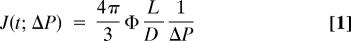Physical plasticity of the nucleus in stem cell differentiation (original) (raw)
Abstract
Cell differentiation in embryogenesis involves extensive changes in gene expression structural reorganization within the nucleus, including chromatin condensation and nucleoprotein immobilization. We hypothesized that nuclei in naive stem cells would therefore prove to be physically plastic and also more pliable than nuclei in differentiated cells. Micromanipulation methods indeed show that nuclei in human embryonic stem cells are highly deformable and stiffen 6-fold through terminal differentiation, and that nuclei in human adult stem cells possess an intermediate stiffness and deform irreversibly. Because the nucleo-skeletal component Lamin A/C is not expressed in either type of stem cell, we knocked down Lamin A/C in human epithelial cells and measured a deformability similar to that of adult hematopoietic stem cells. Rheologically, lamin-deficient states prove to be the most fluid-like, especially within the first ≈10 sec of deformation. Nuclear distortions that persist longer than this are irreversible, and fluorescence-imaged microdeformation with photobleaching confirms that chromatin indeed flows, distends, and reorganizes while the lamina stretches. The rheological character of the nucleus is thus set largely by nucleoplasm/chromatin, whereas the extent of deformation is modulated by the lamina.
Keywords: chromatin remodeling, cell mechanics, nuclear plasticity
Developmental “plasticity” generally refers to a cell's ability to modulate its gene expression (1) and often reflects changes in chromatin structure (2). Stem cell nuclei are therefore said to be more plastic than fully differentiated cells (3). Major differences are indeed found in chromatin conformation (4, 5), nuclear protein expression (6–8), and also DNA and histone modification (9). Differentiation further entails immobilization of some nucleoplasmic proteins and is said to functionally “rigidify” the genome, setting a relatively permanent preference for particular expression profiles (10, 11). Motivated by such remodeling, we sought to directly assess collective physical properties of the nucleus in differentiation.
In an embryo, more deformable nuclei could confer a significant advantage to less differentiated cells that must squeeze their way through developing tissues. Remarkably large nuclear deformations are seen, for example, during the migration of stem cell-like progenitor cells in brain tissue (Fig. 1A Upper) (12). Although such data illustrate deformability, we hypothesized that the nuclei of stem cells would prove more deformable, and it is indeed shown by micropipette aspiration that stem cell nuclei deform to a greater extent under a fixed stress compared with typical differentiated cell types. We go on to characterize the time-dependent deformation or creep under a constant applied stress and then also once the stress is removed from the nucleus. In the latter phase, we provide evidence of physical plasticity in which irreversible deformations persist without fracture. Single chromatin fibers in forced extension also exhibit plasticity, based perhaps on dissociation of histones from DNA (13–15). Our recent work on adult stem cells has additionally shown that multipotent cells are highly contractile and can generate significant cytoskeletal stress (16), representing a potential driving force not only for cell motility (e.g., Fig. 1A) but also for nuclear remodeling. Here we assess some of the macromolecular determinants of nuclear deformability by fluorescence-imaged microdeformation (Fig. 1A), combined with quantitative stress measurements, photobleaching, and knockdown of lamin proteins at the periphery of the nucleus.
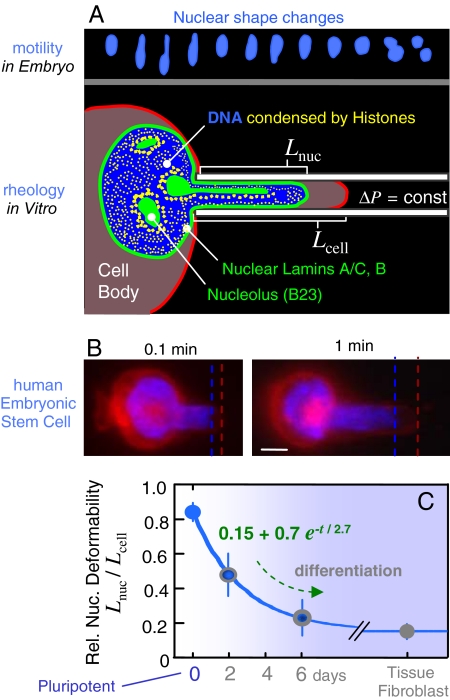
Fig. 1.
