regions and accumulate at the 3′ ends of coding genes (Kim et al. 2004). Rat1 also localizes to snoRNA genes, but is dispensable for termination at these genes (Kim et al. 2006). How XRN2/Rat1 is localized to target genes is unknown, and it is therefore important to identify additional XRN2/Rat1-interacting protein(s), and determine whether they are required for recruitment of XRN2 to the Pol II elongation and/or polyadenylation complexes or for transcription termination.
In this study, we provide new insights into the function of XRN2 in 3′-end formation and termination. We show that recruitment of XRN2 to 3′-processing complexes is necessary for efficient degradation of the 3′ downstream RNA in vitro as well as termination in vivo. Unexpectedly, we found that XRN2 associates with p54nrb–protein-associated splicing factor (PSF), a multifunctional protein complex thought to be involved in transcription, splicing, and polyadenylation (e.g., see Patton et al. 1993; Gozani et al. 1994; Mathur et al. 2001; Rosonina et al. 2005; Liang and Lutz 2006). We establish the functional significance of this interaction by showing that p54nrb/NonO (p54) is also required for degradation of the 3′-cleaved RNA in vitro and for XRN2 recruitment and for efficient termination in vivo.
Results
XRN2 associates with 3′-processing factors
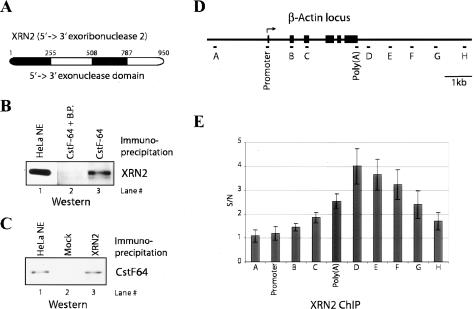
Our recent work showed that human Cdc73, a subunit of the Paf1 complex (Paf1C), a multisubunit protein complex associated with elongating Pol II (Rozenblatt-Rosen et al. 2005), interacts with the cleavage/polyadenylation factors CPSF, CstF, and symplekin (data not shown). Intriguingly, analysis of additional proteins from the same immunopurification by mass spectrometry identified XRN2 (Fig. 1A). To extend these results, and to confirm the association of XRN2 with 3′-processing factors, we generated anti-peptide polyclonal antibodies directed against XRN2 and CstF-64 and performed reciprocal coimmunoprecipitations with HeLa nuclear extracts (NEs). We found that an anti-CstF-64 antibody coimmunoprecipitated XRN2 in the absence but not presence of blocking peptide (Fig. 1B, lanes 2,3), while the anti-XRN2 antibody coimmunoprecipitated CstF-64 (Fig. 1C, lane 3).
Figure 1.
XRN2 is associated with 3′-processing factors. (A) Structure of human XRN2. 5′–3′ exonuclease domains are shown by black boxes. (B) HeLa NE was immunoprecipitated with anti-CstF-64 antibody. (Lane 3) IPs were resolved and blotted with anti-XRN-2 antibody. (Lane 2) Control IP was performed with anti-CstF-64 antibody plus blocking peptide. (Lane 1) Ten percent of NE was loaded to indicate the position of XRN2. (C) Reciprocal IPs with anti-XRN2 antibody show association with CstF-64. NE was immunoprecipitated with anti-XRN2 antibody. (Lane 3) IPs were resolved and visualized by immunoblot with CstF-64 antibody. (Lane 2) XRN2 antibody was omitted in the mock IP. (Lane 1) NE (12.5%) was loaded to indicate the position of CstF-64. (D) Schematic representation of the β-actin locus and the locations of the PCR primers used for ChIP analysis. Each exon is indicated by a black box. The transcription initiation site is indicated by an arrow. (E) XRN2 accumulates after the poly(A) site. ChIP analysis of XRN2 on the β-actin locus is shown. The _Y_-axis shows the signal-to-noise ratio (S/N) of XRN2 IP relative to control IP (anti-XRN2 antibody plus blocking peptide). Error bars show the standard deviation from three independent replicates.
We next wished to determine whether the above interaction might reflect a functional association to ensure that XRN2 is present at the 3′ end of transcribed genes, to function in 3′ processing and/or termination. To this end, we performed chromatin immunoprecipitation (ChIP) experiments utilizing cross-linked HeLa cell extracts and examined XRN2 cross-linking at sites along the length of the β-actin gene (Fig. 1D). Results are presented as the ratio of signals observed in the presence or absence of blocking peptide during the immunoprecipitation (IP) (Fig. 1E; see Materials and Methods). Consistent with similar studies of Rat1 in yeast (Kim et al. 2004; Luo et al. 2006), we found that XRN2 cross-linking was strongest immediately after the poly(A) site and then gradually decreased, consistent with a function in degradation of the 3′-cleaved RNA and/or termination. XRN2 was not detected upstream of or at the promoter, but cross-linking increased gradually along the length of the gene.
XRN2 is required for degradation of 3′ downstream RNA
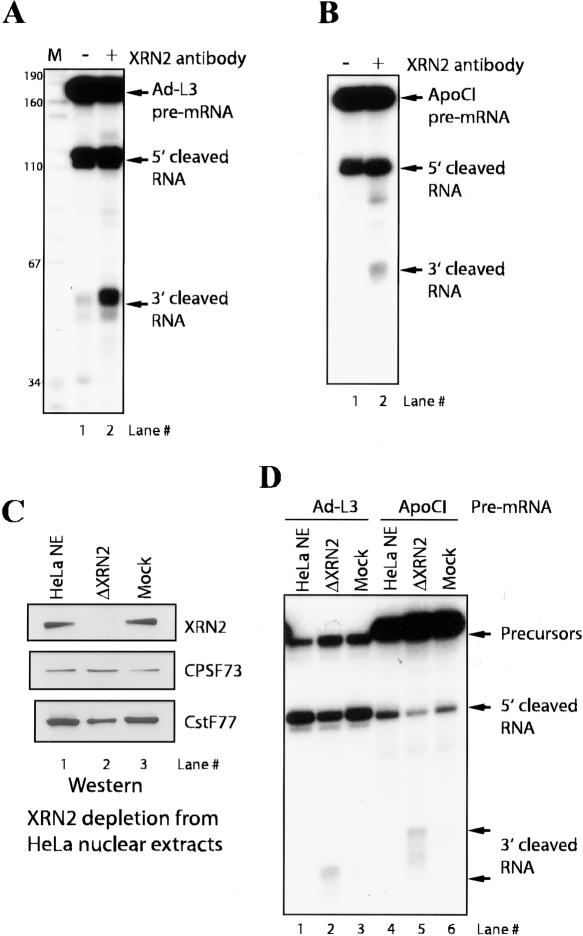
Both Rat1 and XRN1 are capable of cotranscriptional degradation of downstream cleavage products in yeast (Luo et al. 2006). Therefore, a probable function for XRN2 when associated with 3′-processing factors is to degrade the uncapped cleaved RNA following endonucleotic cleavage at the poly(A) site, although a possible role in facilitating the actual cleavage reactions has also been suggested (Luo et al. 2006). To test these possibilities directly, we performed in vitro cleavage assays with an adenovirus L3 pre-mRNA substrate to monitor both 3′-end-processing efficiency as well as degradation of the 3′-cleaved RNA fragment. In an initial experiment, we found that addition of the anti-XRN2 antibody used above into reaction mixtures resulted in the accumulation of 3′-cleaved RNA, while cleavage activity (as judged by accumulation of the upstream 5′ cleavage product) was not affected (Fig. 2A). Identical results were obtained with a poly(A) signal derived from a second RNA, the apolipoprotein CI pre-mRNA (Fig. 2B). Control antibodies affected neither cleavage activity nor degradation of the 3′-cleaved RNA (data not shown).
