p21 transcription is regulated by differential localization of histone H2A.Z (original) (raw)
Abstract
In yeast cells, H2A.Z regulates transcription and is globally associated within a few nucleosomes of the initiator regions of numerous promoters. H2A.Z is deposited at these loci by an ATP-dependent complex, Swr1.com. Here we show that H2A.Z suppresses the p53 → p21 transcription and senescence responses. Upon DNA damage, H2A.Z is first evicted from the p21 promoter, followed by the recruitment of the Tip60 histone acetyltransferase to activate p21 transcription. p400, a human Swr1 homolog, is required for the localization of H2A.Z, and largely colocalizes with H2A.Z at multiple promoters investigated. Notably, the presence of sequence-specific transcription factors, such as p53 and Myc, provides positioning cues that direct the location of H2A.Z-containing nucleosomes within these promoters. Collectively, this study strongly suggests that certain sequence-specific transcription factors regulate transcription, in part, by preferentially positioning histone variant H2A.Z within chromatin. This H2A.Z-centered process is part of an epigenetic process for modulating gene expression.
Keywords: H2A.Z, Myc, p21, p400, p53
Eukaryotic DNA is condensed many fold (e.g., 10,000) into chromatin, the basic unit of which contains 146 base pairs (bp) of DNA and an octamer of histone proteins (H2A, H2B, H3, and H4). Due to the high level of compaction, chromatin typically represses certain cellular DNA transactions, including transcription. For successful transcription, it is argued that nucleosomes need to be remodeled or evicted from promoter regions for the transcriptional machinery to be efficiently recruited to a target gene.
The incorporation of histone variants into specific nucleosomes within a promoter region constitutes a mechanism by which promoter region chromatin can become more permissive to transcription initiation and elongation following receipt of a proper physiological cue. One such histone variant is H2A.Z. In Saccharomyces cerevisiae, it can elicit positive effects on gene expression (Santisteban et al. 2000; Adam et al. 2001; Larochelle and Gaudreau 2003). In addition, H2A.Z regulates genes that are proximal to telomeres and acts as a “buffer” to antagonize the spread of heterochromatin into euchromatic regions (Meneghini et al. 2003). Furthermore, recent reports from our laboratory and others (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005) have shown that H2A.Z is preferentially localized within a few nucleosomes of the initiator regions of multiple promoters in the yeast genome. Interestingly, these H2A.Z-rich loci are largely devoid of transcriptional activity, which suggests that the variant histone prepares genes for activation (Guillemette et al. 2005) and/or operates as a transcriptional repressor. Finally, yeast H2A.Z was shown to regulate nucleosome positioning, which provides mechanistic insight into how its presence can alter promoter transcriptional state (Guillemette et al. 2005).
An ATP-dependent chromatin remodeling complex that specifically loads H2A.Z onto chromatin and exchanges it with H2A exists in yeast (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004). This complex, in which the catalytic subunit is Swr1, also shares essential subunits with the NuA4 histone acetyltransferase complex (Krogan et al. 2003; Kobor et al. 2004). In addition to their importance in gene regulation, the Swr1 complex, H2A.Z, and NuA4 are all involved in the regulation of yeast chromosome stability (Krogan et al. 2004). This is noteworthy because, in mammalian cells, depletion of H2A.Z causes major nuclear and chromosomal abnormalities as witnessed by a high incidence of lagging chromosomes and chromatin bridges (Rangasamy et al. 2004).
There are two homologs of Swr1 in human cells: p400/Domino (herein referred to as p400), and SRCAP (Doyon and Cote 2004; Jin et al. 2005). There are also three uncharacterized p400-type SWI2–SNF2 molecules, including hIno80 (Bakshi et al. 2006). Members of this family of SWI2/SNF2 chromatin remodeling enzymes each contain a spacer region inserted into the SWI2/SNF2 homology region (Smith and Peterson 2005).
p400 was originally isolated as an E1A-associated protein (Fuchs et al. 2001), and it was also shown to interact with p53, Myc, and SV40 large T antigen (Chan et al. 2005). It is also required for E1A to induce p53-mediated apoptosis (Samuelson et al. 2005). SRCAP has been isolated as a CREB-binding protein (Johnston et al. 1999). While one report shows that both p400 and SRCAP constitute part of the same complex (Doyon et al. 2004), a recent study shows that SRCAP and p400 exist in distinct complexes with H2A.Z (Cai et al. 2005; Ruhl et al. 2006). Recently an SRCAP-containing complex was purified, and it was shown to have the ability to exchange H2A–H2B for H2A.Z–H2B in reconstituted mononucleosomes (Ruhl et al. 2006). It remains to be determined whether mammalian homolog(s) of Swr1, such as p400 and SRCAP, also catalyze H2A.Z deposition in vivo.
Depletion of p400 elevates p21 synthesis to initiate premature senescence in primary human fibroblasts (Chan et al. 2005). Senescence was first observed in tissue culture cells as a stable form of cell growth arrest provoked by diverse stresses (Hayflick and Moorhead 1961; Sharpless and DePinho 2005). Recently, oncogene-induced senescence was shown to occur in various precancerous lesions both in humans and mice (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005; Michaloglou et al. 2005), further suggesting that senescence acts as a defense mechanism against malignant cell development (Sager 1991; Hanahan and Weinberg 2000). Importantly, the action of p400 at p21 depends on the function of p53, a key regulator of p21 transcription (el-Deiry et al. 1993).
Given the possibility of a link between p400 and H2A.Z, we questioned whether H2A.Z is also an important regulator of p21 expression. The results of this effort show that H2A.Z depletion induces p21 expression in a p53-dependent fashion, as well as the premature senescence of primary diploid fibroblasts. Similar to senescence induced by p400 depletion, inactivating p53 or p21 blocked the emergence of certain senescent phenotypes following H2A.Z depletion. In a normal setting, H2A.Z is highly enriched at discrete p53-binding sites that lie within the p21 promoter. This distinctive localization pattern depends on the presence of p53, and was detected at other p53 target gene promoters as well. The presence of p400 is required to localize H2A.Z at those loci, and purified recombinant p400 from insect cells can carry out in vitro exchange of H2A.Z–H2B dimers into chromatin. H2A.Z and p400 localization at the p53-binding sites in p21 was severely diminished following p21 induction, and this process was not dependent on active p21 transcription per se. After H2A.Z and p400 eviction from the p53-binding sites in p21, we observed that the Tip60 histone acetyltransferase had been recruited to the distal p53-binding site in the promoter to positively regulate p21 expression. Finally, overexpression of Myc, a known suppressor of p21 synthesis, significantly increased H2A.Z localization at the Myc-binding site in the TATA initiator region of the p21 promoter. This observation is consistent with the view that Myc represses p21 expression by preferentially recruiting H2A.Z-containing nucleosome(s) to this element.
Results
Depletion of H2A.Z in primary fibroblasts induces p21 transcription as well as the p53 → p21 senescence pathway
Considering that p400, a Swr1 homolog, regulates p21 expression (Chan et al. 2005), and that Swr1 can remodel chromatin as well as exchange H2A.Z in the process, we asked whether H2A.Z also regulates p21 transcription. Specifically, multiple retroviral vectors were engineered to encode a short hairpin RNA (shRNA) directed against human H2A.Z, one of which has been previously published (sh-H2A.Z-2) (Rangasamy et al. 2004). When tested, three out of four of these vectors (sh-H2A.Z-2, sh-H2A.Z-3, and sh-H2A.Z-4) suppressed H2A.Z expression to levels equivalent to 5%–20% of control BJ cells transduced with vector alone (Fig. 1A). In vector and sh-H2A.Z-1-transduced BJ cells, we also detected a higher molecular weight band in the H2A.Z Western blot (Fig. 1A). This higher molecular weight species represents a ubiquitinated form of H2A.Z (data not shown), and this form was also depleted following H2A.Z knockdown. H2A.Z mRNA levels decreased in parallel with protein levels (data not shown). The knockdown of H2A.Z was also tested in other human cell lines (i.e., MCF7, HEK293, BJ, WI38, and U2OS) with comparable effects (data not shown).
