Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation (original) (raw)
Abstract
MicroRNAs (miRNAs) are potent post-transcriptional regulators of protein coding genes. Patterns of misexpression of miRNAs in cancer suggest key functions of miRNAs in tumorigenesis. However, current bioinformatics tools do not entirely support the identification and characterization of the mode of action of such miRNAs. Here, we used a novel functional genetic approach and identified miR-221 and miR-222 (miR-221&222) as potent regulators of p27Kip1, a cell cycle inhibitor and tumor suppressor. Using miRNA inhibitors, we demonstrate that certain cancer cell lines require high activity of miR-221&222 to maintain low p27Kip1 levels and continuous proliferation. Interestingly, high levels of miR-221&222 appear in glioblastomas and correlate with low levels of p27Kip1 protein. Thus, deregulated expression of miR-221&222 promotes cancerous growth by inhibiting the expression of p27Kip1.
Keywords: cancer, genetic screen, miRNA, p27
Introduction
The p27Kip1 gene is a member of the Cip/Kip family of cyclin-dependent kinase (CDK) inhibitors that function to negatively control cell cycle progression (recently reviewed in Koff (2006)). It binds to CDK2 and cyclin E complexes to prevent cell cycle progression from G1 to S phase. p27Kip1 also acts as a tumor suppressor and its expression is often disrupted in human cancers. Studies in mice have shown that loss of p27Kip1 increases tumor incidence and tumor growth rate in either specific genetic backgrounds, or when mice are challenged with carcinogens (Fero et al, 1998). Decreased p27Kip1 levels have been correlated with tumor aggressiveness and poor patient survival (Ponce-Castaneda et al, 1995; Loda et al, 1997; Porter et al, 1997; Lu et al, 1999; Mineta et al, 1999; Migita et al, 2002).
Although p27Kip1 is characterized as a tumor suppressor, inactivating point mutations with loss of heterozygosity are rarely observed in human cancer. Therefore, the low levels of p27Kip1 protein observed in many aggressive types of cancer are likely to be mediated by other mechanisms (Ponce-Castaneda et al, 1995). The abundance of p27Kip1 protein is largely controlled through a variety of post-transcriptional regulatory mechanisms (Alessandrini et al, 1997; Kardinal et al, 2006; Chu et al, 2007; Grimmler et al, 2007), among which are sequestration by cyclin D/CDK4 complexes, accelerated protein destruction and cytoplasmic retention (Koff, 2006). In certain types of cancers, such as colorectal cancer, high expression levels of Skp2 and Cks1, specific p27Kip1 ubiquitin ligase subunits, were strongly associated with low p27Kip1 expression and aggressive tumor behavior (Hershko and Shapira, 2006). However, several studies have indicated that genes controlling the stability of p27Kip1 protein might not always account for its lower expression in cancer, and that p27Kip1 can also be regulated at the level of translation (Hengst and Reed, 1996; Millard et al, 1997; Chilosi et al, 2000).
MicroRNAs (miRNAs) are a class of small non-coding RNAs that function to control gene expression through association with the 3′-UnTranslated Region (3′UTR) of protein coding genes and subsequent induction of translation inhibition, which can also be associated with transcript destabilization (Bagga et al, 2005; Giraldez et al, 2006; Wu et al, 2006). miRNAs have been found to be implicated in a large variety of cellular processes, and their aberrant expression has been linked to disease (Kloosterman and Plasterk, 2006). Recently, research has uncovered both the tumor suppressive and oncogenic potential of a number of miRNAs, underscoring their importance in human cancer (Lu et al, 2005; He et al, 2005b; Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Kent and Mendell, 2006; Mayr et al, 2007). In particular, we have constructed and used a library of miRNA expressing vectors (miR-Lib; see Voorhoeve et al, 2006) to identify the oncogenic potential of the miR-372 family using a functional genetic approach (Voorhoeve et al, 2006). As p27Kip1 is mostly controlled at the post-transcriptional level and miRNAs are potent regulators of gene expression, we hypothesized a role for miRNAs in the contribution of cancer progression through the suppression of p27Kip1 expression.
Results
miR-221 and miR-222 are potent suppressors of p27Kip1 expression
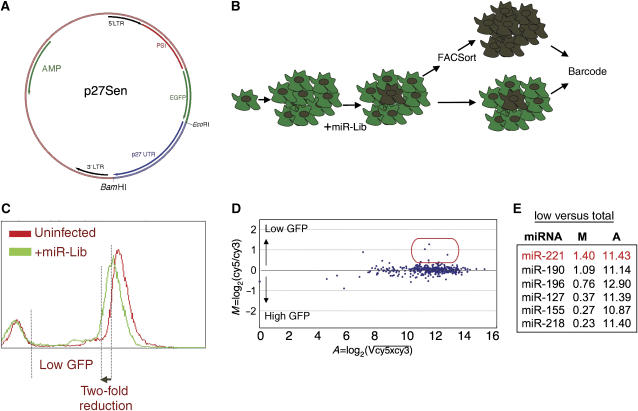
To identify miRNAs that control the expression of p27Kip1 through its 3′UTR, we constructed a retroviral sensor vector containing the GFP coding region upstream of the 3′UTR of p27Kip1 (p27Sen; Figure 1A). We transduced p27Sen into HeLa cells and expanded a single clone expressing GFP-p27-3′UTR (Figure 1B, data not shown). Subsequently, all miRNA expressing vectors from our miRNA expression library (miR-Lib; for details see Voorhoeve et al, 2006) were individually transduced and drug-selected to obtain resistant growing cells, each containing a unique integrated miR-Vec. Around 90% of vectors yielded stable clones. These cells were mixed into one pool and after 2 weeks of culturing, flow cytometry analysis revealed only a slight lower overall GFP signal in the miR-Lib-transduced cells when compared with the untransduced population (Figure 1C). This effect was most likely due to nonspecific promoter competition between miR-Vec and the sensor vector. Subsequently, from the total miR-Lib-transduced population, we sorted out the low-level GFP expressing cells (less than half of the fluorescence peak signal), extracted genomic DNA and compared the abundance of miR-Vec inserts between low and total GFP expressing populations in a barcode experiment using miR-Array (Figure 1D; for details see Voorhoeve et al, 2006). This analysis identified miR-Vec constructs that were enriched in the low GFP expressing cells (Figure 1E). miR-221 vector produced the most pronounced and reproducible effect.
