Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase (original) (raw)
Abstract
The bacterial preprotein translocase is comprised of a membrane-embedded oligomeric SecYEG structure and a cytosolic dimeric SecA ATPase. The associations within SecYEG oligomers and SecA dimers, as well as between these two domains are dynamic and reversible. Here, it is shown that a covalently linked SecYEG dimer forms a functional translocase and a high affinity binding site for monomeric and dimeric SecA in solution. The interaction between these two domains stimulates the SecA ATPase, and nucleotides modulate the affinity and ratio of SecA monomers and dimers bound to the linked SecYEG complex. During the translocation reaction, the SecA monomer remains in stable association with a SecYEG protomer and the translocating preprotein. The nucleotides and translocation-dependent changes of SecA–SecYEG associations and the SecA dimeric state may reflect important facets of the preprotein translocation reaction.
Keywords: ATPase/channel/oligomerization/preprotein translocation/SecA/SecYEG
Introduction
Preproteins containing an N-terminal classical leader peptide cross the membrane by means of a multisubunit translocase. The membrane core of the preprotein translocase is formed by the heterotrimeric SecYEG complex, universally conserved in bacteria, archaea and eukaryotes (Cao and Saier, 2003). The cytosolic part of the bacterial translocase is the dimeric ATPase SecA (Lill et al., 1990; Driessen, 1993). Preproteins bind to the translocase at SecA, triggering multiple rounds of ATP binding and hydrolysis (Hartl et al., 1990). The current model of preprotein translocation suggests that SecA interchanges between membrane-‘inserted’ and ‘deinserted’ states at the membrane-embedded SecYEG complex, thereby allowing the sequential movement of preprotein segments across the membrane (Economou and Wickner, 1994).
SecA is the motor subunit of the translocase, but the precise mechanism by which ATP-derived energy is coupled to the preprotein movement remains elusive. In addition to its high affinity for membrane-embedded SecYEG (Hartl et al., 1990), SecA also interacts with numerous ligands: leader and mature regions of preproteins, acidic phospholipids, SecB, nucleotides, Mg2+, Zn2+ and its own mRNA (Economou, 2002; and references therein). Accordingly, the crystal structure of SecA reveals a complex multidomain protein with numerous binding folds (Hunt et al., 2002). Recent studies suggest that SecA undergoes major rearrangements upon binding to acidic phospholipids or leader peptides, leading to the dissociation or polymerization of dimeric SecA (Or et al., 2002; Benach et al., 2003). An earlier study involving heterodimers with one ATPase-inactive subunit has shown that the SecA dimer is important for translocation (Driessen, 1993), yet the recent findings suggest that SecA monomerization and polymerization may play major roles in translocation (Or et al., 2002; Benach et al., 2003).
Similarly, another current debate concerns the dynamics and stoichiometry of the SecYEG translocation channel. Electron microscopy studies have first revealed that the SecYEG protomer, or its eukaryotic counterpart, assembles into an oligomer with a pore-like structure (Hanein et al., 1996; Meyer et al., 1999; Manting et al., 2000). Except for one study, other evidence for oligomeric SecYEG associations comes from cross-linking, sedimentation analysis, native gel electrophoresis, electron crystallography, genetic and fluorescence resonance energy transfer (FRET) experiments (Manting et al., 2000; Yahr and Wickner, 2000; Collinson et al., 2001; Veenendaal et al., 2001; Bessonneau et al., 2002; Breyton et al., 2002; Matsuo et al., 2003; Mori et al., 2003). Altogether, these different studies suggest that the SecYEG oligomers are dynamic assemblies and different oligomeric states may be involved during translocation.
Knowledge of the subunit dynamics of the SecYEG oligomers, SecA dimers and SecYEG–SecA associations is thus central to understanding the mechanism of protein translocation. This is a difficult question because some of these interactions are transient and occur only at the membrane or during preprotein translocation. Novel experimental tools and methods of analysis need to be developed. The present work reports that the stabilization of the SecYEG dimer via covalent linkage allows the detection of a stable SecYEG–SecA complex in detergent solution. It is shown that nucleotides, which play a critical role in regulating SecA-mediated preprotein movement, also modulate the SecA–SecYEG associations. Furthermore, at the translocation channel, the preprotein is stably engaged with monomeric SecA and monomeric SecYEG. These results support the possibility that changes in the SecA dimeric state play a critical role during the translocation reaction. The oligomeric state of the functional SecYEG translocation channel is discussed.
Results
A tandem of two covalently linked SecY subunits is functional for preprotein translocation
A gene encoding two SecY subunits fused in tandem (secYY) was constructed by linking two secY open reading frames (ORFs), in-frame, without an intervening stop codon. The hybrid was cloned along secE and secG in an expression-controlled plasmid vector (pTrcSecEYYG). The thermosensitive phenotype of the secY mutant strain (CJ107; Wolfe et al., 1985), as well as the cold-sensitive phenotype of the SecDFyajC-depleted strain (BL325; Duong and Wickner, 1997), were suppressed by expression of this plasmid (Figure 1A). Thus, two covalently linked SecY subunits are likely to participate in a functional translocase.
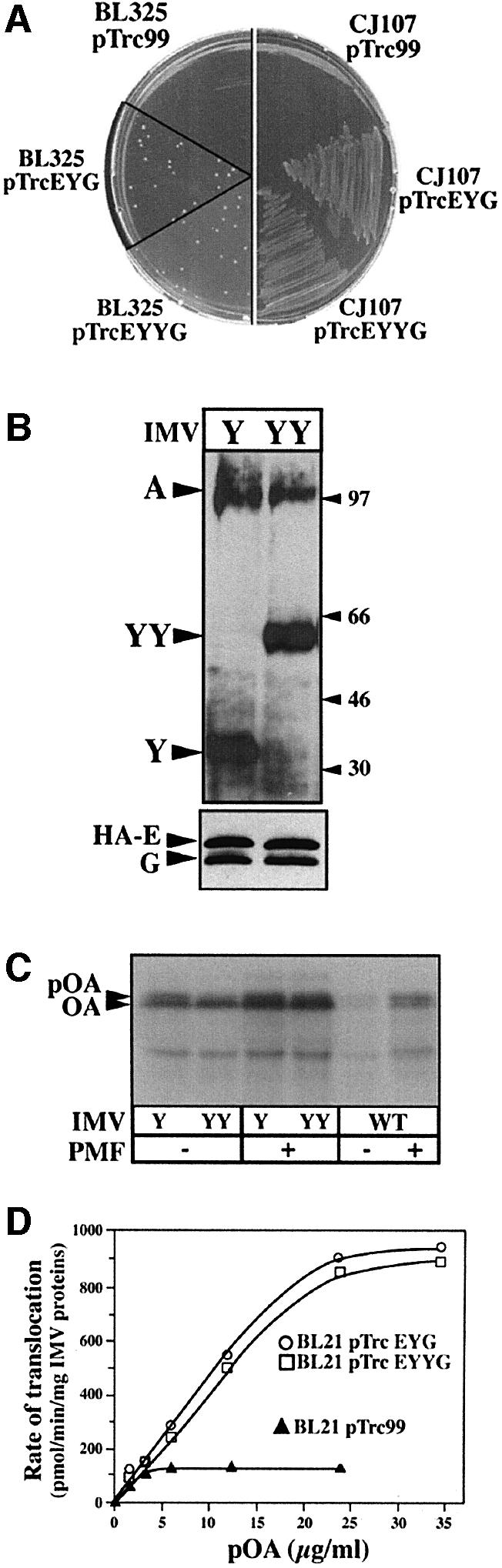
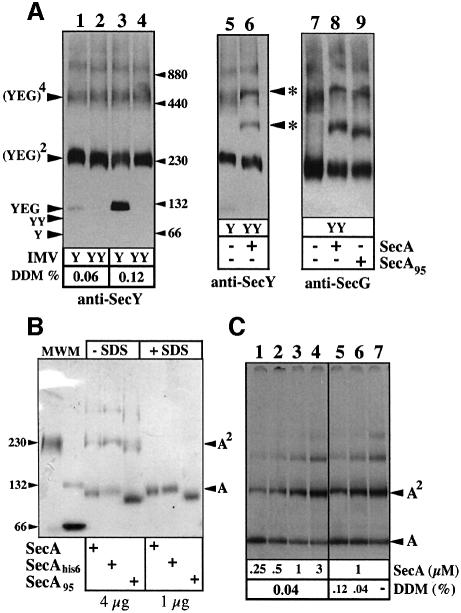
Fig. 1. The covalently linked SecYEG dimer is functional for pre protein translocation. (A) Overproduction of SecYYEG suppresses the SecDFyajC– cold-sensitive and SecY thermo-sensitive phenotypes. Strain BL325 (BL21 tgt::kan-araC+ -PBAD_::yajCsecDF_) or CJ107 (Wolfe et al., 1985) were transformed with the isopropyl-β-d-thio galactopyranoside (IPTG)-inducible plasmid pTrc99 or pTrc99 encoding SecYEHAG (pTrcEYG) or SecYYEHAG (pTrcEYYG). Transformants were plated on LB/ampicillin (50 µg/ml) contain ing 1 mM IPTG and incubated at the restrictive temperature. (B) Immunostaining of IMV preparations. About 1 µg of IMV proteins prepared from E.coli BL21 overexpressing SecYEHAG (IMVs-Y) or SecYYEHAG (IMVs-YY) were analyzed by SDS–PAGE and immunostained with the indicated antibodies. In the presence of SDS, SecY (48.5 kDa) and SecYY (97 kDa) migrate aberrantly. (C and D) Translocase activity of the IMV preparations. Translocation reactions were performed and analyzed as described in Materials and methods.