Nuclei of human stem cells are more deformable than nuclei of differentiated cells. (A Upper) Neural progenitor cell nuclei (12) show large deformations during in vivo migration (SI Fig. 8). Micropipette aspiration mimics such distortions. (B) Human ESCs were aspirated after differentiation in culture, and the ratio of nuclear extension to cytoplasmic extension was measured as _L_nuc/_L_cell. A day-0 pluripotent ESC is shown with fluorescent dyes labeling nuclear DNA (blue) and the cell membrane (red). (Scale bar: 3 μm.) Dead cells were excluded by counterstaining with propidium iodide. (C) As differentiation progresses, ESC nuclei stiffen nearly 6-fold relative to cytoplasm, and the decrease in relative compliance fits an exponential decay. Differentiated cells such as embryonic fibroblasts also have a nucleus that is stiffer than the cytoplasm. Aspiration of at least three cells and two to three time points per cell were analyzed for each “day” (average ± SD).
Results and Analysis
Nuclei in Stem Cells Deform More Readily than in Differentiated Cells.
To controllably assess the deformability of nuclei as stem cells differentiate, live cell microaspiration of nuclei within primary human embryonic stem cells (ESCs) was performed. Pluripotent cells (day-0) with fluorescently labeled nuclei were aspirated at constant pressure and compared with cells at days 2 and 6 after induction to neurectodermal differentiation (17). The ratio of nuclear projection length to cell membrane extension (_L_nuc/_L_cell) after 1 min of fixed pressure aspiration provides a first simple metric of relative nuclear deformability (Fig. 1B); this ratio is near unity for highly deformable nuclei, and it is low for very stiff nuclei. We find that ESC nuclei are highly deformable and that they stiffen over several days in culture, approaching a 6-fold higher relative stiffness of 0.15 that is typical of differentiated cells such as embryonic fibroblasts (Fig. 1C). An empirically fit exponential yields an effective time constant of 2.7 days for differentiation-associated stiffening of the nucleus relative to the cell body; this timescale appears roughly consistent with global changes in nucleoprotein expression such as lamins in ESCs (7, 17). The limiting ratio for _L_nuc/_L_cell (i.e., nuclear:cytoplasmic stiffness) of ≈0.15 is also consistent with past measurements of relative nuclear compliance for other differentiated cells (18).
Hematopoietic stem cells (HSCs) obtained from bone marrow are less multipotent than ESCs, but HSCs can still differentiate into all of the mature blood cell types (19) as well as a few solid tissue cells that include epithelial cells (20). Micropipette aspiration of human-HSC nuclei subjected to a step increase in pressure show progressive flow into the pipette, and these stem cell nuclei again exhibit greater deformation than fibroblast nuclei at the same stress (Fig. 2 B and C): HSC nuclei deform more than twice as much after 200 sec of aspiration. A relative deformability for HSC nuclei of _L_nuc/_L_cell ≈ 0.5 identifies HSCs as roughly equivalent to ESCs at day 3 ± 1 (Fig. 1C). On the basis of this physical measure, HSCs would be rightly viewed as partially differentiated.
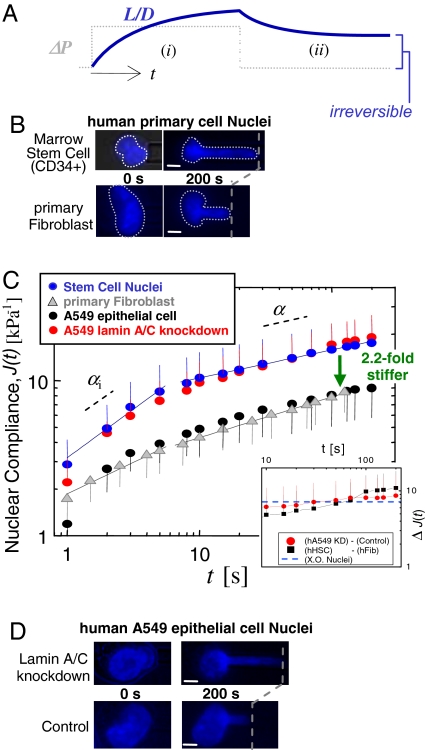
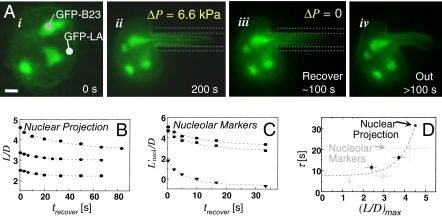
Fig. 2.
Marrow-derived human HSCs have more compliant nuclei than primary human fibroblasts, which may correspond to a lack of Lamin A/C. (A) Constant pressure aspiration into a micropipette (diameter, D) with creep compliance response for relative aspirated length L/D (i). Release of Δ_P_ and incomplete recovery of L/D (ii). (B–D) After 200 sec of aspiration, HSC nuclei (blue) have deformed 2.2-fold more than fibroblast nuclei (gray), indicating that the latter are stiffer. A549 cells (black) are made equally compliant by knockdown of Lamin A/C (red) (average ± SD; n = 15–20 nuclei). (Scale bars: 3 μm.)