Figure 1.
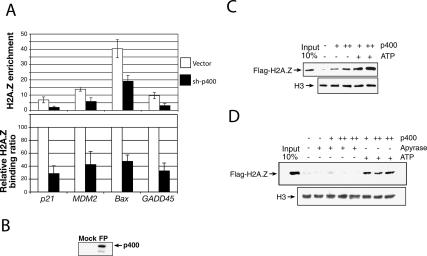
H2A.Z represses the basal expression of p21 in p53+/+ cells, and regulates senescence in human fibroblasts. (A) Western blot showing the knockdown of H2A.Z expression by different H2A.Z hairpins. (B, left panel) Expression levels of p21, cyclin D1, and H2Ai in U2OS cells before and after H2A.Z knockdown using shH2A.Z-2. (Right panel) Expression levels of p21 in HCT116 p53+/+ and p53−/− paired cells before and after H2A.Z knockdown using shH2A.Z-2. mRNA expression levels were normalized against expression levels of the 36B4 ribosomal gene, as measured by Q-PCR. (C) FACs analysis of BJ cells following H2A.z depletion. Results from duplicate samples were presented. (D) Cell growth assay performed in BJ cells. (E) Senescent β-gal staining of WI-38 cells following the indicated perturbations. sh-p400 was included as a positive control. (F) DNA staining by DAPI of WI-38 cells following the indicated treatments.
p400 depletion leads to p21 induction in cells that contain functional p53 (Chan et al. 2005). Here, we made use of U2OS cells, which synthesize wild-type p53, and asked whether depletion of H2A.Z also leads to p21 induction. Indeed, depletion of H2A.Z (using the shH2A.Z-2 vector) resulted in a significant increase of p21 expression (Fig. 1B). In the same experiment, H2Ai and cyclin D1 expression levels were unchanged (Fig. 1B, left panel). Therefore, the increase of p21 expression observed following H2A.Z knockdown is specific. Similar results were also obtained using the shH2A.Z-3 and shH2A.Z-4 vectors (data not shown).
Next, we asked whether H2A.Z depletion has an effect on p21 expression in cells that lack functional p53. When p400 was depleted from cells containing nonfunctional p53, these cells failed to exhibit p21 induction (Chan et al. 2005). Such an experiment was carried out in SaOS2 cells, which are also p53−/−, and the first question was whether p21 promoter activity was affected. Notably, when H2A.Z was depleted from SaOS2 cells, no p21 up-regulation was observed (see Fig. 5E, below). An analogous experiment was carried out in isogenic p53+/+ and p53−/− HCT116 cells, and p53+/+ cells underwent p21 induction while the p53−/− cells did not (Fig. 1B, right panel). Taken together, these results suggest that H2A.Z can down-regulate p21 expression and that p53 contributes to p21 induction in an H2A.Z-depleted setting.
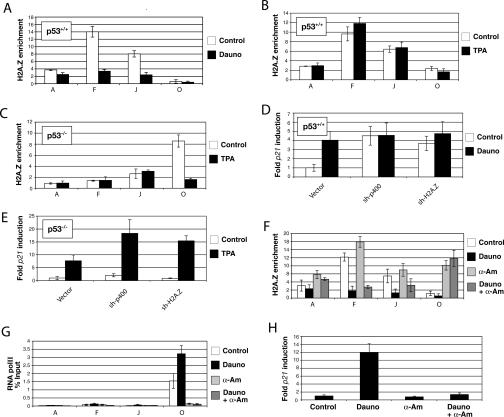
Figure 5.
Local eviction of H2A.Z-containing nucleosomes upon induction of the p21 promoter is p53 dependent. (A) H2A.Z enrichment at specific sites within the p21 promoter region (cf. Fig. 4) before and after treatment with 250 μM daunorubicin for 12 h in p53+/+ cells. (B) ChIP analysis of H2A.Z enrichment at the p21 promoter in p53+/+cells treated with 50 ng/mL TPA. (C) ChIP experiments showing the effect of TPA treatment on the localization of H2A.Z in p53−/− cells. The various segments analyzed are also from Figure 4 (D) Relative expression of p21 before and after daunorubicin treatment of p53+/+ cells depleted for H2A.Z and p400. (E) Expression levels of p21 before and after treatment with TPA for 4 h in p53−/− cells depleted in H2A.Z and p400. (F) H2A.Z enrichment at the p21 promoter in p53+/+ cells treated with 250 μM daunorubicin and/or 100 μM α-amanitin for 12 h. (G) ChIP corresponding to the data shown in F, showing binding of RNA polII (8WG16 antibody). (H) p21 expression levels of U2OS cells following various treatments.
Acute depletion of p400 is known to induce premature senescence of primary fibroblasts in a p53 → p21-dependent manner (Chan et al. 2005). Given that H2A.Z depletion activates p21 expression (Fig. 1B), we asked whether H2A.Z depletion also leads to senescence in primary fibroblasts. BJ cells depleted of H2A.Z using specific shRNA vectors showed a significant decrease of S phase cells (Fig. 1C), accompanied by decreased BrdU incorporation (data not shown). H2A.Z depletion also dramatically inhibited cell proliferation (Fig. 1D), and H2A.Z-depleted BJ cells, much like p400-depleted BJ cells (Chan et al. 2005), displayed prominent β-gal staining, suggesting that they had become senescent (Fig. 1E). Similar findings were observed when H2A.Z was depleted from IMR90 and WI-38, two other primary fibroblastic cell lines (data not shown; see also Fig. 2). When WI-38 cells were depleted of H2A.Z and their DNA staining properties were examined by fluorescence microscopy, a large fraction (>40% in multiple experiments) of cells displayed characteristic, senescence-associated heterochromatic foci (SAHF) (Fig. 1F). Similar features were observed when activated RasV12 was overexpressed in these cells or when they were actively p400 depleted (Fig. 1F). Senescent β-gal staining and SAHF phenotypes were observed 9–11 d after cells were transduced with retroviral H2A.Z shRNA. Taken together, these results suggest that H2A.Z depletion mimics p400 depletion and leads to premature senescence in primary fibroblasts.
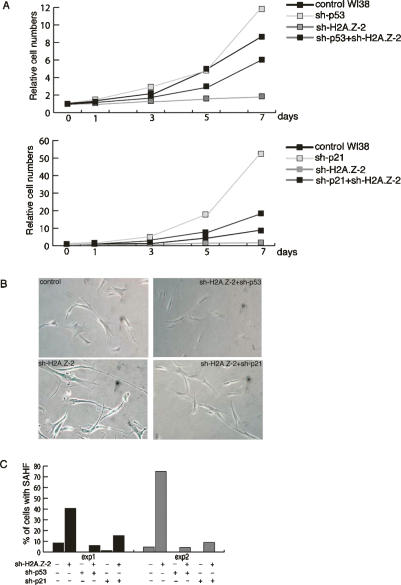
Figure 2.
Inactivation of p53/p21 rescues H2A.Z depletion-induced senescent phenotypes. (A) Cell growth assay in WI-38 cells following the indicated treatment. (B) Senescent β-gal staining of the WI-38 cells following the indicated treatment. (C) Summary of SAHF in WI-38 cells exposed to the indicated perturbation. The numerical values are listed in Supplementary Table S1. Cells were fixed and counted on days 10 and 12 post-infection in Experiments 1 and 2, respectively.