Figure 2.
Immunodepletion of XRN2 results in accumulation of 3′ downstream RNA. (A) Anti-XRN2 antibody prevents degradation of 3′ downstream RNA. In vitro cleavage assays were performed with NE and adenovirus L3 (Ad-L3) pre-mRNA. (Lane 2) Anti-XRN2 antibody was added to the reaction mixture. Ad-L3 pre-mRNA, 5′-cleaved RNA, and 3′ downstream RNA are indicated by arrows. DNA size markers are shown on the left side. (B) In vitro cleavage assay of Apolipoprotein CI (ApoCI) pre-mRNA.(Lane 2) Cleavage reaction with anti-XRN2 antibody was performed. (C) Immunodepletion of XRN2. (Lane 2) XRN2-depleted NE (ΔXRN2) was resolved by SDS-PAGE and blotted with the indicated antibodies. Immunoblots of depleted (lane 3) and untreated (lane 1) NE are shown. (D) Cleavage reactions of Ad-L3 (lanes 1–3) and ApoCI (lanes 4–6) pre-mRNAs were performed with XRN2-depleted (lanes 2,5), mock-depleted (lanes 3,6), and untreated (lanes 1,4) NE.
The above results suggest that XRN2 is required for degradation of 3′-cleaved RNA in vitro but not for cleavage itself. To address this possibility more directly, we performed in vitro cleavage assays using XRN2 immunodepleted NEs, and as a control, mock-depleted NEs. Immunoblots confirmed that XRN2 was specifically depleted from the extracts (Fig. 2C, top panel), while under the IP conditions used, the amounts of CPSF-73 and CstF-77 were reduced at most only slightly (Fig. 2C, middle and bottom panels). Consistent with the above data, we found that the 3′-cleaved RNA accumulated in the XRN2 depleted NEs (Fig. 2D, lanes 2,5), while this RNA was essentially undetectable in mock-depleted and untreated NEs (Fig. 2D, lanes 1,3,4,6). Cleavage activity was only minimally reduced, likely reflecting the modest codepletion of CstF. These data indicate that XRN2 is required in vitro for degradation of the 3′ cleavage product but not for cleavage activity itself.
Coupling XRN2 to 3′ processing stimulates exonuclease activity in NE
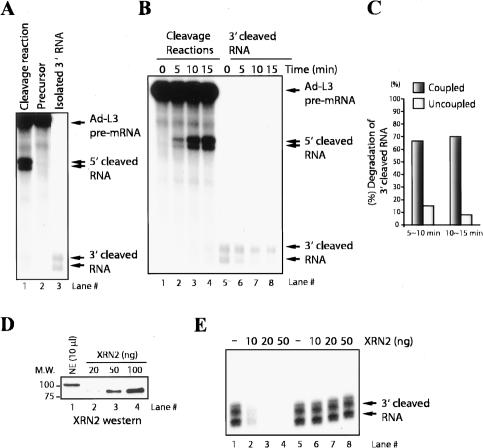
We next wished to investigate whether the association of XRN2 with 3′-processing factors has a functional significance. For example, our data indicate that the presence or absence of XRN2 does not affect processing efficiency, but perhaps coupling 3′ cleavage to degradation of the downstream product enhances the efficiency of the latter reaction. To investigate this possibility, we monitored the kinetics of exonuclease activity both coupled and uncoupled with 3′-end cleavage. To this end, we isolated 3′-cleaved RNA (Fig. 3A, lane 3) and reincubated it in NE under conditions identical to those in typical cleavage assays (Fig. 3B; Kaneko and Manley 2005). Degradation of the downstream RNA when coupled to cleavage was quantitated by determining the ratio of the amount of downstream RNA to the expected amount of this species, calculated from the amount of 5′ cleavage product. We found that only 10% ∼ 15% of isolated 3′-cleaved RNA was degraded at time intervals of 5–10 and 10–15 min (Fig. 3B [lanes 5–8], C). Significantly, time-course assays of authentic cleavage reactions showed 65% ∼ 70% of 3′-cleaved RNA was degraded during these time intervals (Fig. 3B [lanes 1–4], C).
Figure 3.
Degradation of 3′ downstream RNA is coupled with 3′ processing. (A) Preparation of RNA substrates for degradation assays. The 3′-cleaved RNA (lane 3) was gel purified from cleaved Ad-L3 pre-mRNA (lane 1). (B, lanes 1–4) Time-course analysis of in vitro cleavage reaction was performed with NE and Ad-L3 pre-mRNA. (Lanes 5–8) The identical reaction was performed with 3′ downstream RNA instead of Ad-L3 pre-mRNA. The 3′ downstream RNA was prepared by gel purification of cleaved Ad-L3 pre-mRNA. (C) Degradation of 3′ downstream RNA was quantified at a time interval between 5 ∼ 10 min and 10 ∼ 15 min of the reactions. (D) Quantitation of XRN2 protein in NE. Indicated amounts of NE (lane 1) and recombinant, C-terminal-truncated XRN2 (lanes 2–4) were resolved and visualized by immunoblot with XRN2 antibody. (E) XRN2 ribonuclease reactions were performed under typical ribonuclease conditions (lanes 1–4) or 3′ cleavage conditions (lanes 5–8) with 3′ downstream RNA and increasing amounts (0, 10, 20, and 50 ng) of XRN2.
