Social cheating in Pseudomonas aeruginosa quorum sensing (original) (raw)
Abstract
In a process termed quorum sensing, bacteria use diffusible chemical signals to coordinate cell density-dependent gene expression. In the human pathogen Pseudomonas aeruginosa, quorum sensing controls hundreds of genes, many of which encode extracellular virulence factors. Quorum sensing is required for P. aeruginosa virulence in animal models. Curiously, quorum sensing-deficient variants, most of which carry a mutation in the gene encoding the central quorum sensing regulator lasR, are frequently isolated from acute and chronic infections. The mechanism for their emergence is not known. Here we provide experimental evidence suggesting that these lasR mutants are social cheaters that cease production of quorum-controlled factors and take advantage of their production by the group. We detected an emerging subpopulation of lasR mutants after ≈100 generations of in vitro evolution of the P. aeruginosa wild-type strain under culture conditions that require quorum sensing for growth. Under such conditions, quorum sensing appears to impose a metabolic burden on the proliferating bacterial cell, because quorum-controlled genes not normally induced until cessation of growth were highly expressed early in growth, and a defined lasR mutant showed a growth advantage when cocultured with the parent strain. The emergence of quorum-sensing-deficient variants in certain environments is therefore an indicator of high quorum sensing activity of the bacterial population as a whole. It does not necessarily indicate that quorum sensing is insignificant, as has previously been suggested. Thus, novel antivirulence strategies aimed at disrupting bacterial communication may be particularly effective in such clinical settings.
Keywords: acyl-homoserine lactone, bacterial communication, evolution, lasR, virulence
The diverse quorum-sensing (QS) systems found in bacteria can be considered the paradigm for prokaryotic cooperation and communication. In QS, bacteria produce diffusible signal molecules that allow the coordination of group behavior. In many Gram-negative bacteria, these signals are acyl-homoserine lactones (acyl-HSL). One of the best-described acyl-HSL signaling systems is that governing gene expression in the opportunistic pathogen Pseudomonas aeruginosa. There are two acyl-HSL signals in P. aeruginosa, _N_-(3-oxododecanoyl)-HSL (3OC12-HSL) and _N_-butyryl-HSL (C4-HSL), produced by the LasI and RhlI enzymes, respectively. These signals accumulate intra- and extracellularly as the cells grow. Above a certain threshold level, the signals bind to their cognate transcription factors, LasR and RhlR, respectively, to activate expression of target genes (1–4). The two QS circuits are arranged hierarchically because the LasR-LasI system controls the RhlR-RhlI system (5). Transcriptome studies have shown that both systems together control the expression of hundreds of genes (6–8).
More than 30 quorum-controlled genes encode virulence factors, such as extracellular enzymes (LasA protease, elastase, alkaline protease, lipase, phospholipase C), secondary metabolites (hydrogen cyanide, pyocyanin), and toxins (exotoxin A). These virulence factors are thought to contribute to both acute and chronic P. aeruginosa infections in immunocompromised individuals, including those affected by the genetic disorder cystic fibrosis. In several animal models, QS mutants are significantly attenuated in their virulence compared with the wild type (4). Nevertheless, QS-deficient variants have been isolated from a variety of different infections and other environments (9). Most of the isolates possess mutations in the central regulatory gene lasR. For example, in one study, 12 of 66 clinical and environmental P. aeruginosa isolates had insertion, missense, or nonsense mutations in lasR (10). To reconcile both findings, it has been proposed that QS may not be important for the particular infection from which the deficient strains were isolated (10). An alternative explanation is that QS-deficient variants may be social cheaters (9, 11, 12).
Cheating is considered a major problem in the evolution of cooperation (13–15). Cheaters are individuals that reap the benefit of a social trait (for example, the production of “public goods”) while contributing less than average to the cost. They have been shown to arise in several microbial systems (16–18), including P. aeruginosa. In mixed populations, strains of P. aeruginosa that do not produce extracellular, iron-scavenging siderophores outcompete the wild type (19). Likewise, it is conceivable that a QS deficient subpopulation might be able to exploit production of extracellular quorum-controlled products by the slower-growing wild type. However, although QS controls the expression of ≈6% of all P. aeruginosa genes, QS mutants do not exhibit a faster growth rate under standard culture conditions in the laboratory (20, 21). As shown recently, P. aeruginosa lasR mutants can have a selective advantage after cessation of growth (21). When cultured in unbuffered complex medium, lasR mutants are more resistant to cell lysis and death than the wild type in stationary phase at high cell densities and alkaline pH. The ecological implications of this observation remain to be determined.