Loss of either p53 or p21 rescues certain phenotypes that arise during p400 depletion-dependent senescence (Chan et al. 2005). To investigate whether H2A.Z and p400 act in the same genetic pathway to induce senescence, we asked whether the inactivation of p53 or p21 rescues H2A.Z-induced senescence. In this regard, similar to the case of p400 depletion, shRNA-mediated depletion of either p53 or p21 suppressed the growth arrest (Fig. 2A), senescent β-gal staining (Fig. 2B), and SAHF (Fig. 2C; Supplementary Table S3) induced by H2A.Z knockdown. The efficiency of sh-p53 and sh-p21 have been described previously (Chan et al. 2005), and both reagents can efficiently inhibit p21 expression (Supplementary Fig. S1). It was apparent in these experiments that a large fraction of H2A.Z-depleted cells revealed intense p21 staining (∼80% in H2A.Z depleted vs. 30% in control WI38). The intense p21 staining was largely suppressed in cells that had been transduced with shRNA vectors that target either p53 or p21 (Supplementary Fig. S1). Moreover, depleting either p53 or p21 accelerated the growth of naïve WI-38 cells (Fig. 2A). Notably, when WI-38 cells were codepleted of H2A.Z and either p53 or p21, the cells proliferated more slowly than when the cells were singly depleted of p53 or p21 (Fig. 2A). Taken together, these results suggest that senescence induced by H2A.Z depletion is dependent on the integrity of the p53 → p21 pathway. Since knocking down p53 or p21 expression failed to fully restore the proliferation rate of WI-38 cells when H2A.Z was codepleted, perhaps H2A.Z depletion also impedes WI-38 proliferation in a p53/p21-independent manner.
H2A.Z is enriched at p53-binding sites in chromatin
To gain insight into how H2A.Z regulates p21 expression, chromatin immunoprecipitation (ChIP) and tiled quantitative PCR (Q-PCR) assays were used to search for regions of preferential localization of H2A.Z within or near the p21 promoter. Figure 3A provides a graphic representation of the p21 promoter that includes the location of specific p53-binding sites in the transcription initiator region, and the nature of the tiled Q-PCR amplicons used in these experiments. Figure 3B depicts a ChIP experiment using anti-H2A.Z antibodies both in p53+/+ (U2OS; black bars) and in p53−/− cells (SaOS2; white bars). H2A.Z enrichment was calculated as the ratio of anti-H2A.Z to canonical anti-H2A ChIP. The latter is an indication of local nucleosome density. Raw data for H2A.Z and H2A binding are shown in Supplementary Figure S2. Similar enrichment ratios were obtained when normalizing H2A.Z to histone H3 density by ChIP analysis (data not shown). Surprisingly, we found that H2A.Z, in p53+/+ cells, was strongly enriched at the distal and proximal p53-binding sites, albeit more intensely at the former than the latter. Notably, the H2A.Z localization pattern was significantly different in SaOS2 cells (p53−/−), where the p53-distal binding site peak was lost, and binding appeared to be redistributed over the TATA initiator region. The disappearance of the H2A.Z peak at the p53-binding sites was also observed in p53−/− HCT116 cells, using p53 wild-type HCT116 as a control (see Fig. 6, below; Supplementary Fig. S3).
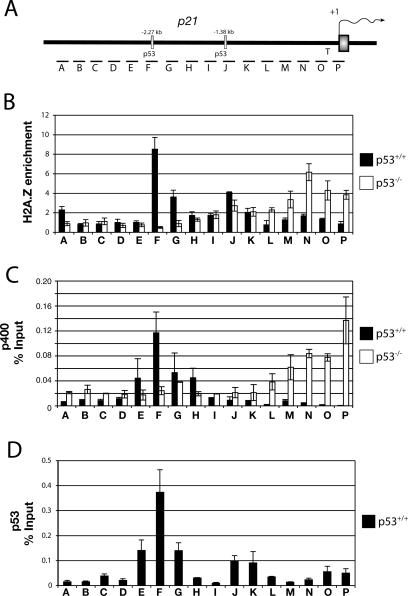
Figure 3.
H2A.Z colocalizes with p53 and p400 at the p21 promoter. (A) Diagram of the p21 promoter region and its indigenous p53 response elements, and segments (columns A–P) used for Q-PCR. (B) ChIP analysis of H2A.Z enrichment (percent input of H2A.Z/H2A) at the p21 promoter in U20S (p53+/+) and SAOS2 (p53−/−) cells. (C) ChIP assays for p400 on the p21 promoter in U2OS and SAOS2 cells. (D) ChIP experiments determining the recruitment of p53 at the p21 promoter region in U2OS cells.
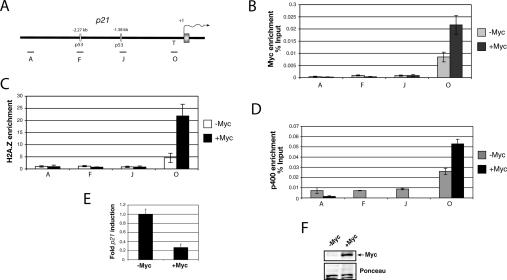
Figure 6.
c-Myc directs H2A.Z recruitment at the p21 TATA region in p53−/− cells. (A) Diagram of the p21 promoter region with the amplicons used for Q-PCR. (B) ChIP assay showing c-Myc binding at the p21 promoter in HaCat-tetMyc cells that overexpress c-Myc under the control of tetracycline. (C) ChIP analysis of H2A.Z enrichment before and after c-Myc overexpression. (D) ChIP analysis of p400 binding before and after c-Myc overexpression. (E) Basal expression level of p21 in HaCat-tetMyc. (F) Immunoblot assay showing c-Myc expression in the HaCat-tetMyc cells cultured for 24 h with or without tetracycline.
Interestingly, in multiple p53 wild-type-producing cell lines, including U20S, the H2A.Z localization pattern resembled that of p400, which was determined by low-resolution ChIP (Chan et al. 2005). The high-resolution ChIP assay results in U2OS cells reconfirmed the published results, showing that p400 is preferentially enriched at the distal, but not the proximal p53-binding site (Fig. 3C; Chan et al. 2005). Moreover, in SaOS2 (p53−/−) cells, p400 was concentrated at the TATA initiator region of p21, much like H2A.Z (Fig. 3C). Figure 3D depicts the results of a ChIP assay performed in U20S, revealing that both the distal and proximal sites are occupied by p53 under the experimental conditions that were employed. The results suggest that H2A.Z, p400, and p53 colocalize at the distal binding site in the p21 promoter. Furthermore, the presence of p53 is required to localize H2A.Z at the p53-binding sites within the p21 promoter. The fact that p400 did not appear to be significantly enriched at the proximal p53-binding site could be due to the fact that the p400 antibody used in our experiment is less efficient than either the p53 or the H2A.Z antibodies that were used in the experiment. Alternatively, the presence of H2A.Z at the proximal p53-binding site is a p400-independent event.
Since H2A.Z, p400, and p53 colocalized at the distal p53-binding element on the p21 promoter, we asked whether this was also the case at other p53-responsive genes. The GADD45, Bax, and MDM2 promoters were surveyed by ChIP to assess H2A.Z- and p400-binding profiles. In all cases, both H2A.Z and p400 were tightly focused at known, cognate p53-binding sites (Supplementary Fig. S4). Overall, these results suggest that H2A.Z and p400 colocalize with p53 at multiple, p53-responsive promoters.
p400 is required for the preferential localization of H2A.Z to p53-binding sites
Since p400 is a human ortholog of yeast Swr1 and p400 colocalizes with H2A.Z at p21 and other p53-responsive genes, we asked whether p400 is required to incorporate H2A.Z at p53–promoter-binding sites. Specifically, we used an shRNA vector directed at p400 (Chan et al. 2005) to knock down p400 expression. Under those conditions, H2A.Z binding was monitored by ChIP at the p53-binding sites of p21, GADD45, Bax, and MDM2. Figure 4A shows that, upon p400 depletion, H2A.Z levels bound at all p53 target genes that were investigated fell. Results are either represented as actual H2A.Z enrichment at these loci (Fig. 4A, top panel), or as a relative binding ratio to better visualize the contribution of p400 in H2A.Z localization (Fig. 4A, bottom panel). H3 binding was also monitored as a nucleosome density control, and there was no significant reduction at any of the sites that were interrogated (data not shown).