To confirm that the above results reflect the behavior of XRN2, we examined the activity of a purified, recombinant XRN2 derivative in in vitro assays. Using quantitative Western blotting, we first determined that NE contains ∼10 ng/μL XRN2 (Fig. 3D). As our reaction mixtures typically contain 2 μL of NE, we examined the fate of purified 3′-cleaved RNA (Fig. 3B) when incubated with 10, 20, or 50 ng of XRN2 under conditions optimal for exonuclease activity or as used in 3′-processing assays (Fig. 3E; see Materials and Methods). While as expected the RNA was efficiently degraded under optimal nuclease conditions (Fig. 3E, lanes 2–4), it was not degraded significantly under 3′-processing conditions (Fig. 3E, lanes 6–8). Together, these results indicate that coupling XRN2 to 3′ processing stimulates its exonuclease activity in NEs. Possible mechanisms for this enhancement are discussed below.
The C-terminal region of XRN2 is necessary and sufficient for protein interactions
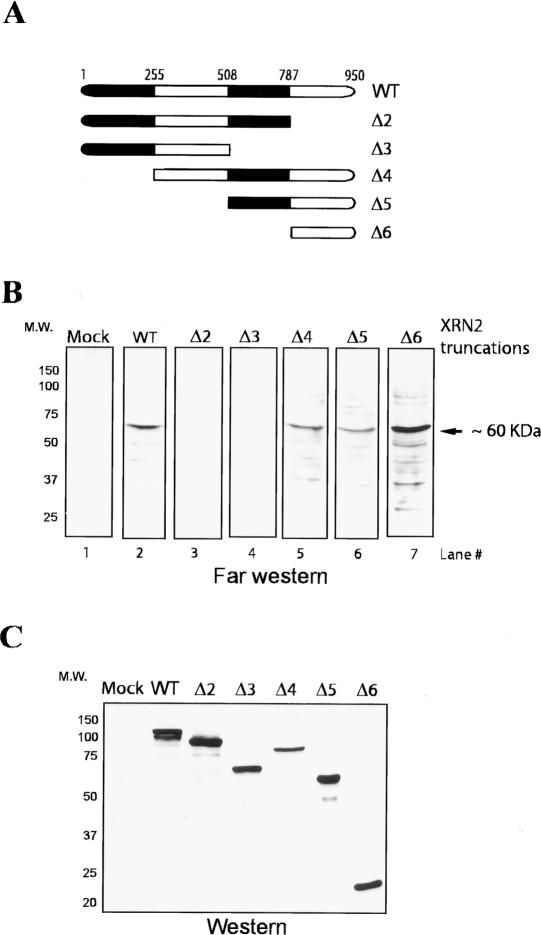
The above experiments indicate that XRN2 associates with the 3′-processing machinery and that this facilitates efficient degradation of 3′ RNA products. We therefore wished to investigate the interactions involved in recruitment of XRN2 to the 3′-processing machinery. To this end, we first set out to define the regions of XRN2 required for interactions with other proteins. An initial set of experiments was performed by employing a Far Western blotting assay. NEs were resolved by SDS-PAGE and transferred to nitrocellulose membrane. To probe for interactions that may be indirect as well as direct, we generated a series of Flag-tagged XRN2 constructs (Fig. 4A) and transfected them into HeLa cells. Cell extracts were prepared to probe the membranes, and XRN2-interacting protein(s) were detected with anti-Flag antibodies. We found a number of XRN2-interacting proteins, including a major one of ∼60 kDa, with wild-type XRN2 (Fig. 4B, lane 2). Significantly, XRN2 deletions of the C-terminal region encompassing 786–950 amino acids (XRN2 786–950) and further deletions eliminated the interactions (Fig. 4B, lanes 3,4), while none of the N-terminal deletions affected them (Fig. 4B, lanes 5–7). These experiments indicate that XRN2 residues 786–950 are necessary and sufficient for interactions with a number of HeLa nuclear proteins.
Figure 4.
The C-terminal region (786–950) of XRN2 is necessary and sufficient for protein–protein interactions. (A) Schematic representation of Flag-tagged wild-type and XRN2 truncations. After transient transfection of XRN2 expression vectors into HeLa cells, cell lysates were used as probes for Far Western assay. (B) NE was resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with either wild-type XRN2 (lane 2) or XRN2 truncations (XRN2 Δ2–Δ6, lanes 3–7). (Lane 1) A lysate of mock-transfected HeLa cells is used as a negative control. (C) Immunoblots of XRN2-truncated proteins. Proteins were detected by anti-Flag antibody.
Identification of XRN2-associating proteins
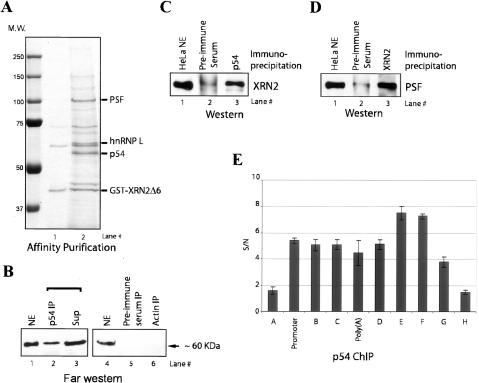
We next wished to identify the proteins capable of interacting with the XRN2 CTD. To this end, an XRN2 (786–950)-GST fusion protein was expressed in Escherichia. coli and purified from extracts on glutathione beads. The beads carrying GST-XRN2 (786–950) and beads bound to GST alone were then incubated with NE. After extensive washing, proteins were eluted with glutathione and separated by SDS-PAGE. We detected three major species with GST-XRN2 (786–950) that were not present in the GST alone beads (data not shown) or beads with GST-XRN2 (786–950) before incubating with NE (Fig. 5A). Analyses by MALDI-TOF MS sequencing revealed that these proteins were heterogeneous ribonucleoprotein L (hnRNP L), polypyrimidine tract-binding PSF and p54. PSF and p54 are related proteins, showing close homology within a central region that contains two RNA recognition motifs and a homo–heterodomierization domain (Dong et al. 1993; Yang et al. 1993). The two proteins have been shown to bind each other and, as mentioned above, have been implicated in transcription, splicing, and polyadenylation. It thus seemed feasible that these proteins might also function in conjunction with XRN2, and we therefore investigated this interaction further.
Figure 5.