Western blot analysis of inner membrane vesicles (IMVs) prepared from the Escherichia coli strains carry ing plasmids pTrcSecEYG (termed IMVs-Y) or pTrcSecEYYG (termed IMVs-YY) showed that SecYY/SecY, SecE and SecG were correctly synthesized and overproduced (Figure 1B). The level of endogenous SecA, which normally co-purifies with IMVs, was similar in these preparations (Figure 1B). An in vitro translocation assay showed that both IMVs-Y and IMVs-YY support efficient proOmpA translocation (Figure 1C), each stimulated by the proton-motive force. A quantitative translocation assay using an increasing concentration of preprotein substrate (Figure 1D) confirmed that the translocation capacity is increased ∼10 times in IMVs-Y or IMVs-YY compared with wild-type IMVs (i.e. without Sec overproduction). Hence, it can be concluded that the SecYY hybrid molecule can participate in an active preprotein translocase.
The SecYY hybrid molecule forms a stable complex with SecE and SecG
Blue native gel electrophoresis (BN–PAGE; Schägger and von Jagow, 1991) of solubilized IMV proteins was performed to probe the oligomeric association of SecYY with SecE and SecG. From the IMVs-Y detergent extracts, SecYEG monomers, dimers and tetramers were immunodetected (Figure 2A, lanes 1 and 3). The SecYEG monomer:dimer ratio depends on the detergent concentration, while SecYEG tetramers and higher oligomers appear with the Sec overproduction, as previously shown (Bessonneau et al., 2002). From the IMVs-YY detergent extracts, oligomers of molecular weight similar to those seen with IMVs-Y were detected, with the preponderance of a (SecYEG)2-like assembly (Figure 2A, lanes 2 and 4). All the assemblies cross-reacted with anti-SecY or anti-SecG antibodies (lane 7), while the SecYY-containing complex was purified by affinity chromatography using a His6-tagged version of SecE (Figure 3A). Thus, the SecYY molecule forms stable associations with both SecE and SecG. Furthermore, the native SecYEG dimer migrated at the same position as the purified or membrane-solubilized SecYY-containing complex (Figures 2A and 3A). The relative migration of these native and SDS-dissociated assemblies strongly suggest that SecYY is associated with two SecE and two SecG (referred to as SecYYE2G2 or linked SecYEG dimer, hereafter), although a different stoichiometry cannot be excluded. The theoretical molecular weight of these complexes is given in the legend of Figure 3.
Fig. 2. Oligomeric behavior of covalently-linked SecYEG dimer and SecA. (A) Immunodetection of SecYEG oligomers in membranes enriched with SecYEG (IMVs-Y) or SecYYE2G2 (IMVs-YY). IMVs were solubilized in TSG buffer at the indicated detergent concentration. Protein aliquots (∼0.2 µg) were analyzed by linear gradient BN–PAGE (4–12%) and immunostained. Where indicated, IMVs were incubated with SecA or SecA95 (4 µg; 0.8 µM protomer). The asterisks indicate the probable complex of SecYYE2G2 with monomeric and dimeric SecA. (B) SecA, His6-tagged SecA or His6-tagged SecA95 in TSG buffer (±SDS) were analyzed on a native gel followed by Coomassie blue staining. Molecular weight markers are BSA and catalase. (C) [35S]SecA (∼20 000 c.p.m., ∼100 ng) was mixed at room temperature with unlabeled SecA to reach the desired concentration. Samples were diluted in TSG buffer containing DDM and analyzed by BN–PAGE (4–10%) and autoradiography. The final SecA and DDM concentrations in each sample are indicated.
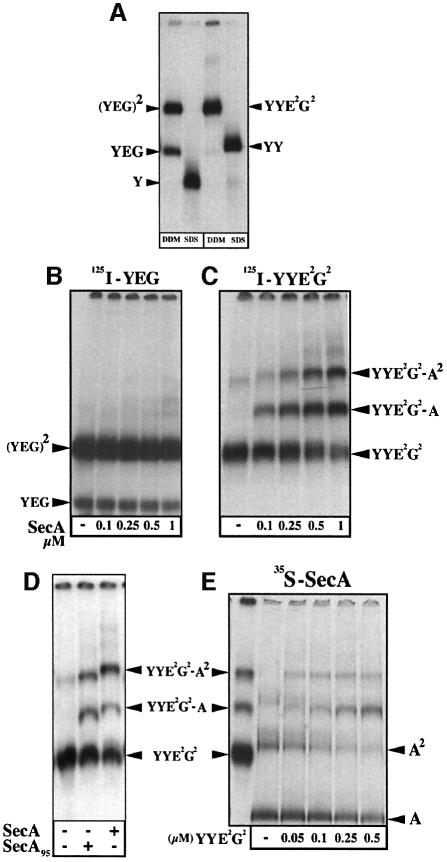
Fig. 3. The stabilized SecYEG dimer forms a stable binding site for SecA monomers and dimers. (A) Oligomeric state of the purified SecYEG complexes. [125I]SecYEG and [125I]SecYYE2G2 purified complexes (∼30 000 c.p.m., ∼20 ng, in TSG buffer 0.03% DDM or 0.1% SDS) were analyzed by BN–PAGE (5–13%) and autoradio graphy. (B–D) Complexes between radiolabeled SecYEG and unlabeled SecA. [125I]SecYEG and [125I]SecYYE2G2 purified complexes (∼30 000 c.p.m., 0.7 µM) were incubated (5 min, 22°C) with SecA or SecA95 at the indicated concentration in TSG buffer containing 0.04% DDM. The resulting complexes were separated by BN–PAGE (4–12%) and revealed by autoradiography. (E) Complexes between radiolabeled SecA and unlabeled SecYYE2G2. [35S]SecA (∼20 000 c.p.m., 1 µM) was incubated in TSG buffer containing 0.04% DDM with the indicated concentration of the purified SecYYE2G2 complex. The resulting complexes were analyzed by BN–PAGE (4–12%) and autoradiography. For _M_r reference, a sample of the [125I]SecYYE2G2 complex incubated with unlabeled SecA is loaded in the left lane of the gel. Calculated molecular weights of the Sec proteins and Sec complexes: SecY (48.5 kDa); SecYY (97 kDa); His-SecE (14 kDa); SecG (11 kDa); His-SecA (103 kDa); His-SecA95 (95 kDa); SecYEG (73.5 kDa); SecY2E2G2 (147 kDa); SecY4E4G4 (294 kDa); SecY2E2G2–SecA (250 kDa); SecY2E2G2–SecA2 (353 kDa).