Figure 1.
A genetic screen to identify miRNA suppressors of p27Kip1. (A) A schematic representation of p27Sen, a retroviral vector for stable expression of GFP under the control of p27-3′UTR. (B) A clonal population of GFP-p27-3′UTR expressing cells was subjected to transduction with the entire miRNA library (miR-Lib) in a single-well format, selected and then mixed for further analysis. Two independent experiments were performed. Subsequently, low expressing GFP cells were sorted and the abundance of miRNA inserts was compared to the total population using miR-Array. (C) A GFP expression profile of control and miR-Lib-transduced cells. The region marked ‘low GFP' was sorted out of the entire population of cells. (D) A representative MA plot showing the signal and change of signal of each miRNA insert. (E) The top six hits found enriched in the low expressing GFP cells.
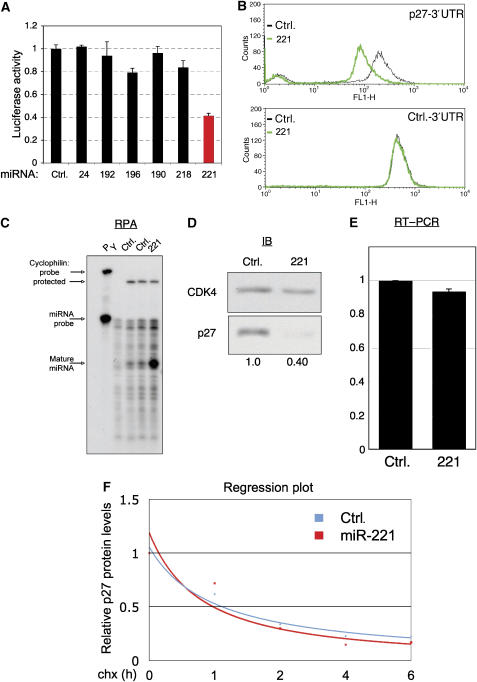
To verify and quantify the effect of each miRNA vector in the list of enriched miRNA vectors, we subcloned the p27-3′UTR downstream of luciferase. Analysis of transiently transfected cells revealed that only the miR-221 expressing vector significantly suppressed p27-3′UTR activity around 2.5-fold (Figure 2A). To further substantiate the specificity of p27-3′UTR-mediated suppression by miR-221, we stably introduced it into cells containing p27Sen or Control-Sen vectors. Consistent with the results above, we found that miR-221 expression suppressed GFP expression in p27Sen-transduced cells by 2.5-fold (Figure 2B). As the expression of GFP in the control-Sen-transduced cells remained unchanged, we concluded that miR-221 is a potential regulator of the 3′UTR of p27.
Figure 2.
miR-221 inhibits translation of p27Kip1. (A) Luciferase reporter experiments were performed with _Firefly_-luciferase-p27-3′UTR, control _Renilla_-luciferase and the indicated miRNA constructs. The luciferase ratio between the Firefly and Renilla of the control sample was adjusted to 1. A summary of three independent experiments is shown. (B) A stable p27Sen-HeLa cell line was transduced with miR-221 and control expressing vectors, and drug-selected for a week. Polyclonal cell populations were analyzed by flow cytometry a week later. (C) HeLa cells were transduced with control or miR-221 expressing vectors and drug-selected for a week. Subsequently, total RNA was extracted from the stable cells and RPA was performed with miR-221 and control cyclophilin probes. The lane P contains probes without RNase treatment, Y is a lane where yeast RNA was used as control. (D) An immunoblot analysis with p27 and CDK4 antibodies on the same cell populations as in panel B. Quantification was performed using Tina 2.0 software. (E) QRT-PCR was performed on the same RNA extracts used in panel C. (F) HeLa cells stably expressing control or miR-221 vectors were treated with 100 μg/ml cyclohexamide. At the indicated time points, whole-cell extracts were prepared and analyzed by immunoblot with p27 and control tubulin antibodies. Band intensities were quantified using Tina 2.0 software, and the resulting p27-tubulin ratios plotted in a regression plot.
We next examined the expression of miR-221 and its effect on endogenous p27Kip1 expression. By RNase protection assay (RPA), we detected low miR-221 level in HeLa cells and found potent expression of miR-221 from its vector (Figure 2C). By immunostaining, we found the endogenous p27Kip1 protein level to be 2.5-fold lower in HeLa cells stably expressing miR-221, compared with control cells (Figure 2D). Intriguingly, by quantitative RT–PCR, we found that the mRNA level of p27 remained unaltered in the miR-221-transduced cells, indicating that miR-221 controls p27Kip1 translation but not mRNA stability (Figure 2E). To rule out any miR-221-mediated effect on p27 protein stability, a cyclohexamide experiment was performed (Figure 2F; Supplementary Figure S3). Although miR-221 expressing HeLa cells have lower p27 protein levels, the half-life of p27 was comparable to control cells. Therefore, we conclude that miR-221 expression does not change the rate of p27 decay.
p27Kip1 protein level changes during cell cycle progression, accumulating when cells progress through G1 and sharply decreasing just before cells enter S phase (Kaldis, 2007). Additionally, p27Kip1 protein levels rise when cells exit cell cycle to G0, and decreases when cells enter the cell cycle again (Kaldis, 2007). These alterations in p27Kip1 levels are mainly caused by regulation at the protein degradation level (Alessandrini et al, 1997; Kardinal et al, 2006; Chu et al, 2007; Grimmler et al, 2007). To examine the effect of miR-221 on p27Kip1 levels during the cell cycle, we blocked cells in mitosis and then released them to enter G1 and S phases. Immunoblot analysis revealed that while a global reduction in p27Kip1 was observed in miR-221 cells compared with control, still, a similar relative increase in p27 levels during G1 and decrease just before S phase were seen in both cell types (Supplementary Figure S1A). As expected from p27 function in G1, its lower levels in miR-221 expressing cells resulted in a faster entry of cells into S phase, while entering into G1 was unaffected (Supplementary Figure S1B). Similarly, the exit of cells from the cell cycle and the relative accumulation of p27 during this process were not affected by miR-221 in primary human cells (data not shown). Thus, we identified miR-221 as a suppressor of endogenous p27Kip1 expression.