PSF–p54nrb is associated with XRN2. (A) Affinity purification of glutathione S-transferase GST-XRN2Δ6-bound proteins. (Lane 1) Bacterially expressed GST-XRN2Δ6 was incubated with NE. (Lane 2) After extensive washing, GST-XRN2Δ6-bound proteins were eluted with glutathione and developed by SDS-PAGE. (B) Far Western analysis shows that the IP of p54 contains XRN2-interacting proteins. Immunoprecipitated NEs with anti-p54nrb (lane 2), preimmune serum (lane 5), and anti-actin (lane 6) were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with Flag-tagged XRN2Δ6. (C) NEs were immunoprecipitated with anti-p54 (lane 3) and preimmune serum (lane 2). IPs were resolved and visualized by Western blot with anti-XRN2 antibody. (Lane 1) Ten percent of NE was loaded to indicate the position of XRN2 protein. (D) Reciprocal IPs with anti-XRN2 revealed interaction with p54/PSF. NEs were immunoprecipitated with anti-XRN2 (lane 3) and preimmune serum (lane 2). IPs were resolved and blotted with anti-PSF antibody. (Lane 1) Ten percent of NE was loaded to indicate the position of PSF. (E) p54 localizes throughout an active gene. ChIP analysis of p54 on the β-actin locus is shown (see Fig. 1D). The _Y_-axis shows the signal-to-noise ratio (S/N) of p54 IP relative to control IP (p54 antibody plus blocking peptide).
p54/PSF associates with XRN2 and localizes throughout an active gene
We next set out to characterize the association between p54/PSF and XRN2 in more detail. We first wished to determine if p54 is one of the XRN2-interacting proteins identified by Far Western blotting (see Fig. 4B). To this end, we immunopurified p54 from NE and performed the Far Western blotting assay as above. We found an ∼60-kDa protein was detected in the p54 IP (Fig. 5B, lane 2), while no proteins were detected in the control IPs (Fig. 5B, lanes 5,6). These experiments indicate that p54 likely corresponds to the major XRN2-interacting protein detected in NE. To obtain evidence that p54/PSF associates with XRN2 in vivo, we performed coimmunoprecipitates with NEs. We found that anti-p54 antibodies coimmunoprecipitated XRN2 while IP with preimmune serum displayed no significant signal (Fig. 5C, lanes 2,3). Furthermore, anti-XRN2 antibodies, but not preimmune serum, coimmunoprecipitated PSF (Fig. 5D, lanes 2,3).
To determine whether p54/PSF and XRN2 have the potential to associate on active genes, we performed ChIP assays along the length of the β-actin gene (see Fig. 1D for the position of primers pairs). We found that p54 was cross-linked at the promoter and throughout the coding region (Fig. 5E), consistent with its proposed roles in transcription and RNA processing. Significantly, p54 remained cross-linked to regions downstream from the poly(A) site (regions D–F), while we did not detect significant signals upstream of the promoter or far downstream from the poly(A) site (regions A and H). Taken together, these experiments suggest that p54–PSF associates with XRN2, in a manner consistent with a role in coupling 3′ processing and transcription termination.
Immunodepletion of p54nrb–PSF results in accumulation of 3′-cleaved RNA in vitro
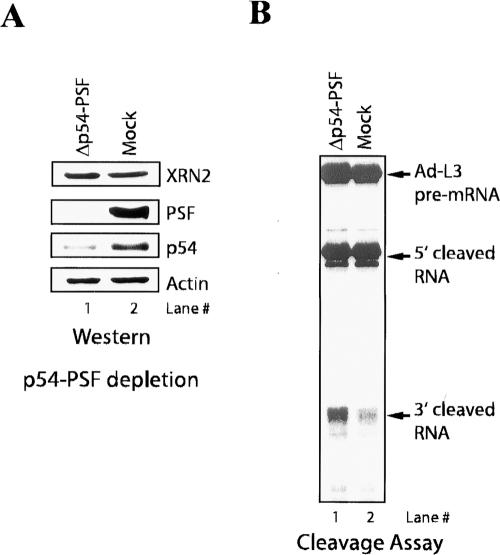
We next wished to investigate the functional significance of the p54–PSF association with XRN2. The data presented above provide evidence that XRN2 associates with the 3′-processing machinery and that this functions to facilitate efficient degradation of 3′ cleavage products. We therefore reasoned that p54–PSF might play a role in coupling 3′ cleavage to degradation of the downstream products, perhaps by recruiting XRN2 to the processing complex. To test this, we immunodepleted both p54 and PSF from NEs under stringent conditions to deplete p54–PSF but not XRN2 (see Materials and Methods), and used these NEs for in vitro cleavage assays. Immunoblots confirmed that p54–PSF was efficiently depleted from NEs, while the amount of XRN2 remained unchanged (Fig. 6A). Strikingly, 3′-cleaved RNA accumulated in the p54–PSF-depleted NE but not in the mock-depleted NE (Fig. 6B). Depletion of p54–PSF did not affect cleavage activity, as no effect on accumulation of 5′ cleavage products was detected. These data suggest that p54–PSF, like XRN2, is required for degradation of 3′-cleaved RNA but not for cleavage activity in vitro.
Figure 6.
Immunodepletion of p54/PSF results in accumulation of 3′-cleaved RNA. (A) Immunodepletion of p54/PSF. (Lane 1) p54/PSF-depleted NE (Δp54–PSF) was resolved by SDS-PAGE and blotted with the indicated antibodies. (Lane 2) Mock depletion (Mock) is also shown. (B) Cleavage reactions of Ad-L3 pre-mRNA were performed with p54–PSF-depleted (lane 1) and mock-depleted (lane 2) NE.
RNAi knockdown of p54 causes a termination-related defect in vivo
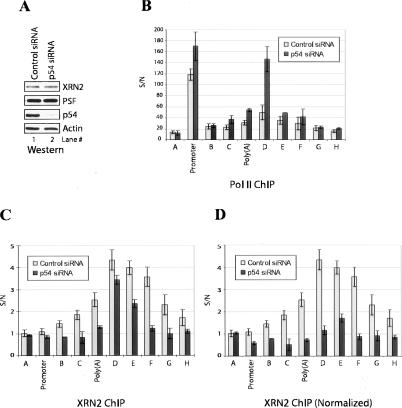
The above experiments indicate that p54–PSF associates with XRN2 and is required for efficient degradation of downstream cleavage products. Given the requirement of XRN2 for degradation of downstream RNA and efficient termination, we hypothesized that p54/PSF might play a role in coupling 3′ processing to termination. To test this idea, we performed ChIP assays to measure Pol II density along the the β-actin gene using p54 small interfering RNA (siRNA)-depleted cells. To this end, HeLa cells were treated with a p54-specific siRNA or with a nontargeting siRNA as a negative control. (Comparable experiments with PSF were not feasible because PSF RNAi resulted in cell death [results not shown].) Immunoblots confirmed that the amount of p54 was significantly decreased in the p54 siRNA-treated cells while the amount of XRN2 (and PSF) remained unchanged (Fig. 7A). Cross-linked extracts were immunoprecipitated with N20, an antibody that recognizes an epitope in the body of the Pol II largest subunit. While the nontargeting siRNA did not affect distribution of Pol II compared with untreated cells (data not shown), significant effects were observed in the p54-depleted cells. Most notably, Pol II accumulation increased markedly immediately downstream from the poly(A) site (Fig. 7B, region D). A smaller increase was detected at the poly(A) site itself, but Pol II levels decreased normally further downstream (regions E and F). Similar results were obtained with a second anti-Pol II antibody (8WG16, which recognizes an unphosphorylated CTD epitope) (data not shown). Significantly, recent studies have suggested that Pol II pausing at a site within region D plays an important stimulatory role in XRN2-mediated termination in the β-actin gene (Gromak et al. 2006), and our results suggest that p54 also plays a role in this process.