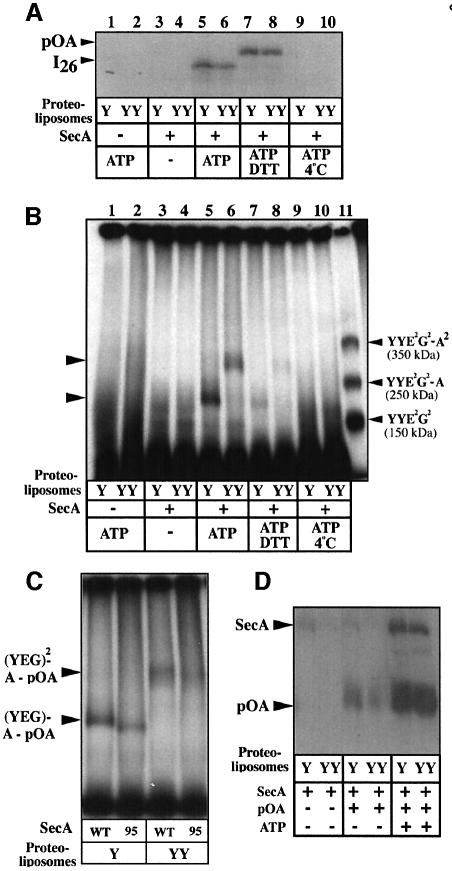
Interestingly, when SecA was added in excess prior to IMV solubilization, two additional complexes were detected from the IMVs-YY detergent extracts (Figure 2A, labeled *). A C-terminal truncated version of SecA (SecA95; Breukink et al., 1995) shifted these two complexes to lower molecular weight positions (Figure 2A, lane 9). Thus, these two complexes probably represent stable associations of SecA with the linked SecYEG dimer. Before further analysis, the oligomeric property of SecA was studied by BN–PAGE. In solution, SecA presents complex oligomeric behavior with a stoichiometry depending on protein concentration, detergent, temperature and composition of the buffer (Or et al., 2002; Woodbury et al., 2002; Benach et al., 2003). In this study, the comparison between the migration of molecular weight markers and SDS-dissociated SecA indicated that SecA exists as monomers and dimers, although higher oligomers were also present (Figure 2B). The SecA95 protein, which is active for preprotein translocation but displays slightly higher endogenous ATPase activity (Breukink et al., 1995), also formed dimers. A monomer–dimer equilibrium was detected using increasing concentrations of SecA mixed with a constant amount of radiolabeled SecA (Figure 2C, lanes 1–4). At higher concentrations, SecA formed higher order associations (Figure 2C). These polymers may resemble those previously identified by cross-linking analysis and analytical ultracentrifugation (Hirano et al., 1996; Dempsey et al., 2002; Benach et al., 2003). Incubation of SecA with dodecyl-maltoside (DDM) triggered the progressive dissociation of the SecA oligomers (Figure 2C, lanes 5–7; Or et al., 2002). At the DDM and SecA concentrations used in this study (such as lanes 1–3), dimeric SecA was still detected.
The linked SecYEG dimer binds monomeric and dimeric SecA
A previous BN–PAGE analysis did not detect stable SecYEG–SecA associations or oligomerization of the SecYEG complex upon incubation of IMVs-Y with SecA (Bessonneau et al., 2002; and Figure 2A, lane 5). Similarly, the purified and radiolabeled native SecYEG complex did not form obvious associations with SecA (Figure 3B). Faint bands migrating above the SecYEG dimer were detected at higher SecA concentrations (Figure 3B, right lanes), suggesting that a transient or unstable interaction between native SecYEG and SecA may occur. In contrast, incubation of SecA with the purified and radiolabeled SecYYE2G2 complex readily revealed two higher molecular weight assemblies (Figure 3C; labeled SecYYE2G2–A and SecYYE2G2–A2, respectively). Furthermore, the truncated SecA95 shifted these two bands to lower molecular weight positions, attesting to the presence of SecA in these complexes (Figure 3D). In the absence of SecA, a low-intensity band migrating slightly below SecYYE2G2–A2 was detected (Figures 2A, and 3A, C and D). This band migrates at the same position as the (SecYEG)4 assembly (Figure 2A) and probably corresponds to an association of two SecYYE2G2 complexes [i.e. (SecYEG)4]. However, the two complexes that appear upon addition of SecA migrate at positions distinct from this band and depend on both the molecular weight and concentration of SecA (Figure 3C and D). An association between (SecYYE2G2)2 and monomeric SecA would be expected to migrate at a higher position and the shift induced by a single SecA95 (–8 kDa) to be less obvious. In the converse experiment, radiolabeled SecA was incubated with increasing concentrations of SecYYE2G2 (Figure 3E). The same two complexes were also detected, concomitantly with the progressive disappearance of the SecA monomers and dimers. Thus, it is very likely that the SecYYE2G2 complex associates with monomeric and dimeric SecA.
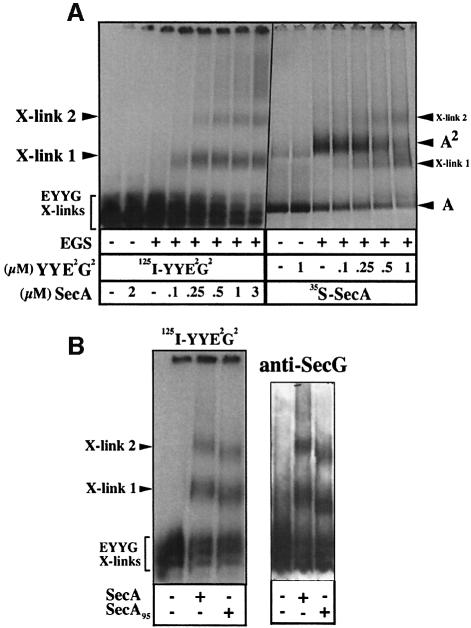
This conclusion was also reached by cross-linking analysis. The radiolabeled SecYYE2G2 complex was incubated with SecA, then with an amine-reactive cross-linking reagent. Two major cross-linked products were formed in a SecA-dependent manner (labeled X-link1 and X-link2; Figure 4A, left panel). At high SecA concentrations, higher molecular weight cross-links were also detected. In the converse experiment, radiolabeled SecA was incubated with SecYYE2G2, and cross-linked products of molecular weight similar to X-link1 and X-link2 were identified (Figure 4A, right panel). Smears and aggregates on top of the gel appeared with the increase in the SecYYE2G2 concentration. Finally, incubation of the truncated SecA95 with the linked SecYEG dimer shifted the X-link1 and X-link2 bands to lower molecular weight positions, attesting to the presence of SecA in these cross-links (Figure 4B). These two cross-linked products were also detected with anti-SecG antibodies (Figure 4B) or anti-SecY antibodies (not shown). Thus, the X-link1 and X-link2 products probably correspond to cross-links of SecYYE2G2 with monomeric and dimeric SecA, respectively.