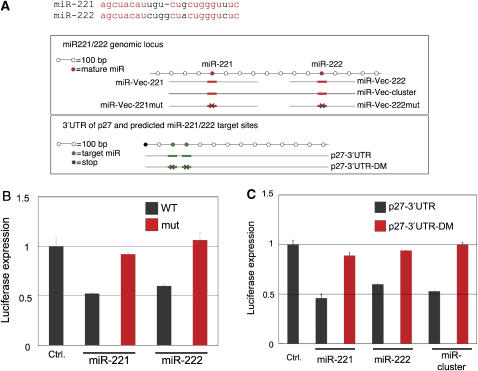
miR-221 is part of a gene cluster also expressing miR-222, a close homologue of miR-221. Both miRNAs share an identical seed sequence and are predicted (by PicTar and TargetScanS) to bind to p27-3′UTR at two sites (Figure 3A). Our library already contained a vector designed to express miR-222. However, due to a low virus titer, this vector was among the 10% of unsuccessful stable clones in our screen (data not shown). To examine the effectiveness of miR-222, as well as the specificity of both miR-221 and miR-222, we constructed seed-mutated miR-221 and miR-222, as well as a cluster vector. RPA analysis verified the respective expression and lack of expression of wild type and mutated miR-221 and miR-222 constructs (data not shown). As expected from their seed sequence identity, luciferase reporter assays revealed a similar suppressive activity of miR-222 toward p27-3′UTR as miR-221 (Figure 3B). Importantly, mutating the seed sequence of miR-221 and miR-222 completely abolished their suppressive activity, indicating that the expression of the miRNAs is responsible for p27 suppression. In line with these experiments, the introduction of a cluster containing vector that directs the expression of both miRNAs gave a similar suppressive activity (Figure 3C).
Figure 3.
Specificity of p27Kip1 inhibition by miRNA-221 and miR-222. (A) A schematic representation showing the mature miR-221 and miR-222 sequences (in red, identical sequences). In addition, the genomic locus of miR-221&222 cluster with the inserts used in the miRNA vectors is also shown. Red boxes represent miRNA precursor positions. Also, the 3′UTR of p27Kip1 is shown. The green boxes represent the two predicted miR-221&222 targeting sequences (as predicted by Pictar and TargetScanS software). (B) Luciferase reporter experiments were performed as in Figure 2A. 221SM and 222SM are constructs in which the seed sequence of the miRNAs was altered. (C) Luciferase reporter experiments were performed as in Figure 2A. p27-3′UTR-DM (double mutant) is a construct where both predicted miR-221&222 targeting sequences were modified (see Materials and methods). (B, C) The histograms show a summary of the results of three independent experiments.
The 3′UTR of p27 contains two predicted miR-221 and 222 target sequences. We have mutated these predicted sites in order to examine their requirement for miR-221 and miR-222 (miR-221&222) function, using luciferase reporter assays (Figure 3A). We found that the mutated p27-3′UTR (DM) was completely refractory to the miR-221- and miR-222-suppressive effect (Figure 3C). As each single mutant retained at least some sensitivity to miRNAs 221&222 (data not shown), this result indicates that both predicted sites are required for miR-221&222 effects. In summary, we have identified and verified miR-221&222 as potent suppressors of p27Kip1 expression.
miR-221&222 activity is required for cancer cell proliferation
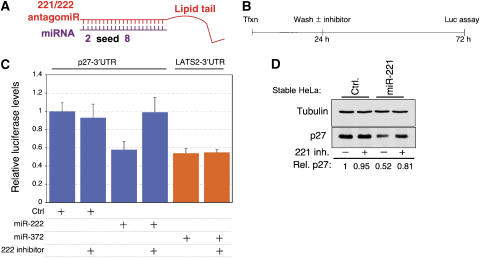
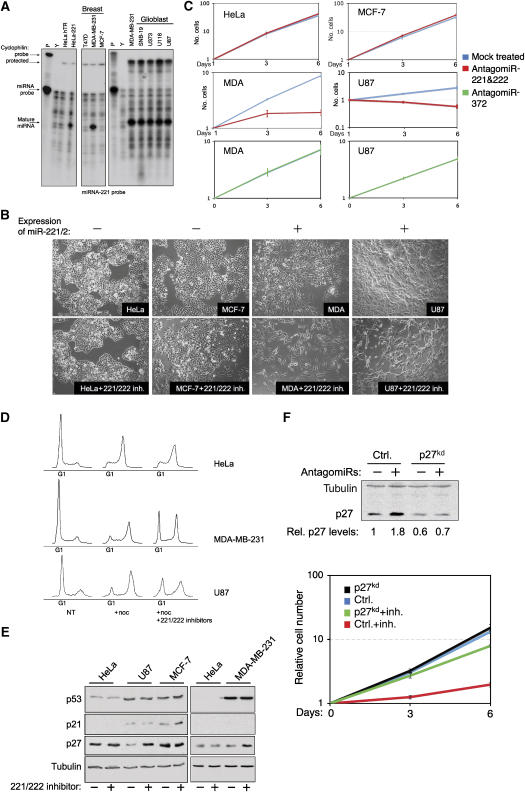
Intriguingly, three papers reporting the differential expression of miRNAs in primary glioblastomas, papillary thyroid carcinomas and pancreas tumors, describe the upregulation of miR-221 as part of a cancer signature (Ciafre et al, 2005; He et al, 2005a; Pallante et al, 2006; Lee et al, 2007). In accordance with their findings, our data suggest that these miR-221&222 expressing tumors might depend on p27Kip1 downregulation for their survival. To examine this issue, we generated antagomiR-221&222 molecules, antisense RNA oligos containing a cholesterol molecule at their 5′ end that are 2′-_O_-methylated at every nucleotide (Figure 4A, see also Materials and methods for details). These molecules were previously shown to be capable of passing through cellular membranes to inhibit miRNA action by sequestering it from its targets (Krutzfeldt et al, 2005). To test antagomiR function, we added antagomiR-222 to the culture medium of cells that were cotransfected 24 h earlier with luciferase-p27-3′UTR and either miR-222 or control expressing constructs (Figure 4B). Figure 4C shows that while the expected reduction (∼2-fold) in luciferase expression was seen when miR-222 was cotransfected with p27-3′UTR, the luciferase level remained as high as in the control cells in the presence of antagomiR-222, indicating a complete blockade of miR-222 activity. To confirm specificity, we also added antagomiR-222 to cells transfected with constructs expressing miR-372 and its target luciferase-LATS2-3′UTR (see Voorhoeve et al, 2006). In this case, miR-372 remained fully active in the presence of the antagomiR, indicating the specific inhibitory effect of antagomiR-222 on miR-222 (Figure 4C). Similar results were obtained with antagomiR-221 (data not shown). To support these data, we also tested the effect of antagomiR-221 on HeLa cells that have stable expression of miR-221. Figure 4D shows that the addition of antagomiR-221 to cells containing miR-221 restored p27Kip1 protein level to almost its normal level. As a result of antagomiR-221&222 administration, we detected lower levels of each of the mature miRNAs (Supplementary Figure S5). Thus, these results establish the specific and powerful inhibitory effect exerted by antagomiR-221 and antagomiR-222 on their corresponding miRNAs. Equally important, these observations also demonstrate the tight interaction between miR-221&222 activity and p27Kip1 gene expression.
