RHAMM Is a Centrosomal Protein That Interacts with Dynein and Maintains Spindle Pole Stability (original) (raw)
Abstract
The receptor for hyaluronan-mediated motility (RHAMM), an acidic coiled coil protein, has previously been characterized as a cell surface receptor for hyaluronan, and a microtubule-associated intracellular hyaluronan binding protein. In this study, we demonstrate that a subset of cellular RHAMM localizes to the centrosome and functions in the maintenance of spindle integrity. We confirm a previous study showing that the amino terminus of RHAMM interacts with microtubules and further demonstrate that a separate carboxy-terminal domain is required for centrosomal targeting. This motif overlaps the defined hyaluronan binding domain and bears 72% identity to the dynein interaction domain of Xklp2. RHAMM antibodies coimmunprecipitate dynein IC from Xenopus and HeLa extracts. Deregulation of RHAMM expression inhibits mitotic progression and affects spindle architecture. Structure, localization, and function, along with phylogenetic analysis, suggests that RHAMM may be a new member of the TACC family. Thus, we demonstrate a novel centrosomal localization and mitotic spindle-stabilizing function for RHAMM. Moreover, we provide a potential mechanism for this function in that RHAMM may cross-link centrosomal microtubules, through a direct interaction with microtubules and an association with dynein.
INTRODUCTION
The receptor for hyaluronan (HA)-mediated motility (RHAMM, intracellular hyaluronan binding protein [IHABP], CD168), first described by Turley and colleagues (Turley, 1992;Turley and Torrance, 1985; Turley et al., 1987,1991), is a multifaceted protein with both intracellular and extracellular functions (Hardwick et al., 1992; Assmann et al., 1999; Pilarski et al., 2001). RHAMM mediates the HA-specific motility of a variety of cell types, including transformed cell lines and ex vivo malignant leukocytes (Masellis-Smith et al., 1996; Pilarski_et al_., 2001). RHAMM is the founding member of the B(X)7B hyaladherin protein family and interacts with HA through basic (B) 9–11 amino acid (aa) B(X)7B motifs (Yang et al., 1994). Encoded by 18 exons, RHAMM localizes to chromosome 5q33.2-qter (Spicer et al., 1995) and encodes an 85-kDa protein with extensive coiled coil structure and a basic amino-terminal globular domain (Hardwick_et al_., 1992; Assmann_et al_., 1999). In addition to its role in cell migration, RHAMM expression, and overexpression, have been linked to ras transformation, tumor progression, and metastasis (Hall_et al_., 1995). Moreover, RHAMM has been identified as a microtubule-associated protein that also interacts with the actin cytoskeleton (Assmann et al., 1999). Overexpression of RHAMM in B lymphoid malignancies, and other cancers, suggests that it may play a role in human oncogenesis and disease progression (Wang et al., 1998; Crainie et al., 1999). Two RHAMM splice variants, RHAMM-exon 4 and RHAMM-exon13, have been identified in myeloma patients and cancer cell lines (Assmann et al., 1999; Crainie et al., 1999); all variants localize to the mitotic spindle, although loss of exon 4 inhibits interaction with interphase microtubules (Assmann et al., 1999). The localization of RHAMM isoforms at the mitotic spindle provides a putative mechanism through which RHAMM expression regulates mitotic integrity, ploidy, and possibly carcinogenesis.
In animal cells, the duplication of centrosomes, and their subsequent polar separation, initiate the establishment of a bipolar microtubule array. Many centrosomal and noncentrosomal proteins, including microtubule motor complexes, coordinate early mitotic events. Kinesin-like protein 2 family members (Xklp2, Hklp2, and KRP180) are essential for centrosomal separation and the maintenance of spindle bipolarity (Boleti et al., 1996). The carboxy-terminal leucine zipper of Xklp2 is required for its centrosomal and spindle pole localization, via an interaction with targeting protein for Xklp2 (TPX2) and the dynein/dynactin motor complex (Wittmann et al., 1998). TPX2 has recently been shown to mediate Ran-GTP–dependent microtubule assembly and to target Aurora A kinase to the spindle pole (Gruss et al., 2002; Kufer et al., 2002). A recurring mechanism in spindle pole formation and stability is dynein/dynactin-mediated recruitment of structural proteins to the spindle pole (reviewed in Zimmerman and Doxsey, 2000). Pericentrin, a 220-kDa coiled coil protein, plays an integral role in centrosomal stability by acting as a molecular scaffold and interacting with numerous proteins and protein complexes, including dynein (Purohit et al., 1999; Zimmerman and Doxsey, 2000). Although centrosomal localization of pericentrin is microtubule independent (Gillingham and Munro, 2000), based on the observations that nocodazole disruption fails to abolish pericentrin centrosomal localization, recruitment of pericentrin to centrosomes requires an association with the dynein motor complex (Young et al., 2000; Zimmerman and Doxsey, 2000). Nuclear mitotic apparatus protein (NuMA), a highly coiled coil 240-kDa protein, is also concentrated at spindle poles, where it cross-links microtubule minus ends, through an association with the dynein/dynactin complex (Gaglio et al., 1996). Recently, a direct microtubule interaction has been localized to the carboxy terminal tail domain of NuMA (Haren and Merdes, 2002). Thus, NuMA may function in microtubule sliding and spindle stability by directly binding with microtubules at the carboxy terminus, complexing with the dynein/dynactin motor complex and anchoring one microtubule relative to another sliding microtubule (Gaglio et al., 1996).
The transforming acidic coiled coil (TACC) proteins, a family of proteins that concentrate to centrosomes through a conserved coiled coil carboxy-terminal domain called the TACC domain, also play a role in organizing centrosomal microtubules (Gergely et al.,2000a,b;Lee et al., 2001). The TACC domain of _Drosophila_-TACC, D-TACC, has been shown to interact with minispindles, msps, the Drosophila homolog of human colonic and hepatic tumor overexpressed (TOGp) protein (Cullen and Ohkura, 2001;Lee et al., 2001). The TOG family of proteins (TOGp, XMAP215, and Msps) can bind directly to microtubules and promote their polymerization. In Drosophila, Msps is transported to microtubule minus ends by the kinesin-like protein Ncd and anchored to centrosomes by an association with d-TACC (Cullen and Ohkura, 2001;Lee et al., 2001); centrosomal targeting of D-TACC is regulated by Aurora A kinase, which, in turn, is dependent on TPX2-mediated targeting (Giet et al., 2002;Kufer et al., 2002). Like RHAMM, TACC proteins have been intimately linked to carcinogenesis. TACC1 overexpression transforms mouse fibroblasts (Still et al., 1999a). A TACC2 isoform has been identified as the tumor suppressor protein Azu-1 in breast carcinoma lines and TACC3 is up-regulated in multiple cancer lines (Still et al., 1999b; Chen et al., 2000). Moreover, TACC3 is essential for hematopoietic stem cell function and may play a primary role in the regulation of p53 function within hematopoietic cells (Piekorz et al., 2002).
This article investigates the involvement of RHAMM at the mitotic spindle with emphasis on its localization in nonadherent cell lines derived from human lymphocytes. We demonstrate that RHAMM shares structural and functional similarity with proteins that are essential for the maintenance of the mitotic spindle. RHAMM localizes to the centrosome during interphase and to the spindle poles during mitosis. Consistent with sequence similarity to the Klp2 family, we demonstrate that RHAMM interacts with the dynein complex in vivo. The centrosomal targeting domain of RHAMM localizes to the previously characterized hyaluronan binding domain (Yang et al., 1994); we show this motif is phylogenetically related to the TACC and Klp2 centrosomal targeting motifs. We find that overexpression of GFP-RHAMM leads to mitotic delays and apoptosis. We also find that disruption of RHAMM mitotic function, through microinjection of purified anti-RHAMM antibodies, affects spindle integrity and results in the formation of tripolar and tetrapolar spindles. We have shown that in addition to its other functions, RHAMM is a centrosomal protein that interacts with the dynein microtubule motor complex, functions in the maintenance of spindle integrity, and is chromosomally located proximal to the putative FGFR4-TACC4 gene cluster on chromosome 5qter.
MATERIALS AND METHODS
Plasmids and Antibodies
The RHAMMFL, RHAMM-exon 4, and RHAMM-exon13 were all amplified with the 5′_Xho_I-RHAMM and 3′ _Kpn_I-RHAMMFL primers by using platinum high-fidelity Taq (Invitrogen Life Technologies, Carlsbad, CA) under standard conditions as recommended by the manufacturer. Polymerase chain reaction products were cloned into pCR2.1-TOPO (Invitrogen). The RHAMM679, RHAMM623, RHAMM525 deletion constructs were all amplified from the pCR2.1-TOPO-RHAMMFL vector, after sequencing, with 5′ _Xho_I-RHAMM and the respective deletion primer. These products were cloned into pCR2.1-TOPO. The respective RHAMM constructs were then cloned into pEGFP-C1 (BD Biosciences Clonetech, Palo Alto, CA) by using _Xho_I and _Kpn_I. All GFP-RHAMM constructs were prepared for transfection using the EndoFree maxi kit (QIAGEN, Valencia, CA). The green fluorescent protein (GFP)-tubulin construct was purchased from BD Biosciences Clonetech.
Primer Sequences. The following primer sequences were used:
- 5′ XhoI-RHAMM cGCtcgagAt_ATG_TCCTTTCCTAAG
- 3′ KpnI-RHAMMFL cc_gGTacc_CTTCCATGATTCTTG
- 3′ KpnI-RHAMM679 TGGTGTTTGgTACCTAGAAC_TTa_ATTCAAT
- 3′ KpnI-RHAMM623CAATAAggTAcCATA_TtA_ATCTCTTATTTTATT
- 3′ KpnI-RHAMM525GAAACTGTGgTacCTTTAAT_TTa_TGTTTCCTTTAG
- 5′ XhoI-RHAMM500 C_ctcgag_atGCAACTGAGAGCTC
- 5′ XhoI-RHAMM624 AA_ctcGAG_ATTCATATGCTAAATT
- 3′ KpnI-RHAMMstop GggtaCcGT_tta_CTTCCATGATTCTTG
Primer sequences are shown 5′ to 3′, lowercase letters denote mispriming from de novo mRNA, bold sequences denote restriction endonuclease recognition sites, and italic letters denote start and stop codons.
Antibodies. α-Tubulin (clone B-5-1-2), γ-tubulin (clone GTU-88), and dynein (clone 70.1) were purchased from Sigma-Aldrich (St. Louis, MO); pericentrin was from Babco (Richmond, CA); and the mouse monoclonal NuMA antibodies were identified in a monoclonal antibody (mAb) screen for mitotic chromosome scaffold proteins (Compton et al., 1991). The polyclonal RHAMM antibody CM1 was produced by Washington Biotechnology (Baltimore, MD) to the following carboxy-terminal peptide sequence: G681IKHFDPSKAFHHESK696. The polyclonal serum was affinity purified over a peptide-loaded NHS-activated Sepharose 4B column (Amersham Biosciences, Piscataway, NJ), washed, eluted with glycine pH 2.5, neutralized, and quantified by OD280. Fractions were pooled and concentrated with Ultrafree-MC (Millipore, Bedford, MA) to >10 mg/ml. The specificity of this serum was tested by immunoblot, immunoprecipitation, and immunofluorescent analysis (Figure 5). A second anti-RHAMM serum was as described previously (Assmann et al., 1999); this antiserum is a pan-specific anti-RHAMM sera raised against a bacterially expressed GST-RHAMM fusion protein (RHAMM exons 10–13, aa307–498) (Assmann et al., 1999). The anti-TOGp serum was as described previously (Dionne et al., 2000). Secondary antibodies were from Molecular Probes (Eugene, OR).
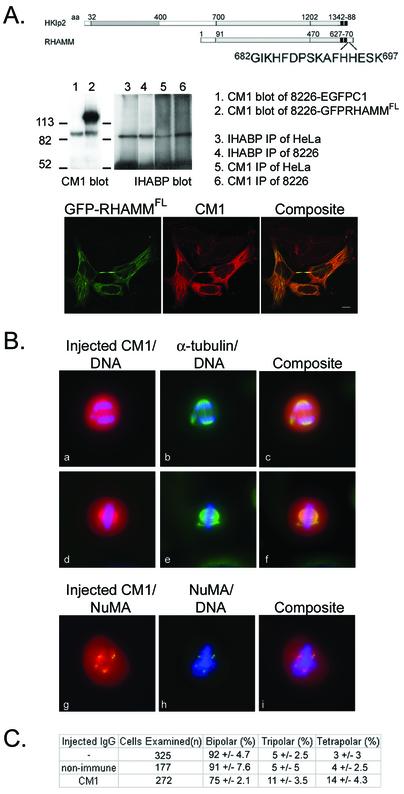
Figure 5.
Microinjection of anti-RHAMM antibodies induces multipolar spindles. (A) Representation of the location, relative to Hklp2, and sequence of the peptide from which a polyclonal anti-RHAMM serum (CM1) was derived. The specificity of the purified CM1 serum was determined by immunofluorescence, immunoblotting (lane 1 and 2), immunoprecipitation (lane 3–6), and reactivity with GFP-RHAMMFL fusion proteins. Lane 1 and 2 demonstrates that CM1 specifically blots endogenous RHAMM and GFP-RHAMM in RPMI 8226 cells transfected with GFP-RHAMM (lane 2) or vector control (EGFP-C1, lane 1). Lanes 3–6 demonstrates that CM1 immunoprecipitates from HeLa and RPMI 8226 lysates an equivalent-sized protein as the previously validated IHABP antibody. Bar, 5 μm. (B) HeLa cells were plated onto gridded coverslips and were synchronized at the G1/S boundary by a double thymidine block. Cells were injected 1–2 h after they were released from the G1/S boundary. Asymmetrical and symmetrical tripolar and tetrapolar spindles were observed. DAPI staining revealed that all spindles in these abnormal mitotic figures segregated DNA. Microinjected RHAMM antibody (first column) reveals that the injected antibody localizes to the spindle poles. NuMA staining (g–i) confirms that the abnormal spindle poles contain NuMA. (C) Graphical representation of microinjection results. Four separate experiments were performed and spindles were scored as bipolar, tripolar, tetrapolar, and hexapolar spindles (our unpublished data). The results are given as mean percentage and SD of the mean for each spindle phenotype.
Cell Culture, Transient Transfection, Nocodazole Treatment, and Immunofluorescence
Cells from RPMI 8226, a human lymphoblastic cell line derived from the peripheral blood of a multiple myeloma patient, and Raji, a human lymphoblastic cell line derived from a Burkitts lymphoma patient, were grown in suspension in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen), at 37°C in 5% CO2. HeLa cells, a human adherent epithelial cell line derived from a patient with cervical adenocarcinoma, were grown in DMEM medium supplemented with 10% fetal bovine serum (Invitrogen), at 37°C in 5% CO2. Cells were passaged 24 h before transfection. Suspension cells were transfected by electroporation (270 mV, 960 μF, 47–53 ms), and stable GFP-α-tubulin transfectants were selected for in 600 μg/ml G418. HeLa cells were transfected with LipofectAMINE 2000 (Invitrogen) following manufacturers' protocols. For nocodazole experiments, HeLa cells were incubated for 90 min in 25 μM nocodazole (Sigma-Aldrich) before immunofluorescence. Suspension cells and HeLa were fixed and permeabilized in cold MeOH at defined time points posttransfection. Cells were washed with phosphate-buffered saline (PBS)-0.5% Triton X-100 (Sigma-Aldrich) before immunofluorescence. Primary and secondary antibodies were diluted in PBS-0.1% Tween (Sigma-Aldrich) + skim milk powder (blocking buffer), and all antibody incubations were for 30 min at room temperature. For double-staining experiments, antibodies were added sequentially. Cells were washed three times in PBS-0.5% Tween before and after incubations. Cells were mounted in 90% glycerol/PBS + 4,6-diamidino-2-phenylindole (DAPI) and images were acquired using a confocal LSM 510 or multiphoton microscope (Carl Zeiss, Thornwood, NY). Images were processed using MetaMorph software (Universal Imaging, Downingtown, PA) and Photoshop 5.02 software (Adobe Systems, Mountain View, CA).
Immunoprecipitations
HeLa cells were transfected using LipofectAMINE 2000 (Invitrogen) and the manufacturer's protocol. After transfection, cells were incubated for 12 to 16 h, in the presence of OptiMEM (Invitrogen), whereas untransfected cells were incubated in fresh DMEM-10% fetal bovine serum. After incubation, cells were released from plates with 1× trypsin, washed three times with PBS, and lysed at 5 × 106–107 cells/ml in 1% 3-[(3-cholamidopropy-l)dimethylammonio]propanesulfonate (CHAPS) plus 10 μg/ml leupeptin, 10 μg/ml antipain, and 1 mM phenylmethylsulfonyl fluoride (all from Sigma-Aldrich). For some experiments, cells were washed and lysed in situ with lysis buffer. All immunoprecipitation procedures were performed at 4°C. Lysates were precleared with protein A-Sepharose beads (Amersham Biosciences) with rotation for 30 min. Precleared lysates were incubated with antibodies (3 μl/150 ml of lysate) for 2 h at 4°C with rotation, and then with protein A beads (40 μl of a 1:1 slurry in CHAPS+) for 1 h at 4°C with rotation. Beads were collected with centrifugation and washed four times with CHAPS+ buffer. Immunoprecipitated proteins were eluted with boiling SDS buffer and analyzed by a 5% stacking/8% separating SDS-PAGE. Precleared lysates (25 μl) and postimmunoprecipitation fractions were analyzed to determine efficiency and relative quantity of the immunoprecipitations.
Microinjection
HeLa cells were plated onto coverslips and synchronized at the G1/S boundary by double thymidine block (2.5 mM thymidine). Cells were injected 1 h after release from thymidine block. Microinjection was performed with a semiautomatic microinjector (model 5412; Brinkman Eppendorf, Westbury, NY). For each coverslip, 200–300 cells were injected with each antibody mix over the course of ∼30 min. Affinity-purified antibodies were kept in calcium- and magnesium-free PBS and were filtered through a 0.22-μm microfiltration cup (Millipore) before microinjection. Cells were released from the G1/S block by washing with PBS and replaced with fresh media before microinjection. Cells were fixed with 3.5% paraformaldehyde ∼12 h after release from the G1/S boundary. Cells were permeabilized with 0.2% Triton X-100 in KB (10 mM Tris, pH 7.5, 0.15 M NaCl, 0.1% bovine serum albumin) for 5 min and washed with KB before antibody incubation. To minimize loss of loosely attached mitotic cells, the coverslips were centrifuged at 200 × g for 2 min in a clinical centrifuge (GPKR; Beckman Coulter, Fullerton, CA). The injected antibodies were detected by Cy5-conjugated anti-rabbit secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA).