Fig. 4. Cross-linking production between SecYYE2G2 and SecA. (A) [125I]SecYYE2G2 (0.1 µM) or [35S]SecA (1 µM) were incubated in CL buffer (5 min, 22°C) with the indicated concentration of unlabeled SecYYE2G2 or unlabeled SecA, respectively. After cross-linking by EGS and dissociation of the non-covalent protein complexes by SDS, samples were analyzed by BN–PAGE (4–12%) and autoradiography. (B) [125I]SecYYE2G2 or unlabeled SecYYE2G2 were incubated as in (A) with SecA or SecA95, and cross-links were revealed by autoradiography or immunostaining.
Nucleotide-dependent SecYEG–SecA interactions
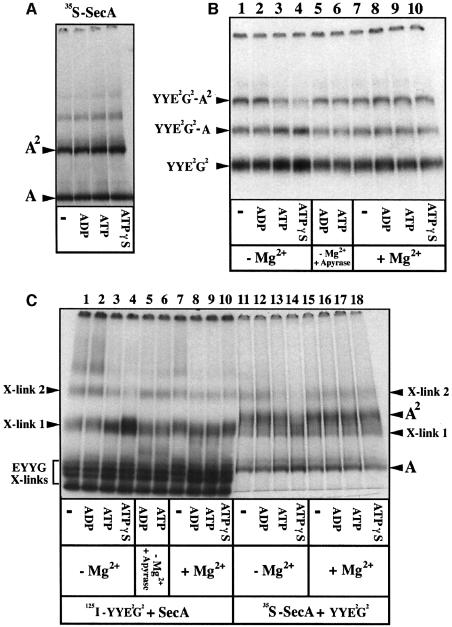
Experiments such as cross-linking, FRET, small angle X-ray scattering (Driessen, 1993; Shilton et al., 1998; Or et al., 2002) and BN–PAGE (Figure 5A) showed that ADP, ATP or ATPγS do not significantly change the SecA monomer–dimer equilibrium. However, these nucleotides were found to affect the SecA–SecYEG interactions profoundly (Figure 5B). In the presence of ADP, the SecA dimer was stably associated with the SecYYE2G2 complex (lane 2). In contrast, the SecA dimer dissociated from the SecYYE2G2 complex upon addition of ATP (lane 3 compared with lane 1). This dissociation was even stronger in the presence of ATPγS (lane 4). Rapid depletion of hydrolyzable nucleotides upon addition of apyrase restored the initial level of SecA dimers bound to SecYYE2G2 (lanes 5 and 6). Unexpectedly, the effect of ATP or ATPγS was strongly reduced when magnesium was included in the incubation buffer (lanes 7–10). Similar results were obtained by cross-linking analysis (Figure 5C). Only low amounts of X-link2, corresponding to a covalent complex of SecYYE2G2 with SecA2, were obtained in the presence of ATP or ATPγS, compared with ADP (Figure 5C, lanes 3 and 4 compared with lane 2). This ATP- or ATPγS-dependent decrease in the amount of X-link2 was also seen in the converse experiment using radiolabeled SecA (compare lanes 13 and 14 with lane 12). The ATP-dependent decrease of X-link2 production was abolished upon addition of apyrase (lanes 5 and 6). Again, the effects of ATP or ATPγS on the X-link2 production were reduced when magnesium was present in the incubation buffer (lanes 8–10 and 16–18).
Fig. 5. Nucleotide-dependent SecA–SecYEG associations. (A) Nucleotides do not change the oligomeric state of SecA in solution. [35S]SecA (∼20 000 c.p.m., 1 µM) was incubated (10 min, 22°C) in TSG buffer containing 0.04% DDM with the indicated nucleotides (1 mM). Samples were analyzed by BN–PAGE (4–12%) and autoradiography. (B) Complexes between radiolabeled SecYEG and unlabeled SecA after addition of nucleotides. [125I]SecYYE2G2 (0.7 µM) was mixed with SecA (1 µM) in TSG buffer (0.04% DDM; ±5 mM MgCl2). After incubation (5 min, 22°C), nucleotides were added and the incubation was prolonged for 5 min. Samples (lanes 5 and 6) were treated with potato apyrase (grade VII; 20 U/ml) for an additional 2 min. All samples were returned to ice before analysis by BN–PAGE (4–12%) and autoradiography. (C) Cross-link production between SecYYE2G2 and SecA after addition of nucleotides. [125I]SecYYE2G2 (0.1 µM) or [35S]SecA (1 µM) were incubated in CL buffer (±5 mM MgCl2) with unlabeleled SecA (2 µM) or unlabeled SecYYE2G2 (0.5 µM), respectively. After incubation with nucleotides (as described in B), samples were treated by EGS, then dissolved by SDS and analyzed by BN–PAGE (4–12%) and autoradiography.
Thus, together, the data show that ATP binding to SecA tends to destabilize the association between the SecA dimer and the linked SecYEG dimer. Interestingly, both the BN–PAGE analysis and cross-linking experiments indicate that the level of SecYYE2G2–A (or X-link1) increases upon addition of ATP or ATPγS (Figure 5B and C, lanes 3 and 4). To some extent, this increase is in relation to the decrease of the level of SecYYE2G2–SecA2 (or X-link2). This last observation may suggest that only one SecA protomer dissociates from the SecYYE2G2–A2 complex upon binding of ATP. Alternatively, it may reflect an increased affinity of the SecA monomer for the SecYEG complex.
Activation of the SecA ATPase by the SecYEG complex in solution
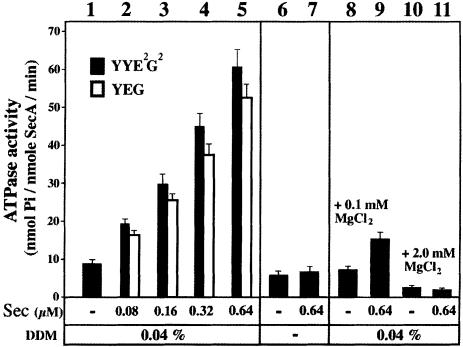
The SecA ATPase activity is stimulated by acidic lipids containing membranes and by membrane-embedded SecYEG (Lill et al., 1990; Hendrick and Wickner, 1991). The nucleotide-dependent SecA–SecYEG associations (Figure 5) suggest that a SecYEG-dependent SecA activation may also occur in solution. Indeed, SecA was found to hydrolyze large amounts of ATP during incubation with the SecYYE2G2 complex, and the release of inorganic phosphate augmented with the increase of SecYYE2G2 present in the reaction (Figure 6, lanes 1–5). In the absence of detergent, the SecYYE2G2 complex aggregates and the SecYYE2G2-dependent ATPase activity was lost (lanes 6 and 7). Upon addition of magnesium, the SecYYE2G2-dependent ATPase activity was strongly repressed (lane 11). The endogenous ATPase activity of SecA was also diminished by this cation (lane 10). The native SecYEG complex stimulated the SecA ATPase activity to a similar extent (Figure 6), which was also repressed by addition of magnesium (not shown). Therefore, in the absence of added magnesium and in the presence of the native or linked SecYEG complex, the SecA enzyme is stimulated and consumes ATP. Activation of the SecA ATPase by the native SecYEG complex suggests that an interaction between these two domains, at least transient, must also occur in solution. Furthermore, both the SecYEG-dependent SecA ATPase and the nucleotide-dependent SecA–SecYEG associations (Figure 5) are reduced by magnesium, suggesting that a relationship between these two events may exist.