Figure 7.
Knockdown of p54 leads to defective termination and XRN2 recruitment. (A) siRNA knockdown of p54. Whole-cell lysates of HeLa cells transfected with p54 (lane 2) and control (lane 1) siRNA were resolved by SDS-PAGE and blotted with the indicated antibodies. (B) ChIP analysis of Pol II on the β-actin locus is shown (see Fig. 1D). The _Y_-axis shows the signal-to-noise ratio (S/N) of Pol II IP relative to control IP (Pol II antibody plus blocking peptide). (C) p54 is required for recruitment of XRN2 to Pol II EC. ChIP analysis of XRN2 was performed as described in Figure 1E. (D) The XRN2 ChIP signals are presented normalized to the Pol II ChIP signals with or without p54 depletion.
The above data suggest that p54/PSF may be involved in recruitment of XRN2 to the Pol II EC. To test this, we again performed ChIP experiments using p54-depleted cells and examined the distribution of XRN2. XRN2 association was considerably reduced, most notably at a region encompassing the poly(A) site itself and just downstream from region D (Fig. 7C). Given that the Pol II ChIP profile was altered by p54 depletion (Fig. 7B), an alternative means of presenting the XRN2 ChIP signals is to normalize them to the Pol II signals. When presented in this manner, the decrease in XRN2 cross-linking was considerably more striking, especially at regions downstream from the poly(A) site (Fig. 7D). We conclude that depletion of p54 reduced XRN2 recruitment while increasing Pol II termination-related pausing downstream from the poly(A) site. It is possible that these results may underestimate the role of p54 in these processes, due to incomplete depletion and/or possible redundancy with PSF. Our results are similar to but distinct from those observed when XRN2 itself was depleted by siRNA treatment (Gromak et al. 2006), and together with our other data support the idea that p54/PSF plays a role in enhancing XRN2-mediated exonuclease activity in 3′ processing and transcription termination.
Discussion
In this study, we demonstrated that XRN2 is recruited to the polyadenylation machinery to facilitate degradation of the 3′ downstream cleaved RNA. We also found that XRN2 associates with p54nrb/NonO (p54)–PSF, multifunctional factors implicated in transcription, splicing, and polyadenylation and other nuclear processes (e.g., see Zhang and Carmichael 2001). Significantly, p54/PSF is required for efficient degradation of the downstream RNA in vitro and for efficient XRN2 recruitment and proper Pol II pausing/termination in vivo. Together, our data extend previous results regarding the role of XRN2 in 3′ processing and termination (Kim et al. 2004; West et al. 2004; Luo et al. 2006) by showing that p54/PSF plays an important role in coupling 3′ processing and termination in mammalian cells. Below we discuss how XRN2 is recruited to 3′-processing machinery in conjunction with p54–PSF, the mechanism for stimulation of exonuclease activity coupled to 3′-end processing, and the new role(s) of p54–PSF in 3′ processing and termination.
We initially identified XRN2 by virtue of its apparent association with the Paf1 complex. Paf1C was originally identified as an RNA Pol II-associated complex in yeast, and is required for transcription of several genes. It associates with the Spt4–Spt5 and yeast FACT chromatin elongation factors (e.g., see Krogan et al. 2002). More recent reports have demonstrated that Paf1C affects 3′-end processing of both mRNAs and snoRNAs (Penheiter et al. 2005; Sheldon et al. 2005). Paf1C is therefore thought to coordinate interaction of 3′-processing factors and nuclear factors that function in transcription and chromatin modification (for reviews, see Hampsey and Reinberg 2003; Rosonina and Manley 2005). Given our ChIP experiments indicating that XRN2 accumulates toward the 3′ end of the β-actin gene, it is possible that Paf1C participates in cotranscriptional recruitment of XRN2. Consistent with this, PSF was also detected in the immunopurified Paf1C in which we detected XRN2 (our unpublished data).
Our data showed that XRN2 is not required for efficient 3′ cleavage in vitro and functions only in degradation of the downstream RNA. These results are consistent with studies in yeast and human that found that loss of Rat1/XRN2 did not have a detectable effect on 3′-end formation in vivo (Kim et al. 2004; West et al. 2004). However, a recent study suggested that inactivation of Rat1 in yeast reduced recruitment of certain polyadenylation factors to 3′ end of genes and altered usage of poly(A) sites in transcripts that undergo alternative polyadenylation (Luo et al. 2006). Our data suggest a parsimonious explanation for these disparate results. Specifically, we propose that XRN2 has no effects on interactions involving the core polyadenylation machinery. However, we suggest that it alters factor recruitment and/or interactions that occur in vivo during transcription-coupled 3′ processing. This could affect the rate of processing of specific poly(A) sites and thus influence relative utilization of alternative sites, while not affecting accumulation of steady-state levels of most mRNAs in which the polyadenylation step is not rate limiting. The increased pausing of Pol II downstream from the β-actin poly(A) site that we detected is consistent with a possible delay in initiation of processing.
An interesting question is how the coupling of 3′ cleavage to degradation of the downstream RNA enhances the efficiency of exonuclease activity. It may simply be that recruitment of XRN2 to an RNA substrate promotes its degradation. Alternatively, Rai1 is known to enhance Rat1 exonuclease activity in yeast (Xue et al. 2000), and XRN2-interacting proteins, perhaps p54/PSF, might stimulate XRN2 activity. Still another possibility is that integration of 3′ processing and degradation could prevent the downstream RNA from binding nuclear hnRNPs, which might in turn prevent or slow degradation. In any event, our data suggest that XRN2 is recruited to the 3′-processing machinery to facilitate efficient degradation of 3′ downstream RNA and not to enhance assembly or efficiency of the basal polyadenylation machinery.