Figure 4.
p400 incorporates H2A.Z into chromatin around selected p53-binding sites. (A) ChIP experiments showing the relative binding of H2A.Z after p400 knockdown at different p53 target genes. The primers used in these experiments correspond to the p53-binding sites for each gene (fragment F [cf. Fig. 3], p21; fragment E, GADD45; fragment F, BAX; fragment E, MDM2). (B) Immunoblot assay with antibody against p400 demonstrating the effect of Flag-specific purification (FP). (C) In vitro exchange of H2A.Z–H2B dimers by p400. Flag-H2A.Z was detected by immunoblotting using an anti-Flag antibody. The H3 immunoblot was used here as a loading control. (D) Similar in vitro exchange of H2A.Z–H2B dimers by p400 but with the addition of apyrase to show dependency of ATP for the reaction.
SRCAP, another Swr1 ortholog, can carry out cell-free H2A.Z–H2B exchange in chromatin (Ruhl et al. 2006). Thus, we tested the possibility that SRCAP might incorporate H2A.Z at the p53-binding site of p21. Supplementary Figure S5C also shows that, upon SRCAP depletion, H2A.Z levels dropped, but to a significantly smaller degree than following p400 depletion. Furthermore, SRCAP depletion did not alter p21 expression (Supplementary Fig. S5A). Taken together, these and the above-noted results suggest that p400 makes a more significant contribution than SRCAP to localizing H2A.Z at those p53-binding sites that were investigated.
In order to further study the involvement of p400 in the specific chromatin loading of H2A.Z, in vitro exchange assays were carried out, using in vitro-assembled mononucleosomes and affinity-purified human p400 (Fig. 4B). Highly purified human histone octamers were assembled on a 5S 196-bp DNA fragment (Simpson et al. 1985) that was end-labeled with biotin. Purified recombinant H2A.Z–H2B (Flag-tagged H2A.Z) dimers were added to the reaction with or without ATP. Streptavidin-captured templates were washed, and the efficiency of the exchange reaction was monitored by immunoblotting directed at Flag-H2A.Z. The addition of increasing quantities of p400 catalyzed the replacement of H2A–H2B with H2A.Z–H2B, particularly after addition of ATP (Fig. 4C). The purified yeast H2A.Z deposition complex, the Swr1 complex, is known to contain endogenous ATP (Mizuguchi et al. 2004), a condition that may account for background levels of exchange observed in the presence of p400 but in the absence of exogenous ATP. Thus, in order to further characterize the ATP-dependency of the p400-mediated exchange reaction, we made use of apyrase to degrade residual ATP that might still have been associated with the purified p400 (Fletcher et al. 2002) that was added to the above-noted reaction mixtures (Fig. 4D). In the presence of apyrase, no significant H2A.Z–H2B exchange was observed, thus showing the ATP-dependency of the reaction. The ATP-dependency of the p400-catalyzed reaction was also verified using ATPγS, a nonhydrolyzable source of ATP that also prevented H2A.Z–H2B dimer exchange (data not shown). These results imply that the p400 complex-driven exchange reaction is ATP dependent, and that the purified complex contains endogenous ATP.
Although affinity-isolated p400 from insect cells was highly purified, we cannot exclude the possibility that it copurified with one or more insect Domino complex subunits essential for carrying out the exchange reaction. Mock purification from Sf9 cells without recombinant p400 expression did not yield any H2A.Z deposition activity, again implying that the observed activity was, at a minimum, clonal p400 dependent (data not shown). In conclusion, the aforementioned experiments strongly suggest that p400 (and, perhaps, a complex containing this protein) can exchange H2A.Z–H2B dimers for H2A–H2B within specific chromatin loci, in a process that requires ATP.
H2A.Z is evicted from the p21 promoter in a transcription-independent fashion following p53-dependent p21 activation
Since shRNA-mediated removal of H2A.Z from the p53-binding site leads to p21 induction, it became important to understand whether H2A.Z is also naturally evicted from the p53-binding sites following the receipt of physiological signal(s) that activate p21 expression. To activate p21, we utilized daunorubicin, a DNA damaging agent known to strongly induce p21 via the p53 pathway (Seoane et al. 2002). The p21 regions that were surveyed in the relevant ChIP experiments are illustrated in Supplementary Figure S3A. Figure 5A depicts the results of a ChIP assay carried out with anti-H2A.Z. The data show that, as expected, there was enrichment of the variant histone at the relevant p53-binding sites in U2OS cells. However, upon induction of the p21 gene with daunorubicin, H2A.Z binding was markedly diminished. Notably, p400 binding to the p53-distal site was also diminished in this setting (Supplementary Fig. S6). These data underscore the notion that p400 has a repressive effect on p21 transcription mediated at least in part by H2A.Z (Chan et al. 2005).
In order to determine whether H2A.Z operates solely by the p53 pathway, we asked whether H2A.Z disappears from the p21 promoter upon its activation by a p53-independent pathway. For this purpose, we used the phorbol ester and protein kinase C activator, tetradecanoyl-phorbol acetate (TPA), which can induce p21 expression in a setting wherein the p53 gene is missing (Fig. 5E; Seoane et al. 2002), presumably by engaging proximal promoter-binding factors that activate p21 independently of p53. When TPA was added to U2OS cells (p53+/+), it induced p21 expression, as expected (Supplementary Fig. S7). Interestingly, when anti-H2A.Z ChIP was performed on TPA treated U2OS, H2A.Z was not lost from the relevant p53-binding sites (Fig. 5B). This result differs from the outcome in daunorubicin-treated U2OS cells (Fig. 5, cf. B and A). Taken together, these two data sets suggest that H2A.Z is only evicted from its p53-binding sites when p21 is induced in a p53-dependent manner.
Notably, in p53+/+ cells, the degree of activation of p21 by daunorubicin was comparable to that triggered by depletion of either p400 or H2A.Z (Fig. 5D). This suggests that the contribution of p53 to p21 induction via this pathway is to counteract the repressive effect of p400–H2A.Z. Therefore, one can argue that H2A.Z and p400 regulate p21 gene expression by preventing p53 from directly activating this target gene when cells are not stressed, and thereby helps to prepare p21 for physiological, p53-mediated activation.
As observed previously (Fig. 3), H2A.Z preferentially associated with the initiator region of the p21 promoter in p53−/− SaOS2 cells (site O). Here, given the absence of wild-type p53, one wonders whether a different H2A.Z remodeling scheme is employed to activate p21. While H2A.Z or p400 depletion did not affect basal p21 expression in these cells, these manipulations did increase the extent of p21 induction following TPA treatment (Fig. 5E). Moreover, by ChIP assay, H2A.Z was evicted from the initiator region of the p21 promoter after TPA exposure (Fig. 5C). Thus, in this context, H2A.Z does not act primarily as a repressor of p21 expression; rather, it controls the extent of p21 induction by TPA.
We also wanted to know whether H2A.Z eviction following daunorubicin-mediated p21 induction was directly linked to the active transcriptional state of p21. To test this, α-amanitin, an inhibitor of RNA polymerase II (RNA pol II) activity, was used to block p21 transcription. In its presence, we asked whether H2A.Z was still evicted from the p53-binding sites upon gene induction. Figure 5F shows that upon addition of α-amanitin, H2A.Z levels at p53-binding sites were comparable to those observed in the uninduced p21 gene (sites F and J). Interestingly, H2A.Z also accumulated at the non-p53-binding site-containing initiator region (site O) in this setting (Fig. 5F). Similar to Figure 5A, the addition of daunorubicin evicted H2A.Z from p53-binding sites. When daunorubicin was added together with α-amanitin, H2A.Z was still evicted from the relevant p53-binding sites. H2A.Z was evicted from the p53-binding sites specifically, since the α-amanitin-generated H2A.Z peak at site O was not affected by daunorubicin treatment. A ChIP experiment using RNA pol II antibodies showed that, as expected, RNA pol II levels were detected at the TATA initiator region of p21 and, upon daunorubicin addition, these levels increased further (Fig. 5G), a result consistent with those obtained by others (Espinosa et al. 2003; Gomes et al. 2006). However, as previously shown, RNA pol II levels were indistinguishable from background when α-amanitin was added (Fig. 5G). Quantification of p21 expression levels revealed that, whenever α-amanitin was present, p21 induction was abolished (Fig. 5H). Taken together, these results suggest that H2A.Z is quickly remodeled upon activation of p21 via the p53 pathway, and this remodeling can occur even in the absence of active p21 transcription. Furthermore, α-amanitin appears to prevent RNA pol II from being prebound to p21, and, under these conditions, H2A.Z-containing nucleosomes replaced RNA pol II at the TATA initiator region of the gene. This is much like what is observed in p53−/− cells; i.e., in the absence of p53. RNA polII is not prebound to the p21 promoter (Espinosa et al. 2003) and, therefore, cannot prevent H2A.Z from associating with the TATA initiator region.