Fig. 6. Activation of the SecA ATPase by SecYEG. SecA (0.8 µM) was incubated at room temperature with the purified SecYEG or SecYYE2G2 complexes in TSG buffer (±0.04% DDM; ±MgCl2) containing 2 mM of ATP. The Sec concentration refers to that of the SecYEG protomer (SecYYE2G2 = 2 × SecYEG). After 10 min incubation at 22°C, the inorganic phosphate produced by the SecA ATPase was measured by the photocolorimetric method of Lanzetta et al. (1979).
Monomeric SecA and preprotein form a stable complex at the SecYEG translocation channel
We previously reported that a preprotein arrested during its translocation (intermediate I26) forms a sufficiently stable complex with the SecYEG translocation channel to be analyzed by BN–PAGE (Bessonneau et al., 2002). The same technology was employed here. First, the purified SecYEG and SecYYE2G2 complexes were reconstituted into liposomes and then incubated with SecA, bovine serum albumin (BSA), ATP and [125I]proOmpA conjugated to the globular BPTI molecule. Both SecYEG- and SecYYE2G2-proteoliposomes are functional for protein translocation, and the I26 intermediate was generated in both cases, although with less efficiency in the latter case (Figure 7A, lanes 5 and 6). In the absence of SecA, ATP or at 4°C, the intermediate I26 was not formed (Figure 7A, lanes 1–4, and 9 and 10). In the presence of dithiothreitol (DTT), which freed proOmpA from BPTI, the arrested intermediate I26 completed its translocation across the membrane (lanes 7 and 8).
Fig. 7. The SecA monomer is engaged with preprotein and the SecYEG channel during translocation. (A) [125I]ProOmpA–BPTI (∼100 000 c.p.m.; 4 µg/ml) was incubated (10 min, 37°C) in TL buffer (50 µl final volume) with SecYEG- or SecYYE2G2-proteoliposomes (15 µg/ml), BSA (200 µg/ml), SecA (80 µg/ml; 0.8 µM) and ATP (2 mM) or DTT (2 mM) where indicated. Reactions were terminated by addition of apyrase (20 U/ml). Aliquots of the translocation reactions (1/10 vol.) were treated with proteinase K and analyzed by 12% SDS–PAGE and autoradiography. (B) The remainder of the translocation reaction performed in (A) was solubilized with an equal volume of TSG buffer containing 0.12% DDM. Aliquots were analyzed by BN–PAGE (4–10%) and autoradiography. For _M_r reference, [125I]SecYYE2G2 complexed to SecA was loaded on the same gel (lane 11). (C) As in (B) lanes 5 and 6, but using the truncated SecA95 or wild-type SecA to drive translocation in SecYEG- or SecYYE2G2-proteoliposomes. (D) The translocation reactions described in (A) were supplemented with [125I]SecA (∼100 000 c.p.m.; 4 nM). At the end of the incubation (10 min, 37°C), proteoliposomes were solubilized with 300 µl of TSG buffer, 0.1% DDM (10 min, 4°C). After centrifugation (10 min, 100 000 g), the membrane extract was incubated with anti-SecG IgG coupled to protein A–Sepharose beads. Immuno precipitates were washed (3 × 0.3 ml of TSG buffer) and analyzed by 10% SDS–PAGE and autoradiography. Theoretical molecular weight of the SecYEG–SecA–pOA complexes: pOA (37 kDa); SecYEG– pOA (112 kDa); (SecYEG)2–pOA (187 kDa); SecYEG–SecA–pOA (212 kDa); (SecYEG)2–SecA (250 kDa); (SecYEG)2–SecA–pOA (287 kDa); (SecYEG)4–pOA (343 kDa).
In parallel, the translocation reactions described above were treated with DDM and the solubilized materials were analyzed by BN–PAGE (Figure 7B). The molecular weight of the potential complexes is given in the legend of Figure 7. With the SecYYE2G2-proteoliposomes, proOmpA migrated as a complex of high molecular weight (lane 6), in between SecYYE2G2–A and SecYYE2G2–A2 (250 and 350 kDa, respectively). The size of this complex (lane 6) is too large to correspond to an association of SecYYE2G2 and proOmpA (187 kDa). It is also too small to correspond to an association of proOmpA with a (SecYYE2G2)2 complex (343 kDa). Given the affinity of the SecYYE2G2 complex for SecA, a likely composition of the complex observed in lane 6 (Figure 7B) is an association of SecYYE2G2–proOmpA with monomeric SecA (293 kDa). With the SecYEG-proteoliposomes, proOmpA was trapped with the SecYEG complex (lane 5; and Bessonneau et al., 2002) and migrated in between the SecYEG dimer (150 kDa) and the SecYYE2G2–SecA complex (250 kDa). This complex (lane 5) was proposed to correspond to an association of proOmpA with dimeric SecYEG (187 kDa; Bessonneau et al., 2002), but it may also correspond to an association of proOmpA with monomeric SecA and monomeric SecYEG (212 kDa).
To analyze further the composition of these translocation complexes, preprotein translocation was driven with truncated SecA95 instead of wild-type SecA (Figure 7C). With the SecYYE2G2 proteoliposomes, SecA95 shifted the SecYYE2G2–proOmpA complex to a lower molecular weight position (lanes 3 and 4). With the SecYEG-proteoliposomes, SecA95 also decreased the molecular weight of the SecYEG–proOmpA complex (lanes 1 and 2). Thus, both the linked SecYEG dimer and the native SecYEG complex are engaged with SecA during translocation. This conclusion was confirmed by co-immunoprecipitation (Figure 7D). Anti-SecG antibodies efficiently co-immunoprecipitated a complex of SecYEG and translocation-arrested proOmpA (Bessonneau et al., 2002), but this study shows that SecA is also present in the immunoprecipitate (Figure 7D). In the absence of a preprotein intermediate, the complex between SecA and SecYYE2G2 may have been dissociated during the washes of the immunoprecipitates (Figure 7D). Thus, our previous observation about the composition of the translocation complex was mistaken (Bessonneau et al., 2002). This study show that the translocation complex is actually formed by the ternary association of SecYEG–SecA–proOmpA. With the SecYYE2G2 proteoliposomes, the second SecYEG protomer remains associated with the translocation complex because of the covalent linkage.