Nuclei Exhibit Power Law Creeping Flow.
Aspiration of the various nuclei with a step in pressure shows an increase in L_nuc = L(t) over long times (200 sec) and also, upon release of the applied stress, a permanent deformation (Fig. 2A). The response in the recovery phase provides clear evidence of plasticity during the aspiration phase, but first we characterize the flow behavior, or rheology (21), of the aspiration phase. L(t) is scaled by pipette diameter D and aspiration pressure Δ_P to obtain a creep compliance.
The constant Φ = 2.1 (22). On a log-log scale, the data for HSC and fibroblast nuclei exhibits regime(s) of power law behavior that fit the following form.
The prefactor A has units of inverse pressure or stiffness [kPa−1] and, to facilitate comparisons to past experiments on nuclei conducted on approximately second timescales (22, 23), _t_o ≡ 1 sec. This _t_o conforms to the convention for rheological coefficients (having units: Pa secα) and is also the earliest measurable timescale in our experiments. Eq. 2 captures a wide range of mechanical responses, from simple to complex: α = 0 indicates a simple elastic solid and α = 1 indicates a simple fluid. An intermediate power law is indicative of a viscoelastic or viscoplastic response. Cytoskeletal rheology often yields α ≈ 0.2–0.3 (24, 25), which is understood to reflect a broad spectrum of characteristic timescales and is typical of complex macromolecular structures. Additionally, while fractal organization of chromatin density within the nucleus (26) provides a possible structural basis for a wide range of characteristic times, theory for simple polymer gels gives α = df/(df + 2), with df the fractal dimension of the gel (27). Such a relation suggests that a higher α corresponds to a higher df, which is consistent with a significantly higher df of decondensed chromatin after treatment with a histone deacetylase inhibitor (26).
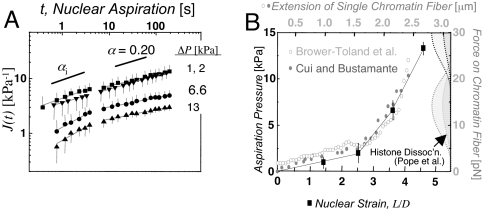
We find that fits of Eq. 2 to the HSC results as well as to the Lamin A/C-knockdown results split into two phases below and above ≈10 sec [Fig. 2C and supporting information (SI) Fig. 9]. For consistency we also separate the remaining aspiration data into two regimes (see Fig. 5A and SI Tables 1 and 2). In the initial phase, HSC results fit with αi ≅ 0.60, indicating greater fluidity than fibroblasts with αi ≅ 0.41 (SI Table 1). Beyond 10 sec, the creep slows for both types of nuclei, and α ≅ 0.2 indicates a more solid-like behavior. Relating these power law exponents to specific fractal dimensions is beyond the scope here and is a general challenge even in simple systems (27).
Fig. 5.
Scale-free creep of nuclei, and nonlinear stress-strain mapping onto chromatin fiber data. (A) Nuclear creep in TC7 cells as a function of pressure (average ± SD; n = 10–15 nuclei). At large Δ_P_, J shows a crossover at τplastic ≅ 8–10 sec. (B) Overlay of nuclear aspiration results and single chromatin fiber extension data (13, 14). The data from Brower-Toland et al. is normalized to the initial length of the Cui and Bustamante fiber, and stretched by a factor of 1.75. The histograms on the right indicate force-dependent frequency of histone dissociation (15).
Network of Lamin A/C Modulates Nuclear Compliance.
Differentiation of ESCs entails changes in multiple structural components of the nucleus, including altered expression of Lamin proteins (A/C and B) (7), as well as changes in the extent of heterochromatinization (10). Neither ESCs nor HSCs have detectable levels of Lamin A/C (28) (confirmed by immunofluorescence and blotting), and so we hypothesized that observed changes in nuclear rheology reflect altered Lamin A/C expression. To mimic this aspect of the stem cells with the human-derived epithelial cell line A549, we knocked down Lamin A/C by >85% with plasmid-based small hairpin RNA (shRNA) mediated interference (SI Fig. 10_A_). While immunofluorescence detected little to no change in either Lamin B expression or key chromatin modification markers of the cell's differentiated state (SI Fig. 10_B_), the A549 Lamin A/C knockdowns exhibit a nuclear rheology that is indistinguishable from that of HSCs (Fig. 2C), whereas the rheological properties of untreated A549 cells prove very similar to those of committed fibroblasts (Fig. 2C, black and gray symbols). Changes in Lamin A/C are therefore sufficient to cause a ≈2-fold difference in stiffness (Fig. 2 B–D). Also, although the extent of deformation is dramatically altered by changes in Lamin A/C expression, the characteristic power law in creeping flow of nuclei is unaffected by Lamin A/C knockdown. Underlying rheological properties of nuclei might therefore be lamin-independent.