c-Myc directs H2A.Z to the TATA initiator region of the p21 promoter
c-Myc is known to repress p21 transcription by binding to the TATA initiator region of the p21 promoter (Seoane et al. 2002). This was proposed to explain how c-Myc can influence p53 responses, favoring apoptosis over cytostasis after DNA damage (Seoane et al. 2002). Interestingly, c-Myc is also known to bind to p400 (Fuchs et al. 2001). Given that p53 can direct H2A.Z at the p21 promoter, we wondered whether c-Myc directs H2A.Z to the TATA initiator region of the p21 promoter. In order to test this hypothesis, we made use of a HaCaT cell line that induces c-Myc overexpression using a “Tet-off” system (Seoane et al. 2002). This cell line is p53−/−, and was used to show that c-Myc overexpression represses p21 expression (Seoane et al. 2002). c-Myc expression was induced when tetracyclin (Tet) was removed from the culture medium. First, ChIP was carried out using antibody against c-Myc. Figure 6B shows that c-Myc preferentially associates with the p21 TATA initiator region, as previously reported, but not to other regions tested, including the p53-binding sites (Seoane et al. 2002). When c-myc was induced 24 h after Tet removal (Fig. 6F), a threefold increase of c-Myc binding to the TATA initiator region was observed (Fig. 6B). Consistent with reported observations (Seoane et al. 2002; Wu et al. 2003) overexpressed c-Myc repressed p21 expression (Fig. 6E). When an H2A.Z ChIP assay was performed before and after c-Myc induction, we observed that H2A.Z was preferentially localized to the initiator region of the p21 promoter in both settings. Importantly, the level of H2A.Z binding was enriched by approximately fourfold following c-Myc overexpression (Fig. 6C). This enrichment was specific, since we did not observe increased H2A.Z binding to other regions of the p21 promoter. Importantly, similar results were observed with p400 (Fig. 6D), thus connecting p400 to increased H2A.Z binding at p21 in this experimental setting. Given that HaCaT cells express a mutated form of p53 (two point mutations in both alleles), we did not detect H2A.Z or p400 bound at the p53-binding sites (Fig. 6C,D), a result consistent with the findings in SaoS2 and the p53−/− HCT116 cells. Taken together, these results suggest that c-Myc recruits p400 or a complex thereof to the initiatot region of p21, which in turn directs H2A.Z to that region as part of its repression of p21 expression.
Tip60 and H2A.Z are independently recruited to the p53-binding sites at the p21 promoter to regulate p21 expression
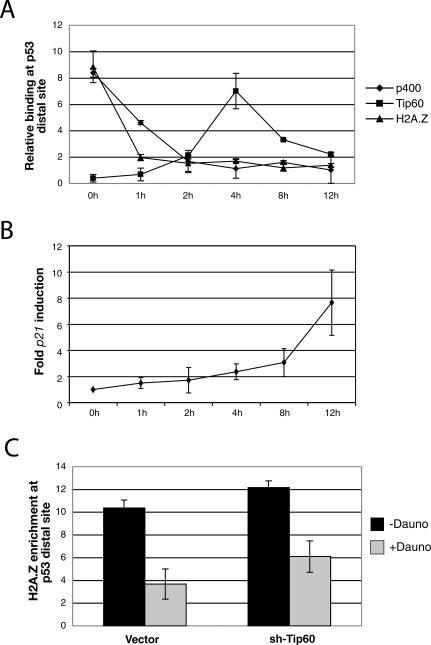
Tip60 and p400 are proposed to be components of the same complex (Fuchs et al. 2001; Doyon et al. 2004), yet they elicit opposite effects on p21 transcription. Tip60 serves as a transcription coactivator to activate p53-dependent p21 transcription (Berns et al. 2004; Legube et al. 2004; Tyteca et al. 2006) and p400, together with H2A.Z, represses p53 → p21 transcription. A recent report also demonstrates that Tip60 depletion by small interfering RNA (siRNA) can reverse p21 induction following p400 depletion (Tyteca et al. 2006). In order to investigate how Tip60 counteracts p400/H2A.Z to activate p21 expression, ChIP was performed with antibodies against H2A.Z, p400, and Tip60 at the distal p53-binding site on the p21 promoter in U2OS cells that were induced to express p21 by daunorubicin (Fig. 7A). In unstressed cells (0 h), H2A.Z and p400, but not Tip60, showed preferential accumulation at this site (Fig. 7A). As expected, H2A.Z and p400 quickly disappeared from the distal p53-binding site following daunorubicin treatment. The H2A.Z peak had disappeared after 1 h of daunorubicin treatment, and p400 binding dropped to background levels by 2 h of treatment (Fig. 7A). Interestingly, Tip60 began to accumulate at the distal p53-binding site after 1 h of daunorubicin treatment, reaching a peak at 4 h. It decreased significantly thereafter. We next monitored p21 expression after daunorubicin treatment in an effort to correlate the p21 expression profile with that of p400, Tip60, and H2A.Z binding. The results in Figure 7B indicate that p21 expression commences at the onset of p400 and H2A.Z eviction, and correlates with active Tip60 recruitment. These results suggest a model wherein Tip60 and p400 act as independent complexes at the p21 promoter to regulate its function. After DNA damage, H2A.Z/p400 first vacate the p53-binding sites, followed by recruitment of Tip60, which serves as a coactivator of p21 expression. This model explains why Tip60 and p400 exert opposing effects on p21 transcription, and why depleting Tip60 by siRNA counteracts p400 depletion to inhibit p21 transcription.
Figure 7.
p400-independent recruitment of Tip60 at the p53-distal site of the p21 promoter. (A) ChIP assay showing the kinetics of H2A.Z, p400, and Tip60 occupancy during p21 promoter activation after daunorubicin treatment in U2OS cells. Q-PCR was performed using primers corresponding to the distal p53 site of the p21 promoter (fragment F). (B) p21 mRNA expression levels after daunorubicin treatment of U2OS cells. (C) ChIP experiment showing H2A.Z enrichment at the distal p53 site after Tip60 knockdown before and after daunorubicin treatment.
Recently, Tip60 was shown to acetylate ATM, thereby triggering a fully activated ATM-dependent DNA-damage signaling cascade (Sun et al. 2005). Therefore, one wonders whether Tip60 participates in a signaling cascade to evict H2A.Z and p400 from the p53-binding sites following daunorubicin treatment. Moreover, since Tip60 can exist in the same molecular complex as p400, it is possible that Tip60 also affects p400-mediated H2A.Z deposition. In these cases, one possibility, given results described in this report, is that depleting Tip60 from cells would affect basal H2A.Z deposition and/or H2A.Z eviction from the p21 promoter following DNA damage. To investigate these possibilities, we carried out H2A.Z ChIP at the distal p53-binding site of the p21 promoter in control U2OS cells, and in U2OS cells depleted of Tip60 by shRNA. Figure 7C shows that Tip60 knockdown does not significantly affect H2A.Z localization at the distal p53 site whether or not p21 is induced with daunorubicin. The Tip60 shRNA was functional, because Tip60 knockdown reduced p21 expression following daunorubicin treatment (data not shown). These data suggest that H2A.Z/p400 and Tip60 act in two separate steps to regulate p21 expression. The binding of H2A.Z and p400 to the p53-binding site is repressive toward p21 expression. H2A.Z and p400 need to be removed from the p53-binding site, perhaps as a necessary step to allow subsequent recruitment of Tip60, which then activates p21.