Discussion
Our previous work suggested that the dimeric stoichiometry of the SecYEG channel bore the translocation intermediate (Bessonneau et al., 2002), but we missed the possible participation of SecA. Here, it is shown that the translocation complex bearing the preprotein is actually formed by monomeric SecYEG, as previously reported (Yahr and Wickner, 2000), in association with monomeric SecA. Remarkably, higher concentrations of detergent further destabilized the SecYEG–SecA–preprotein complex (Bessonneau et al., 2002), but the intermediate of dissociation formed by monomeric SecYEG and the translocation-arrested preprotein was not detected. This suggests that the translocating polypeptidic chain may be weakly associated with the SecYEG protomer or that SecA may be structurally necessary to maintain this association during detergent solubilization. A site-specific photocross-linking study has shown that SecY and SecA simultaneously contact the translocating chain, suggesting that SecA contacts the polypeptide conduit intimately (Joly and Wickner, 1993). However, while the ternary complex SecYEG–SecA–preprotein appears as the most stable part of the translocation complex upon detergent solubilization and analysis by BN–PAGE, this result does not exclude the participation of other SecYEG protomers, either for the formation of the channel or for its activity. In fact, most current studies suggest that the SecYEG complex, in its non-translocating state, exists as oligomers. These studies include cysteine cross-linking, sedimentation analysis, native gel electrophoresis, electron microscopy, electron crystallography, genetic and FRET experiments (Meyer et al., 1999; Manting et al., 2000; Collinson et al., 2001; Veenendaal et al., 2001; Bessonneau et al., 2002; Breyton et al., 2002; van der Sluis et al., 2002; Matsuo et al., 2003; Mori et al., 2003). The SecYEG monomer exists in solution but readily reforms dimers upon dilution with detergent and reconstitution into lipids (Bessonneau et al., 2002; Breyton et al., 2002; Mori et al., 2003). Thus, a SecYEG protomer may form the central core of the translocation channel, but it is likely that the SecYEG oligomers are actively involved. The presence of SecA and an arrested preprotein in the translocation pathway may have weakened the interactions between the SecYEG protomers, thus becoming unstable in detergent solution.
The current model for post-translational translocation proposes that SecA inserts with the first loop of the preprotein into the SecYEG channel to become both shielded from the lipid phase and partly exposed to the other side of the membrane (Economou and Wickner, 1994; Eichler and Wickner, 1997; Ramamurthy and Oliver, 1997). A single membrane-embedded SecYEG protomer may not be sufficient to allow the SecA insertion, whereas the lipid bilayer surrounding the protomer may further oppose this event. In contrast, the SecYEG protomers within the dimer are weakly associated and their interface forms a cytosolic cavity (Bessonneau et al., 2002; Breyton et al., 2002). The prl mutations further destabilize the SecYEG associations and increase the binding affinity of SecA and the translocation efficiency (van der Wolk et al., 1998; Bessonneau et al., 2002). The non-essential SecG subunit facilitates translocation and also stabilizes SecYEG tetramers and higher order oligomers when the SecYEG complex is overexpressed in the membrane (Bessonneau et al., 2002). In such a scenario, the interface of the SecYEG dimers or other oligomers may favor the initial step of the translocation reaction such as SecA binding, insertion and transfer of the first loop of the preprotein into the translocation channel. One of the SecYEG protomers would bind to preprotein and SecA, thereby forming the most stable part of the translocation complex upon detergent solubilization. Oligomeric formation of SecYEG and its eukaryotic homolog, Sec61p, is stimulated in membranes by association with SecA and ribosomes, respectively (Hanein et al., 1996; Manting et al., 2000). Different oligomeric structures may be essential for the initial steps of translocation and may specialize according to the mode of translocation, the energy source or the protein substrate.
This study also provides information about the stoichiometry and dynamics of the SecYEG–SecA association. This association was demonstrated previously by indirect assays, such as affinity binding measurements, in vivo cross-linking and genetic analysis (Manting et al., 1997; Matsumoto et al., 1997; Snyders et al., 1997; Lill et al., 1990). The current work shows that a linked SecYEG dimer readily forms a complex with monomeric and dimeric SecA in solution. Higher order SecYEG oligomers do not seem necessary for this interaction. In support of this finding, analytical ultracentrifugation and gel filtration experiments show that a SecYEG dimer stabilized with an antibody can bind dimeric SecA (I.Collinson, personal communication). Thus, the antibody or the covalent linkage may have imposed a single conformational state on the SecYEG dimer, thereby forming a high affinity SecA-binding site. In contrast, the native SecYEG complex forms dynamic assemblies in detergent solution and exists as monomers, dimers and higher order oligomers at high concentration (Manting et al., 2000; Collinson et al., 2001; Bessonneau et al., 2002). This oligomeric variability may prevent the stabilization and detection of the SecA-binding site. Nonetheless, the present study does not exclude other transient SecA–SecYEG associations, nor the possibility that the linkage between two SecYEG protomers induces an abnormal conformation, resulting in an artificial SecA affinity effect. However, the linked SecYEG dimer is functional for preprotein translocation, complements Sec-defective strains and stimulates the SecA ATPase activity. Furthermore, a cysteine cross-linking study has shown that both the N- and C-termini of neighboring SecY are in close proximity. The cysteine-linked dimers possess translocase activity (van der Sluis et al., 2002). The projection structure of dimeric SecYEG shows two monomers related to each other by an antiparallel symmetry (Breyton et al., 2002). Accordingly, the cytosolic loop formed by the covalent linkage between the N- and C-termini of two SecY molecules may allow sufficient conformational freedom to respect the native SecYEG dimeric structure, both in membrane and in solution. It is thus conceivable that the various analysis performed with this hybrid reflect, at least partially, the intra- and inter-molecular interactions that normally occur in the native SecYEG dimer.
These analysis show that nucleotides modulate the SecYEG–SecA associations (Figure 5). A previous work has revealed that the binding affinity of SecA for IMVs is reduced in the presence of ATP (van der Wolk et al., 1998). Here, it is shown that in the presence of ADP, both monomeric and dimeric SecA are stably associated with the linked SecYEG complex. Upon addition of ATP or non-hydrolyzable ATPγS, the SecA dimer tends to dissociate from the SecYEG-binding site. In contrast, in the same circumstances, the SecA monomer remains stably associated. Furthermore, an increase in the amount of the SecA monomer bound to the linked SecYEG complex is detected in the presence of ATPγS. This increase may correspond to the monomerization of the SecYEG-bound SecA dimer or to an increased affinity of the SecA monomer upon binding of the nucleotide. Clearly, nucleotides modulate the binding affinity and binding stoichiometry of SecA at the SecYEG dimer. Further studies will be required to probe the dynamics of the SecYEG–SecA interaction, but the observed nucleotide-dependent SecYEG–SecA stoichiometry may reflect important aspects of this interaction during ATP-driven preprotein translocation.
Accordingly, a link may exist between the modulation of SecYEG–SecA associations by nucleotides and the SecYEG–SecA ATPase (Figure 7) since both are diminished by magnesium in solution. However, a repression of the endogenous SecA ATPase by Mg2+ also takes place in solution (Kim et al., 2001; this study) and the lipid-stimulated SecA ATPase occurs at low Mg2+ concentration (Lill et al., 1990). The molecular mechanism of ATP binding and hydrolysis is not yet well defined, but it appears that magnesium increases the energy barrier for the SecYEG-dependent SecA activation. It is suggested from the SecA crystal structure that a low Mg2+ concentration favors rapid nucleotide exchange at the ATP-binding folds and contributes to the so-called SecA domain dissociation reaction (Hunt et al., 2002). In solution, the interaction of SecA and SecYEG may further contribute to the SecA domain dissociation, thereby leading to the activation of the SecA ATPase. The reconstitution of the SecYEG–SecA ATPase in detergent solution provides a novel in vitro assay to assess the mechanism of the SecA activation, without the contribution of the lipid bilayer and the SecA lipid ATPase (Lill et al., 1990).