Our recent compliance measurements of the Xenopus oocyte (XO) nucleus provide a useful comparison because the mechanics of XO nuclei are dominated by the lamin network that surrounds dilute chromatin (29). We find here that J_XO approximates the difference in nuclear compliance (Δ_J) between HSCs and fibroblasts as well as between A549 controls and A549-Lamin-knockdowns (Fig. 2C Inset). A stiff lamina thus seems likely to explain nuclear stiffening in late differentiation.
Chromatin Is Stiff When Condensed but Otherwise Flows.
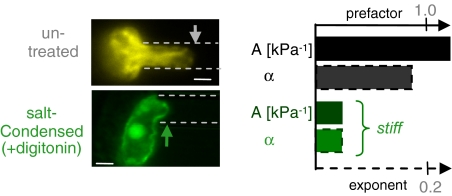
The remarkable compliance of nuclei in pluripotent ESCs (Fig. 1A) seems likely to reflect high accessibility of chromatin (10). Some terminally differentiated nuclei have, in contrast, highly condensed chomatin that might be expected to be stiff (30). Divalent cations such as Ca2+ and Mg2+ will condense nuclei (22, 31), and so we introduced these ions into nuclei via selective permeabilization of the cell membrane by the detergent digitonin, which does not permeabilize the nuclear membrane. Condensation by divalent cations results in extremely stiff nuclei with small values for the creep compliance factors A and α (Fig. 3). Similar effects were seen with the ionophore A23187, which more selectively permeabilizes the plasma membrane to Ca2+.
Fig. 3.
Divalent salts condense chromatin and stiffen the nucleus. Compared with untreated nuclei, quantification reveals decreases in both the creep prefactor, A, and the creep exponent, α, due to cation-induced condensation. (Scale bar: 3 μm.)
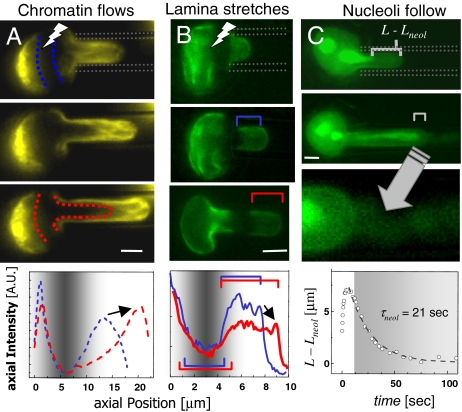
The lamin-independent nature of nuclear flow (i.e., α ≈ 0.2), combined with the strong effects of condensation, implicates chromatin in the physical plasticity of the nucleus. We therefore tracked chromatin with GFP-histones (H2B), as well as GFP Lamin A in TC7 epithelial cells. A dark stripe of GFP-H2B was made by laser photobleaching (FRAP/FIMD, fluorescence recovery after photobleaching/fluorescence-imaged microdeformation). The parabolic flow profile of fluorescence proves consistent with a strong but sheared linkage between chromatin and the lamina (Fig. 4A and SI Fig. 11). Axial intensity profiles show that chromatin is progressively compacted toward the tip as the bleached stripe convects during aspiration. In some nuclei, chromatin bundles align and visibly extend within the pipette, indicative of physical remodeling (SI Fig. 11, asterisk).
Fig. 4.
Distinctive mechanics of nuclear components. (A) FRAP of GFP-H2B chromatin (pseudocolored yellow) reveals chromatin flow within the pipette. Chromatin compaction at the tip is quantified in the axial intensity profiles, and the arrow indicates an increase in intensity with compaction. (B) The lamina (green) is stably stretched into the pipette, and the arrow indicates a decrease in intensity with dilation. (C) Nucleoli slowly follow chromatin toward the nuclear tip, as GFP permeates the intact envelope (arrow into magnified view in bottom panel). (Scale bar: 3 μm.)
GFP-Lamin A stretches during aspiration (Fig. 4B), with widening of the bleached stripe and lengthening of the unbleached tip area, which also thins as it stretches. The lack of recovery of the photobleached region provides direct evidence that large, sustained strains do not significantly increase Lamin A incorporation into the nuclear lamin over the experimental timescale. In unbleached nuclei, the observed gradient in density is similar to that of other elastic networks, particularly the red cell spectrin network (32) and the nuclear lamina of both HeLa cells and mouse embryonic fibroblasts (23).