Figure 4.
Inactivation of miR-221&222 with corresponding antagomiRs. (A) A diagram showing the design of antagomiR-221 and antagomiR-222. (B, C) MCF-7 cells were transfected with the indicated luciferase reporter constructs, as in Figure 2A. Twenty-four hours following transfection, antagomiR-222 (25 μg) was added to the culture medium. Luciferase activity was measured 48 h later. Results show a summary of three independent experiments. (D) Stable control and miR-221 expressing HeLa cell populations were treated with antagomiR-221 for a period of 24 h. Whole cell extracts were prepared and analyzed by immunoblot with p27 and control tubulin antibodies. Bands were quantified using Tina 2.0 software.
Using antagomiR-221&222, we set out to test the relationship between miR-221&222, p27Kip1 and cell proliferation. We first examined several breast and glioblastoma cancer cell lines for miR-221&222 expression (Figure 5A). To explore the effect of miR-221&222 antagomiRs on cellular proliferation, we selected two cell lines that showed endogenous expression of miR-221&222 (U87 and MDA-MB-231), and two negative cell lines (MCF-7 and HeLa). Interestingly, treatment of U87 and MDA-MB-231 with both antagomiR-221&222 resulted in a clear proliferation arrest phenotype, which was accompanied by a significant reduction in detectable miRNA levels (Supplementary Figure S5). However, no significant effect on proliferation was observed in treated MCF-7 and HeLa cell lines (Figure 5B). We verified and quantified the proliferation block using a 3T3 cell growth protocol (Figure 5C). Here, we observed that while treated HeLa cells and MCF-7 cells continued to proliferate indistinguishably from mock treated cells, both U87 and MDA-MB-231 cells ceased proliferating almost completely and very rapidly following treatment. The arrest observed in U87 and MDA-MB-231 required simultaneous addition of both antagomiRs, as the addition of only one antagomiR (221 or 222) or a control antagomiR designed to inhibit miR-372 was not sufficient to affect cellular growth (Figure 5C and data not shown). This indicates a functional overlap between miR-221 and miR-222 in controlling proliferation. Flow cytometry analysis showed that the antagomiR-221&222-induced proliferation arrest observed in the miR-221&222 expressing cell lines was due to a block in G1, which is consistent with the role of p27Kip1 in controlling progression through G1 phase of the cell cycle. As observed above, miR-221&222 are regulators of p27Kip1. Interestingly, when p27Kip1 levels were examined by immunostaining, only the cells containing miR-221&222 (U87 and MDA-MB-231) showed increased levels of the cell cycle inhibitor p27Kip1 after administration of the antagomiRs (Figure 5E). Furthermore, we found that regulation of p27 by miR-221&222 is a general feature, as inhibition of endogenously expressed miR-221&222 in BJ human primary fibroblasts by antagomiRs also resulted in elevation of p27 protein levels (Supplementary Figure S4). The effect on p27Kip1 was specific, since no change was observed in p21Cip1, a stress-associated cell cycle inhibitor and p27's closest homologue, or in p53, a major tumor suppressor gene. Moreover, the induction of p27 by antagomiRs-221&222 was direct, as this was observed as early as 4 h following treatment, and preceded any detectable changes in cell cycle profile (Supplementary Figure S2). To examine whether the inhibition of p27Kip1 is essential for the effect of miR-221&222 on cell proliferation, we studied U87 cells treated with antagomiR-221&222 when p27 levels were suppressed using an shRNA vector targeting p27Kip1 (p27kd). Figure 5F shows that while the proliferation of control U87 cells was efficiently blocked when miR-221&222 activity was inhibited, p27kd cells continued to proliferate to a comparable extent as untreated cells (green line). No significant effect of p27kd was seen in the mock-treated cells (compare black and blue lines). These results demonstrate the causal relationship between miR-221&222 and cell proliferation. In certain cancer cell lines, the expression of miR-221&222 is required for maintaining low p27Kip1 protein levels and continuous cellular growth.