Discussion
In this study, we show that H2A.Z is a negative regulator of p21 expression. Depletion of this variant histone in human cells leads to p21 activation and premature senescence. Strikingly, H2A.Z colocalizes with p53 within the promoter region of p21 and several other p53 target genes, and does so in a p53-dependent fashion. Importantly, in response to certain cellular stresses, such as DNA damage, cells trigger a p53-dependent H2A.Z remodeling event to activate a p21 transcriptional response. Notably, in p53+/+ U2OS cells, elements of the transcription machinery (i.e., RNA Pol II) are prebound to the p21 promoter—contrary to what was observed in p53−/− cells (see Espinosa et al. 2003)—and may well prepare the p21 promoter for transcription activation (Espinosa et al. 2003). In this setting, the data presented here indicate that in normally proliferating p53+/+ U2OS cells, H2A.Z constitutes a repressive barrier to p53-dependent p21 activation, and that it needs to be evicted from its p53-binding sites in order to activate p21 transcription. The level of p21 induction following daunorubicin treatment was no greater than what was observed in cells that were artificially depleted of H2A.Z by shRNA. Also, in cells that were depleted of H2A.Z, further treatment with daunorubicin did not cause any greater level of p21 expression. Therefore, we conclude that p53 likely overcomes the repressive effect of H2A.Z to fully activate p21 in response to cellular stress. The remodeling event that leads to H2A.Z eviction from the distal p53-binding site is independent of the p21 transcription status, since p21 transcription could be inhibited, yet H2A.Z eviction from the promoter still occurred.
Apart from p53, a different growth-affecting protein with transcription modulating effects, c-Myc, also directs H2A.Z to a c-myc-bound site in the p21 promoter in the vicinity of the TATA initiator region. The recruitment of H2A.Z to the c-Myc-binding region also correlated with c-Myc-mediated repression of p21 transcription. Overall, our data demonstrate that H2A.Z likely represses p21 transcription by being differentially localized within chromatin by the action of at least two different DNA-binding transcription-modulating factors, p53 and c-Myc.
We also show that the human homolog of the yeast Swr1, p400, is required for the localization pattern of H2A.Z at the p53-binding sites within multiple promoters. p400 colocalized with H2A.Z at multiple p53-binding sites, and they also colocalize at other loci, including a number of ER-responsive promoters (data not show). Knockdown of p400 expression led to severe reduction in H2A.Z binding to all p53-binding sites tested. Moreover, purified human p400 catalyzed H2A.Z–H2B dimer exchange on in vitro-assembled nucleosomes. Importantly, cells depleted of H2A.Z by shRNA largely recapitulated the phenotypes of p400 depletion, leading to p53 → p21 activation and premature senescence. In both cases, the resulting senescent phenotypes were prevented when p53 or p21 was codepleted. Intriguingly, p53 levels are modestly elevated following H2A.Z depletion, though no significant levels of p53 S15 phosphorylation was observed (Chan et al. 2005; H.M. Chan and D.M. Livingston, unpubl.), suggesting that activation of p53 may not be through the classical DNA damage type response. Depletion of two, other p400 complex components, Gas41 (Park and Roeder 2006), and YL1 (data not shown), also activated p53 → p21 transcription. Taken together, these data suggest that the p400 complex plays a role in specific deposition of H2A.Z in chromatin. Proper localization of H2A.Z by the p400 complex at the p21 promoter forms a repressive barrier to prevent aberrant p53 → p21 activation, and suppresses senescence in normally proliferating primary fibroblasts.
We also investigated a possible role for SRCAP-containing complexes in regulating H2A.Z positioning. One SRCAP-containing complex was reported to support cell-free H2A.Z–H2B exchange in chromatin (Ruhl et al. 2006). Thus far, our analysis indicates that p400 plays a more significant role than SRCAP in localizing H2A.Z at those chromatin loci that were investigated. It remains to be determined whether these observations also hold true on a genome-wide scale.
Since promoter depletion of H2A.Z as well as certain p400 complex members correlates with p21 activation, we investigated the mechanism underlying these phenomena by studying the Tip60 histone acetyltransferase. It is a component of the p400 complex (Fuchs et al. 2001), yet it behaves as a coactivator of p53 to activate p21 (Berns et al. 2004; Legube et al. 2004; Tyteca et al. 2006). Furthermore, it was shown that depleting Tip60 by RNA interference (RNAi) counteracts p21 induction following p400 RNAi treatment (Tyteca et al. 2006). Our ChIP analyses reveal that p400 and Tip60 act as two separate complexes in a temporal manner to regulate p21 transcription. In normally cycling cells, H2A.Z and p400, but not Tip60, occupy the distal p53-binding site at the p21 promoter. Upon DNA damage, H2A.Z and p400 are cleared from the p21 promoter, a situation that possibly allows the recruitment of Tip60 to the distal p53-binding site where it activates p21. Here, we considered the possibility that Tip60 also participates in H2A.Z deposition within chromatin, or participates in an ATM-dependent DNA damage signaling cascade (Sun et al. 2005) to activate p21. However, our H2A.Z ChIP analyses in Tip60-depleted U2OS cells have not yielded evidence to support these possibilities. In conclusion, our data suggest that the recruitment of Tip60 to the p53-binding site following H2A.Z and p400 eviction is a necessary step for p53 → p21 transcription activation.
Data presented here indicate that sequence-specific transcription factors, such as p53 and c-Myc, define the positioning of H2A.Z within certain genomic segments. H2A.Z tightly colocalizes with p53, Myc, and ER (data not shown) within multiple sites in chromatin. In cells that carry p53 mutations, H2A.Z is delocalized from the known p53-binding sites. Since both p53 and c-Myc interact with p400, we favor a model where transcription factors can recruit a p400-associated chromatin remodeling complex, which in turn incorporates H2A.Z locally. Genome-wide studies in yeast cells have revealed that H2A.Z preferentially associates with regions that lie adjacent to translational start sites (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005). Nonetheless, there is no evidence that transcription factors generally promote H2A.Z chromatin deposition in yeast. It remains to be determined whether, as a general matter, mammalian cells adopt a different strategy for H2A.Z deposition.
While H2A.Z and p400 repress p21 transcription, they do not generally serve as negative regulators of gene transcription. H2A.Z and p400 colocalize with p53 on multiple p53-responsive promoters surveyed in this study (including GADD45 and Bax). However, H2A.Z or p400 knockdown did not activate the basal transcription of these genes (data not shown). In yeast, there is abundant evidence that H2A.Z regulates gene expression both positively and negatively (Allis et al. 1980; Santisteban et al. 2000; Adam et al. 2001; Meneghini et al. 2003). No clear mechanism exists that explains how H2A.Z exerts its influence on transcription. Therefore, the behavior of H2A.Z on gene expression may be context dependent. For example, in SaOS2 cells that lack p53, the transcription machinery is not prerecruited to p21, and H2A.Z preferentially accumulates at the TATA initiator region of the promoter. In this state, H2A.Z does not have a repressive effect on basal p21 transcription. However, H2A.Z-containing nucleosomes at the TATA initiator region are still remodeled upon TPA-mediated activation, and the presence of H2A.Z determines the extent of p21 induction. This finding underscores the importance of chromatin structure in the regulation of p21 expression, and it also suggests that accurate targeting of H2A.Z within defined chromatin subregions may be vital to the control of certain gene expression patterns and their related physiological responses.
Lastly, transcription factors, like p53 and c-Myc, are known to regulate the expression of a large group of potential genes. One model to explain such a widespread gene expression regulatory effect is that the c-Myc oncoprotein may behave as a general chromatin regulator (Knoepfler et al. 2006). Our finding that p53 and c-Myc can dictate histone variant H2A.Z positioning at certain promoters suggests one general mechanism of how transcription factors regulate chromatin dynamics and gene expression. In this scenario, H2A.Z positioning is part of an epigenetic regulatory process, together with specific histone post-translational modifications to modulate gene expression profiles in a local, context-dependent manner.