Our discovery that XRN2 functionally associates with p54 and PSF provides unexpected new insight into how termination and 3′ processing are coupled. Numerous studies have suggested that splicing and 3′-end formation are coupled to transcription (for reviews, see Hirose and Manley 2000; Proudfoot et al. 2002; Zorio and Bentley 2004). This function is at least partially mediated by the Pol II CTD, which appears to recruit specific spliceosome and polyadenylation components cotranscriptionally (e.g., see McCracken et al. 1997). Intriguingly, p54 and PSF were purified by both hypo- and hyperphosphorylated CTD (Emili et al. 2002) and identified as part of a large transcription/splicing complex (Kameoka et al. 2004). Furthermore, p54 (but not XRN2) has been found to copurify with active cleavage/polyadenylation complexes bound to an RNA substrate (Y. Shi, D. DiGiammartino, and J.L. Manley, unpubl. data) and to associate with a complex capable of modulating polyadenylation in vitro (Liang and Lutz 2006).
Our data provided evidence that p54/PSF functions in 3′-end formation and transcription termination by recruiting XRN2 to the transcription/polyadenylation complex. But given that p54 (and presumably PSF) is present along the length of the gene, why is XRN2 only recruited to the 3′ end? One possibility is that a conformational change occurs in the Pol II EC—for example, at the downstream pause site—that allows a surface of p54/PSF that binds XRN2 to be exposed. Another not mutually exclusive possibility is that phosphorylation of one or both proteins facilitate binding of XRN2. Indeed, p54 and PSF are both known to be phosphorylated, and multiple kinases have been found to be capable of phosphorylating one or both and altering their protein interactions or functional properties (e.g., see Proteau et al. 2005; Huang et al. 2007). An interesting candidate to influence 3′-proximal events such as XRN2 recruitment is Cdk9/cyclinT, or P-TEFb (Marshall and Price 1995). P-TEFb is known to phosphorylate the CTD at the Ser2 position toward the 3′ end of transcribed genes and, importantly, to enhance transcription-coupled 3′-end formation (Ahn et al. 2004; Ni et al. 2004). Although the possibility that p54 or PSF is a P-TEFb target has not been investigated, both proteins contain multiple potential cdk phosphorylation sites (S/T-P), and p54 is in fact known to be phosphorylated by at least two cdks during M phase (Proteau et al. 2005).
We also identified hnRNP L as an XRN2-associated protein. hnRNP L is an abundant nuclear protein and has been found to associate with both the mRNA export factor TAP and the exon junction complex protein Aly/REF (e.g., see Guang et al. 2005). Several studies have suggested that hnRNP L is multifunctional, with roles in splicing, export, mRNA stability, and translation (e.g., see Hahm et al. 1998; Shih and Claffey 1999; Hui et al. 2003). Significantly, hnRNP L is also capable of stimulating polyadenylation (Guang et al. 2005). It will be intriguing to investigate if and how hnRNP L contributes to the function of XRN2.
The mechanism by which Pol II pausing promotes termination has not been clearly demonstrated. Pausing facilitates cleavage/polyadenylation (Yonaha and Proudfoot 1999, 2000), likely allowing CTD-mediated interactions between the RNA and polyadenylation factors, and thereby perhaps indirectly promoting termination. Pausing may also allow other changes in the Pol II EC that are important for termination, such as recruitment and activation of XRN2. Consistent with this, pausing was found to correlate with degradation of the downstream cleaved RNA by XRN2 (Gromak et al. 2006). Our ChIP experiments demonstrated an accumulation of Pol II at or near the β-actin pause site in the absence of p54, and a concomitant decrease in XRN2, notably just upstream at the poly(A) site. We suggest that one function of Pol II pausing is to allow XRN2 to associate with the EC and to initiate exonuclease activity. When this process is defective, pausing is enhanced.
In summary, our data provided evidence that XRN2 recruitment to the 3′-processing machinery is required for efficient degradation of 3′-cleaved RNA and critical for proper Pol II termination. Surprisingly, p54/PSF was also found to be required for optimal XRN2 activity and termination, likely affecting a function in recruitment of XRN2 to the Pol II EC and/or polyadenylation machinery. While many questions remain regarding the function of p54/PSF in 3′ processing and transcription termination, our results provided new insight into the complex coupling between RNA processing and transcription.
Materials and methods
Antibodies, blocking peptides, and siRNA sequences
Antibodies were obtained from Bethyl Laboratories (anti-XRN2 [BL-2039, BL-2041], anti-CPSF-73 [BL-1906], anti-CstF-77 [BL-1894], CstF-64 [BL-1889], and anti-p54), Sigma (anti-actin), Santa Cruz Biotechnology (anti-Pol II #N-20), and Stratagene (anti-Flag M2). Anti-CstF-64 monoclonal antibody (αCstF-64 mAb) was described previously (Takagaki et al. 1990). Rabbit polyclonal antibody specific for PSF was a gift from Dr. J. Patton (Vanderbilt University, Nashville, TN). Blocking peptides were obtained from Bethyl Laboratories (CstF-64, XRN2, and p54) and Santa Cruz Biotechnology (Pol II #N-20). The siRNA for p54 mRNA targeted 5′-GCAUUCCUGAAGUCUCUAA-3′. siCONTROL nontargeting siRNA #1 was from Dharmacon.
Immunodepletion of XRN2 and p54/PSF
HeLa NE was prepared as described previously (Kaneko and Manley 2005). To prepare ΔXRN2 NE, NE was first diluted into an equal volume of 1× dilution buffer (25 mM Tris at pH 7.9, 125 mM NaCl, 0.25% NP-40) and then mixed twice with protein A beads (GE Healthcare) coated with or without anti-XRN2 antibody for 2 h at 4°C. An identical procedure was carried out to prepare Δp54/PSF NE by using anti-p54 and PSF antibodies, except a 2× dilution buffer was used.
In vitro 3′ cleavage assays
Cleavage assays were performed in 12.5-μL reaction mixtures containing NEs (2 μL), ∼1 ng labeled pre-mRNA substrates, 9.6 mM HEPES-NaOH (pH 7.9), 9.6% glycerol, 24 mM (NH4)2SO4, 0.24 mM DTT, 4% polyvinyl alcohol, 0.24 mM PMSF, 0.12 mM EDTA, and 0.5 mM 3′ dATP at 30°C (Kaneko and Manley 2005). To isolate 3′-cleaved RNA, the cleavage assay also contained 2 mM EDTA. 3′-Cleaved RNA was then purified from a 6% polyacrylamide gel.
Ribonuclease assay
Exoribonuclease assays were performed in reaction mixtures containing 20 mM Tris (pH 7.9), 50 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 2 μg BSA, purified 3′ Ad-L3 downstream RNA, and indicated amounts of recombinant XRN2 (gift of S. Xiang and L. Tong, Columbia University, New York, NY). Reaction mixtures were incubated at 37°C for 15 min. RNA products were isolated and fractionated by 6% PAGE in 8.3 M urea.