Figure 5.
Oncogenic addiction of certain cancer cell lines to miR-221&222 activity. (A) RPA was performed with miR-221 probe using total RNA from the indicated cell lines. Sizes of probes and mature miR-221 are indicated. (B) The indicated cancer cell lines were grown in the presence or absence of a mixture of antagomiR-221 and anagomiR-222 oligos. Images were obtained 4 days after treatment. (C) The growth of the indicated cancer cell lines, either exposed or not to antagomiR treatment, was measured using a 3T3 protocol. (D) The indicated cell lines were either treated or not (NT, non-treated) with a mixture of antagomiR-221 and antagomiR-222 oligos. Forty-eight later, cells were split and nocodazole was added (+Noc). Flow cytometry analysis was performed 24 h later. (E) Cells were treated as in panel B, whole cell lysate was extracted 3 days after treatment and subjected to immunoblot analysis with antibodies to detect the indicated proteins. (F) U87 cells were transfected with either control or a p27-shRNA expressing construct. Efficiency of transfection was at least 80%, as determined by GFP treatment (data not shown). After 24 h, cell populations were treated with a mixture of antagomiR-221 and antagomiR-222 and proliferation was determined as in panel C. An immunoblot analysis for p27Kip1 and control tubulin proteins was performed on wild-type and p27kd-transfected U87 cells. Bands were quantified using Tina 2.0 software.
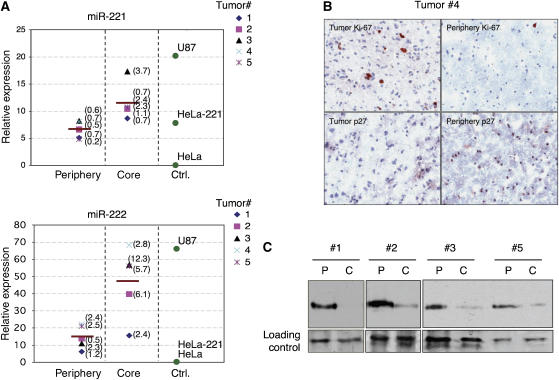
Next, we searched for evidence for this mechanism in human cancer. It was recently shown that high level of miR-221&222 is part of a signature for poor prognosis in glioblastomas (Ciafre et al, 2005). We therefore compared the expression levels of miR-221&222 and p27Kip1 in grade 3 tumors and peripheral tissues from 5 glioblastoma patients. In quantitative RT–PCR, five tumors (#1–5) showed miR-221&222 expression that was significantly higher in the core tumor region when compared with the peripheral normal region (Figure 6A). As controls, we used U87 and HeLa, which are miR-221&222-positive and -negative cell lines, respectively, as well as HeLa-221, a cell line that was engineered to express miR-221 only. Interestingly, the elevated level of miR-221&222 in tumors #1, 2, 3, 4 and 5 was correlated with reduced expression of p27Kip1. In immunohistochemistry, p27Kip1 staining was high and nuclear in peripheral tissue, whereas only a very weak staining was seen in the core of the tumor (Figure 6A). As control, we stained for Ki-67, a marker for cell proliferation expressed in many tumors. Additionally, using immunoblot analysis of whole-cell extracts, we detected lower levels of p27Kp1 protein in the core of tumors #1, 2, 3 and 5 compared to their periphery controls (Figure 6C). These results suggest that in a large proportion of glioblastoma tumors, elevated levels of miR-221&222 are correlated with low levels of its target p27Kip1. Altogether, our results propose a mechanism by which deregulated expression of miR-221&222 promotes cancerous growth by inhibiting the expression of p27Kip1 in human tumors.
Figure 6.
Deregulated miR-221 and miR-222 expression in glioblastomas correlate with p27Kip1 levels. (A) QRT-PCR for the expression of miR-221 and miR-222 was performed on RNA extracted from five tumor frozen tissues and five frozen peripherial tissue (0.5 cm from core tumor). As controls, we used the miR-221&222-negative HeLa cells, HeLa with miR-Vec-221 (HeLa-221) and U87, which are positive for both miRNAs. In parentheses are the standard deviations of three experiments. (B) Core and periphery material of tumor #4 was stained brown with Ki-67 and p27 antibodies. Nuclei were stained blue. (C) Whole-cell extracts were obtained from core (C) and periphery (P) material of tumors #1, 2, 3 and 5. A 20 μg weight of each sample was loaded on gel and subjected to either immunostaining with p27 antibody or silver staining, to demonstrate equal loading.
Discussion
Perhaps the main challenge in the identification and characterization of miRNA function is the gain of a comprehensive understanding of their effect on the expression of target mRNAs and the resulting outcome on relevant cellular pathways and phenotypes. Until now, only a handful of comprehensive studies on miRNA function have been completed. One major reason for this is the prominent effect of miRNAs on protein translation, an effect that still cannot be quantified genome wide. A second reason is that each cellular phenotype exhibits a different sensitivity to changes in protein levels and pathway activity, due to complex and robust network connections. This individual protein-to-phenotype behavior prompted us to test the effectiveness and activity of miRNAs on a single gene of interest in an unbiased manner. We therefore designed a novel genetic screen to experimentally identify potent miRNA effectors for a single gene. We exploited a miRNA expression library to select for miRNAs that target p27Kip1, a gene whose tumor suppressor activity correlates with its cellular protein levels. In contrast to genes such as p53 and pRb, whose complete loss is required to inactivate the pathway, the tumor suppressor activity of p27Kip1 is haplo-insufficient. Our results uncover miR-221 and miR-222 as endogenous regulators of p27Kip1 and demonstrate the importance of its genetic interaction for the proliferation of certain cancer cells. Notably, this genetic screening method can be used for the identification and characterization of miRNAs targeting other tumor suppressors and oncogenes.