Labeling nucleoli with GFP-B23 (nucleophosmin) reveals slow nucleolar extrusion into the pipette during aspiration (Fig. 4C), as quantitated by the decreased distance from nucleolus-to-tip (L − _L_neol). Further kinematic analyses (SI Figs. 12 and 13) suggest nucleoli are stiffer than nucleoplasm, consistent perhaps with higher electron density in EM imaging as well as reduced accessibility of intranuclear dextran (33). Visible loss of soluble GFP from the nucleus (Fig. 4C, arrow) indicates slow flow through the nucleus into the pipette, but the intact appearance of the lamina suggests permeation through nuclear pores, consistent with nuclear export processes (34).
Plastic Flow of Nuclei and Nucleoli Maps into Chromatin Fiber Compliance.
Distortions seen in chromatin and nucleoli are large and occur on both short timescales, as viscoelastic deformation, and on long timescales under small stress, consistent with plastic flow (21). To quantify these regimes and to also relate chromatin deformation within the nucleus to nonlinear extension forces known for isolated chromatin (13–16), four different aspiration stresses were applied to individual nuclei (Fig. 5A). At low Δ_P_ of 1–2 kPa, the creep of differentiated cell nuclei is fit relatively well by a single power law. However, at larger stresses (≥6.6 kPa), a crossover time in the creep compliance is seen at τplastic ≅ 8–10 sec. This is denoted as plastic because the small deformations (L/D ∼ 1) held for short times ≤ τplastic lead to essentially full recovery, contrasting with results below for long-time aspiration. For t > τplastic, the nuclear creep exponent α proves independent of applied stress with α ≅ 0.2 ± 0.04 (see SI Table 2), which is in agreement with results above as well as data on fully isolated nuclei in buffers low in divalent cations (21). At short times, t ≤ τplastic, the creep exponent αi is distinctly higher and tends to increase with increasing stress, indicative of a more fluid-like character in this initial regime. Stem cells also display biphasic behavior around τplastic ≅ 10 sec (Fig. 1C). We thus observe a stress-sensitive “fluidization” of all nuclei that is characterized by an increase in the initial creep exponent (αi) with increasing stress. Stem cell nuclei, which are overall more compliant, are fluidized at lower stress (2 kPa) than the stiffer differentiated nuclei (6.6 kPa).
Nuclear compliance measurements and visualization of H2B reorganizations indicate a central role for chromatin mechanics, but additional analyses make a stronger case. J(t, Δ_P_) decreases strongly with pressure, indicating that the nucleus stiffens with increasing stress similar to experiments on single chromatin fibers. We plot the large strain L/D at 200 sec versus the applied stress (Fig. 5B), spanning the same experimental timescale for extensional studies of individual chromatin fibers: hundreds of nanometers per second for several micrometers over 15–30 sec (13, 14). Overlaying the force-extension behavior of chromatin fibers on the nucleus experiments presented here, we determined an effective chromatin fiber diameter d_chrom from the nuclear aspiration pressure Δ_P and the force f per chromatin fiber, i.e., _d_chrom ∼ (f Δ_P_−1)1/2. The best fit yields _d_chrom ≅ 45 nm. This estimation should not be overinterpreted because it makes many naive assumptions, such as homogeneous stress in chromatin and aligned fibers, but the result is surprisingly close to the most recent measurements of bimodal chromatin fiber diameters of 33 nm and 44 nm (35). We speculate that the nonlinear nature of nuclear rheology might therefore be attributable to chromatin.
Irreversible Nuclear Deformation.
Flow and irreversibility at the molecular scale often correspond to disruption and/or entanglement of macromolecular structures. Some of the past studies of isolated chromatin postulate the dissociation of nucleosomes in forced extension (13–15). To quantify changes in histone mobility in stressed nuclei, FRAP was applied to GFP-H2B on the highly extended chromatin within the pipette. Stressed or not, the mobile fraction of H2B remained low at ≈8% (SI Fig. 11 C and D), which agrees with results from other cell lines (36). Fluorescent bundles of GFP-H2B also appear well defined without loss of resolution or intensity inside the pipette where stresses are clearly high (Fig. 4A and SI Fig. 11). The creeping flow in aspiration is therefore supported in part by changes in chromatin conformation.