Here, we show that P. aeruginosa QS imposes a metabolic burden on the growing bacterial cell under conditions that require QS for growth. Under such conditions, lasR mutants have a growth advantage compared with the wild type, providing a compelling explanation for their enrichment in QS-dependent infections. Thus, lasR mutants can be considered social cheaters.
Results
In Vitro Evolution of P. aeruginosa Under Conditions That Require QS.
We set out to test the hypothesis that QS deficient variants of P. aeruginosa are social cheaters. We reasoned that they have a selective advantage in mixed populations, and that this advantage manifests itself particularly in environments that require bacterial communication of the population as a whole. Our experimental system was as follows: We grew P. aeruginosa under batch culture conditions in minimal medium containing sodium caseinate as the sole carbon source. Growth on casein requires the production of QS-dependent extracellular proteases (22). As opposed to other biopolymers that could serve as QS-dependent substrates, such as mucin, chitin, or casein itself, caseinate (the sodium salt of the milk protein casein) is soluble and therefore facilitates handling of cultures and accurate quantitation of bacterial growth. P. aeruginosa was inoculated from an LB starter culture, grown in caseinate minimal medium for 24 h, diluted into fresh medium, and incubated again for 24 h. This cycle was continued for 20 days.
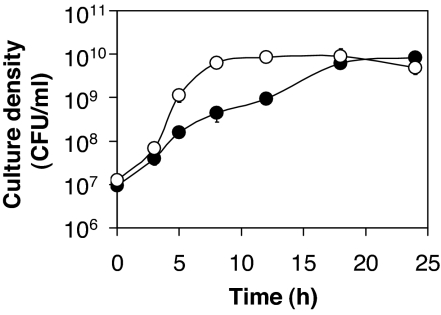
P. aeruginosa was actively growing for virtually the entire 24-h cycle (Fig. 1). Growth appeared slightly biphasic, which might be attributed to the utilization of different breakdown products (oligopeptides and amino acids) during proteolytic growth. Extracellular proteases carried over from the previous culture probably also accelerated growth initially. After 24 h of growth, cultures appeared bright blue-green, indicative of high-level pyocyanin production. Cultures never reached late stationary phase, and their pH was always ≤8. Thus, the accumulation of lasR mutants would not be the result of their increased resistance to cell lysis and death at pH >9 after termination of growth, as reported in ref. 21. P. aeruginosa grew significantly slower on caseinate than on its breakdown products, casamino acids (CAA) (Fig. 1).
Fig. 1.
Growth of P. aeruginosa. P. aeruginosa PAO1 was grown in M9 minimal medium supplemented with 1% (wt/vol) caseinate (filled circles) and 0.5% (wt/vol) CAA (open circles). Bacteria were inoculated from 24-h cultures grown in the same media. Depicted are representative growth curves during day 3 of subculturing.
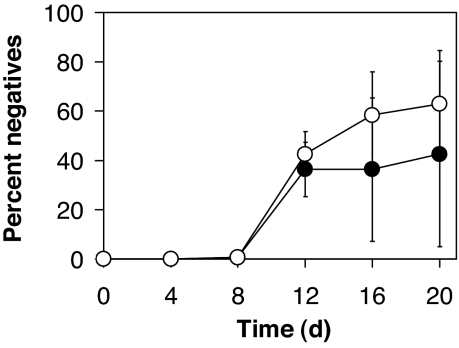
After 4, 8, 12, 16, and 20 days, aliquots were appropriately diluted, and 100 colonies were scored for protease production on skim milk plates and for growth on adenosine. Whereas the skim milk plate assay is a general indicator for the production of extracellular (QS-dependent) proteases, growth on adenosine is a more specific indicator of LasR function. This assay is based on the observation that lasR mutants do not grow on adenosine as the sole carbon source because the expression of a key degradative enzyme, nucleoside hydrolase (Nuh), depends on LasR (21). After 12 days of culturing, protease- and Nuh-negative variants emerged (Fig. 2). Most variants were both protease- and Nuh-negative. Not a single variant from any of the days isolated was protease-negative and Nuh-positive. The fraction of protease-positive and Nuh-negative variants was low initially (6% average on day 12), and increased as culturing progressed (22% and 20% average on days 16 and 20, respectively).
Fig. 2.
In vitro evolution of P. aeruginosa. Shown are the percentages of Nuh-negative (open circles) and protease-negative (filled circles) variants during 20 days of P. aeruginosa PAO1 growth in M9 minimal medium supplemented with 1% (wt/vol) caseinate. Cultures were subcultured into fresh medium every 24 h.