Materials and methods
Cell culture, retroviral infections, and shRNA design
U2OS, SaOS2, HCT116 cells (a gift from Dr. Bert Vogelstein, Howard Hughes Medical Institute, Baltimore, MD), and WI-38 cells (American Type Culture Collection) were cultured in a 10% CO2-containing atmosphere in DMEM (Invitrogen) containing 10% fetal bovine serum (FBS) and antibiotics. BJ cells were maintained in DMEM supplemented with 20% M199 (Invitrogen), 15% serum, and antibiotics. The HaCaT-tetMyc cells (gift from Joan Massagué, Sloan-Kettering Cancer Center, New York) were cultured in DMEM containing 10% FBS, 500 μg/mL G418, and 300 μg/mL hygromycin. Daunorubicin (Sigma), TPA (Sigma), and α-amanitin oleate (Sigma) were used at concentrations of 250 μM, 50 ng/mL, and 100 μM, respectively. Retroviral infections were performed as described in Chan et al. (2005). Methods of cell cycle analysis, cell growth assay, and senescent β-gal assay were described previously (Chan et al. 2005). ShRNA sequence against p400 (GGACTTGGTTCTCATCGAC), H2A.Z (#1, TTCGAAATGGCTGGCGGTA; #2, GAAGAAAGGACA ACAGAAGACT; #3, TCTAGGACGACCAGTCATG; #4, ATA CTCTAACAGCTGTCCA), and p21 (CTTCGACTTTGTCAC CGAG) were cloned into pSuperRetro-(Puro) backbone (oligoengine) for stable suppression of p400 and p21, respectively.
Cell proliferation assay
The cell proliferation assay has been described previously, with slight modification (McCurrach and Lowe 2001; Chan et al. 2005). Briefly, 1 × 104 cells were seeded in a 24-well plate. Twelve hours after plating, one plate was fixed (time zero) with glutaraldehyde. The cell was washed with PBS twice, and the plate was kept at 4°C until the experiment was complete. At the indicated time point, other assay plates were also fixed and stored at 4°C. When the experiment was completed, all wells were stained with 500 μL of 0.1% crystal violet for 30 min at room temperature, and washed twice with PBS; plates were washed thoroughly in distilled water. Plates were dried at room temperature, and the attached crystal violet was solublized by adding 200 μL of 10% acetic acid to each well. The solution was pipetted up and down several times to mix well, the solublized dye was transferred to a 96-well plate, and optical density was measured at 590 nm. The values were normalized to those of the time zero plate. Staining was proportional to the relative cell number. Each data point presented is the average value of triplicate samples.
RNA analysis
Total RNA was isolated using the RNeasy kit (Qiagen). RNA was reverse-transcribed into first-strand cDNA using AMV reverse transcriptase (Promega). Samples were subjected to Q-PCR using MX3000P (Stratagene). Q-PCR for 36B4 mRNA (which encodes a ribosomal protein subunit) was used as an internal control. The relative abundance of p21 mRNA (or other test genes) was calculated after normalization using 36B4 mRNA, where relative expression levels were calculated as 2−ΔCT where ΔCT = CT test gene − CT 36B4. Primers used are listed in Supplementary Table S1.
ChIP assays
ChIP assays were performed essentially as described previously (Gevry et al. 2003) with a panel of specific polyclonal antibodies generated in-house or from commercial sources, as well as preimmune and no antibody controls. Samples were sonicated to generate DNA fragments <500 bp. PCR was performed using a set of primers relevant to the promoter regions of the p21, GADD45, MDM2, and BAX genes, and analyzed by Q-PCR. The primers used in real-time PCR are listed in Supplementary Table S2.
Antibodies
The p21 polyclonal (C-19), RNA pol II antibodies (N20), and Myc(N-262) were purchased from Santa Cruz Biotechnology, and the p21 monoclonal antibody is from Cell Signaling Technology. Polyclonal antibodies to Histone H2A (07-146) and Tip60 (07-038) were from Upstate Biotechnology. Histone H3 (ab 1791) was purchased from Abcam, and the p53 (Ab-2) monoclonal antibody was from Oncogene. Antibody to p400 was raised against a C-terminal p400 peptide (MRVPAVRLKTPTKP PCQ) and affinity-purified, and has been described (Chan et al. 2005). The H2A.Z antibody used for immunoblotting was a kind gift from Dr. Pat Nakatani (Dana Farber Cancer Institute, Boston, MA), and the H2A.Z antibody used for ChIP assays was raised against an N-terminal H2A.Z peptide (CSLIGKKGQQKT).
Mononucleosome assembly and exchange assays
A 196-bp DNA fragment containing a 5S positioning element was generated by EcoRI digestion of pIC-2085S (gift from Jacques Côté, Université Laval, Québec, Canada). The DNA fragment was biotinylated using biotin-14-dATP and Klenow fragment. Mononucleosomes were assembled by salt gradient dialysis using HeLa core histones (ratio 1:80) (Owen-Hughes et al. 1999). Exchange assays were performed as described by Ruhl et al. (2006) with minor modifications. Briefly, the equivalent of 30 ng of DNA immobilized on avidin-coupled dynabeads was preincubated for 15 min at 30°C with or without apyrase (1 U) and human p400-Flag purified from SF9 insect cells. After a preincubation step, exchange reactions were assayed for 60 min at room temperature with 300 ng of Flag-H2A.Z/H2B dimer in exchange buffer (25 mM HEPES-KOH at pH 7.6, 0.1 mM EDTA, 5 mM MgCl2, 10% glycerol, 0.02% NP-40, 1 mM DTT, 0.1 mg/mL BSA, 70 mM KCl) in the absence or presence of 1 mM ATP. Beads were washed twice with exchange buffer containing 400 mM KCl and once with exchange buffer containing 70 mM KCl and were eluted using SDS gel loading buffer. The incorporation of Flag-H2A.Z was analyzed by immunoblot using an anti-Flag M2 antibody (Sigma). The membrane was stripped and reprobed with anti-histone H3 antibody (Abcam) as a loading control for chromatin.
Acknowledgments
We thank Amy Svotelis for critical comments on the manuscript, and Drs. Pat Nakatani and Jacques Côté for sharing antibodies, reagents, and advice. We are also grateful to Dr. Joan Massagué for the gift of HaCat cells, and Dr. Bert Vogelstein for the HCT116 cells. This work was supported by grants from the Cancer Research Society, Inc., of Canada and the Canadian Cancer Society awarded to L.G., and from the U.S. National Cancer Institute to D.M.L. L.G. holds a Canada research chair on mechanisms of gene transcription, and N.G. was a recipient of a post-doctoral fellowship from NSERC. H.C. was a recipient of a Human Frontiers Fellowship.