A recent study shows that a monomerized SecA mutant retains some translocation activity while the SecA dimer dissociates upon contact with acidic phospholipids (Or et al., 2002). During translocation, only monomeric SecA is found tightly associated with the translocation complex upon detergent solubilization (Figure 7). Altogether, the findings indicate that the SecA monomer has a key role during the translocation reaction but do not exclude the participation of the SecA dimer or other multimers. Hydrophobic peptide alters the dimeric conformation of SecA, toward dissociation but also polymerization (Or et al., 2002; Benach et al., 2003). The cross-linking experiments also suggest that polymeric SecA may be associated with the linked SecYEG dimer when SecA is present at high concentration (Figures 4 and 5). A high concentration of SecA allows proton-motive force-independent translocation (Yamada et al., 1989). Recently, electron microscopy studies and crystal structure analysis have revealed that the interface of the SecA dimer, and possibly higher order oligomers, forms a pore-like structure which may be important for the function of SecA (Sharma et al., 2003; Wang et al., 2003). These observations further complicate the current understanding of the SecA-dependent translocation mechanism. The results presented in this study strongly suggest that changes in the oligomeric associations within and between SecA and SecYEG protomers play a critical role during the translocation reaction. The reconstitution of the SecA–SecYEG complex in solution amenable to biochemical analysis provides us with a novel tool to analyze these molecular interactions further.
Materials and methods
Plasmids
Plasmids pTrcEHAYG and pBadEhisYG were described previously (Duong and Wickner, 1997; Collinson et al., 2001). The _Bam_HI–_Sal_I DNA fragment encoding His6-SecE was replaced in pTrcEHAYG to give plasmid pTrcEhisYG. To construct pTrcEHAYYG and pTrcEhisYYG, a 1.3 kb _Sal_I–_Sac_I DNA fragment encoding the secY ORF with the _Sac_I restriction site replacing the stop codon was generated by PCR and inserted in pTrc99a to give pTrcY′. Another 1.3 kb _Sac_I–_Xba_I DNA fragment encoding the secY ORF but with the _Sac_I restriction site replacing the initiation codon was inserted into pTrc99a to give pTrc′Y. The secY gene carried by pTrcY′ and pTrc′Y was sequenced. The _Sal_I–_Sac_I and _Sac_I–_Xba_I DNA fragments extracted from pTrcY′ and pTrc′Y were co-inserted into pTrcEHAYG and pTrcEhisYG digested with _Sal_I and _Xba_I to excise the secY gene. To construct C-terminal His6-tagged SecA, a 1.3 kb 5′_Nde_I–3′_Eco_RV DNA fragment was generated by PCR. Another 1.3 kb 5′_Eco_RV–3′_Xho_I DNA fragment encoding the second half of the secA ORF, but with the _Xho_I restriction site replacing the stop codon, was generated by PCR. The two PCR fragments were co-inserted into pET-21a (Novagen). The 2.5 kb _Nco_I DNA fragment encoding almost the entire secA gene was replaced by the same fragment extracted from pT7-SecA2 (generous gift from Dr D.Oliver) to give pET-SecAhis. A stop codon was introduced at Val831 to give pET-SecA95his.
Protein purification
SecB, proOmpA and proOmpA–BPTI were purified as described (Hartl et al., 1990). The purification of the SecYEhisG and SecYYE2hisG2 complexes was achieved by Ni2+-chelating chromatography, according to the procedure described by Collinson et al. (2001). The purified Sec complexes were stored at –80°C in TSG buffer (50 mM Tris–HCl pH 7.9, 150 mM NaCl, 10% glycerol, 1 mM DTT) containing 0.04% DDM. His-tagged SecA was purified by Ni2+-chelating chromatography, then by gel filtration on a pre-packed Sephacryl S-200 HR column equilibrated in HSG buffer (50 mM K-HEPES pH 7, 50 mM NaCl, 10% glycerol, 1 mM DTT). His-tagged SecA is fully active and was used throughout this study.
Radiolabeling
Cell growth and 35S metabolic labeling of SecAhis were performed as described by Eichler and Wickner (1997). [35S]SecAhis was purified by Ni2+-chelating chromatography and desalted on a G-25 spin column (Bio-Rad) equilibrated in TSG buffer. The specific activity of the purified [35S]SecA was ∼2 × 105 c.p.m./µg. 125I-labeling of the SecYEG and SecYYE2G2 complexes, proOmpA and proOmpA–BPTI was as previously described (Duong and Wickner, 1997; Bessonneau et al., 2002). The specific activity of the 125I-labeled SecYEG complexes was ∼1.5 × 106 c.p.m./µg.
ProOmpA translocation assay
ProOmpA translocation assays were performed in 50 µl of TL buffer (50 mM Tris–HCl pH 7.9, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT) containing SecAhis (40 µg/ml), SecB (40 µg/ml), BSA (200 µg/ml), IMVs (50 µg/ml), ATP (2 mM) and [125I]proOmpA (∼60 000 c.p.m.; 2 µg/ml). The transmembrane proton gradient was abolished by CCCP (carbonyl cyanide _m_-chlorophenylhydrazone; 20 µM) when necessary. After 5 min incubation at 37°C, translocation reactions were stopped on ice and treated with proteinase K (1 mg/ml, 15 min), trichloroacetic acid (TCA)-precipitated and analyzed by 12% SDS–PAGE and autoradiography. Quantitative proOmpA translocation was performed as described above, except that [125I]proOmpA was pre-mixed with unlabeled proOmpA (2–34 µg/ml). Reactions were incubated 15 min at 37°C with 2 mM ATP. Translocated [125I]proOmpA was visualized by autoradiography and analyzed by scanning densitometry using the ImageQuant software. The data were quantified by comparison with a [125I]proOmpA standard curve. The [125I]proOmpA–BPTI translocation assay is described in the legend of Figure 7.
Cross-linking assay
Both SecA and SecYYE2G2 were desalted a Sephadex G-25 column equilibrated in CL buffer (50 mM K-HEPES pH 7, 150 mM NaCl, 10% glycerol, 0.04% DDM). Cross-linking reactions were performed in 20 µl of CL buffer with 1 mM of EGS (ethylene glycol bis-succinimidylsuccinate; Sigma) for 15 min at 22°C. Reactions were terminated by addition of 1/10 vol. of quenching buffer (2 M glycine, 1 M Tris–HCl pH 8.0). The samples were incubated with SDS (0.1%, 10 min) under vigorous mixing followed by gel electrophoresis and autoradiography.
Other methods
Preparation of linear gradient blue native gels, electrophoretic conditions and electroblotting were as described by Schägger and von Jagow (1991). BN–gels were calibrated with high molecular weight markers (ferritin, 440 kDa/880 kDa; catalase, 232 kDa; BSA, 66 kDa/132 kDa). Immunoblots were visualized using the ECL reagents (AP Biotech). Protein concentrations were determined using the Bradford reagent (Biorad). The ATPase activity was measured by the colorimetric method of Lanzetta et al. (1979). Absorption was measured at 660 nm and compared with a standard curve prepared with a phosphate salt. SecA ATPase assays were performed in duplicate and the graph in Figure 6 represents an average of two independent experiments. Reconstitution of the SecYEG or SecYYE2G2 complex into liposomes was as previously described (Collinson et al., 2001).
Acknowledgments
Acknowledgements
I am grateful to Drs J.Eichler, I.Collinson and D.Belin for critical reading of the manuscript, and V.Besson for technical assistance during this work. This work was supported by the CNRS and by the Fondation pour la Recherche Médicale.