Fifty colonies obtained randomly from days 12, 16, and 20 could all be functionally complemented for growth on adenosine and protease production (if the respective variant was also protease-negative) by a plasmid carrying lasR. Twelve candidates, four each from days 12, 16, and 20, were chosen for further characterization. All of the candidates showed impaired pyocyanin production, and all but two candidates also showed greatly reduced LasA activity as well as 3OC12-HSL and C4-HSL levels (Table 1). The secreted protease LasA and the secondary metabolite pyocyanin are additional quorum-controlled factors in P. aeruginosa (23, 24). DNA sequencing revealed that all 12 candidates were mutated in lasR (Table 1). Nine candidates had point mutations (transitions) in lasR that resulted in single substitutions of conserved, semiconserved, or nonconserved amino acids (25). Three candidates had 37 bp deletions extending from the promoter region into the 5′ coding region of lasR, thereby removing translational start sequences and the N-terminal four amino acids of LasR.
Table 1.
Characterization of QS cheater variants
| Variant | Experiment* | lasR mutation† | Change‡ | Skim milk proteolysis§ | Growth on adenosine§ | LasA activity§ | 3OC12-HSL, %¶ | C4-HSL, %¶ | Pyocyanin, %¶ |
|---|---|---|---|---|---|---|---|---|---|
| lasR1 | 1 (12) | Δ (−25 to +12) | Truncation | − | − | − | 4.0 ± 0.2 | <20 | 5.6 ± 2.3 |
| lasR2 | 1 (12) | Δ (−25 to +12) | Truncation | − | − | − | 4.7 ± 0.9 | <20 | 4.8 ± 1.7 |
| lasR3 | 1 (12) | C→T (+221) | P74L | + | − | − | 3.5 ± 0.4 | <20 | 3.6 ± 0.8 |
| lasR4 | 1 (16) | Δ (−25 to +12) | Truncation | − | − | − | 4.3 ± 0.7 | <20 | 3.6 ± 1.6 |
| lasR5 | 2 (12) | C→T (+683) | A228V | ++ | − | + | 100 ± 10 | 230 ± 50 | 16 ± 2 |
| lasR6 | 2 (16) | G→A (+541) | E181K | − | − | − | 3.9 ± 0.8 | <20 | 2.9 ± 0.7 |
| lasR7 | 2 (20) | G→A (+541) | E181K | − | − | − | 4.6 ± 0.5 | <20 | 1.6 ± 1.3 |
| lasR8 | 2 (20) | C→T (+683) | A228V | ++ | − | + | 100 ± 10 | 110 ± 40 | 11 ± 4 |
| lasR9 | 3 (12) | G→A (+541) | E181K | − | − | − | 4.0 ± 0.6 | <20 | 1.2 ± 0.8 |
| lasR10 | 3 (16) | G→A (+541) | E181K | − | − | − | 4.2 ± 0.5 | <20 | 4.7 ± 2.1 |
| lasR11 | 3 (20) | C→T (+344) | T115I | + | − | − | 4.1 ± 0.5 | <20 | 4.0 ± 1.8 |
| lasR12 | 3 (20) | C→T (+683) | A228V | − | − | − | 4.0 ± 0.4 | <20 | 7.0 ± 2.9 |
| lasR::TcR | N/A | Insertion | N/A | − | − | − | 5.1 ± 1.1 | <20 | 2.7 ± 1.7 |
| Wild type | N/A | None | None | ++ | ++ | ++ | 100 ± 10 | 100 ± 20 | 100 ± 20 |
Interestingly, one of three LasR mutants with the nonconserved A228V substitution (lasR12) was impaired in all QS-dependent phenotypes tested, whereas two (lasR5 and lasR8) displayed near-wild-type levels of acyl-HSL production, increased proteolysis, and slightly elevated production of pyocyanin. All three A228V mutants were Nuh-negative. These discrepant phenotypes probably resulted from a second site gain-of-function mutation in lasR5 and lasR8 rather than a second site loss-of-function mutation in lasR12, because all of the quorum-controlled phenotypes could be restored fully in lasR12 by complementation with lasR (data not shown).
It has been reported that in certain environments, such as those in the cystic fibrosis lung, hypermutable strains with >20-fold elevated mutation rates emerge (26). Our lasR mutants do not appear to be such mutators. Three selected candidates, lasR1, lasR8, and lasR10, exhibited mutation frequencies after exposure to the antibiotic rifampin that were similar to those of the PAO1 parent, the defined lasR mutant [supporting information (SI) Table 3], and values reported in ref. 27.
In summary, we conclude that lasR mutants can emerge under culture conditions that require QS-dependent extracellular factors for growth.
Cheater-Load.