Plasmids
Human XNR2 cDNA, obtained by RT–PCR of oligo(dT)-selected HeLa cell RNA, was inserted into p3xFlag-CMV-7 (Sigma). XRN2 truncation mutants were generated by PCR. For bacterial expression, XRN2-truncated cDNA was inserted into pGEX-2T (GE Healthcare).
Far Western blot analyses
Flag-tagged XRN2 expression vectors were transfected into HeLa cells. After 24 h, cell extracts were prepared and used as bait proteins. HeLa NE (∼50 μg) was loaded onto 10% SDS-PAGE and transferred to nitrocellulose. Filters were blocked in PBST containing 5% nonfat dry milk for 12–16 h at 4°C. Filters were then washed in PBST and in binding buffer (20 mM Tris at pH 7.9, 100 mM NaCl, 0.1% Tween-20, 10% glycerol, 1 mM MgCl2, 0.1 mM ZnSO4, 1 mM PMSF), then incubated with bait proteins in binding buffer for 2 h at 4°C. Blots were washed three times for 10 min in PBST and detected with anti-Flag M2 antibody.
Affinity purifications
Undialyzed HeLa NE (10 mL) was first diluted in 30 mL of AP buffer (20 mM Tris at pH 7.9, 0.5% Triton X-100, 10% glycerol, 1 mM MgCl2, 0.1 mM ZnSO4, 1 mM PMSF, proteinase inhibitor) containing 100 mM NaCl. Diluted NE was incubated with bacterially expressed and purified GST-XRN2Δ6 (∼50 μg) and glutathione beads (300 μL) for 2 h at 4°C. Beads were washed three times in AP buffer containing 0.5 M NaCl. Bound proteins were eluted with 25 mM reduced glutathione, and loaded onto a 10% SDS-PAGE gel. CBB-stained proteins were excised, digested by trypsin, then analyzed by MALDI-TOF.
IP and immunoblot analysis
HeLa NE (50 μL) was incubated with indicated antibodies (∼2 μg) or preimmune serum bound to protein A beads. IPs were carried out for 3 h at 4°C in 200 μL of Buffer D (Dignam et al. 1983). Beads were collected by centrifugation and washed three times with Buffer D. Aliquots of the beads were analyzed by SDS-PAGE and immunobloting with indicated antibodies. In Figure 1B, IPs with an anti-CPSF-64 antibody (Bethyl) with or without blocking peptide were analyzed. In Figure 1C, IPs with an anti-XRN2 antibody were analyzed by using an anti-CstF-64 mAb.
ChIP assays
HeLa cells were grown in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum. Transfections of siRNA were performed by using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s guidelines. After 48 h, transfected cells were analyzed by Western blotting and ChIP assays. ChIP analysis was carried out as described (Nelson et al. 2006) with minor modifications. Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature. When ChIP assays using anti-Pol II antibody were carried out, cells were cross-linked with 0.4% formaldehyde. Cross-linking was stopped by addition of glycine (pH 7.0) to 125 mM final concentration. Cells were washed three times with cold PBS and harvested in IP buffer (150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris at pH 7.9, 1 mM PMSF, protease inhibitors). Samples were sonicated to generate DNA fragments of ∼500 base pairs. After adding each antibody preincubated with or without blocking peptide for 30 min at 4°C, the shared chromatin was incubated in an ultrasonic water bath for 30 min at 4°C. After centrifugation, supernatants were incubated with protein A beads for 45 min at 4°C. Beads were washed five times with IP buffer. Immunocomplexes were boiled for 10 min and incubated with Proteinase K for 30 min at 55°C. After boiling for 10 min, DNA was extracted by phenol/chloroform and precipitated with ethanol. Real-time PCR was carried out as described (Kaneko and Manley 2005). Sequences of primers are given in Supplemental Material.
Acknowledgments
We thank J. Patton for anti-PSF antibody, S. Xiang and L. Tong for recombinant XRN2 protein, and I. Boluk for help with the manuscript. We thank E. McIntush (Bethyl Laboratories) for antibody design and Bethyl Laboratories for preparation of polyclonal antibodies. This work was supported by NIH grant GM28983 to J.L.M., and gifts from the Caring for Carcinoid Foundation and from Drs. Raymond and Beverly Sackler to M.M.
Footnotes
References
- Ahn S.H., Kim M., Buratowski S., Kim M., Buratowski S., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Barilla D., Lee B.A., Proudfoot N.J., Lee B.A., Proudfoot N.J., Proudfoot N.J. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V.I., Scherthan H., Solinger J.A., Buerstedde J.M., Heyer W.D., Scherthan H., Solinger J.A., Buerstedde J.M., Heyer W.D., Solinger J.A., Buerstedde J.M., Heyer W.D., Buerstedde J.M., Heyer W.D., Heyer W.D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J., Lee B.A., Keller W., Proudfoot N.J., Keller W., Proudfoot N.J., Proudfoot N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti C., Tollervey D., Presutti C., Tollervey D., Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J.L., Manley J.L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Sadowski M., Hubner W., Weiser S., Keller W., Blank D., Sadowski M., Hubner W., Weiser S., Keller W., Sadowski M., Hubner W., Weiser S., Keller W., Hubner W., Weiser S., Keller W., Weiser S., Keller W., Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G., Lebovitz R.M., Roeder R.G., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Horowitz D.S., Kobayashi R., Krainer A.R., Horowitz D.S., Kobayashi R., Krainer A.R., Kobayashi R., Krainer A.R., Krainer A.R. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A., Shales M., McCracken S., Xie W., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J., Shales M., McCracken S., Xie W., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J., McCracken S., Xie W., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J., Xie W., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J., Kobayashi R., Blencowe B.J., Ingles C.J., Blencowe B.J., Ingles C.J., Ingles C.J. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N., Bentley D.L., Bentley D.L. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Patton J.G., Reed R., Patton J.G., Reed R., Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N., West S., Proudfoot N.J., West S., Proudfoot N.J., Proudfoot N.J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Felthauser A.M., Mertz J.E., Felthauser A.M., Mertz J.E., Mertz J.E. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 2005;25:6303–6313. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B., Kim Y.K., Kim J.H., Kim T.Y., Jang S.K., Kim Y.K., Kim J.H., Kim T.Y., Jang S.K., Kim J.H., Kim T.Y., Jang S.K., Kim T.Y., Jang S.K., Jang S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M., Reinberg D., Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L., Manley J.L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hollingworth D., Noble C.G., Taylor I.A., Ramos A., Noble C.G., Taylor I.A., Ramos A., Taylor I.A., Ramos A., Ramos A. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA. 2006;12:555–560. doi: 10.1261/rna.2304506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.J., Tang Z., Lin R.J., Tucker P.W., Tang Z., Lin R.J., Tucker P.W., Lin R.J., Tucker P.W., Tucker P.W. Phosphorylation by SR kinases regulates the binding of PTB-associated splicing factor (PSF) to the pre-mRNA polypyrimidine tract. FEBS Lett. 2007;581:223–232. doi: 10.1016/j.febslet.