Footnotes
References
- Adam M., Robert F., Larochelle M., Gaudreau L., Robert F., Larochelle M., Gaudreau L., Larochelle M., Gaudreau L., Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 2001;21:6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C.D., Glover C.V., Bowen J.K., Gorovsky M.A., Glover C.V., Bowen J.K., Gorovsky M.A., Bowen J.K., Gorovsky M.A., Gorovsky M.A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Bakshi R., Mehta A.K., Sharma R., Maiti S., Pasha S., Brahmachari V., Mehta A.K., Sharma R., Maiti S., Pasha S., Brahmachari V., Sharma R., Maiti S., Pasha S., Brahmachari V., Maiti S., Pasha S., Brahmachari V., Pasha S., Brahmachari V., Brahmachari V. Characterization of a human SWI2/SNF2 like protein hINO80: Demonstration of catalytic and DNA binding activity. Biochem. Biophys. Res. Commun. 2006;339:313–320. doi: 10.1016/j.bbrc.2005.10.206. [DOI] [PubMed] [Google Scholar]
- Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Madiredjo M., Nijkamp W., Weigelt B., Nijkamp W., Weigelt B., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Dorken B., Jenuwein T., Schmitt C.A., Jenuwein T., Schmitt C.A., Schmitt C.A. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Cai Y., Jin J., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Jin J., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W., Washburn M.P., Conaway R.C., Conaway J.W., Conaway R.C., Conaway J.W., Conaway J.W. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- Chan H.M., Narita M., Lowe S.W., Livingston D.M., Narita M., Lowe S.W., Livingston D.M., Lowe S.W., Livingston D.M., Livingston D.M. The p400 E1A-associated protein is a novel component of the p53 → p21 senescence pathway. Genes & Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., Scher H.I., Ludwig T., Gerald W., Ludwig T., Gerald W., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Zaballos A., Flores J.M., Barbacid M., Flores J.M., Barbacid M., Barbacid M., et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Cote J., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Selleck W., Lane W.S., Tan S., Cote J., Selleck W., Lane W.S., Tan S., Cote J., Lane W.S., Tan S., Cote J., Tan S., Cote J., Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B., Mercer W.E., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Espinosa J.M., Verdun R.E., Emerson B.M., Verdun R.E., Emerson B.M., Emerson B.M. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Fletcher T.M., Xiao N., Mautino G., Baumann C.T., Wolford R., Warren B.S., Hager G.L., Xiao N., Mautino G., Baumann C.T., Wolford R., Warren B.S., Hager G.L., Mautino G., Baumann C.T., Wolford R., Warren B.S., Hager G.L., Baumann C.T., Wolford R., Warren B.S., Hager G.L., Wolford R., Warren B.S., Hager G.L., Warren B.S., Hager G.L., Hager G.L. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 2002;22:3255–3263. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M., Lane W.S., Nakatani Y., Livingston D.M., Nakatani Y., Livingston D.M., Livingston D.M. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Gevry N.Y., Lalli E., Sassone-Corsi P., Murphy B.D., Lalli E., Sassone-Corsi P., Murphy B.D., Sassone-Corsi P., Murphy B.D., Murphy B.D. Regulation of niemann-pick c1 gene expression by the 3′5′-cyclic adenosine monophosphate pathway in steroidogenic cells. Mol. Endocrinol. 2003;17:704–715. doi: 10.1210/me.2002-0093. [DOI] [PubMed] [Google Scholar]
- Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Szostek S.A., Emerson B.M., Espinosa J.M., Emerson B.M., Espinosa J.M., Espinosa J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L., Adam M., Blanchette M., Robert F., Gaudreau L., Blanchette M., Robert F., Gaudreau L., Robert F., Gaudreau L., Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hayflick L., Moorhead P.S., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Jin J., Cai Y., Li B., Conaway R.C., Workman J.L., Conaway J.W., Kusch T., Cai Y., Li B., Conaway R.C., Workman J.L., Conaway J.W., Kusch T., Li B., Conaway R.C., Workman J.L., Conaway J.W., Kusch T., Conaway R.C., Workman J.L., Conaway J.W., Kusch T., Workman J.L., Conaway J.W., Kusch T., Conaway J.W., Kusch T., Kusch T. In and out: Histone variant exchange in chromatin. Trends Biochem. Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Johnston H., Kneer J., Chackalaparampil I., Yaciuk P., Chrivia J., Kneer J., Chackalaparampil I., Yaciuk P., Chrivia J., Chackalaparampil I., Yaciuk P., Chrivia J., Yaciuk P., Chrivia J., Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Zhang X.Y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Zhang X.Y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Gafken P.R., McMahon S.B., Eisenman R.N., McMahon S.B., Eisenman R.N., Eisenman R.N. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Jennings J.L., Link A.J., Madhani H.D., Rine J., Link A.J., Madhani H.D., Rine J., Madhani H.D., Rine J., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Haw R.A., Pootoolal J., Tong A., Canadien V., Pootoolal J., Tong A., Canadien V., Tong A., Canadien V., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Davey M.G., Pootoolal J., Hughes T.R., Pootoolal J., Hughes T.R., Hughes T.R., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M., Gaudreau L., Gaudreau L. H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 2003;22:4512–4522. doi: 10.1093/emboj/cdg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legube G., Linares L.K., Tyteca S., Caron C., Scheffner M., Chevillard-Briet M., Trouche D., Linares L.K., Tyteca S., Caron C., Scheffner M., Chevillard-Briet M., Trouche D., Tyteca S., Caron C., Scheffner M., Chevillard-Briet M., Trouche D., Caron C., Scheffner M., Chevillard-Briet M., Trouche D., Scheffner M., Chevillard-Briet M., Trouche D., Chevillard-Briet M., Trouche D., Trouche D. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 2004;279:44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- Li B., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Chen J., Seidel C., Gerton J., Workman J.L., Seidel C., Gerton J., Workman J.L., Gerton J., Workman J.L., Workman J.L. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurrach M.E., Lowe S.W., Lowe S.W. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 2001;66:197–227. doi: 10.1016/s0091-679x(01)66010-2. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D., Wu M., Madhani H.D., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Shay J.W., Mooi W.J., Peeper D.S., Mooi W.J., Peeper D.S., Peeper D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C., Shen X., Landry J., Wu W.H., Sen S., Wu C., Landry J., Wu W.H., Sen S., Wu C., Wu W.H., Sen S., Wu C., Sen S., Wu C., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T., Utley R.T., Steger D.J., West J.M., John S., Cote J., Havas K.M., Workman J.L., Utley R.T., Steger D.J., West J.M., John S., Cote J., Havas K.M., Workman J.L., Steger D.J., West J.M., John S., Cote J., Havas K.M., Workman J.L., West J.M., John S., Cote J., Havas K.M., Workman J.L., John S., Cote J., Havas K.M., Workman J.L., Cote J., Havas K.M., Workman J.L., Havas K.M., Workman J.L., Workman J.L. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol. Biol. 1999;119:319–331. doi: 10.1385/1-59259-681-9:319. [DOI] [PubMed] [Google Scholar]
- Park J.H., Roeder R.G., Roeder R.G. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol. Cell Biol. 2006;26:4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Schreiber S.L., Rando O.J., Madhani H.D., Rando O.J., Madhani H.D., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D., Greaves I., Tremethick D.J., Greaves I., Tremethick D.J., Tremethick D.J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- Ruhl D.D., Jin J., Cai Y., Swanson S., Florens L., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Jin J., Cai Y., Swanson S., Florens L., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Cai Y., Swanson S., Florens L., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Swanson S., Florens L., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Florens L., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Washburn M.P., Conaway R.C., Conaway J.W., Chrivia J.C., Conaway R.C., Conaway J.W., Chrivia J.C., Conaway J.W., Chrivia J.C., Chrivia J.C. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Sager R. Senescence as a mode of tumor suppression. Environ. Health Perspect. 1991;93:59–62. doi: 10.1289/ehp.919359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson A.V., Narita M., Chan H.M., Jin J., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., Narita M., Chan H.M., Jin J., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., Chan H.M., Jin J., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., Jin J., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W., Fuchs M., Livingston D.M., Lowe S.W., Livingston D.M., Lowe S.W., Lowe S.W. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 2005;280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova T., Smith M.M., Kalashnikova T., Smith M.M., Smith M.M. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Seoane J., Le H.V., Massague J., Le H.V., Massague J., Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., DePinho R.A., DePinho R.A. Cancer: Crime and punishment. Nature. 2005;436:636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- Simpson R.T., Thoma F., Brubaker J.M., Thoma F., Brubaker J.M., Brubaker J.M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Smith C.L., Peterson C.L., Peterson C.L. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 2005;25:5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Chen S., Fernandes N., Price B.D., Jiang X., Chen S., Fernandes N., Price B.D., Chen S., Fernandes N., Price B.D., Fernandes N., Price B.D., Price B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyteca S., Vandromme M., Legube G., Chevillard-Briet M., Trouche D., Vandromme M., Legube G., Chevillard-Briet M., Trouche D., Legube G., Chevillard-Briet M., Trouche D., Chevillard-Briet M., Trouche D., Trouche D. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 2006;25:1680–1689. doi: 10.1038/sj.emboj.7601066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Cetinkaya C., Munoz-Alonso M.J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.G., Cetinkaya C., Munoz-Alonso M.J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.G., Munoz-Alonso M.J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.G., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.G., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.G., Beuger V., Eilers M., Leon J., Larsson L.G., Eilers M., Leon J., Larsson L.G., Leon J., Larsson L.G., Larsson L.G. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- Zhang H., Roberts D.N., Cairns B.R., Roberts D.N., Cairns B.R., Cairns B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]