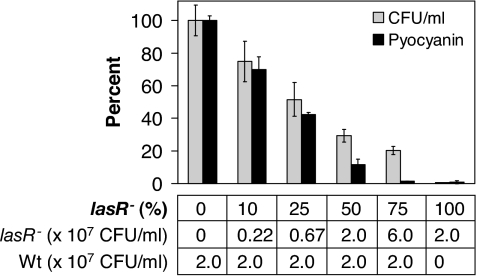
During our long-term experiment, P. aeruginosa culture densities did not decrease substantially after lasR mutants arose (CFU/ml were 3–4 × 109 after the initial 24-h cycle, and 0.8–1.1 × 1010 after subsequent cycles). One might have expected that the emergence of QS-deficient variants or cheaters that do not contribute to the degradation of caseinate would slow growth. At a critical cheater threshold, the entire population might even cease to grow. To investigate the cheater-load in our experimental system, i.e., the potential decrease in productivity as a result of a cheating subpopulation, we quantified growth of mixed populations of a defined, in frame lasR deletion mutant and its isogenic PAO1 parent after 24 h of culture. We inoculated each coculture with the same amount of wild-type cells, and added increasing amounts of lasR mutant cells. This approach allowed us to specifically evaluate the metabolic cost of cheating independent of inoculum effects. A decrease in wild-type inoculum size itself delays growth (data not shown), not only because cell numbers are reduced but presumably also because acyl-HSL signals and extracellular enzymes carried over from the previous culture are diluted.
With increasing amounts of lasR mutant cells, culture density and pyocyanin production decreased successively (Fig. 3). Thus, the presence of cheaters does incur a significant cost to the bacterial population as a whole. The fact that we did not observe this trend in our in vitro evolution experiment indicates that the emergence of cheaters gradually changes the population dynamics during long-term culture, which in turn imposes selective pressure for compensatory adaptation.
Fig. 3.
Cheater-load. Cocultures of a defined unmarked P. aeruginosa lasR mutant and its parent were grown in M9 minimal medium supplemented with 1% (wt/vol) caseinate. Shown are percentage growth (light bars) and percentage pyocyanin production (dark bars) after 24 h of coculture compared with a PAO1 pure culture. As indicated, the inoculum size of the PAO1 parent (in coculture and by itself) was kept constant at 2 × 107 CFU/ml, and increasing amounts of lasR mutant cells were added as “cheater-load.”
Consistent with this notion, we found that the fraction of Nuh-negative but protease positive mutants increased over time, indicating that in several instances protease production was restored in _lasR_-mutant backgrounds. When the results from each individual replicate experiment are considered separately, this phenomenon is particularly apparent in Replicates 1 and 2 (SI Fig. 6). Such a trend was not observed in Replicate 3, suggesting that in this case a different compensation strategy evolved (which also explains the large standard deviations for the averaged data in Fig. 2).
Enrichment of lasR Mutants in Coculture.
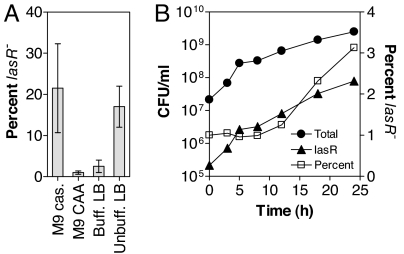
The emergence of lasR mutants during long-term culture suggests that these mutants have a growth advantage compared with the wild type. Because growth rates cannot be assessed in pure culture (the lasR mutant alone does not grow in caseinate), we cocultured an unmarked lasR deletion mutant and its PAO1 parent in M9 minimal medium with caseinate or CAA as the sole carbon source. The initial lasR mutant to wild-type ratio was 1:100. After three 24-h cycles, there was significant enrichment of the lasR mutant in M9 caseinate but not in M9 CAA (Fig. 4A). The pH of both cultures was similar (7.5–8.0 after 24 h). To compare these findings to those reported for the pH-dependent enrichment of lasR mutants in complex medium after termination of growth (21), we also cocultured both strains in buffered and unbuffered LB. Consistent with previous results, there was no enrichment in buffered LB (pH of 8.0–8.5 after 24 h), but there was enrichment in unbuffered LB (pH of 9.0–9.5 after 24 h).
Fig. 4.
Enrichment cocultures. P. aeruginosa cocultures were inoculated with a PAO1 parent to lasR mutant ratio of 100:1. The inoculum size of the PAO1 parent was 2 × 107 CFU/ml. (A) Enrichment after growth of the wild type and the unmarked lasR mutant for three consecutive 24-h cycles in the indicated media. (B) Growth of the wild type and the lasR::TcR mutant during one 24-h cycle. Shown are CFU/ml of the lasR mutant and of the entire culture as determined by plating on selective and nonselective media (Left), and the percentage of lasR mutant cells (Right). Enrichment was significant (P < 0.05) for 12-, 18-, and 24-h time points, as determined by Student's t test. Error bars are too small to be seen.