In a final set of experiments focused again on plasticity, nuclei aspirated at the low stress of 2 kPa for 200 sec were allowed to recover after aspiration (Figs. 2A and 6). Even at this pressure, large and irreversible deformation is evident, with recovery of no more than 20% of the maximum L/D. The timescale of recovery increases with the maximum L/D imposed, implying progressive loss of physical memory (Fig. 6B). Irreversible distortion is also evident in nucleoli stretched along the nuclear projection (Fig. 6C). Timescales for partial recovery fit an exponential in strain (L/D)max and extrapolate to a minimum at small extensions of τrecover ≅ 8 sec (Fig. 6D). This value appears consistent with τplastic for crossover to slow plastic creep under stress (Figs. 2C and 5A) and appears to correspond roughly to a crossover from viscoelastic to viscoplastic deformation. Because Lamin A/C knockdown cells behave similarly, chromatin seems responsible for much of nuclear plasticity.
Fig. 6.
In situ irreversibility of stretched nuclei. (A) The undeformed nucleus (i) is aspirated for ≈200 sec (ii). Recovery within the micropipette for ≈100 sec (iii) is followed by ejection from the pipette (iv). (Scale bar: 3 μm.) Plasticity is evident in the persistently deformed shapes of nuclei and nucleoli, and the time required for even partial recovery increases for both (B and C) at large deformations. (D) Extrapolation of the data to the small strain limit yields τrecover ≅ 8 sec.
Discussion
Nuclear Rheology Is Modulated by Chromatin “Stemness” and Lamin A/C Density.
Transcriptional plasticity of stem cells is reflected in part in the high molecular mobility of some key nucleosomal proteins (10, 37), and here we show that such genetic plasticity is highly collective and measurable as an overall physical plasticity of nuclei. Physical malleability could be important to cell function. Very large nuclear deformations observed in vivo would facilitate migration through solid tissues (12)—a process that would benefit from low Lamin A/C expression. The highly specialized, differentiated neutrophil also makes its way through small tissue openings (out of blood vessels), and this cell's distensible multilobed nucleus is known to lack Lamin A/C (38).
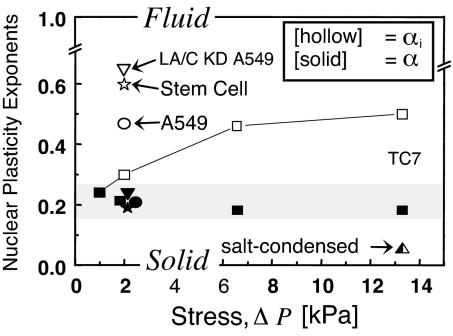
Because of the many differences between stem cells and their progeny lineages (30, 39, 40), we took a model approach to study the changes in nuclear ultrastructure. The human epithelial cell line A549 was knocked down for Lamin A/C, and, for generality, another common epithelial cell line (TC7 derived from kidney of an African green monkey) was stressed at different pressures to assess changes in creep relaxation (Fig. 7). On timescales shorter than τplastic and at stresses low enough for cells to generate themselves through cytoskeletal forces (16), αi indicates strikingly fluid-like behavior for nuclei. Nuclei of undifferentiated stem cells and Lamin A/C knockdown cells also appear more fluid (α > 0.5) than solid and are the most compliant states here.
Fig. 7.
Modulation of nuclear plasticity. Initial deformation response at small stress is more fluid for stem cells and Lamin A/C knock down cells, but differentiated nuclei also display increasingly fluid responses at larger stress. At longer times, all nuclei are more solid-like, with a weak power law exponent of ≈0.2 that is typical of macromolecular networks.
Under sustained stress, nuclei display more solid-like, weak power law rheology, as has been measured in many cytoskeletal networks. Microdeformation imaging of mammalian nuclei expressing GFP-Lamin A indeed suggest a solid-like lamin network (23), whereas chromatin and nucleoli flow, shear, and compact locally in physical rearrangements typical of a complex fluid. Consistent with a dominant fluidic contribution of nucleoplasm, the stem cells lacking Lamin A/C, as well as the A549-knockdowns all display power law rheology similar to differentiated cell nuclei with a full complement of lamins.
Physical Memory Through Conformational Remodeling.
In the mechanical life of the nucleus, the character of the viscoelastic response seems to be set by the nucleoplasm and perhaps its interconnection to the lamina, whereas the lamina substantially increases the durability of the nucleus, bearing up to 50% of the applied stress. This model is consistent with recent studies of _lmna_−/− mouse fibroblasts (41, 42) that suggest mechanical contributions of Lamin A/C of a similar magnitude. However, these past studies were complicated by the fact that laminopathies perturb differentiation (43–45), and differentiation processes are seen here to change the physical plasticity of the nucleus.
Irreversible realignment of chromatin fiber bundles and nucleoli are indicative of the types of conformational changes (Fig. 4 A and C, and SI Fig. 10) that might result in gene colocalization (46) and rearrangement of expression-affecting chromosome territories (47–49). Local changes in nuclear lamina density (Fig. 4B) also illustrate possibilities for lamin-modulated changes in gene expression (50), and the longevity of such distortions compared with molecular timescales provides an opportunity for sustained influence. Indeed, the time dependence of much of the physical remodeling seen here exhibits a recurring and surprisingly small characteristic time of τplastic ≅ τrecover ≅ 8–10 sec (Fig. 6 B_—_D), consistent with particle-tracking microrheology experiments in which a crossover from “elastic trapping” of beads in chromatin to “diffusive behavior” of the beads is also observed to be of the order of seconds (51, 52). Under stress, memory of the initial state is increasingly lost as nuclei relax into a new state, which may be relevant to the physical and genetic underpinnings of dysmorphic nuclear structures. Normalizing stresses should likewise be required to maintain spheroidal nuclei, as processes such as cell spreading, contraction, and migration would all be expected to perturb nuclear shape. Thus, on timescales of biochemical reprogramming of gene expression, the chromatin proves highly malleable and is physically plastic.
Materials and Methods
Human ESC Culture and HSC Isolation.
Human ESCs, line WA07 (H7, WiCell) were maintained in DMEM high glucose with 20% knockout serum replacer, and grown on mitomycin-treated mouse embryo fibroblasts. Culture on low-density feeder cells (1/3 normal) produced a gradual, controlled differentiation to neurectodermal lineages (17). Normal human bone marrow cells were obtained by the Stem Cell Core at the University of Pennsylvania after informed consent. Mononuclear cells were purified by Ficoll gradient centrifugation, and CD34+ cells (HSCs) were purified by using a Miltenyi Biotec AutoMacs magnetic cell sorter. HSCs were kept on ice for same day study.
Lamin A/C Knockdown.
Lentiviral shRNA against the human lmna gene, targeting the sequence CGACTGGTGGAGATTGACAAT (Sigma, TRCN0000061833) was infected into A549 cells. Cells were plated near confluency in 24-well dishes and treated with viral particles at a multiplicity of infection of ≈10 in conjunction with 2 μl of the transduction agent ViroMag R/L (OZ Biosciences, Marseille, France). Transduced cells were selected with the antibiotic puromycin (8 μg/ml), and subjected to a second round of infection. This procedure resulted in cells with Lamin A/C levels of <20% compared with nontransduced cells.
Sample Preparation and Buffers.
ESCs were collected by treatment for 5 min with 1 mM EDTA in Ca2+/Mg2+ free Hanks' balanced salt solution, followed by detachment from the substrate with a glass needle, and aspiration in culture media. All other adherent cells were trypsinized and resuspended in aspirating buffer (135 mM NaCl/5 mM KCl/5 mM Hepes/1.8 mM CaCl2/2 mM MgCl2/10% BSA, and the viability dye propidium iodide). At least 30 min before aspiration, differentiated cells were treated with 10 μM latrunculin A to depolymerize the actin cytoskeleton. Cells were loaded into a sample chamber that allows micropipette access from the side. Only nonphase, lucid viable cells that excluded propidium iodide were aspirated. Nuclei were visualized via Hoechst 33342 DNA dye (Invitrogen), and ESC plasma membranes were visualized by a fluorescently conjugated wheat germ agglutinin. All chemicals were purchased from Sigma–Aldrich unless indicated. Further details of labeling and imaging are provided in SI Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank D. M. Gilbert (State University of New York, Syracuse), M. Olson (University of Mississippi, Jackson), and T. Misteli (NCI, Bethesda) for respective gifts of GFP-Lamin A, B23, and H2B plasmids. We also thank S. Sen for helpful discussion. This work was supported by an Ashton Fellowship (to J.D.P.) and funding from the National Institutes of Health (to K.N.D. and D.E.D.) and the National Science Foundation and Muscular Dystrophy Association (to D.E.D.).
Abbreviations
ESC
embryonic stem cell
HSC
hematopoietic stem cell
shRNA
small hairpin RNA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Blau H, Pavlath G, Hardeman E, Chiu C, Silberstein L, Webster S, Miller S, Webster C. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 2.Slack JMW, Tosh D. Curr Opin Genet Dev. 2001;11:581–586. doi: 10.1016/s0959-437x(00)00236-7. [DOI] [PubMed] [Google Scholar]
- 3.Szutorisz H, Dillon N. BioEssays. 2005;27:1286–1293. doi: 10.1002/bies.20330. [DOI] [PubMed] [Google Scholar]
- 4.Labrador M, Corces VG. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 5.West AG, Fraser P. Hum Mol Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 6.Worman HJ, Courvalin J-C. Int Rev Cytol. 2005;246:231–279. doi: 10.1016/S0074-7696(05)46006-4. [DOI] [PubMed] [Google Scholar]
- 7.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 8.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Proc Natl Acad Sci USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li E. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 10.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosak ST, Groudine M. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- 12.Tsai J-W, Chen Y, Kriegstein AR, Vallee RB. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Y, Bustamante C. Proc Natl Acad Sci USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Proc Natl Acad Sci USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pope LH, Bennink ML, van Leijenhorst-Groener KA, Nikova D, Greve J, Marko JF. Biophys J. 2005;88:3572–3583. doi: 10.1529/biophysj.104.053074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 17.Ozolek JA, Jane EP, Krowsoski LA, Sammak PJ. Stem Cells Dev. 2007;16(3):403–412. doi: 10.1089/scd.2006.0109. [DOI] [PubMed] [Google Scholar]
- 18.Maniotis AJ, Chen CS, Ingber DE. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 20.Krause DS, Theise ND, Collector MI, Henegariu O, Hwang S, Gardner R, Neutzel S, Sharkis SJ. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 21.Larson RG. The Structure and Rheology of Complex Fluids. New York: Oxford Univ Press; 1999. [Google Scholar]
- 22.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowat AC, Foster LJ, Neilsen MM, Weiss M, Ipsen JH. J R Soc Interface. 2004;2:63–69. doi: 10.1098/rsif.2004.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Phys Rev Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman BD, Massiera G, Van Citters KM, Crocker JC. Proc Natl Acad Sci USA. 2006;103:10259–10264. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toth KF, Knoch TA, Wachsmuth M, Frank-Stohr M, Stohr M, Bacher CP, Muller G, Rippe K. J Cell Sci. 2004;117:4277–4287. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- 27.Scanlan JC, Winter HH. Macromol. 1991;24:47–54. [Google Scholar]
- 28.Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 29.Dahl KN, Kahn SM, Wilson KL, Discher DE. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 30.Krauss SW, Lo AJ, Short SA, Koury MJ, Mohandas N, Chasis JA. Blood. 2005;106:2200–2205. doi: 10.1182/blood-2005-04-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelhardt M. Biochem Biophys Res Commun. 2004;324:1210–1214. doi: 10.1016/j.bbrc.2004.09.175. [DOI] [PubMed] [Google Scholar]
- 32.Discher DE, Mohandas N, Evans EA. Science. 1994;266:1032–1035. doi: 10.1126/science.7973655. [DOI] [PubMed] [Google Scholar]
- 33.Gorisch SM, Lichter P, Rippe K. Histochem Cell Biol. 2005;123:217–228. doi: 10.1007/s00418-005-0752-y. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, Weber JD. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura H, Cook PR. J Cell Biol. 2001;153:1341–1354. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yellajoshyula D, Brown DT. Proc Natl Acad Sci USA. 2006;103:18568–18573. doi: 10.1073/pnas.0606478103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yabuki M, Miyake T, Doi Y, Fujiwara T, Hamazaki K, Yoshioka T, Horton AA, Utsumi K. Physiol Chem Phys Med NMR. 1999;31:77–84. [PubMed] [Google Scholar]
- 39.Shav-Tal Y, Lee B-C, Bar-Haim S, Hadas S, Zipori D. J Cell Biochem. 2001;81:379–392. doi: 10.1002/1097-4644(20010601)81:3<379::aid-jcb1052>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 40.Tamada H, Thuan NV, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, Wakayama T, Kikyo N. Mol Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broers JLV, Peeters EAG, Kuijpers HJH, Endert J, Bouten CVC, Oomens CWJ, Baaijens FPT, Ramaekers FCS. Hum Mol Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 42.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong LG, Ng JK, Lammerding J, Vickers TA, Meta M, Cote N, Gavino B, Qiao X, Chang SY, Young SR, et al. J Clin Invest. 2006;116:743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boguslavsky RL, Stewart CL, Worman HJ. Hum Mol Genet. 2006;15:653–663. doi: 10.1093/hmg/ddi480. [DOI] [PubMed] [Google Scholar]
- 45.Favreau C, Higuet D, Courvalin J-C, Buendia B. Mol Cell Biol. 2004;24:1481–1492. doi: 10.1128/MCB.24.4.1481-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. J Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manders EMM, Kimura H, Cook PR. J Cell Biol. 1999;144:813–822. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremer T, Cremer C. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 50.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Proc Natl Acad Sci USA. 2004;101:13221–13226. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. J Cell Sci. 2004;117:2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information