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Stangl K., Lane W.S., Bindereif A., Stangl K., Lane W.S., Bindereif A., Lane W.S., Bindereif A., Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M.M., Duque P., Konarska M.M., Konarska M.M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Manley J.L., Manley J.L. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol. Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Nedea E., Greenblatt J.F., Buratowski S., Greenblatt J.F., Buratowski S., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M., Vasiljeva L., Rando O.J., Zhelkovsky A., Moore C., Buratowski S., Vasiljeva L., Rando O.J., Zhelkovsky A., Moore C., Buratowski S., Rando O.J., Zhelkovsky A., Moore C., Buratowski S., Zhelkovsky A., Moore C., Buratowski S., Moore C., Buratowski S., Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F., Shilatifard A., Buratowski S., Greenblatt J.F., Buratowski S., Greenblatt J.F., Greenblatt J.F. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Lutz C.S., Lutz C.S. p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA. 2006;12:111–121. doi: 10.1261/rna.2213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen E., Darnell J.E., Shenk T., Falck-Pedersen E., Darnell J.E., Shenk T., Darnell J.E., Shenk T., Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl. Acad. Sci. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Bentley D., Bentley D. A ribonucleolytic rat torpedoes RNA polymerase II. Cell. 2004;119:911–914. doi: 10.1016/j.cell.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Luo W., Johnson A.W., Bentley D.L., Johnson A.W., Bentley D.L., Bentley D.L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes & Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.F., Price D.H., Price D.H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Mathur M., Tucker P.W., Samuels H.H., Tucker P.W., Samuels H.H., Samuels H.H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L., Patterson S.D., Wickens M., Bentley D.L., Wickens M., Bentley D.L., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Sova P., Bomsztyk K., Denisenko O., Sova P., Bomsztyk K., Sova P., Bomsztyk K., Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T., Schwartz B.E., Werner J., Suarez J.R., Lis J.T., Werner J., Suarez J.R., Lis J.T., Suarez J.R., Lis J.T., Lis J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Osheim Y.N., Proudfoot N.J., Beyer A.L., Proudfoot N.J., Beyer A.L., Beyer A.L. EM visualization of transcription by RNA polymerase II: Downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- Patton J.G., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B., Galceran J., Tempst P., Nadal-Ginard B., Tempst P., Nadal-Ginard B., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes & Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Penheiter K.L., Washburn T.M., Porter S.E., Hoffman M.G., Jaehning J.A., Washburn T.M., Porter S.E., Hoffman M.G., Jaehning J.A., Porter S.E., Hoffman M.G., Jaehning J.A., Hoffman M.G., Jaehning J.A., Jaehning J.A. A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Petfalski E., Dandekar T., Henry Y., Tollervey D., Dandekar T., Henry Y., Tollervey D., Henry Y., Tollervey D., Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proteau A., Blier S., Albert A.L., Lavoie S.B., Traish A.M., Vincent M., Blier S., Albert A.L., Lavoie S.B., Traish A.M., Vincent M., Albert A.L., Lavoie S.B., Traish A.M., Vincent M., Lavoie S.B., Traish A.M., Vincent M., Traish A.M., Vincent M., Vincent M. The multifunctional nuclear protein p54nrb is multiphosphorylated in mitosis and interacts with the mitotic regulator Pin1. J. Mol. Biol. 2005;346:1163–1172. doi: 10.1016/j.jmb.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger A., Dye M.J., Furger A., Dye M.J., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rigo F., Kazerouninia A., Nag A., Martinson H.G., Kazerouninia A., Nag A., Martinson H.G., Nag A., Martinson H.G., Martinson H.G. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol. Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Manley J.L., Manley J.L. From transcription to mRNA: PAF provides a new path. Mol. Cell. 2005;20:167–168. doi: 10.1016/j.molcel.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Ip J.Y., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Ip J.Y., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Tucker P., Ingles C.J., Blencowe B.J., Ingles C.J., Blencowe B.J., Blencowe B.J. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol. Cell. Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E., Kaneko S., Manley J.L., Kaneko S., Manley J.L., Manley J.L. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Hughes C.M., Nannepaga S.J., Shanmugam K.S., Copeland T.D., Guszczynski T., Resau J.H., Meyerson M., Hughes C.M., Nannepaga S.J., Shanmugam K.S., Copeland T.D., Guszczynski T., Resau J.H., Meyerson M., Nannepaga S.J., Shanmugam K.S., Copeland T.D., Guszczynski T., Resau J.H., Meyerson M., Shanmugam K.S., Copeland T.D., Guszczynski T., Resau J.H., Meyerson M., Copeland T.D., Guszczynski T., Resau J.H., Meyerson M., Guszczynski T., Resau J.H., Meyerson M., Resau J.H., Meyerson M., Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K.E., Mauger D.M., Arndt K.M., Mauger D.M., Arndt K.M., Arndt K.M. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.C., Claffey K.P., Claffey K.P. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Manley J.L., MacDonald C.C., Wilusz J., Shenk T., Manley J.L., MacDonald C.C., Wilusz J., Shenk T., MacDonald C.C., Wilusz J., Shenk T., Wilusz J., Shenk T., Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes & Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J., Gromak N., Proudfoot N.J., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Whitelaw E., Proudfoot N., Proudfoot N. α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α 2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Bai X., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W., Bai X., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W., Ho J., Brown J., Stevens A., Johnson A.W., Brown J., Stevens A., Johnson A.W., Stevens A., Johnson A.W., Johnson A.W. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Hanke J.H., Carayannopoulos L., Craft C.M., Capra J.D., Tucker P.W., Hanke J.H., Carayannopoulos L., Craft C.M., Capra J.D., Tucker P.W., Carayannopoulos L., Craft C.M., Capra J.D., Tucker P.W., Craft C.M., Capra J.D., Tucker P.W., Capra J.D., Tucker P.W., Tucker P.W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M., Proudfoot N.J., Proudfoot N.J. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- Yonaha M., Proudfoot N.J., Proudfoot N.J. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 2000;19:3770–3777. doi: 10.1093/emboj/19.14.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carmichael G.G., Carmichael G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu J., Gilmour D.S., Fu J., Gilmour D.S., Gilmour D.S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A., Bentley D.L., Bentley D.L. The link between mRNA processing and transcription: Communication works both ways. Exp. Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]