To determine growth rates in a more direct way, we cocultured a lasR insertion mutant containing a tetracycline resistance cassette with the wild type (1:100 ratio) in M9 caseinate and followed growth during the initial 24-h cycle (Fig. 4B). The fraction of lasR mutant cells, as quantified by plate counts on selective media, increased between 8 and 24 h of growth. The doubling times during this phase of near-logarithmic growth were 331 ± 18 min for the entire culture, 335 ± 19 min for the wild type alone, and 206 ± 8 min for the lasR insertion mutant alone. The same enrichment pattern was observed in subsequent cycles of coculturing (data not shown). When cultured individually in M9 CAA, the doubling times of the unmarked mutant, the insertion mutant, and the PAO1 wild type were indistinguishable (29.6 ± 0.6 min, 29.9 ± 0.4 min, and 29.6 ± 0.8 min, respectively).
Expression of QS Genes.
Compared with the parent strain, lasR mutants have a growth advantage in mixed culture under conditions that require QS for growth. We predict that this is so because quorum-controlled genes are highly expressed in the wild-type strain under these conditions. Such high-level expression would impose a metabolic burden on the wild-type cell and provide lasR mutants with a selective advantage. To test our prediction, we used real-time PCR to compare the expression of selected QS genes under closely related conditions that do and do not require QS for growth (M9 caseinate and M9 CAA, respectively).
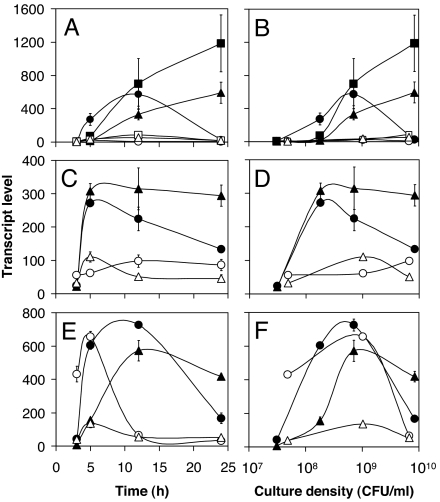
The transcription of quorum-controlled genes aprA (encoding alkaline protease), lasA (encoding LasA protease), and rhlA (encoding rhamnosyltransferase) was much higher in M9 caseinate than in M9 CAA, and was induced at low cell density during growth (Fig. 5). These extracellular factors were expressed maximally when the lasR mutant showed enrichment in coculture (Fig. 4B). Similar gene expression patterns were observed for the acyl-HSL receptor genes lasR and rhlR, and for the acyl-HSL synthase gene rhlI. Interestingly, the transcription of the other acyl-HSL synthase gene, lasI, was similar under both conditions. The expression patterns of both lasI and rhlI correlated well with the levels of 3OC12-HSL and C4-HSL in culture (Table 2). LasRI and RhlRI-dependent target genes were equally affected because aprA is controlled primarily by LasRI, rhlA is controlled primarily by RhlRI, and lasA is controlled by both LasRI and RhlRI (7).
Fig. 5.
QS gene expression. Transcription of selected genes during P. aeruginosa PAO1 growth in M9 minimal medium supplemented with 1% (wt/vol) caseinate (filled symbols) and 0.5% (wt/vol) CAA (open symbols) was measured by real-time PCR. Transcript levels are given in picograms of a genomic DNA standard and were plotted versus time (Left) and culture density (Right). (A and B) aprA (circles), lasA (squares), and rhlA (triangles). (C and D) lasR (circles) and rhlR (triangles). (E and F) lasI (circles) and rhlI (triangles). Measurements were made during day 3 of subculturing.
Table 2.
Acyl-HSL production of P. aeruginosa PAO1
| Carbon source* | 3OC12-HSL, μM† | C4-HSL, μM† |
|---|---|---|
| Caseinate | 0.29 ± 0.03 | 2.2 ± 0.2 |
| CAA | 0.43 ± 0.04 | 0.49 ± 0.03 |
Discussion
We have shown that long-term culture of P. aeruginosa in defined medium that requires QS for growth selects for the emergence of QS-deficient variants. Adaptation probably also involved other changes undetectable by our screen. All of the variants we characterized were mutated in lasR, and these mutations are sufficient to account for the loss-of-function phenotypes observed (Table 1). Point mutations in lasR affected conserved (Pro-74 and Glu-181), semiconserved (Thr-115), and nonconserved (Ala-228) amino acid residues. LasR P74L has previously been shown to be defective in multimerization and consequently failed to activate transcription (25). In the crystal structure of the LasR homolog TraR, the residue corresponding to Glu-181 forms a salt bridge with another conserved residue that stabilizes the primary DNA-recognition helix (28, 29). Thr-115 is adjacent to two other residues that are important for LasR multimerization, Gly-113 and Pro-117 (25), and Ala-228 is located in a region of LasR that is involved in DNA binding (25).