miR-221&222 and p27Kip1
From about 400 miRNAs in our library, we have identified one family that significantly controls p27Kip1 expression. As many predicted p27Kip1-targeting miRNAs showed no significant effect on p27Kip1 protein level, and since no other miRNA vector was reproducibly enriched in our screen, this suggests that targeting p27Kip1 is specific to the miR-221 family. However, since endogenously expressed miRNAs might obscure effects on the reporter gene induced by our miR-Vectors, it does not rule out other miRNAs targeting p27. Moreover, additional miRNAs regulating p27 might not be detected by the developed method, due to either insensitivity to detect weak regulation of the miRNA toward the target, or inefficient processing of the overexpressed miRNA. Still, the finding that p27 is regulated by miR-221&222 proves that this method is a powerful tool for identifying target-specific miRNAs.
An additional interesting point is how many other relevant targets do miR-221&222 have in our cell system? Previously, it was shown that c-kit is a target of miR-221&222 (He et al, 2005a). However, c-kit expression could not be detected in the cell lines we used. At the moment, there are no genome-wide methods that are sensitive and quantitative enough to identify miRNA effects on the protein level. Thus, how many additional targets miR-221&222 have in our cell system and which of them is required for their oncogenic function are interesting questions that remain to be explored.
Relationship between miRNAs and mRNA
Using target prediction algorithms and large-scale mRNA expression arrays, the current view of miRNA function suggests that miRNAs possess the potential to affect the expression of hundreds of target genes. Our results indicate a highly specific and influential effect of very few miRNAs on a protein-coding gene of interest. These views are not contradictory, as very few miRNA-targets, once downregulated, have the capacity to influence a certain phenotype of interest in a given cellular environment. Second, we should not exclude the possibility that many predicted miRNA targets are indifferent to miRNA activity simply because their gene expression is robust due to negative feedback loops. Thus, we speculate that a given phenotype can be affected by the activity of at least some miRNAs through the suppression of only few targets.
Oncogenic addiction of several cancer cell lines to miR-221&222
Based on our data, we can conclude that regulation of p27 by miR-221&222 is a common phenomenon, as p27 protein levels are regulated by these miRNAs in both cancerous and primary cells. Therefore, we would predict that the upregulated expression of the miR-221&222 cluster in human tumors would decrease p27 levels and as such will correlate with poor prognosis. Indeed, aberrant expression of miR-221&222 was observed in poor-prognosis miRNA signatures in tumors such as papillary thyroid carcinoma (He et al, 2005a; Pallante et al, 2006), pancreatic adenocarcinoma and glioblastoma (Ciafre et al, 2005). Our results show that persistent expression of miR-221&222 is required for the abnormal cellular proliferation of at least some tumor cell lines, in particular glioblastoma. We find that in high proportion of glioblastoma cancers, high level of miR-221&222 in the tumor is associated with lower level of p27kip1. In combination with our tissue culture results, our data suggest that miR-221&222 function as oncogene by controlling cell cycle progression through inhibition of p27kip1. The requirement of miR-221&222 for tumor survival may suggest that it might be possible to use antagomiR-221&222 as a form of cancer therapy.
Materials and methods
Constructs and inhibitors
Genomic DNA was isolated using the QIAamp DNA mini kit (Qiagen), according to the manufacturer's instructions.
P27Sen-GFP was constructed by deleting the MCS, PGK-promoter and blasticidin resistance marker from pMSCV-blast, followed by insertion of GFP-MCS from pEGFP-C1 (Clontech). The 3′UTR of p27 was synthesized from genomic DNA using forward GCGAATTCttaaACAGCTCGAATTAAGAATATGTTTCC (which includes an in-frame stop codon for GFP) and reverse GCGGATCCGCTATGGAAGTTTTCTTTATTGATTACTTAA TGTG primers. The resulting PCR product was cloned downstream of the GFP gene.
P27Sen-Luc was prepared by cloning the 3′UTR of p27 from the GFP sensor vector into pGL3 (Promega) downstream of the luciferase gene. For creating pGL3-p27-3′UTR-DM (double mutant), a PCR was performed using the following primer: CCCCAAAGTTTATGTGGGATCCAAAAGGTAAAAACTATA TACACAGGTAGTACAATGAAGCAAATAAGGAAAAACCTA ATTGCATAATGGGATCCCCAACGCTTTTAGAGGC. In red are the _Bam_HI sites that substituted the two seed sequences.
The miR-Vec-221/222 and MSCV-hTR constructs were prepared as previously described (Kedde et al, 2006).
The miR-Vec-221/222 seed mutants were prepared in the same manner as described above for creating the double mutated p27 3′UTR. In this case, the 221/222 seed sequences were replaced by _Hin_dIII sites (underlined):
221: GAAACCCAGCAGACAAAGCTTTGTTGCCTAACGAAC
222: GAGACCCAGTAGCCAGAAGCTTTGCTGATTACGAAAG
Sequences of the miR-221 and miR-222 antagomiRs (Dharmacon):
221: Chl-AGCCUGAAACCCAGCAGACAAUGUAGCUGUUGCC
222: Chl-CAUCAGAGACCCAGUAGCCAGAUGUAGCUGCUGA
The p27kd was designed to target the sequence gtacgagtggcaagaggtg (Le et al, 2003), and was prepared as described in Brummelkamp et al (2002).
RPA
RPA was performed using mirVana miRNA probe construction and detection kits (Ambion), according to the manufacturer's instructions. A 2.5 μg weight of total RNA was used for each reaction. The primer sequence of the 221 RPA probe was GCAACAGCTACATTGTCTGCTGGGTTTCAGGCTcctgtctc
The antisense cyclophilin probe contained nucleotides 46–149 with accession no. BC013915 (Voorhoeve et al, 2006).