References
- Benach J. et al. (2003) Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J. Biol. Chem. 278, 3628–3638. [DOI] [PubMed] [Google Scholar]
- Bessonneau P., Besson,V., Collinson,I. and Duong,F. (2002) The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J., 21, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E., Nouwen,N., van Raalte,A., Mizushima,S., Tommassen,J. and de Kruijff,B. (1995) The C terminus of SecA is involved in both lipid binding and SecB binding. J. Biol. Chem., 270, 7902–7907. [DOI] [PubMed] [Google Scholar]
- Breyton C., Haase,W., Rapoport,T.A., Kuhlbrandt,W. and Collinson,I. (2002) Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature, 418, 662–665. [DOI] [PubMed] [Google Scholar]
- Cao T.B. and Saier,M.H. (2003) The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys Acta, 1609, 115–125. [DOI] [PubMed] [Google Scholar]
- Collinson I., Breyton,C., Duong,F., Tziatzios,C., Schubert,D., Or,E., Rapoport,T. and Kuhlbrandt,W. (2001) Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J., 20, 2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey B.R., Economou,A., Dunn,S.D. and Shilton,B.H. (2002) The ATPase domain of SecA can form a tetramer in solution. J. Mol. Biol., 315, 831–843. [DOI] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1997) Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J., 16, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1999) The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J., 18, 3263–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A.J. (1993) SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry, 32, 13190–13197. [DOI] [PubMed] [Google Scholar]
- Economou A. (2002) Bacterial secretome: the assembly manual and operating instructions. Mol. Membr. Biol., 19, 159–169. [DOI] [PubMed] [Google Scholar]
- Economou A. and Wickner,W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell, 78, 835–843. [DOI] [PubMed] [Google Scholar]
- Eichler J. and Wickner,W. (1997) Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc. Natl Acad. Sci. USA, 94, 5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J., Brunner,J. and Wickner,W. (1997) The protease-protected 30 kDa domain of SecA is largely inaccessible to the membrane lipid phase. EMBO J., 16, 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Matlack,K.E., Jungnickel,B., Plath,K., Kalies,K.U., Miller,K.R., Rapoport,T.A. and Akey,C.W. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Lecker,S., Schiebel,E., Hendrick,J.P. and Wickner,W. (1990) The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E.coli plasma membrane. Cell, 63, 269–279. [DOI] [PubMed] [Google Scholar]
- Hendrick J.P. and Wickner,W. (1991) SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J. Biol. Chem., 266, 24596–24600. [PubMed] [Google Scholar]
- Hirano M., Matsuyama,S. and Tokuda,H. (1996) The carboxyl-terminal region is essential for SecA dimerization. Biochem. Biophys. Res. Commun., 229, 90–95. [DOI] [PubMed] [Google Scholar]
- Hunt J.F., Weinkauf,S., Henry,L., Fak,J.J., McNicholas,P., Oliver,D.B. and Deisenhofer,J. (2002) Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science, 297, 2018–2026. [DOI] [PubMed] [Google Scholar]
- Joly J.C. and Wickner,W. (1993) The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J., 12, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Ahn,T., Ko,J., Park,C. and Kim,H. (2001) Effect of divalent cations on the ATPase activity of Escherichia coli SecA. FEBS Lett., 493, 12–16. [DOI] [PubMed] [Google Scholar]
- Lanzetta P.A., Alvarez,L.J., Reinach,P.S. and Candia,O.A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem., 100, 95–97. [DOI] [PubMed] [Google Scholar]
- Lill R., Dowhan,W. and Wickner,W. (1990) The ATPase activity of SecA is regulated by acidic phospholipids, SecY and the leader and mature domains of precursor proteins. Cell, 60, 259–269. [DOI] [PubMed] [Google Scholar]
- Manting E.H., van der Does,C. and Driessen,A.J. (1997) In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J. Bacteriol., 179, 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manting E.H., van Der Does,C., Remigy,H., Engel,A. and Driessen,A.J. (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J., 19, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G., Yoshihisa,T. and Ito,K. (1997) SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. EMBO J., 16, 6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo E., Mori,H. and Ito,K. (2003) Interfering mutations provide in vivo evidence that Escherichia coli SecE functions in multimeric states. Mol. Gen. Genet., 268, 808–815. [DOI] [PubMed] [Google Scholar]
- Meyer T.H., Ménétret,J.-F., Breitling,R., Miller,K.R., Akey,C.W. and Rapoport,T.A. (1999) The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic sec61p complex. J. Mol. Biol., 285, 1789–1800. [DOI] [PubMed] [Google Scholar]
- Mori H., Tsukazaki,T., Masui,R., Kuramitsu,S., Yokoyama,S., Johnson,A.E., Kimura,Y., Akiyama,Y. and Ito,K. (2003) Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J. Biol. Chem., 278, 14257–14264. [DOI] [PubMed] [Google Scholar]
- Or E., Navon,A. and Rapoport,T. (2002) Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J., 21, 4470–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V. and Oliver,D. (1997) Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J. Biol. Chem., 272, 23239–23246. [DOI] [PubMed] [Google Scholar]
- Schägger H. and von Jagow,G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem., 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Sharma V., Arockiasamy,A., Ronning,D.R., Savva,C.G., Holzenburg,A., Braunstein,M., Jacobs,W.R.,Jr and Sacchettini,J.C. (2003) Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl Acad. Sci. USA, 100, 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilton B., Svergun,D.I., Volkov,V.V., Koch,M.H., Cusack,S. and Economou,A. (1998) Escherichia coli SecA shape and dimensions. FEBS Lett., 436, 277–282. [DOI] [PubMed] [Google Scholar]
- Snyders S., Ramamurthy,V. and Oliver,D. (1997) Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J. Biol. Chem., 272, 11302–11306. [DOI] [PubMed] [Google Scholar]
- van der Sluis E.O., Nouwen,N. and Driessen,A.J. (2002) SecY–SecY and SecY–SecG contacts revealed by site-specific crosslinking. FEBS Lett., 527, 159–165. [DOI] [PubMed] [Google Scholar]
- van der Wolk J.P., Fekkes,P., Boorsma,A., Huie,J.L., Silhavy,T.J. and Driessen,A.J.M. (1998) PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA–SecY interaction during the initiation of translocation. EMBO J., 17, 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal A.K., van der Does,C. and Driessen,A.J. (2001) Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J. Biol. Chem., 276, 32559–32566. [DOI] [PubMed] [Google Scholar]
- Wang H.W., Chen,Y., Yang,H., Chen,X., Duan,M.X., Tai,P.C. and Sui,S.F. (2003) Ring-like pore structures of SecA: implication for bacterial protein-conducting channels. Proc. Natl Acad. Sci. USA, 100, 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe P.B., Rice,M. and Wickner,W. (1985) Effects of two sec genes on protein assembly into the plasma membrane of Escherichia coli. J. Biol. Chem., 260, 1836–1841. [PubMed] [Google Scholar]
- Woodbury R.L., Hardy,S.J. and Randall,L.L. (2002) Complex behavior in solution of homodimeric SecA. Protein Sci., 11, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T.L. and Wickner,W.T. (2000) Evaluating the oligomeric state of SecYEG in preprotein translocase. EMBO J., 19, 4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Matsuyama,S., Tokuda,H. and Mizushima,S. (1989) A high concentration of SecA allows proton motive force-independent translocation of a model secretory protein into Escherichia coli membrane vesicles. J. Biol. Chem., 264, 18577–18581. [PubMed] [Google Scholar]