We found a correlation between the enrichment of lasR mutants in coculture and QS gene expression in the parent strain (Figs. 4 and 5). When P. aeruginosa was cultured in M9 caseinate, the transcription of QS genes was highly induced during growth, and lasR mutants had a selective advantage. The expression of individual QS genes has also been shown to be elevated during growth under microaerobic conditions, which has been proposed to reflect the cystic fibrosis lung environment (30), and during growth in mucin, a glycoprotein that P. aeruginosa degrades and may use as a nutrient source during colonization of mucosal surfaces (31, 32). Consequently, lasR mutants are predicted to have a selective advantage under such conditions. On the other hand, in M9 CAA medium, QS genes were expressed at low levels and lasR mutants were not enriched. Similarly, in buffered complex medium, most QS-regulated genes are not induced until the cessation of growth (7, 33), and lasR mutants did not have a selective advantage (Fig. 4A). lasI was the only gene among those characterized that did not show higher expression in M9 caseinate than in M9 CAA. lasI is also not induced significantly during stationary phase growth in complex medium (7), which appears to be the result of a homeostatic control mechanism (34). Consistent with these results is the finding that a basal level of lasI transcription is sufficient for the full induction of most quorum-controlled genes, i.e., that the level of 3OC12-HSL is not the limiting factor (7, 33, 35).
When quorum-controlled genes were highly expressed in M9 caseinate coculture, lasR mutants grew ≈60% faster than the parent strain (Figs. 4B and 5). This finding is remarkable given the current estimate that up to 6% of all P. aeruginosa genes are quorum-controlled (6–8). Thus, under conditions that require QS for growth, the bacterial cell appears to devote an unproportionally large fraction of its metabolic energy to the high-level production of quorum-controlled factors, resulting in high selective pressure for genetic adaptation. In this respect, a mutation in the central regulatory gene lasR, which is atop the QS hierarchy and controls all quorum-controlled functions, seems logical. A mutation in the cognate acyl-HSL synthase gene, lasI, would not amount to the same effect, because such mutants would still respond to the signals generated by the wild-type ancestor. Our mutational analysis did not provide any evidence for mutations in other global regulatory pathways that affect QS gene expression, such as the GacAS/RsmAZ cascade or the catabolite repressor homolog Vfr (3). Because these pathways affect many different cellular functions beyond QS, such mutations would presumably result in reduced overall fitness. Mutations in lasR would also be expected to arise under conditions in which QS was dispensable and not induced during growth. However, in the absence of selective pressure, their accumulation would be dictated by random mutation rates (because there is currently no evidence for a mutational hotspot) and thus be orders of magnitude slower.
We propose that the high-level induction of quorum-controlled genes in the wild-type strain is a general response to slow-growth conditions, and hence, a response to starvation. The stringent response may be implicated in this process. Overexpression of the stringent response protein RelA has been shown to prematurely activate QS in P. aeruginosa, including the expression of lasR, rhlR, and lasB, as well as the production of acyl-HSL (36). P. aeruginosa is likely to encounter starvation conditions in its natural habitats, such as in oligotrophic soils or during colonization of an infection site. Starvation would be a trigger to highly induce QS gene expression, including extracellular enzymes to break down biopolymers as sources of carbon or nitrogen. We suggest that the lasR mutants that are frequently isolated from clinical and environmental samples emerge from this process. The result would be a heterogeneous community of QS proficient and QS deficient (cheater) variants. Intriguingly, in the only study to date that has examined the diversity of QS phenotypes of a large number of isolates from individual patients, such mixed populations have been found to exist (37). We suspect that cheating might also provide P. aeruginosa with a selective advantage in natural mixed-species environments in which other bacteria secrete extracellular degradative enzymes that generate the necessary nutrients for growth.
P. aeruginosa lasR mutants also emerge in alkalized complex medium after cessation of growth (21). These mutants do not have a growth advantage, but they are more resistant to cell lysis and death than the wild type. Although clinically relevant environments such as human airway surface liquid, cystic fibrosis sputum, and _in vitro_-grown P. aeruginosa biofilms commonly are of slightly acidic to neutral pH (38–40), it is conceivable that QS mediated proteolysis and concomitant ammonification result in a localized increase in pH. Another recent study reported that several _lasR_-deficient cystic fibrosis isolates and a defined lasR mutant of P. aeruginosa strain PA14, but not of PAO1, showed an increased growth yield on certain carbon and nitrogen sources (41). These mechanisms, which appear unrelated to the one we describe here, might contribute to the selection of lasR mutants in nutrient-rich environments.
We have shown that cheating also comes with a cost. In M9 caseinate, wild-type cultures exhibited reduced growth and pyocyanin production in the presence of mutant cells with a defined lasR mutation (Fig. 3), which consume nutrients generated by the proteolytic activity of the wild type. In principle, QS provides a mechanism for the bacterial population of assessing this cheater load (42). If the frequency of cheaters became too high, the population would fail to reach a quorum, cease to produce quorum-controlled factors, and consequently, cease to grow. During our in vitro evolution experiment, however, compensatory mutations appeared to emerge before the cheater load became detrimental to the entire population, essentially converting cheaters into cooperators, as has been observed in Myxococcus xanthus social development (43). In several cases, these compensatory mutations restored protease production in lasR mutant strains (Fig. 2, Table 1, and SI Fig. 6). This has been reported to occur during starvation selection of defined P. aeruginosa lasR mutants and involved up-regulation of rhlI, suggesting compensation by the rhl system (22). Consistent with this finding, one of our protease-positive lasR mutants, lasR5, exhibited elevated production of C4-HSL (Table 1).
Because there is ample experimental evidence for a role of P. aeruginosa QS in acute and chronic infections, the frequent isolation of lasR mutants from diverse environments appeared paradoxical (10, 44). Our results suggest a mechanism for the emergence of such lasR mutants and that these mutants are social cheaters whose presence implies high QS activity of the bacterial population as a whole, thus reconciling seemingly discordant findings. This reinforces the utility of QS as a target for novel antimicrobial strategies, although the capacity for compensatory mutation should be taken into consideration.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains were P. aeruginosa PAO1, a PAO lasR::TcR insertion mutant (45), and the in-frame lasR deletion mutant, PAO6395 (21). Bacteria were grown in Lennox LB broth buffered with 50 mM 3-(_N_-morpholino) propanesulfonic acid (pH 7.0) and in M9 minimal medium [0.68% (wt/vol) Na2HPO4/0.3% KH2PO4/0.05% NaCl/0.1% NH4Cl/0.01% MgSO4/0.001% CaCl2/nonchelated trace element solution, pH ≈7.0] supplemented with 1% (wt/vol) sodium caseinate or 0.5% (wt/vol) CAA as carbon source. Bacterial cultures were routinely grown in an incubator/shaker at 37°C in glass tubes containing 4 ml of media.
Experimental cultures were started from overnight (17 h) buffered LB cultures that had been inoculated with a freshly grown single colony of P. aeruginosa PAO1. The initial densities (OD600) of the first and all subsequent cultures were 0.005–0.01 (corresponding to 1–2 × 107 CFU/ml). Cocultures were inoculated at the indicated lasR mutant-to-wild-type ratios. Cocultures and long-term cultures were grown for 24 h and, where appropriate, subcultured into fresh medium every 24 h. At the indicated times, culture aliquots were removed for further analysis. To determine CFU/ml, cells were appropriately diluted in M9 salts and plated on LB agar or on LB agar supplemented with 50 μg/ml tetracycline. For some experiments, 100 individual colonies were subsequently patched on skim milk agar [1/4-strength LB broth/4% (wt/vol) skim milk/1.5% (wt/vol) agar] and on adenosine agar [M9 medium/0.1% (wt/vol) adenosine/1.5% (wt/vol) agar]. To determine the lasR mutant-to-wild-type ratio in enrichment cocultures, bacteria were scored on skim milk plates only. This method is appropriate because spontaneous lasR mutants are not detectable within the duration of the coculturing experiment. All experiments were performed at least in triplicate, and error bars in figures indicate standard deviations of the mean.
Complementation and Sequencing of lasR Mutants.
Complementation was carried out with the low-copy number plasmid pJN105.lasR, which contains lasR under control of the arabinose-inducible araBAD promoter (46). l-arabinose was added to culture media at a final concentration of 0.75% (wt/vol). Arabinose alone, without the complementing lasR allele, had no effect on _lasR_-dependent phenotypes. All 50 candidates were tested for complementation of protease and Nuh-phenotypes. Mutants lasR1 to lasR12 were further tested for complementation of acyl-HSL and phenazine production. Mutations in lasR were identified as follows: Chromosomal DNA from lasR mutants and the PAO1 parent as a control was used in a PCR with primers las1 and las2 (21) to amplify a 1.24-kb lasR region. Two independently obtained PCR products from each strain were completely sequenced.
Mutation Frequencies of lasR Mutants.
The mutation frequencies after exposure to rifampin were determined as described (27).