Western blot analysis and antibodies
Proteins were analyzed by SDS–PAGE and Western blotting was performed using Immobilon-P transfer membrane (Millipore). Before staining, unspecific sites were blocked in 5% non-fat milk at room temperature for 1 h. Antibodies used were p21 (sc-6246, Santa Cruz Biotechnology), p53 (sc-126, Santa Cruz Biotechnology), p27 (K25020, BD Transduction Laboratories), CDK4 (sc601, Santa Cruz Biotechnology) and tubulin (YL1/2, ECACC). Secondary antibody was goat-anti-mouse-HRP (DakoCytomation). Proteins were detected using ECL (Amersham Biosciences) and film (Kodak).
Luciferase assay
MCF-7 cells were transfected using PEI (Polysciences Inc.). For reporter assays, cells were cultured in 24-well plates and transfected with 5 ng pGL3-p27-3′UTR (or DM), 5 ng Renilla and 0.5 μg of the miR-Vec of interest (WT or SM). Luciferase activity was measured 72 h after transfection using the Dual-luciferase reporter assay system (Promega).
Tissue procurement
Tissue samples were obtained after informed consent from adult patients diagnosed with glioblastoma de novo, freshly resected during surgery and immediately frozen in liquid nitrogen for subsequent total RNA extraction. In order to yield a very specific, case by case matching pair of tumor and control sample, for each patient, we resected a central tumor area (core), surgically and histopathologically recognized as frankly tumoral, and one or more samples from a peripheral glial area (periphery), at an average distance of 0.5 cm from the border of the enhanced tumor, which did not show any evidence of tumor presence, by macroscopical surgeon's evaluation.
Tissue lysates were prepared with ELB lysis buffer (0.1% NP-40, 125 mM NaCl, 50 mM HEPES (pH 7.4) supplemented with protease inhibitor cocktail set (Roche Diagnostics GmbH). A 20 μg weight of total protein was used to detect p27 levels.
Cell culture
HeLa, MCF-7, MDA-MB-231 and U87 were cultured in Dulbecco's modified Eagle's medium (41966, Invitrogen) supplemented with 10% FCS and antibiotics (complete medium). For miR-221/222 inhibitor treatment, cells were grown to 50% confluency and the tissue culture medium was replaced with medium containing either 25 μM (24 wells) or 75 μM (six wells) of each of the inhibitors.
All miRNA transfection and virus harvesting steps were carried out on a Hamilton ML STAR with 96-channel and eight-channel pipetting systems (Hamilton Bonaduz AG, Bonaduz, Switzerland). Protocols were developed at the NKI using Hamilton STAR Software 3.2. Plates were incubated in a Cytomat 2C450 automated tissue culture plate incubator (Thermo Electron Corporation, Asheville, NC, USA). After transfection, the medium was removed using Biotek ELX405 Select plate washer (Biotek Instruments Inc., Winooski, VT, USA). The methods were completely automated; a Hamilton SWAP robotic arm was used to transfer plates between the various instruments.
Immunoperoxidase staining
Serial frozen sections (5 μm) were fixed in acetone for 10 min. Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide/100% methanol for 20 min. Sections were washed in phosphate-buffered saline (PBS) and blocked with 4% BSA in PBS for 30 min. Primary antibodies were diluted (p27 (BD Transduction Laboratories), 1:500; Ki-67 (Dako), 1:4000) in PBS containing 1% BSA, and 100 μl was added to each section. Incubation was carried out for 2 h at room temperature in a humidified chamber. The slides were then washed in PBS. For p27, sections were incubated with biotinylated goat anti-mouse secondary antibody (Dako) at 1:50 in PBS containing 1% BSA for 30 min at room temperature. After washes in PBS-Tween, ABC biotinylation complex (1:200, Dako) was added for 1 h at room temperature. Then, AEC (Sigma) was added to the slide for 20 min. For Ki67, sections were incubated with envision goat-anti-rabbit (1:200, Dako) for 30 min at room temperature. Then, AEC (Sigma) was added to the slide for 20 min.
3T3 cell growth protocol
A total of 3 × 104 cells were plated in triplicates in 24-well plates and propagated in complete medium with or without 221/222 inhibitors (2.5–25 μM) for a week. After 3 days, cells were counted and replated at 30 000 cells to grow for an additional 3 days.
RT–PCR and real-time TaqMan PCR
Total RNA was extracted from HeLa cells stably expressing either miR-221 or a control vector using TRIzol reagent (Life Technologies), according to the manufacturer's instructions. RNA was resuspended in DEPC-treated H2O. Synthesis of cDNA with Superscript III reverse transcriptase (Invitrogen) was primed with oligo(dT). Primers for p27 (Set 1: Fwd AGCGGAGCAATGCGCAGG and Rev TCTTCTGAGGCCAGGCTTCT Set 2: Fwd ACGATTCTTCTACTCAAAACAAAAGAGC and Rev ATTTGGGGAACCGTCTGAAA) and _β_-actin (Fwd CCTGGCACCCAGCACAAT and Rev GGGCCGGACTCGTCATACT) were designed to amplify 100–200 bp fragments. Analyses were carried out using SYBR Green PCR master mix (Applied Biosystems) and ABI Prism 7000 (Amersham-Pharmacia). Results were normalized with respect to _β_-actin expression. _C_t values for gene expression were calculated according to the _C_t method.
TaqMan® microRNA assays (Applied Biosystems) that include RT primers and TaqMan probes were used to quantify the expression of mature miRNA-221 (AB: 4373077) and miRNA-222 (AB: 4373076), in both tissue samples and cell lines. The mean _C_t was determined from triplicate PCRs. Gene expression was calculated relative to 18S rRNA (AB: 4333760F) and multiplied by 104 to simplify data presentation.