RESULTS
RHAMM Localizes to Centrosomes, Spindle Poles, and Midzone Microtubules. The Carboxy-terminal Structure of RHAMM Is Similar to the Klp2 Family
The subcellular localization of RHAMM to interphase microtubules and the mitotic spindle has been demonstrated in adherent lines (Assmann et al., 1999). Because RHAMM overexpression and function also characterizes human lymphocytes and B lymphoid malignancies (Masellis-Smith et al., 1996; Gares et al., 1998; Crainie et al., 1999), we investigated RHAMM localization in nonadherent, suspension cell lines derived from lymphocyte malignancies (RPMI 8226 and Raji). The subcellular localization of endogenous RHAMM was investigated by indirect immunofluorescence by using two polyclonal antisera. The first, termed IHABP, was previously characterized as a pan-specific anti-RHAMM antiserum (Assmann et al., 1999), whereas the second, termed CM1, is characterized below (Figure 5A). The adherent line HeLa was used to confirm RHAMM localization in cells derived from solid tumors (our unpublished data).
Examination of RHAMM localization within interphase, nonadherent cell lines revealed pronounced centrosomal localization in addition to microtubule association (Figure 1A). Although less pronounced in adherent HeLa cells, centrosomal localization was identified within interphase and prophase cells (our unpublished data). We speculate that the more polymerized, filamentous microtubule arrays of adherent lines has prevented previous observation of RHAMM centrosomal localization. During prophase, in both adherent and suspension lines, RHAMM concentrated at the center of microtubule asters and associated with microtubules growing between the two asters (Figure 1B). During prometaphase and metaphase, RHAMM intensified at the spindle pole and extended along spindle microtubules during metaphase. Additionally, RHAMM localized to the spindle midzone during anaphase and telophase. During telophase, RHAMM redistributed from the centrosomes and concentrated to the spindle midzone (Figure 1B). To compare the localization of RHAMM in fixed and live cells and to control for possible fixation and permeabilization artifacts, the distribution of full-length RHAMM (RHAMMFL)-GFP was examined in transiently transfected RPMI 8226, Raji, and HeLa cells. Consistent with the localization of endogenous RHAMM in fixed samples, RHAMMFL-GFP proteins localized to centrosomes, interphase microtubules, the mitotic spindle pole, and midzone microtubules in suspension and in adherent cells. Although microtubules are suboptimally visualized in nonadherent cells, RHAMM antibodies colocalized with microtubules in all interphase cells examined, including methanol fixed suspension cells (our unpublished data).
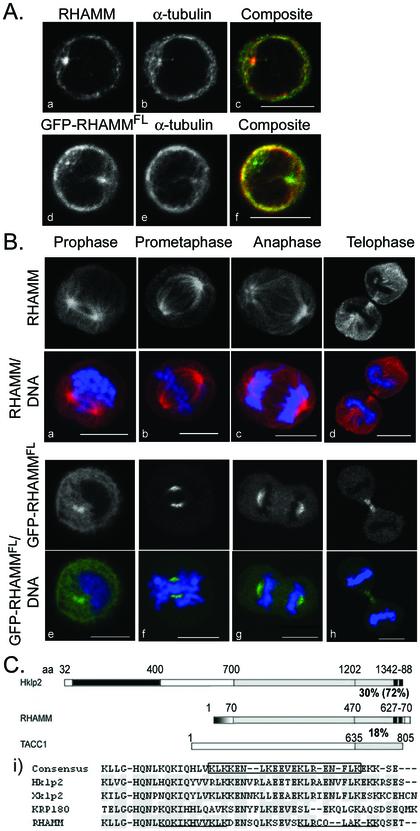
Figure 1.
Structural analysis and localization of RHAMM in suspension cell lines. (A) Interphase localization of RHAMM (left column) by indirect immunofluorescence (a–c) and GFP-RHAMMFL fusion proteins (d–f) and colocalization with α-tubulin (middle column) are shown. Exponentially growing RPMI 8226 cells were transiently transfected with GFP-RHAMMFL (d–f) or methanol treated and sequentially stained with antibodies against RHAMM and α-tubulin. In the composite image c, RHAMM is red and microtubules are green; in composite image d, GFP-RHAMMFL is green, whereas microtubules are red. (B) Mitotic localization of RHAMM by indirect immunofluorescence (a–d) and GFP-RHAMMFL fusion proteins (e–h) are shown. Exponentially growing RPMI 8226 cells were transiently transfected with GFP-RHAMMFL (d–f) or methanol treated and sequentially stained with antibodies against RHAMM and α-tubulin. DNA was visualized with DAPI. Cells in prophase, prometaphase, anaphase, and telophase are shown. Bars, 10 μm. (C) Schematic diagram of RHAMM-predicted secondary structure and functional domains with members of the Klp2 and TACC protein families. The molecular motor domain of Hklp2 (aa 32–400) is identified in black, the amino-terminal microtubule-binding domain of RHAMM (aa 1–70) is in gradient, predicted coiled coil structure of the proteins is in gray, the carboxy-terminal B(X)7B domain of RHAMM (aa 627–70) and basic leucine zipper of Hklp2 (aa 1342–88) are shown in hatched black. Percentage of identity between the COOH-terminal 200 aa is shown; percentage of identity within the basic leucine zipper/HA binding domain is shown in parentheses. (i) RHAMM COOH-terminal sequence was aligned with Klp2 family members by using the PepTool software program. Identical amino acids are outlined in gray. The Klp2 consensus COOH-terminal leucine zipper is boxed. The HA-binding B(X)7B domains of RHAMM are underlined.
Previous analysis of RHAMM established the existence of three major structural domains: the amino-terminal head (aa 1–69; pI = 10.72), which interacts with microtubules; an extensive coil coiled stalk (aa 70–680; pI = 5.06); and a short carboxy-terminal tail (aa 681–724; pI = 8.12) (Assmann et al., 1999). Sequence alignments and structural prediction reveal a relationship between RHAMM and the Klp2 protein family (Figure 1C). Structure prediction for RHAMM, like Xklp2, predicts extensive coiled coil structure within the carboxy-terminal 600 amino acids (Figure 1C) (Boleti et al., 1996;Assmann et al., 1999). Moreover, BLAST 2 sequence analysis (Tatusova and Madden, 1999) of RHAMM primary structure against the Klp2 family reveals significant homology (e-28 for Hklp2), including conservation of the carboxy-terminal leucine zipper, which exhibits 72% identity (Figure 1C, i); this domain is vital to Klp2 centrosomal localization and function through an indirect interaction with the dynein/dynactin complex (Wittmann et al., 1998). The coiled coil terminus of RHAMM, however, bears little sequence identity to the centrosomal targeting TACC domain of TACC1. The carboxy-terminal leucine zipper of RHAMM (boxed inFigure 1C, i) overlaps the defined HA-binding domains (underlined inFigure 1C, i); this region is conserved among RHAMM proteins in human, mouse and rat (Lynn et al., 2001). RHAMM lacks the highly conserved amino-terminal kinesin motor domain but contains a microtubule-binding domain in its place (aa 1–70;Figure 1C). Based on these similarities, we tested whether the conserved carboxy-terminal leucine zipper directed RHAMM to centrosomes and spindle poles.
The Amino Terminus of RHAMM Is Required for Interaction with Interphase Microtubules, whereas the Carboxy-terminal Leucine Zipper Targets RHAMM to the Centrosome
To define the domains in RHAMM responsible for centrosomal targeting, we constructed GFP fusion proteins that correspond to known RHAMM splice variants (RHAMM-exon4 and RHAMM-exon13) as well as RHAMM carboxy-terminal deletion mutants (RHAMM1–525, RHAMM1–623, and RHAMM1–679) and carboxy-terminal fragments (RHAMM625–725 and RHAMM500–725) (Figure 2A).
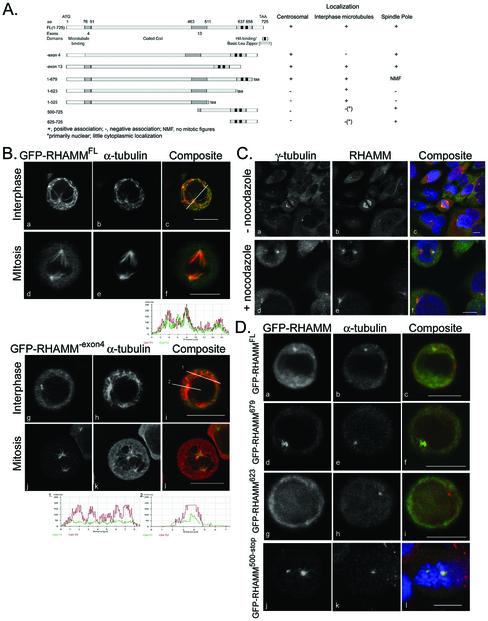
Figure 2.
RHAMM interacts with microtubules at the amino terminus and is targeted to centrosomes by the carboxy terminus. (A) Cartoon diagram of RHAMMFL and the RHAMM splice variants along with the deletion mutants used in the following experiments. The table outlines results of centrosomal localization experiments. For GFP-RHAMM679 variant, no mitotic figures (NMF) were observed for five transfection experiments. (B) EGFP-C1 (our unpublished data), EGFPC1-RHAMMFL (top box), EGFPC1-RHAMM-exon13 (our unpublished data), and EGFPC1-RHAMM-exon4 (bottom box) were transfected into RPMI 8226 and stained for α-tubulin 6 to 8 h posttransfection (middle lanes). α-Tubulin stain allows identification of microtubule colocalization and microtubule-organizing center (centrosome) localization. Cells were visualized in PBS/glycerol (a–f) or cytospun at 200 rpm and visualized (g–l). Line (c and I, inset) profile analysis of channel intensities was used to demonstrate colocalization, or lack thereof, between GFP-RHAMM isoforms and α-tubulin. Representative profiles are shown. Bars, 10 μm. (C) HeLa cells were treated with, or without, nocodazole for 90 min, and sequentially stained for RHAMM, or pericentrin (our unpublished data), and γ tubulin, a centrosomal marker. In the composite images, RHAMM is red, γ-tubulin is green, and DNA is blue. DNA was visualized with DAPI. Bars, 10 μm. (D) Deletion constructs were transfected into RPMI 8226, and cells were stained for γ-tubulin 6 to 8 h posttransfection to indicate centrosomal localization. DAPI channel is included for the GFP-RHAMM625-stop variant to demonstrate prometaphase stage of transfected cell. Interphase cells transfected with GFP-RHAMM500-stop or GFP-RHAMM625-stop demonstrated nuclear accumulation but not centrosomal localization (our unpublished data). Bars, 10 μm.
To investigate the centrosomal localization of RHAMM in nonadherent cells, GFP-RHAMM fusion constructs were transiently transfected into RPMI 8226 and Raji lines. Consistent with previous observations in HeLa cells (Assmann et al., 1999), RHAMM variants that contain exon 4 (i.e., RHAMMFL and RHAMM-exon13) colocalized with interphase microtubules (Figure 2B; our unpublished data). The degree of colocalization is illustrated by the intensity profile along the inset line withinFigure 2B, c. Although the low cytoplasm to nucleus ratio of the suspension cells makes microtubule colocalization experiments difficult, occasional cells with well-defined microtubule networks were observed. Within these cells, GFP-RHAMM variants, containing exon 4, demonstrated precise colocalization with microtubules (our unpublished data). Loss of exon 4 (GFP-RHAMM-exon4) disrupts interphase microtubule interactions as demonstrated by the lack of colocalization along the inset line 1 withinFigure 2B, i. In fact, GFP-RHAMM-exon4 demonstrated diffuse, relatively invariant, cytoplasmic localization (Figure 2B, line 1). However, GFP-RHAMM-exon4 localized to centrosomes (see line 2 profile) and mitotic spindle poles (Figure 2B). Interestingly, whereas GFP-RHAMMFL localized to the spindle pole and along chromosome-contacting microtubules, GFP-RHAMM-exon4 localized primarily to the spindle pole. Nocodazole inhibition of microtubule polymerization was used to determine whether polymerized microtubules were required for the association of RHAMM with centrosomes and the mitotic spindle pole. Pericentrin was used as a positive control because it is known to localize at the centrosome in a microtubule-independent manner (Gillingham and Munro, 2000). Centrosomal localization was analyzed by γ-tubulin staining, and DAPI staining was used to determine the mitotic status of the cells. In the presence of polymerized microtubules (-nocodazole), RHAMM localizes at centrosomes and the mitotic spindle (Figure 2C). As demonstrated by the RHAMM staining within metaphase cells (Figure 2C, e), nocodazole treatment affected cellular architecture and disrupted metaphase mitotic spindles. Consistent with previous reports, pericentrin localization at centrosomes was microtubule independent (our unpublished data); in the absence of polymerized microtubules, RHAMM maintained an association with interphase centrosomes and the mitotic spindle pole (Figure 2C). It is important to note that microtubule-independent centrosomal localization is distinct from microtubule-independent centrosomal targeting. Although RHAMM and other centrosomal proteins such as pericentrin do not require microtubules to remain localized at the centrosome, it is likely that RHAMM centrosomal targeting is facilitated through an interaction, either direct or indirect, with a minus-end directed microtubule molecular motor (see below).
The carboxy-terminal coiled coil domain of RHAMM shares 72% identity with the basic leucine zipper (BZIP) of the Klp2 family (Figure 1C). To test the importance of the conserved leucine zipper in centrosomal targeting, deletion constructs of RHAMM, lacking the carboxy-terminal 200, 102, and 47 aa, were constructed (Figure 2A). Transient transfection of empty vector EGFPC1 was used as a negative control for centrosomal and spindle pole localization; both the centrosome and spindle pole showed slight amplification of transfected EGFP-C1 (our unpublished data). Deletion of the carboxy-terminal 200 aa (RHAMM525; our unpublished data) or 102 aa (RHAMM623;Figure 2D), inhibited the centrosomal localization of the fusion proteins. However, RHAMM proteins that contained the leucine zipper, RHAMMFL (Figure 2D), RHAMM-exon4 (Figure 2B), RHAMM-exon13 (our unpublished data), and RHAMM679 (Figure 2D), localized to the centrosome. The BZIP motif was also essential for localization of RHAMM constructs to the mitotic spindle pole; after examination of five transfection experiments, we were unable to identify a mitotic figure within GFP-RHAMM679-transfected cells. This observation was tested and quantitated in HeLa cells. Transient transfectants of GFP-RHAMM679 and GFP-RHAMMFL were examined for mitotic stage at 12, 13, 14 and 20 h posttransfection and compared with neighboring untransfected cells. GFP-RHAMM679 transfection resulted in only prophase transfectants (n = 47; our unpublished data), whereas GFP-RHAMMFL transfection led to accumulation of prometaphase and metaphase cells (58.6% [34/58] of transfected cells vs. 26.2% [101/386] within the untransfected population; Figure 4). Moreover, the majority of GFP-RHAMM679 transfectants demonstrated large aggregation of GFP fluorescence (our unpublished data). Microtubule associations and centrosomal localization were maintained in transfectants with low levels of fluorescence (our unpublished data). Interestingly, 704TPLK707, a consensus cdc2 phosphorylation site conserved in mouse and rat, falls within the deleted region of RHAMM679. To test the sufficiency of the BZIP motif for centrosomal targeting, we constructed GFP-tagged, carboxy-terminal RHAMM fragments consisting of the terminal 100 and 225 aa, respectively. When transiently transfected into RPMI 8226, these constructs localized to the interphase nucleus and mitotic spindle pole. Although the carboxy terminus does not include a predicted (predictNLS analysis) simple or bipartite nuclear localization signal, characterized by a short stretch of basic aa or two interdependent positively charged clusters separated by a short linker region, it is highly basic (Cokol et al., 2000). Given their basic nature, the nuclear localization of the small carboxy-terminal fragments may be the result of transport, and not simple diffusion, into the nucleus. Both fragments also showed slight localization to the centrosome although not greater than GFP alone. Therefore, the carboxy-terminal basic leucine zipper of RHAMM, which overlaps the defined B(X)7B HA binding domains and is homologous to the Klp2 family, is essential for centrosomal targeting and sufficient for spindle pole localization.
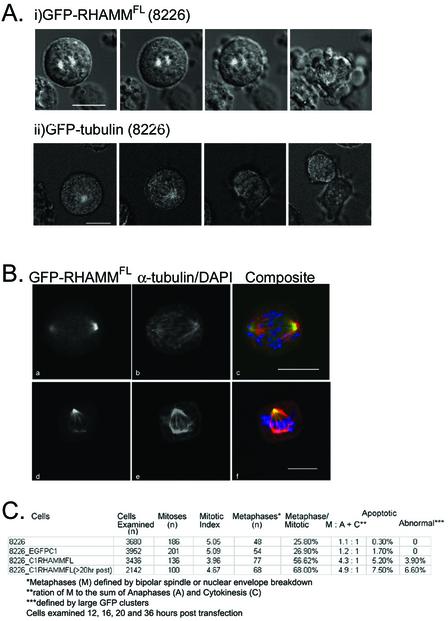
Figure 4.
GFP-RHAMMFLoverexpression inhibits mitotic progression in RPMI 8226 and HeLa (our unpublished data). RPMI 8226 or HeLa cells were transfected with EGFPC1-RHAMMFL, incubated at 37°C for 6 h, and visualized in the presence or absence of Hoescht 33342 at 5 μg/ml. (i) Overexpression of EGFPC1-RHAMMFL within RPMI 8226 and HeLa (our unpublished data) during metaphase induced a block in chromosome alignment and spindle separation, leading to spindle breakdown and cell death. Transfected cells were incubated at 37°C, 5% CO2 and visualized every 5 min for 10 h. (ii) Overexpression of EGFPC1-tubulin within RPMI 8226 did not affect progression through the cell cycle. Within this population, anaphase was completed (∼40 min) ∼25 min after metaphase (14 min). Bars, 10 μm. (B) Overexpression of GFP-RHAMMFL in RPMI 8226 led to the accumulation of mitotic cells in prometaphase (a–c) and metaphase (d–f) with unaligned chromosomes. Cells were transfected, methanol treated at defined time points, and stained with monoclonal antibodies against α-tubulin. DNA was visualized with DAPI. Bars, 10 μm. (C) Graphical representation for time line analysis of mitotic and apoptotic status of untransfected RPMI 8226, EGFP C1 (vector control)-transfected RPMI 8226, and GFP-RHAMMFL-transfected RPMI 8226 at 12, 16, 20, and 36 h posttransfection. Transfected cells were sorted based on viability and fluorescence at 8–10 h posttransfection. Transfected cells were identified as interphase, prophase, prometaphase, metaphase, anaphase, cytokinesis, and apoptotic based on the state and condensation of their DNA. Transfected cells were classified as abnormal if they contained large aggregates of GFP-RHAMM (our unpublished data); the microtubule staining of these cells seemed more diffuse and less organized than cells lacking these aggregates. However, DAPI staining did not indicate that these cells were apoptotic.
RHAMM Interacts with the Dynein Motor Complex In Vivo
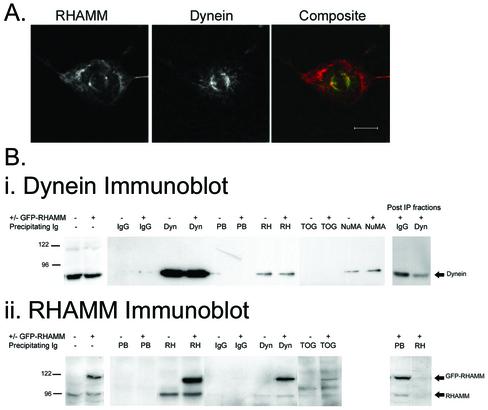
Because the carboxy-terminal leucine zipper is conserved between RHAMM and the Klp2 family and this domain mediates the indirect interaction of Xklp2 and dynein (Wittmann et al., 1998), we investigated whether RHAMM interacts with the dynein motor complex in cells. We first determined the spatial relationship of endogenous RHAMM and dynein IC in HeLa cells. Double immunofluorescence by confocal microscopy demonstrated colocalization of dynein and RHAMM at the spindle pole within mitotic HeLa cells (Figure 3A). Because a large fraction of intracellular RHAMM, and dynein, does not localize to the spindle pole, it is likely that only a subset of the total RHAMM protein interacts with the dynein/dynactin motor complex, and vice versa.
Figure 3.
RHAMM interacts with the dynein motor complex. (A) Endogenous RHAMM and dynein colocalize at the spindle pole in mitotic HeLa cells. HeLa cells were methanol treated and stained sequentially with antibodies against RHAMM (polyclonal) and dynein light IC (monoclonal). Bar, 10 μm. (B) HeLa lysates were prepared as described in MATERIALS AND METHODS. Immunoblot analysis of RHAMM and dynein levels indicates equivalent amounts within transfected and untransfected lysates. An additional band at 120 kDa within the transfected populations represents the GFP-RHAMMFL species. Immunoprecipitation experiments were separately performed on 150 μl of precleared lysates by using a polyclonal RHAMM antibody, the prebleed immune sera, a polyclonal TOGp antibody, a monoclonal NuMA antibody, a monoclonal dynein IC antibody (Sigma-Aldrich), or a IgG2a isotype control antibody (Southern Biotechnology Associates, Birmingham, AL) as described in MATERIALS AND METHODS. Twenty-five microliters (one-fifth of the immunoprecipitation volume) of precleared lysate and postprecipitation (Post-IP) fractions were analyzed to determine relative quantity and efficiency of the precipitations. The resulting immunoprecipitates were blotted with dynein IC monoclonal antibodies (I, dynein immunoblot) or a RHAMM polyclonal serum (ii, RHAMM immunoblot). The expected band size for dynein (dynein immunoblot) as well as endogenous RHAMM and GFP-RHAMM (RHAMM immunoblot) are shown with arrows. Transfected (+GFP-RHAMM) and untransfected (-GFP-RHAMM) lysates are indicated above the blot as are the precipitating antibodies for the experiment. Post-IP fractions for the transfected lysates are shown to indicate the relative efficiency of precipitation for IgG and dynein (dynein immunoblot) or prebleed and RHAMM (RHAMM immunoblot). For dynein immunoblot, all lanes exposed for 30 s. For RHAMM immunoblot, lanes 2–6 were exposed for 15 s, whereas lanes 1, 2, and 7–14 were exposed for 60 s. Precipitating antibodies are indicated as: isotype matched IgG2a (IgG), monoclonal antidynein IC (Dyn), prebleed (PB) antiserum, RHAMM (RH) antiserum, TOGp (TOG) antiserum, and monoclonal NuMA (NuMA).
To extend the confocal colocalization data, we investigated the ability of RHAMM antibodies to coimmunoprecipitate dynein IC from interphase and mitotic_Xenopus_ extracts. A cell cycle-dependent NuMA–dynein–dynactin complex has been identified in immunoprecipitates by using either anti-dynein or anti-NuMA antibody within the Xenopus system (Merdes et al., 1996; Merdes et al., 2000). However, the NuMA–dynein–dynactin complex seems to be less stable in higher vertebrate cells with some, but not others, able to coimmunoprecipitate these protein complexes (Zeng, 2000). This raised the possibility that it may also be difficult to detect a RHAMM–dynein complex in HeLa cells. Thus, we analyzed RHAMM–dynein interactions in_Xenopus_ extracts. Immunoprecipitation of mitotic and interphase_Xenopus_ extracts with RHAMM antibodies revealed a major species at 95 kDa as well as minor bands around 150, 113, and 85 kDa (our unpublished data). These sizes are consistent with those published for RHAMM species in various mammalian tissues (Hall et al., 1995; Assmann et al., 1999). RHAMM antibodies coimmunoprecipitated dynein, with slight amplification within M phase extracts, but some dynein was found to interact with RHAMM during interphase (our unpublished data). Interestingly, the monoclonal dynein IC antibody was unable to coimmunoprecipitate RHAMM protein, consistent with our observation that only a fraction of dynein was associated with RHAMM, and vice versa.
To confirm this association in mammalian cells, HeLa extracts were transiently transfected with GFP-RHAMMFL. CHAPS-soluble lysates from transfected, or untransfected, populations were separately immunoprecipitated with an anti-intermediate chain dynein mAb, IgG2a control antibody, a polyclonal RHAMM antibody, a polyclonal TOGp antibody, a monoclonal NuMA antibody, and a nonimmune serum. As demonstrated inFigure 3C, lanes 1 and 2, transfected and untransfected lysates contained comparable amounts of endogenous proteins with the transfected populations overexpressing a GFP-RHAMM species of ∼120 kDa (lane 2, RHAMM immunoblot). Recently, dynein has been coimmunoprecipitated from HeLa extracts with NuMA antiserum; for this reason, we included a NuMA immunoprecipitation as a positive control for association with dynein (Bhattacharya_et al_., 2002). Also, because the drosophila homolog of TOGp, msps, uses the Ncd motor, and not the dynein complex, to localize to centrosomes (Cullen and Ohkura, 2001), TOGp immunoprecipitation of dynein served as a negative control. RHAMM antibodies coimmunoprecipitated dynein from transfected and untransfected lysates. Comparison of the amount of coprecipitated dynein with the Western blot signal seen in the total lysates indicates that, under the conditions used herein, only a fraction of dynein is recovered in a complex with RHAMM. Although the level of coprecipitated dynein was low, the amount was similar to that precipitated by NuMA antibodies. The specificity of the RHAMM–dynein interaction is demonstrated by the inability of TOGp antibodies, control IgG2a, and nonimmune serum to coimmunoprecipitate dynein. The reciprocal coimmunoprecipitation of RHAMM with dynein IC antibodies was achieved in transfected and untransfected HeLa lysates; again, comparison with the western blot signal indicates that only a fraction of endogenous RHAMM is precipitated by dynein. TOG antibodies also coimmunoprecipitated GFP-RHAMMFL from transfected lysates; this result is consistent with live cell observation, within RPMI 8226, of accumulation of GFP-RHAMMFL aggregates at the cell periphery and vectoral movement of GFP-RHAMMFL toward the cell body (our unpublished data). Thus, the dynamic redistribution of GFP-RHAMMFL in live suspension cells is consistent with minus-end directed motion, mediated by the dynein motor complex, and plus-end association, likely mediated through an interaction with TOG.
Overexpression of GFP-RHAMM Fusions Initiates a Mitotic Block in RPMI 8226 and HeLa Cells
Because GFP-RHAMMFL does not generate stable transfectants within RPMI 8226 and HeLa cells, time-lapse confocal microscopy was used to follow the kinetics of mitosis within these transfectants. Transiently transfected GFP-RHAMMFL cells arrested in mitosis (Figure 4A, i). These transfected cells failed to divide, mitotic spindles broke down, and the cells underwent apoptosis (Figure 4A, i). To demonstrate that the visualization techniques were not causing the mitotic block and that the GFP tag was not influencing the kinetics of mitosis, tubulin-GFP fusions were stably transfected into 8226 cells and followed through mitosis. These cells demonstrated normal division (Figure 4A, ii).
To quantitate the level of GFP-RHAMMFL overexpression and its effect on mitosis, RPMI 8226 cells were transiently transfected with GFP-RHAMMFL or EGFP-C1 vector and sorted 8 h later. An aliquot of transfected cells was examined by immunoblot for overexpression at 16 h posttransfection (Figure 5A). The 8226 cells transfected with EGFPC1 (Figure 5A, lane 1) express an endogenous RHAMM species of ∼90 kDa, whereas the 8226 cells transiently transfected with GFP-RHAMMFL (Figure 5A, lane 2) express endogenous RHAMM and a GFP-RHAMM protein at ∼120 kDa. Band intensities, measured by digital imaging quantitation, indicated that the GFP-RHAMM intensity was approximately five times that of the endogenous RHAMM. The level of overexpression in these sorted populations of transiently transfected RPMI 8226 cells is greater than that in the unsorted population of HeLa cells used in the immunoprecipitation experiments. This difference is due to the sorting of populations (i.e., many unsorted, lysed HeLa cells may not be expressing GFP-RHAMM) and likely gives a more definitive representation of the level of GFP-RHAMM, relative to endogenous RHAMM, within transfected cells. Transfected cells were fixed at 12, 16, 20 and 36 h posttransfection and stained with α-tubulin, to identify the spindle, and with DAPI, to determine mitotic status. Untransfected RPMI 8226 and EGFPC1 (control vector)-transfected RPMI 8226 served as controls. GFP-RHAMMFL overexpression resulted in an increase in the frequency of prometaphase cells; ∼70% of mitotic GFP-RHAMMFL cells were prometaphase arrested 20 h posttransfection (Figure 4, B and C). Interestingly, true metaphase alignment of chromosomes was not achieved in GFP-RHAMMFL-transfected cells. Probably due to the prometaphase arrest, few GFP-RHAMMFL-transfected cells were observed in anaphase or cytokinesis as demonstrated by the increasing metaphase: anaphase + cytokinesis ratio (Figure 4C). In combination with the prometaphase arrest, GFP-RHAMMFL overexpression resulted in a dramatic increase in apoptotic cells as well as the presence of cells with large GFP-RHAMMFL clusters (our unpublished data). It is unclear whether these large GFP-RHAMMFL clusters precede apoptosis; these clusters did not colocalize with α-tubulin and were phenotypically similar to those identified by overexpression of GFP-TACC domain fusions (Gergely et al., 2000a).
Inhibition of Endogenous RHAMM Results in Abnormal Mitotic Figures
To further investigate the mitotic functions of RHAMM, a polyclonal peptide-specific antibody was created against the carboxy-terminal peptide sequence G682-K697, of RHAMM (Figure 5A). This sequence is outside the conserved basic leucine zipper motif to avoid inhibiting Hklp2 function. This serum was affinity purified and the specificity of the antibody was tested by immunoblotting endogenous RHAMM and GFP-RHAMMFL, immunoprecipitating endogenous RHAMM from HeLa and 8226, and colocalization with GFP-RHAMMFL in HeLa and 8226 (Figure 5A).
To determine whether spindle integrity was affected by disruption of RHAMM function, RHAMM peptide antibodies were microinjected into the cytoplasm of cells arrested at the G1/S boundary. The microinjected cells were released from the G1/S boundary and allowed to proceed to mitosis before fixation and analysis by immunofluorescence. In four separate experiments, microinjection of anti-RHAMM antibodies into HeLa cells led to the formation of tripolar and tetrapolar spindles within 11 ± 3.4 and 14 ± 4.3% of injected cells, respectively (Figure 5B). Immunoblocking RHAMM function resulted in symmetrical and asymmetrical tripolar and tetrapolar spindles. Asymmetrical tetrapolar spindles (Figure 5B) were phenotypically similar to those observed within NuMA-immunoblocked CFPAC-1 cells (Gordon et al., 2001); however, microinjection of RHAMM antibodies did not significantly affect spindle focusing. Thus, RHAMM does not seem to be essential for spindle focusing but rather for maintenance of spindle integrity. The fact that roughly three-quarters of injected cells have phenotypically normal spindles suggests that other proteins can compensate for loss of RHAMM function. With all abnormal spindles, the injected RHAMM antibodies and NuMA colocalized to the aberrant spindle poles (Figure 5B).
Chromosomal Location and Phylogenetic Analysis of RHAMM Primary Sequence Suggests Membership in the TACC Family
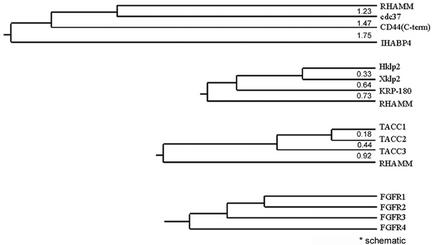
The TACC protein family members are centrosomal proteins that associate with microtubules minus ends through a conserved coiled coil carboxy-terminal TACC domain and are putative regulators of the mitotic apparatus. Despite the low sequence identity within the TACC domain (Figure 1C), RHAMM shares structural and functional similarity to the TACC family. A defining feature of the TACC proteins is an evolutionary conserved relationship with the fibroblast growth factor receptor (FGFR) gene family (Still et al., 1999b). TACC1, 2, and 3 genes map proximal to_FGFR1, 2_, and 3 genes on chromosomes 8p11, 10q26, and 4p16.3, respectively. Currently, no TACC gene has been identified proximal to the FGFR4 locus on chromosome 5q35.1-qter. RHAMM maps to a chromosomal region near to the FGFR4 gene locus at 5q33.2-qter. Given the proximity of RHAMM to FGFR4, we used phylogenetic analysis to compare the carboxy-terminal 240 aa of RHAMM to its current protein family members, the B(X)7B hyaladherins, the TACC family, and the Klp2 family (Figure 6).
Figure 6.
Phylogenetic relationships between members of the BX7B, TACC, Klp2, and FGFR families. Phylogenetic relationships and distances were determined by the MEGA software package by using UPGMA phylogenetic analysis. COOH-terminal sequences were aligned with BLAST and PEPTOOL software, the alignments were entered and analyzed by MEGA software. (A) Relationship between the COOH-terminal 240 aa, which contain the BX7B motif (s), of BX7B family members. (B) Relationship between the COOH-terminal 240 aa, which contain the leucine zipper motif, of klp2 family members and RHAMM. (C) Relationship between the COOH-terminal 240 aa, which contain the TACC domain, of TACC family members and RHAMM. (D) Schematic relationship between FGFR family members as outlined in Still et al., (1999b).
Phylogenetic analysis was performed with the MEGA software program by using carboxy-terminal sequences previously aligned by BLAST and PEPTOOL software (Wishart et al., 1997; Tatusova and Madden, 1999; Kumar et al., 2000). Phylogeny inference was performed using the Unweighted Pair Group Method with Arithmetic Mean and Neighbor-Joining (our unpublished data) distance methods and branch lengths are shown. Theoretically, the evolutionary distance separating two sequences can be defined as the number of mutational events per site underlying the evolutionary history separating these sequences (Brocchieri, 2001). We used the Poisson correction method to approximate these distances.
BLAST analysis of the B(X)7B family members revealed no significant similarity in the B(X)7B-containing domains. CD44, which binds HA with a classical extracellular link domain, contains an intracellular, carboxy-terminal B(X)7B domain and this motif was included in the phylogenetic analysis. As demonstrated by their pairwise evolutionary distances, the B(X)7B family members maintain little evolutionary convergence in their functional domains. The carboxy terminus of RHAMM, currently defined as a B(X)7B domain, shares no significant sequence similarity, and a minimal evolutionary relationship, with other B(X)7B family members.
Comparison of the carboxy terminus of RHAMM with the TACC family reveals that RHAMM diverged before the formation of the TACC3 and TACC1/2 ancestor. This is consistent with the divergence of FGFR4 in the phylogenetic tree of the FGFR family. The carboxy terminus of RHAMM shares minimal identity (∼20%) with TACC1. Therefore, we included the PACT domain of human pericentrin in the phylogenetic analysis to control for the possibility that the RHAMM/TACC relationship is wholly due to RHAMM's coiled coil structure. The PACT domain is an alternative conserved centrosomal targeting domain (Gillingham and Munro, 2000). Pericentrin occupied a separate branch that was 1.10 evolutionary distance units from TACC1 (our unpublished data). Thus, the carboxy terminus of RHAMM is more closely related to the TACC domain than is the PACT domain of human pericentrin.
BLAST analysis of the carboxy-terminal 240 aa of RHAMM reveals significant homology to the Klp2 family (e-13). As there is currently only one human protein member of the Klp2 family (Hklp2), we compared the carboxy-terminal domain of human RHAMM and the sea urchin Klp2-related KRP-180 with those of Hklp2 and Xklp2 to estimate the evolutionary distance between these motifs. As shown in Figure 6, the evolutionary distance between RHAMM (0.73) and Hklp2/Xklp2 approximates that of KRP-180 (0.64). Therefore, the centrosomal targeting motif of RHAMM is more closely related to the Klp2 family than the TACC family. However, the fact that RHAMM lacks the highly conserved molecular motor domain precludes it from membership in the Klp2 family.
DISCUSSION
In this study, we have identified an important functional association between RHAMM, the centrosome, and microtubule-associated proteins. We find that 1) RHAMM is a centrosomal protein that localizes to interphase microtubules, spindle poles, the anaphase midbody and the telophase midzone microtubules; 2) RHAMM contains a centrosome targeting, carboxy-terminal basic leucine zipper and, like Xklp2, interacts with the dynein motor complex; and 3) RHAMM participates in the maintenance of the mitotic spindle. Moreover, RHAMM is an 85-kDa acidic coiled coil protein that is transforming when overexpressed (Hall et al., 1995); functionally related to other TACC family members; and, like TACC 1–3, the RHAMM gene is located near the putative FGFR4-TACC4 gene locus.
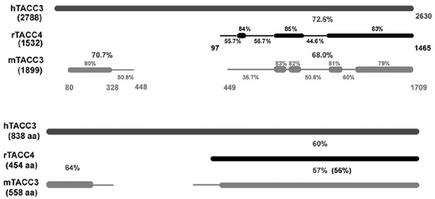
To date, no human TACC4 protein has been identified. A TACC4 protein has been identified in rabbits (rTACC4) (accession no. AAK54244, 454 aa) and shown to directly interact with AKAP350 (Steadman et al., 2002). However, BLAST analysis of the corresponding cDNA (AF372837) against the human genome localized this gene to chromosome 4p16.3 (up to e-29) with exon boundaries that mirror TACC3. This gene product may be the result of a secondary start site in rTACC3 rather than a bone fide rTACC4; in fact, alignment of rTACC4 protein sequence (AAK54244) with an hTACC3 variant (Q9Y6A5) reveals higher identity (60%) than alignment of mTACC3 (XP_132002) with hTACC3 (57%) (Figure 7). Perhaps more convincingly, examination of rTACC4, hTACC3, and mTACC3 cDNA demonstrates that regions of lowest identity between rTACC4 and hTACC3 are also less conserved in mTACC3 (Figure 7). Moreover, the tissue expression of rTACC4 is similar to that defined for mTACC3. MTACC3, but not mTACC2, is highly expressed in the testis, spleen, and lung and absent in the brain, liver, kidney, and muscle (Piekorz et al., 2002); similarly, rTACC4 is highly expressed in the spleen and gut and absent in the brain and liver (Steadman et al., 2002). Interestingly, RHAMM, like TACC3 and rTACC4, is highly expressed in the testis, colon, and stomach but absent in the brain, liver, heart, kidney, and lung (Line et al., 2002).
Figure 7.
Sequence comparison at the cDNA and protein level suggests that rTACC4 is a TACC3 variant initiating at an alternative 5′ start site. Schematic representation of BLAST 2 sequence (nucleotide) analysis of rabbit TACC4 (rTACC4; accession no. AF372837) and murine TACC3 (mTACC3; accession no.NM_011524) against human TACC3 (hTACC3; accession no. NM_006342). The length of the sequences, in base pairs, is bracketed under the sequence name. rTACC4 and mTACC3 were compared with hTACC3 sequence. The overall identities between rTACC4, or mTACC3, and hTACC3 are shown in large font while the identities of smaller segments are shown in small font. The lower levels of identity (≤60%) were insignificant from BLAST analysis and were obtained by PEPTOOL alignment of the sequences. (B) Schematic representation of BLAST 2 sequence (protein) analysis of rabbit TACC4 (rTACC4; accession no. AAK54244) and murine TACC3 (mTACC3; accession no. XP_132002) against human TACC3 (hTACC3; accession no. Q9Y6A5). The length of the sequences, in amino acids (aa), are bracketed under the sequence name. rTACC4 and mTACC3 were compared with hTACC3 sequence. The overall identities between rTACC4, or mTACC3, and hTACC3 are shown, whereas the level of identity between mTACC3 and rTACC4 is bracketed.
Currently, RHAMM is characterized by its HA binding potential through the carboxy-terminal B(X)7B domains (Yang et al.,1993,1994). RHAMM has two additional B(X)7B domains within its NH2-terminal globular head [K40(X)7K48 and R67(X)7K75], which, like the carboxy-terminal domains, contain two internal basic residues. These domains have been identified as binding motifs for interphase and mitotic microtubules (Assmann et al., 1999). We have also shown that the Klp2 family contains putative B(X)7B HA binding domains that are virtually identical to those in RHAMM. For the Klp2 family, these domains are defined as a BZIP motif that allows for centrosomal localization through an indirect interaction with the dynein/dynactin motor complex. We suggest that in addition to their participation in HA binding (Yang et al., 1993), the carboxy-terminal B(X)7B domains of RHAMM are vital components of a consensus BZIP domain that targets this protein to the centrosome and allows for an interaction with the dynein–dynactin complex.
As an acidic (pI = 5.64), extensively coil coiled centrosomal protein that maintains mitotic spindle integrity and is chromosomally located proximal to the FGFR4 locus, RHAMM is a good candidate to be the fourth member of the TACC family. However, RHAMM does not contain the highly conserved, carboxy terminal, centrosomal targeting TACC domain. In fact, RHAMM uses a carboxy-terminal BZIP motif (L-X6-L-X6-L-X6), preceded by lengthy coil coiled structure, to localize to centrosomes. Interestingly, many centrosomal targeting domains may be defined by coiled coil structure terminating in a BZIP motif. Examination of the alignments of two defined centrosomal targeting domains, the TACC and PACT domain protein, reveals carboxy-terminal conservation of leucine residues (our unpublished data). Interestingly, Xenopus and human NuMA, which also interact with the dynein motor complex, contain a stretch of conserved leucines, L209-X9-L-X3-L-X9-L-X3-L-X6-L, that precedes an extensive predicted coiled coil structure (our unpublished data). Recent sequence analysis predicts that NuMA may interact with dynactin through a predicted CH domain (aa 110–210) interaction with Arp 1 (Novatchkova and Eisenhaber, 2002); interestingly, the conserved BZIP motif outlined above lies at the terminus of the predicted CH domain. Moreover, a direct interaction between EB1 and the dynactin subunit p150glued has recently been localized to the carboxy terminus of EB1, which contains a BZIP motif (Askham et al., 2002). Thus, evolutionary pressures may have allowed for divergence among centrosomal targeting domains, and dictated conservation of coiled coil structure terminating, or initiating, in a BZIP motif. RHAMM is unique to the human TACC family in its ability to interact with the dynein motor complex. To date, no association has been demonstrated between TACC1, 2, or 3 and the dynein motor complex, although it has been suggested that D-TACC and Msps may be maintained in the vicinity of the spindle poles by the activity of microtubule–motor complexes such as NuMA–dynein–dynactin (Lee et al., 2001). The divergence of the carboxy-terminal coiled coil domain of RHAMM from the TACC domain may have enabled a RHAMM–dynein interaction, allowing for RHAMM-specific mitotic functions. Thus, RHAMM may be a new member of the TACC family or there may be another, yet to be identified, TACC protein located at 5q35.1. Currently, we conclude that RHAMM is a TACC-like protein and may, in fact, be TACC4.
Because RHAMM can interact with microtubules both directly and indirectly, through an interaction with dynein, RHAMM may function in the maintenance of mitotic spindles by cross-linking centrosomal microtubules. A similar function has been proposed for NuMA, although NuMA has also been shown to affect spindle focusing as well as integrity. Because RHAMM (85 kDa) is much smaller than NuMA (240 kDa), RHAMM–dynein complexes may function proximal to the centrosome compared with NuMA–dynein complexes. Thus, in RHAMM immunoblocked cells, NuMA–dynein complexes may maintain spindle focusing but, in a quarter of injected cells, not spindle integrity. In fact, NuMA is not displaced from the RHAMM-immunoblocked abnormal spindle poles. Endogenous RHAMM function may assist in spindle integrity and microtubule focusing, along with the minus-end directed KIN C motor HSET, in the absence of NuMA but be inadequate to maintain spindle focusing.
To date, it is unclear whether the interaction between RHAMM and the dynein motor complex is direct or indirect; the structural similarity of the carboxy terminus of RHAMM to Xklp2 would suggest that RHAMM, like Xklp2, is targeted to microtubule minus ends through an association with TPX2. The localization of RHAMM, as determined by immunofluorescence and GFP-RHAMMFL, mirrors that of TPX2 throughout mitosis (Gruss et al., 2002). GFP-hTPX2 overexpression, like GFP-RHAMMFL, initiates a prometaphase block and inhibition of TPX2 function, through microinjection of anti-TPX2 antibodies, resulted in abnormal mitoses in 37.8% of injected cells (Gruss et al., 2002). Interestingly, TPX2 has been recently shown to be essential for the spindle targeting of Aurora A kinase (Kufer et al., 2002). Thus, it is possible that RHAMM, like Aurora A kinase and Klp2, is dependent on TPX2 for spindle pole localization.
This work outlines an essential role for RHAMM in the organization and integrity of a bipolar spindle. The role of RHAMM in mitotic stability may partially explain its relationship to cancer. Overexpression of RHAMM, and its variants, characterizes both hematological malignancies and solid tumors (Crainie et al., 1999; Wang et al., 1998); additionally, RHAMM has been identified by SEREX (serological identification of antigens by recombinant expression cloning) analysis to be a tumor-associated antigen in colon cancer, acute myelogenous leukemia, and chronic myeloid leukemia (Greiner et al., 2002; Line et al., 2002). Autoantibodies to centrosomal proteins, such as pericentrin, are a hallmark of autoimmune disease, such as scleroderma (Doxsey et al., 1994;Rattner et al., 1998;Gavanescu et al., 1999). Moreover, NuMA has been investigated as a biomarker for colorectal cancer (Briggman et al., 1999). It is possible that the tumor-specific anti-RHAMM autoantibodies may be indicative of overexpression of RHAMM, or tumor-specific expression of RHAMM variants. Such RHAMM disregulation could potentially affect centrosomal and spindle pole dynamics with consequent dramatic, and possibly oncogenic, effects on cell shape, motility, and ploidy.
Acknowledgments
We thank Dr. V. Assmann for providing RHAMM/IHABP antibodies and Dr. D. Compton for providing TOGp and NuMA antibodies. This research was supported by the Canadian Institutes for Health Research. Funding from the University of Alberta Faculty of Medicine Endowed Ph.D. Studentship, Alberta Heritage Foundation for Medical Research, and the Natural Sciences and Engineering Research Council of Canada studentships supported C.A.M. Funding from the University of Alberta Faculty of Medicine and Dentistry 75th Anniversary Award, the Alberta Heritage Foundation for Medical Research, and the Canadian Institutes for Health Research studentships supported J.J.K. T.J.Y. is supported by National Institutes of Health grant GM-44762, PO1 CA75138, CORE grant CA06927, and an Appropriation from the Commonwealth of Pennsylvania. G.K.C. is supported by the Alberta Cancer Board, Alberta Cancer Foundation New Investigator Award, Petro-Canada Young Innovator Award, and the Canadian Institutes for Health Research.
Abbreviations used: BZIP, basic leucine zipper; FGFR, fibroblast growth factor receptor; GFP, green fluorescent protein; Hklp2, human kinesin-like protein 2; IHABP, intracellular hyaluronan binding protein; KRP180, kinesin-related protein 180; Msps, minispindles; NuMA, nuclear mitotic apparatus protein; NuB1, NuMA binding protein 1; RHAMM, receptor for hyaluronan-mediated motility; TACC, transforming acidic coiled coil; TPX2, targeting protein for Xklp2; TOGp, colonic and hepatic tumor overexpressed protein; Xklp2, Xenopus kinesin-like protein 2.
References
- Askham, J.M., Vaughan, K.T., Goodson, H.V., and Morrison, E.E. (2002). Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell 13, 3627-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, V., Jenkinson, D., Marshall, J.F., and Hart, I.R. (1999). The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J. Cell Sci. 112, 3943-3954. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, N., Wang, Z., Davitt, C., McKenzie, I.F., Xing, P.X., and Magnuson, N.S. (2002). Pim-1 associates with protein complexes necessary for mitosis. Chromosoma 111, 80-95. [DOI] [PubMed] [Google Scholar]
- Boleti, H., Karsenti, E., and Vernos, I. (1996). Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84, 49-59. [DOI] [PubMed] [Google Scholar]
- Briggman, J., et al. (1999). NuMA: evaluation of a new biomarker for the detection of low stage colorectal cancer. Anticancer Res. 19, 2411-2414. [PubMed] [Google Scholar]
- Brocchieri, L. (2001). Phylogenetic inferences from molecular sequences: review and critique. Theor. Popul. Biol. 59, 27-40. [DOI] [PubMed] [Google Scholar]
- Chen, H.M., Schmeichel, K.L., Mian, I.S., Lelievre, S., Petersen, O.W., and Bissell, M.J. (2000). AZU-1: a candidate breast tumor suppressor and biomarker for tumor progression. Mol. Biol. Cell 11, 1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A., Yen, T.J., and Cleveland, D.W. (1991). Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J. Cell Biol. 112, 1083-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crainie, M., Belch, A.R., Mant, M.J., and Pilarski, L.M. (1999). Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood 93, 1684-1696. [PubMed] [Google Scholar]
- Cullen, C.F., and Ohkura, H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637-642. [DOI] [PubMed] [Google Scholar]
- Dionne, M.A., Sanchez, A., and Compton, D.A. (2000). ch-TOGp is required for microtubule aster formation in a mammalian mitotic extract. J. Biol. Chem. 275, 12346-12352. [DOI] [PubMed] [Google Scholar]
- Doxsey, S.J., Stein, P., Evans, L., Calarco, P.D., and Kirschner, M. (1994). Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639-650. [DOI] [PubMed] [Google Scholar]
- Gaglio, T., Saredi, A., Bingham, J.B., Hasbani, M.J., Gill, S.R., Schroer, T.A., and Compton, D.A. (1996). Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135, 399-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gares, S.L., Giannakopoulos, N., MacNeil, D., Faull, R.J., and Pilarski, L.M. (1998). During human thymic development, β1 integrins regulate adhesion, motility, and the outcome of RHAMM/hyaluronan engagement. J. Leukoc. Biol. 64, 781-790. [DOI] [PubMed] [Google Scholar]
- Gavanescu, I., Vazquez-Abad, D., McCauley, J., Senecal, J.L., and Doxsey, S. (1999). Centrosome proteins: a major class of autoantigens in scleroderma. J. Clin. Immunol. 19, 166-171. [DOI] [PubMed] [Google Scholar]
- Gergely, F., Karlsson, C., Still, I., Cowell, J., Kilmartin, J., and Raff, J.W. (2000a). The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA 97, 14352-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely, F., Kidd, D., Jeffers, K., Wakefield, J.G., and Raff, J.W. (2000b). D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19, 241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., McLean, D., Descamps, S., Lee, M.J., Raff, J.W., Prigent, C., and Glover, D.M. (2002). Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156, 437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro, S. (2000). The PACT domain, a conserved centrosomal targeting motif in the coiled–coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, M., Howard, L., and Compton, D. (2001). Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152, 425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner, J., et al. (2002). Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp. Hematol. 30, 1029. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Sillje, H., Karsenti, E., Mattaj, I.W., and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. [DOI] [PubMed] [Google Scholar]
- Hall, C.L., et al. (1995). Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell 82, 19-26. [DOI] [PubMed] [Google Scholar]
- Hardwick, C., Hoare, K., Owens, R., Hohn, H.P., Hook, M., Moore, D., Cripps, V., Austen, L., Nance, D.M., and Turley, E.A. (1992). Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility [published erratum in J. Cell Biol. (1992) 118, 753]. J. Cell Biol. 117, 1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., and Merdes, A. (2002). Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115, 1815-1824. [DOI] [PubMed] [Google Scholar]
- Kufer, T.A., Sillje, H.H., Korner, R., Gruss, O.J., Meraldi, P., and Nigg, E.A. (2002). Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I. B., and Masatoshi, N. (2000). MEGA: Molecular Evolutionary Genetics Analysis, Version 2.0. Pennsylvania State University, University Park, PA, and Arizona State University, Tempe, AZ.
- Lee, M.J., Gergely, F., Jeffers, K., Peak-Chew, S.Y., and Raff, J.W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3, 643-649. [DOI] [PubMed] [Google Scholar]
- Line, A., Slucka, Z., Stengrevics, A., Silina, K., Li, G., and Rees, R.C. (2002). Characterisation of tumour-associated antigens in colon cancer. Cancer Immunol. Immunother. 51, 574-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, B.D., Li, X., Cattini, P.A., Turley, E.A., and Nagy, J.I. (2001). Identification of sequence, protein isoforms, and distribution of the hyaluronan-binding protein RHAMM in adult and developing rat brain. J. Comp. Neurol. 439, 315-330. [DOI] [PubMed] [Google Scholar]
- Masellis-Smith, A., Belch, A.R., Mant, M.J., Turley, E.A., and Pilarski, L.M. (1996). Hyaluronan-dependent motility of B cells and leukemic plasma cells in blood, but not of bone marrow plasma cells, in multiple myeloma: alternate use of receptor for hyaluronan-mediated motility (RHAMM) and CD44. Blood 87, 1891-1899. [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W.C., and Cleveland, D.W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J.D., and Cleveland, D.W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447-458. [DOI] [PubMed] [Google Scholar]
- Novatchkova, M., and Eisenhaber, F. (2002). A CH domain-containing N terminus in NuMA? Protein Sci. 11, 2281-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekorz, R.P., Hoffmeyer, A., Duntsch, C.D., McKay, C., Nakajima, H., Sexl, V., Snyder, L., Rehg, J., and Ihle, J.N. (2002). The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. EMBO J. 21, 653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarski, L.M., Maxwell, C.M., and Gares, S.L. (2001). RHAMM (CD168/IHABP). Protein Rev. 2, 76-84. [Google Scholar]
- Purohit, A., Tynan, S.H., Vallee, R., and Doxsey, S.J. (1999). Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147, 481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner, J.B., Mack, G.J., and Fritzler, M.J. (1998). Autoantibodies to components of the mitotic apparatus. Mol. Biol. Rep. 25, 143-155. [DOI] [PubMed] [Google Scholar]
- Spicer A.P., Roller, M.L., Camper, C.A., McPherson, J.D., Wasmuth, J.J., Hakim, S., Wang, C., Turley, E.A., and McDonald, J.A. (1995). The human and mouse receptors for hyaluronan-mediated motility, RHAMM, genes (HMMR) map to human chromosome 5q33.2-qter and mouse chromosome 11. Genomics 30, 115-117. [DOI] [PubMed] [Google Scholar]
- Steadman, B.T., Schmidt, P.H., Shanks, R.A., Lapierre, L.A., and Goldenring, J.R. (2002). Transforming acidic coiled-coil containing protein 4 interacts with centrosomal AKAP350 and the mitotic spindle apparatus. J. Biol. Chem. 15, 15. [DOI] [PubMed] [Google Scholar]
- Still, I.H., Hamilton, M., Vince, P., Wolfman, A., and Cowell, J.K. (1999a). Cloning of TACC1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene 18, 4032-4038. [DOI] [PubMed] [Google Scholar]
- Still, I.H., Vince, P., and Cowell, J.K. (1999b). The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics 58, 165-170. [DOI] [PubMed] [Google Scholar]
- Tatusova, T.A., and Madden, T.L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174, 247-250. [DOI] [PubMed] [Google Scholar]
- Turley, E.A. (1992). Hyaluronan and cell locomotion. Cancer Metastasis Rev. 11, 21-30. [DOI] [PubMed] [Google Scholar]
- Turley, E.A., and Torrance, J. (1985). Localization of hyaluronate and hyaluronate-binding protein on motile and non-motile fibroblasts. Exp. Cell Res. 1, 17-28. [DOI] [PubMed] [Google Scholar]
- Turley, E.A., Moore, D., and Hayden, J.J. (1987). Characterization of hyaluronate binding proteins isolated from 3T3 and murine sarcoma virus transformed 3T3 cells. Biochemistry 26, 2997-3005. [DOI] [PubMed] [Google Scholar]
- Turley, E.A., Austen, L., Vandeligt, K., and Clary, C. (1991). Hyaluronan and a cell-associated hyaluronan binding protein regulate the locomotion of ras-transformed cells. J. Cell Biol. 12, 1041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Thor, A.D., Moore, D.H., 2nd, Zhao, Y., Kerschmann, R., Stern, R., Watson, P.H., and Turley, E.A. (1998). The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin. Cancer Res. 4, 567-576. [PubMed] [Google Scholar]
- Wishart, D.S., Fortin, S., Woloschuk, D.R., Wong, W., Rosborough, T., Van Domselaar, G., Schaeffer, J., and Szafron, D. (1997). A platform-independent graphical user interface for SEQSEE and XALIGN. Comput. Appl. Biosci. 13, 561-562. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Boleti, H., Antony, C., Karsenti, E., and Vernos, I. (1998). Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143, 673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B., Zhang, L., and Turley, E.A. (1993). Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J. Biol. Chem. 268, 8617-8623. [PubMed] [Google Scholar]
- Yang, B., Yang, B.L., Savani, R.C., and Turley, E.A. (1994). Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J 13, 286-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A., Dictenberg, J.B., Purohit, A., Tuft, R., and Doxsey, S.J. (2000). Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol. Biol. Cell 11, 2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C. (2000). NuMA: a nuclear protein involved in mitotic centrosome function. Microsc. Res Tech. 49, 467-477. [DOI] [PubMed] [Google Scholar]
- Zimmerman, W., and Doxsey, S.J. (2000). Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic 1, 927-934. [PubMed] [Google Scholar]