Assays for 3OC12-HSL, C4-HSL, LasA Protease, and Pyocyanin.
The levels of these factors produced by lasR mutant variants in stationary phase LB cultures were determined according to assays described in refs. 35 and 47–49. For further details, see SI Text.
Real-Time PCR.
Real-time PCR was performed essentially as described in ref. 50. For further details, see SI Text.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dieter Haas (University of Lausanne, Lausanne, Switzerland) for providing P. aeruginosa strain PAO6395, Pete Greenberg (University of Washington, Seattle, WA) for providing purified acyl-HSL, and the Center for Genome Research and Biocomputing at Oregon State University for support with real-time PCR. This work was supported by start-up funds from Oregon State University.
Abbreviations
CAA
casamino acids
CFU
colony-forming units
HSL
homoserine lactone
3OC12-HSL
_N_-(3-oxododecanoyl)-HSL
C4-HSL
_N_-butyryl-HSL
QS
quorum sensing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Fuqua C, Greenberg EP. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 2.Juhas M, Eberl L, Tummler B. Environ Microbiol. 2005;7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 3.Schuster M, Greenberg EP. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 4.Smith RS, Iglewski BH. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 5.Pesci EC, Pearson JP, Seed PC, Iglewski BH. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, et al. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster M, Lohstroh CP, Ogi T, Greenberg EP. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heurlier K, Denervaud V, Haas D. Int J Med Microbiol. 2006;296:93–102. doi: 10.1016/j.ijmm.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. J Bacteriol. 2003;185:7222–7230. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diggle SP, Gardner A, West SA, Griffin AS. Philos Trans R Soc London Ser B. 2007;362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas D. Nat Rev Microbiol. 2006;4:562. doi: 10.1038/nrmicro1466-c1. author reply, 562. [DOI] [PubMed] [Google Scholar]
- 13.Foster KR, Parkinson K, Thompson CR. Trends Genet. 2007;23:74–80. doi: 10.1016/j.tig.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller L, Surette MG. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 15.West SA, Griffin AS, Gardner A, Diggle SP. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 16.Velicer GJ, Kroos L, Lenski RE. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 17.Greig D, Travisano M. Proc R Soc London Ser B. 2004;271(Suppl 3):S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ennis HL, Dao DN, Pukatzki SU, Kessin RH. Proc Natl Acad Sci USA. 2000;97:3292–3297. doi: 10.1073/pnas.050005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin AS, West SA, Buckling A. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 20.An D, Danhorn T, Fuqua C, Parsek MR. Proc Natl Acad Sci USA. 2006;103:3828–3833. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. J Bacteriol. 2005;187:4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Delden C, Pesci EC, Pearson JP, Iglewski BH. Infect Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brint JM, Ohman DE. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 25.Kiratisin P, Tucker KD, Passador L. J Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 27.Ciofu O, Riis B, Pressler T, Poulsen HE, Hoiby N. Antimicrob Agents Chemother. 2005;49:2276–2282. doi: 10.1128/AAC.49.6.2276-2282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez-Ortega C, Harwood CS. Mol Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aristoteli LP, Willcox MD. Infect Immun. 2003;71:5565–5575. doi: 10.1128/IAI.71.10.5565-5575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan K, Surette MG. J Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiteley M, Lee KM, Greenberg EP. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampioni G, Bertani I, Zennaro E, Polticelli F, Venturi V, Leoni L. J Bacteriol. 2006;188:815–819. doi: 10.1128/JB.188.2.815-819.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. J Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Delden C, Comte R, Bally AM. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denervaud V, TuQuoc P, Blanc D, Favre-Bonte S, Krishnapillai V, Reimmann C, Haas D, van Delden C. J Clin Microbiol. 2004;42:554–562. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer H, Widdicombe JH. J Membrane Biol. 2006;211:139–150. doi: 10.1007/s00232-006-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwart H, Moseley WW, Jr, Katz M. Ann NY Acad Sci. 1963;106:709–721. doi: 10.1111/j.1749-6632.1963.tb16679.x. [DOI] [PubMed] [Google Scholar]
- 40.Hunter RC, Beveridge TJ. Appl Environ Microbiol. 2005;71:2501–2510. doi: 10.1128/AEM.71.5.2501-2510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, et al. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Travisano M, Velicer GJ. Trends Microbiol. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Fiegna F, Yu YT, Kadam SV, Velicer GJ. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 44.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberon-Chavez G. Mol Microbiol. 2001;40:708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, Lequette Y, Greenberg EP. Mol Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 47.Essar DW, Eberly L, Hadero A, Crawford IP. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson JP, Passador L, Iglewski BH, Greenberg EP. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuster M, Greenberg EP. BMC Genomics. 2007;8:287. doi: 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information