Flow cytometry
GFP detection For the validation of miRNA hits, HeLa GFP-p27-3′UTR cells were made to stably express these miRNAs. Each of the cell lines was then analyzed for the expression of GFP using FACScan (Becton Dickinson).
GFP sorting Low GFP expressing miR-Lib containing cells were separated by cell sorting using the FACSAria cell sorter from Becton Dickinson.
Cell cycle profile analysis
For the cell cycle experiments, cells were arrested in mitosis using 0.25 μg/ml nocodazole.
Cells were trypsinized, collected by centrifugation and resuspended in PBS containing 0.6% NP-40, 50 μg/ml RNaseA and 50 μg/ml propidium iodide for 10 min. Subsequently, cells were analyzed using FACScan (Becton Dickinson). In each assay, 10 000–100 000 cells were collected by FACScan and analyzed with Cell Quest software (Becton Dickinson).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends
Acknowledgments
We thank Ron Kerkhoven and Mike Heimerikx for technical assistance in using the array facility, Anita Pfauth and Frank van Diepen in assisting with flow cytometry and Roderick Beijersbergen for setting the high-throughput screening facility. This work was supported by grants from the Dutch Cancer Society (KWF) and the European young investigator program (EURYI).
References
- Alessandrini A, Chiaur DS, Pagano M (1997) Regulation of the cyclin-dependent kinase inhibitor p27 by degradation and phosphorylation. Leukemia 11: 342–345 [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Chilosi M, Chiarle R, Lestani M, Menestrina F, Montagna L, Ambrosetti A, Prolla G, Pizzolo G, Doglioni C, Piva R, Pagano M, Inghirami G (2000) Low expression of p27 and low proliferation index do not correlate in hairy cell leukaemia. Br J Haematol 111: 263–271 [DOI] [PubMed] [Google Scholar]
- Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J (2007) p27 phosphorylation by Src regulates inhibition of cyclin E–Cdk2. Cell 128: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351–1358 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ (1998) The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature 396: 177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L (2007) Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 128: 269–280 [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A (2005a) The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102: 19075–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005b) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst L, Reed SI (1996) Translational control of p27Kip1 accumulation during the cell cycle. Science 271: 1861–1864 [DOI] [PubMed] [Google Scholar]
- Hershko DD, Shapira M (2006) Prognostic role of p27Kip1 deregulation in colorectal cancer. Cancer 107: 668–675 [DOI] [PubMed] [Google Scholar]
- Kaldis P (2007) Another piece of the p27Kip1 puzzle. Cell 128: 241–244 [DOI] [PubMed] [Google Scholar]
- Kardinal C, Dangers M, Kardinal A, Koch A, Brandt DT, Tamura T, Welte K (2006) Tyrosine phosphorylation modulates binding preference to cyclin-dependent kinases and subcellular localization of p27Kip1 in the acute promyelocytic leukemia cell line NB4. Blood 107: 1133–1140 [DOI] [PubMed] [Google Scholar]
- Kedde M, le Sage C, Duursma A, Zlotorynski E, van Leeuwen B, Nijkamp W, Beijersbergen R, Agami R (2006) Telomerase-independent regulation of ATR by human telomerase RNA. J Biol Chem 281: 40503–40514 [DOI] [PubMed] [Google Scholar]
- Kent OA, Mendell JT (2006) A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25: 6188–6196 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- Koff A (2006) How to decrease p27Kip1 levels during tumor development. Cancer Cell 9: 75–76 [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M (2005) Silencing of microRNAs in vivo with ‘antagomirs'. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Le XF, Claret FX, Lammayot A, Tian L, Deshpande D, LaPushin R, Tari AM, Bast RC Jr (2003) The role of cyclin-dependent kinase inhibitor p27Kip1 in anti-HER2 antibody-induced G1 cell cycle arrest and tumor growth inhibition. J Biol Chem 278: 23441–23450 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD (2007) Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120: 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda M, Cukor B, Tam SW, Lavin P, Fiorentino M, Draetta GF, Jessup JM, Pagano M (1997) Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat Med 3: 231–234 [DOI] [PubMed] [Google Scholar]
- Lu CD, Morita S, Ishibashi T, Hara H, Isozaki H, Tanigawa N (1999) Loss of p27Kip1 expression independently predicts poor prognosis for patients with resectable pancreatic adenocarcinoma. Cancer 85: 1250–1260 [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita T, Oda Y, Naito S, Tsuneyoshi M (2002) Low expression of p27(Kip1) is associated with tumor size and poor prognosis in patients with renal cell carcinoma. Cancer 94: 973–979 [PubMed] [Google Scholar]
- Millard SS, Yan JS, Nguyen H, Pagano M, Kiyokawa H, Koff A (1997) Enhanced ribosomal association of p27(Kip1) mRNA is a mechanism contributing to accumulation during growth arrest. J Biol Chem 272: 7093–7098 [DOI] [PubMed] [Google Scholar]
- Mineta H, Miura K, Suzuki I, Takebayashi S, Amano H, Araki K, Harada H, Ichimura K, Wennerberg JP, Dictor MR (1999) Low p27 expression correlates with poor prognosis for patients with oral tongue squamous cell carcinoma. Cancer 85: 1011–1017 [DOI] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A (2006) MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer 13: 497–508 [DOI] [PubMed] [Google Scholar]
- Ponce-Castaneda MV, Lee MH, Latres E, Polyak K, Lacombe L, Montgomery K, Mathew S, Krauter K, Sheinfeld J, Massague J (1995) p27Kip1: chromosomal mapping to 12p12-12p13.1 and absence of mutations in human tumors. Cancer Res 55: 1211–1214 [PubMed] [Google Scholar]
- Porter PL, Malone KE, Heagerty PJ, Alexander GM, Gatti LA, Firpo EJ, Daling JR, Roberts JM (1997) Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med 3: 222–225 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends