Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification (original) (raw)
Abstract
Rad18 is a ubiquitin E3 ligase that monoubiquitinates PCNA on stalled replications forks. This allows recruitment of damage-tolerant polymerases for damage bypass and DNA repair. In this activity, the Rad18 protein has to interact with Rad6, the E2 ubiquitin-conjugating enzyme, ubiquitin, PCNA and DNA. Here we analyze the biochemical interactions of specific domains of the Rad18 protein. We found that the Rad6/Rad18 complex forms stable dimers in vitro. Consistent with previous findings, both the Ring domain and a C-terminal region contribute to the Rad6 interaction, while the C-terminus is not required for the interaction with PCNA. Surprisingly we find that the C2HC zinc finger is important for interaction with ubiquitin, apparently analogous to the interactions of classical zinc fingers with ubiquitin such as found in the UBZ and UBM domains in Y-family polymerases. Finally we find that the SAP domain, but not the zinc finger domain, is capable of DNA binding in vitro.
INTRODUCTION
Living organisms are sensitive to DNA damage such as that caused by UV light or chemical mutagens and it is essential for genome stability that the DNA damage is accurately repaired. Many mechanisms are responsible for this repair, but damage that remains during DNA synthesis causes DNA replication to stall. To avoid such stalling there is a specific set of post-replicative repair enzymes that allows bypass of the damaged DNA and continuation of replication. A critical component of this post-replicative repair system is the Rad18 protein. This protein performs one of the earliest steps in the damage recognition and bypass. It acts as an E3 ligase for monoubiquitination of PCNA, together with its cognate E2, Rad6 (1). The monoubiquitination of PCNA allows switching from normal replicative polymerases to Y-family translesion polymerases (2,3). These polymerases are recruited to ubiquitinated PCNA to allow bypass of the damaged lesion (4). Monoubiquitination can also be followed by polyubiquitination by Ubc13/Mms2 as E2 and Rad5 as E3, leading to an error-free repair pathway that involves recombination with the newly synthesized strand (1,5).
The importance of Rad18 is notable, since mice and chicken DT40 cells deficient in Rad18 show sensitivity to various DNA-damaging agents and enhanced genomic instability as seen by increased spontaneous sister chromatid exchange (6). Rad18 is also involved in homologous recombination, cell-type-specific processes such as somatic hypermutation, and in S-phase it plays a role in single-strand break repair (7). In addition, Rad18 was found to play a role in meiosis (8). In agreement with this, the Rad18 protein is found in the nucleus of many different cell types, with the highest abundance in testis (9,10).
Ubiquitin E3 ligases are multidomain proteins that confer the target specificity on a ubiquitin modification system, bringing the ubiquitin conjugating E2 enzyme and the target together. The ubiquitin conjugation activity of Rad18 on PCNA has been reconstituted in vitro (11,12) and was dependent on ubiquitin E1, Rad6 as E2 enzyme, ubiquitin, Mg2+ and ATP, as well as on PCNA loaded onto DNA by the RFC complex. Under these conditions, the enzyme can efficiently monoubiquitinate all three monomers of PCNA. In this process, Rad18 must interact with the E2, Rad6 (13), and the target, PCNA (1) and it was also found to interact with Pol-eta (14) and single-stranded DNA (15). Rad18 utilizes multiple domains for these interactions.
At its N-terminus Rad18 contains a Ring domain, common in ubiquitin E3-ligases, where the Ring domains interact with the cognate E2. In the case of mouse Rad18 (16), it was shown that the E2 Rad6 interacts with the Rad18 Ring domain, and that mutations in this region result in increased sensitivity to DNA-damaging agents. A second region, in the C-terminal domain of Rad18, is also involved in Rad6 binding (5,15,17). Deletion of the peptide 340–395 results in loss of Rad6 interaction in vitro, although localization to damage was not affected (14).
An unusual C2HC Zinc-finger (ZnF, residues 201–225 in human Rad18), has been identified in Rad18, which was suggested to have the potential to bind to DNA (18,19). A sequence that contains this ZnF, between residues 83 and 248, is important for dimer formation according to two-hybrid studies performed in yeast (5) and mammalian Rad18 (16,20). This self-association is disrupted in a mutant, where one of the Zn-binding cysteines is replaced by a phenylalanine (C207F). These experiments (16) gave rise to the suggestion that the ZnF domain is critical for dimerization. The C207F mutation also interferes with auto-monoubiquitination and localization of the Rad18 protein to CPD damage on DNA (21), reinforcing the notion that the ZnF region is of critical importance for Rad18 function.
A SAP domain (243–282 in human Rad18) is located just C-terminal of the ZnF region. This is a domain type named after SAF-A/B, Acinus and Pias, the three proteins where it was first identified (22). In these proteins the SAP domains are involved in DNA interaction. In Rad18, this domain was found to be important for localization to pol-eta-containing foci (21). Regions of the protein that include the ZnF and SAP domain are sufficient for localization to DNA damage even in the absence of DNA replication but mutation of C207F interferes with this localization.
In the C-terminus of human Rad18 Watanabe et al. (14) identified a region (401–445) that is important for Pol-eta binding. Upon UV-damage Rad18 and Rad6 are critical for formation of pol-eta-containing foci on stalled replication forks (14), but other translesion polymerases, such as Rev1 play an additional role in these complexes.
Altogether considerable knowledge on the Rad18 domain structure has been acquired, but relatively few experiments have been performed on the purified mammalian proteins. Here we use biophysical methods to study protein–protein interactions in vitro and map functional domains on mammalian Rad18 proteins. We show that Rad6/Rad18 forms stable dimers of heterodimers and our in vitro ubiquitination reactions and mapping of Rad6 correlates well with published data. Surprisingly we find that only the SAP domain, and not the ZnF domain, is capable of DNA recognition in vitro. In contrast, we find that the ZnF domain binds to ubiquitin, with an affinity similar to other ubiquitin interaction motifs.
MATERIALS AND METHODS
Cloning and expression
Bicistronic PET25b plasmid (Novagen) was constructed containing both genes expressed via one promotor but with two separate ribosome-binding sites (rbs). hRad18 was encoded untagged after the first rbs already present in the plasmid (NdeI/NcoI), while hHR6B was given a His6 N-terminal tag and its own rbs encoded in the reverse Rad18 PCR primer (AflIII/XhoI). A three-point ligation resulted in the dicistronic vector PET25b—hRad18—His6hHR6B. Alternatively, the genes were cloned separately into pET28a (Rad6) and pET22b (Rad18). The isolated Ring domain was cloned into pGEX4T, and the ZnF, SAP and ZnF-SAP domains were cloned into pETM30 (generous gift from Arie Geerlof). Mutagenesis reactions were carried our using a Quikchange kit (Stratagene) according to manufacturer's protocol. Plasmid DNA sequences were confirmed by in-house sequencing.
Protein purification
Overnight pre-cultures were used to inoculate large cultures of LB broth at 37°C. When cell densities reached an absorbance of 1.0 at 600 nm, they were induced with a final concentration of 0.3 mM IPTG and 100 μM ZnCl2 and left to express at 15°C overnight (typically 15–17 h). Cleared cell lysates were incubated with Talon beads (Rad6/Rad18 complexes) or GST beads (GST-ZnF, GST-SAP, GST-ZnF-SAP and GST-RING) (Clontech) and washed with 10-column volumes of 50 mM Tris pH 8.0, 150 mM NaCl, 2 μM ZnCl2 and 5 mM β-ME. Further purification was achieved by Heparin binding for Rad6/Rad18 and finally size exclusion chromatography for all proteins. The single plasmid system PET25b—hRad18—His6hHR6B yielded ∼2 mg of pure protein complex per liter of culture, with an ∼10-fold excess of hHR6B, despite being encoded as the second gene.
Ubiquitination assays
Wheat E1 (1 μM) was pre-incubated with ubiquitin (100 μM), ATP (10 mM) and MgCl2 (10 mM) at room temperature in reaction buffer (25 mM Tris pH 8.0, 150 mM NaCl, 2 μM ZnCl2 and 5 mM β-ME) for 5 min. Rad6/Rad18 (10 μM) and PCNA (35 μM) were added and the reaction incubated at 30°C for 1 hour, or longer when stated. All concentrations are final. Samples were boiled in reducing and denaturing loading buffer and run on 12% SDS–acrylamide gel.
Analytical gel filtration
Forty-two micromolar samples of Rad18/Rad6 were incubated at 4°C for 30 s with a 4-fold excess of PCNA in 25 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-ME, 2 μM ZnCl2. Fifty microliters of this mixture, Rad6/Rad18 or PCNA were analyzed on a 2.4 ml SMART GF Superdex 200 column (S200), PC 3.2/30 (Amersham Biosciences). Alternatively, equivalent experiments were performed with GST-RING and Rad6, alone or in combination, and analyzed on a Superdex 75 (S75), PC 3.2/30 column. Fractions were collected and analyzed by electrophoresis on a 15% SDS-PAGE gel.
Multi-angle static light-scattering (MALLS)
MALLS experiments were performed at 4°C on a Mini-Dawn light scattering detector (Wyatt Technology) online with a Superdex S75 10/30 column in 25 mM Tris pH 8.0, 150 mM NaCl, 2 μM ZnCl2, 5 mM β-ME buffer. Refractive index and light scattering detectors were calibrated against toluene and BSA.
Mass spectrometry to determine ubiquitination sites
Ubiquitination sites were identified from in-gel digested protein bands by online reverse-phase nanoscale liquid chromatography tandem mass spectrometry (RP-nanoLC-MS/MS) using an AB-MDS Sciex QSTAR pulsar quadrupole time of flight mass spectrometer. The extracted tryptic peptide mixtures were auto-sampled onto a RP-C18 column packed in a nanoLC emitter, separated with a 60 min linear gradient from 4–40% MeCN in 0.5% AcOH and eluted directly into the mass spectrometer via a nano-electrospray interface. The mass spectrometer was operated in the data-dependent mode for automatically switching between MS and MS/MS acquisition. Peptide ubiquitination sites were identified via protein database searching of the resulting tandem mass spectra using Mascot.
GST pull-down assay
Twenty-five microliters of glutathione-beads were incubated at 4°C with either buffer (25 mM Tris pH 8.0, 150 mM NaCl, 2 μM ZnCl2, 5 mM β-ME), 500 μg GST or 500 μg GST-ZnF and excess protein was washed out with buffer. The beads were then incubated with 500 μg of ubiquitin for 10 min before washing with buffer. The beads and flow-through solutions were analyzed by electrophoresis on a 15% SDS–PAGE gel.
Electrophoretic mobility shift assay (EMSA) with ubiquitin
Native polyacrylamide gels were run with a final concentration of 200 μM GST-cleaved ZnF with 0, 10, 20, 40, 60, 80, 100, 140 and 200 μM ubiquitin. A control run was performed with final concentration of 100 μM GST with 0, 5, 10, 20, 30, 40, 50, 70 and 100 μM ubiquitin. After a short (30 min) incubation at 4°C, the samples were loaded on a 10% native polyacrylamide gel with a glycerol-based native loading buffer, run at 50 V for 1.5 h at room temperature and stained with Coomassie brilliant blue.
EMSA with DNA
A 500 nM hRad18 + hRad6 was pre-incubated with 0.1 nM 32P-labeled 35T DNA for 7 h at 4°C and either cold 35T or cold ds 20-mer was added in concentrations of 0, 0.01, 0.03, 0.1, 0.3, 1 and 10 μM and left overnight at 4°C. The data shown was with S471A Rad18 as this construct was slightly more stable, although wild-type Rad18 bound with a similar affinity (data not shown). Native loading buffer was added and the samples were loaded on a 4% native polyacrylamide gel at 4°C at 75 V for 45 min. The gel was dried in a slab-dryer and exposed on a phospho-imager plate for 1 h.
Surface plasmon resonance
All experiments were performed at 10°C on a Biacore T-100 instrument (Biacore AB). A specific DNA-binding surface was prepared by binding biotinylated 40-T or gel-filtered 40-mer double-stranded DNA to a SA sensor chip to a density of 20–100RU, while GST-ZnF was immobilized using an amine coupling procedure. GST-ZnF, GST-SAP or ubiquitin in 25 mM Tris, pH 8.0, 125 mM NaCl, 2 μM ZnCl2, 5 mM β-ME and 0.05% [vol/vol] surfactant p20 were injected over the sensor chip at 30 μl min−1 with a 60 or 150 s association phase followed by a 10 min dissociation phase. Binding was also tested at higher flow rates and showed no change in interaction characteristics. The sensor surface was regenerated using a 60 s pulse of 0.2 M glycine-HCl, pH 2.0 followed by a 60 s pulse of 0.05% SDS. Standard double referencing data subtraction methods were used before analysis of kinetics. Curve fitting and other data analyses were performed using Biaevaluation software (Biacore AB).
RESULTS AND DISCUSSION
In order to study the functional domain architecture of the Rad6/Rad18 complex, we have expressed this protein complex in Escherichia coli (Figure 1A, constructs used). We used both the mouse and human versions of the protein in a bicistronic co-expression construct in E. coli (23), with a His-tag on the Rad6 protein for initial purification on Talon beads. The Rad6/Rad18 complex was found to be soluble and could be purified away from a significant excess of Rad6 using heparin affinity and gel-filtration chromatography (Figure 1B). This excess of Rad6 was also seen when expressing Rad6 and Rad18 from independent vectors, although to somewhat lesser extent (23).
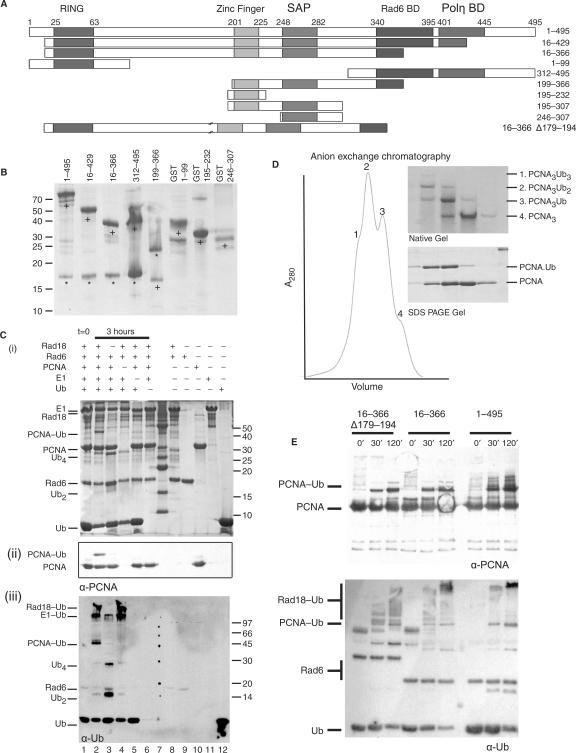
Figure 1.
Rad6/Rad18 complex expression and E3-ligase activity (A) Residue boundaries of the human Rad18 fragments used in the studies (to size with beginning and end residue). (B) A 15% SDS–PAGE gel of the purified fragments; lanes 1–5 coexpression of Rad18 (+) with Rad6 (*). The Rad6 in lane 5 runs at a high molecular weight because of a long linker between the His tag and the protein. (C) Ubiquitination of PCNA. (i) A 15% SDS–PAGE gel, (ii) anti-PCNA western blot and (iii) anti-ubiquitin western blot. Rad6, Rad18, PCNA, ubiquitin E1, ubiquitin, ATP and magnesium incubated for 0 h (lane 1) or 3 h (lanes 2–6), where ubiquitinated PCNA was produced (lane 2). Ubiquitination of PCNA is not evident in reactions lacking Rad18, PCNA, E1 or Ubiquitin (lanes 3–6) or in presence of isolated proteins (lanes 8–12). Ub2 and Ub4 chains were also produced by the Rad6. (D) PCNA trimers are modified with 0, 1, 2 or 3 ubiquitins. Anion exchange chromatography profile shown on native (top) and SDS–PAGE (bottom) gels. (E) Anti-PCNA and anti-ubiquitin western blots showing that Rad18 16–366 Δ179–194, Rad18 16–366 and Rad18 1–495 can all actively ubiquitinate PCNA in the in vitro assay.
To determine whether the complex is functional, we established an in vitro assay for PCNA ubiquitination. In the presence of ubiquitin, ubiquitin-activating enzyme E1, magnesium and ATP, we could detect significant ubiquitination of PCNA as seen by analysis on SDS-PAGE and confirmed on western blot against PCNA and ubiquitin (Figure 1C). The reaction appears to proceed in a stepwise manner, with partially modified trimers more abundant than tri-ubiquitinated PCNA. The activity is presumably suboptimal, since our PCNA is not loaded onto DNA (11,12). Nevertheless, the reaction is dependent on the presence of E1 and E2/E3. Moreover the complex is capable of modifying all three sites on PCNA trimers, as indicated in native PAGE (Figure 1D), while peptide mapping of the ubiquitin site detected K164 as the only modified site (data not shown). Thus our assay recapitulates many of the significant features of the PCNA modification and it can serve as a functional test of enzymatic activity.
We expressed a series of Rad18 deletion constructs as complexes with Rad6 using either the polycistronic system or a two-plasmid system, in order to map the site of interaction of the ligands of Rad18 onto the complex. One complex, Rad18312–495 with Rad6 was produced as a side product of expressing the full-length mouse and human Rad6/Rad18 protein complex. This was the result of a modified Kozak sequence, which apparently acts as an alternative translation start site at methionine 312, as confirmed by N-terminal sequencing. We tested various complexes in the activity assay and we showed that Rad1816–429 is still fully active (data not shown) and Rad1816–366 has maintained substantial activity on PCNA (Figure 1E). Thus the proposed Pol-eta interaction domain (14) is not essential for the PCNA ubiquitination activity by Rad6/Rad18.
N- and C-terminal regions of Rad18 interact with Rad6
The interaction between Rad6 and Rad18 has been studied in detail in yeast, where a C-terminal region of Rad18 was found to be capable of binding to Rad6 (15). In our co-expression studies, we could detect stable complexes in any construct that contained a region ending at Rad18 residue 366. Rad18312–495, which lacked the N-terminal region, still formed a stable complex with Rad6. In Rad18199–366, which was shortened at both the N- and C-terminus, the complex with Rad6 was detectable, but dissociated in high-salt conditions. This is in good agreement with the mapping data from Watanabe et al. (14) who showed that the C-terminal interaction region maps to residues 340–395.
In addition to the C-terminal interaction we could also detect significant interaction of the Ring domain (Rad181–99) of Rad18 with Rad6. Using analytical gel filtration we showed that the peaks of GST-Ring domain could be shifted by Rad6 alone (Figure 2A). This component of the interaction with Rad6 in the Ring domain of Rad18 is in agreement with Tateishi et al. (16) who showed that the interaction was sensitive to mutations in the Ring domain. We also detected a shift of the GST-Ring protein by the Rad6/Rad18312–495 complex (data not shown). This shows that the two Rad6 interaction regions on Rad18 are non-overlapping. Rad18 therefore, interacts with Rad6 through its C- and N-terminus independently. Both these regions are predicted to be partially alpha-helical from primary sequence. In the N-terminal domain predicted helices flank the well-characterized Ring topology, while the C-terminal Rad18-binding domain is predicted to be composed of two β-strands followed by an α-helix. Both of these regions show sequence similarity with Zip3, another proposed ubiquitin E3 ligase (24). Similar E2/E3 interaction is seen in the crystal structure of UbcH7/c-Cbl, where Ubc5 is flanked by two alpha helices from c-Cbl originating from sequentially distant helices (25).
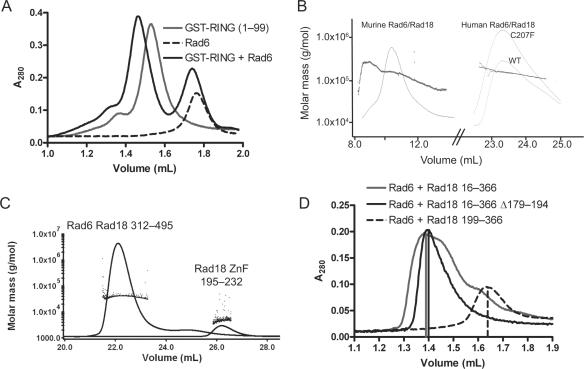
Figure 2.
Rad6 binds to Rad18 and forms a dimer of heterodimers. (A) S75 analytical gel filtration profile showing that mixtures of Rad6 and GST-fused Rad18 (1–99) co-eluted at a higher molecular weight compared with the single proteins. (B) MALLS profile showing the refractive index and scattered light as full-length murine Rad6/Rad18 complexes were eluted from an S75 gel filtration column. The constant scattered light signal across the peak in the gel filtration profile is indicative of a single stable species, while the degree of scattering gives molecular weights consistent with a dimer of heterodimers. Human wild type and C207F Rad6/Rad18 were eluted from an S200 gel filtration column. The MALLS profile revealed them also to be dimers of heterodimers, although the profiles show them to be less monodisperse, probably because the samples were partially degraded. (C) MALLS profile showing the refractive index and scattered light as Rad6/Rad18312–495 complexes and Rad18 ZnF were eluted from an S75 gel filtration column, showing both species to be monomeric. (D) Analytical gel filtration profiles on an S200 analytical gel filtration column. Rad6/Rad18 16–366 and Rad6/Rad18 16–366 Δ179–194 eluted as dimers of heterodimers, while Rad6/Rad18 199–366 eluted as a single heterodimer.
Rad6/Rad18 is a dimer of heterodimers
Our gel-filtration data indicate that the human and mouse Rad6/Rad18 complex forms a dimer of heterodimers. We initially showed the existence of such a complex using chemical cross-linking followed by mass spectrometry (17). To confirm these indirect data, we performed multi-angle static light-scattering (MALLS) online with a gel-filtration setup on the mouse Rad6/Rad18 complex. In this experiment we found that the molecular weight of the complex is 171 kDa, in good agreement with the calculated value of 161.6 kDa for 2xRad6 (19.7 kDa) and 2xRad18 (61.1 kDa) molecules (Figure 2B). Similarly, the human complex showed a molecular weight of 165 kDa compared with a calculated value of 150.5 kDa for 2xRad6 (19.1 kDa) and 2xRad18 (56.2 kDa) molecules (Figure 2B).
In order to detect biochemically which domains are important for dimerization in the Rad6/Rad18 complex, we first analyzed human Rad6/Rad18312–495 and Rad18195–232 (ZnF) by gel filtration and MALLS (Figure 2C) and the data were consistent with monomeric species. Thus, the molecular weight of Rad6/Rad18312–495 was 41.1 kDa compared with 21.0 kDa for Rad18312–495 and 20.0 kDa for Rad6. Similarly the observed molecular weight of the ZnF was 5.1 kDa compared with a predicted molecular weight of 5.6 kDa, although the error was greater at lower molecular weight.
Next we constructed a C207F mutant of human Rad18 and MALLS analysis of the purified complex with Rad6 gave a molecular weight of 171 kDa versus 150.5 kDa for the dimeric species (Figure 2B), showing that in vitro this mutant can still dimerize effectively. Finally we tested a set of deletion constructs in an analytical gel-filtration setup (Figure 2D). All species form complexes that have at least the size of dimers of heterodimers except for the fragment Rad18199–366, which only forms the monomeric Rad6–Rad18 heterodimer.
Dimerization of Rad18 has previously been shown genetically in yeast (5) and human cells (20). In yeast, it was mapped to the region comprising residues 83–248, that contains the ZnF domain, and mutation C207F in the ZnF-domain in human Rad18 could no longer be found to dimerize. Thus the ZnF domain was suggested to be the dimerization domain (20). Our in vitro data do not support this: the monomeric region (199–366) containing the ZnF domain, and the isolated ZnF were monomeric, and the C207F mutation had no effect on the oligomeric state of Rad18 in our assays. The suggested role of the self-association is localization in the cell, with the monomer form mostly found in the cytoplasm (20). In that light the stability of the dimer that we observe is surprising and could indicate that other regulatory steps, such as additional post-translational modifications of the protein are required to create the monomer state.
Taken together, our data are consistent with dimerization being mediated with the N-terminal region of Rad18, but not particularly by the ZnF. We could not directly detect dimerization of the Ring domains, since we could not make soluble Ring domain alone, and GST itself makes dimers so it was not clear if the dimerization was mediated by the GST or the Ring domain. However, the presence of higher order complexes than the GST dimer indicates that Ring/Ring dimerization may occur. Moreover, there is significant precedence for Ring/Ring dimers as shown in the Brca1/Bard1 (26) and Ring1b/Bmi1 (27) heterodimers, while homodimerization was seen in the isostructural U-box complex of Chip (28). At the sequence level, many of the dimeric Ring domains, such as Brca1, Bard1, Ring1b and Bmi1 have significant similarity to the Rad18 Ring domain in the flanking regions that are important for dimerization, and in psi-Blast searches they find each other with more ease than monomeric Ring domains, indicating that Rad18 may well form a similar type of dimer, where the flanking regions of the Ring domain create the dimer interface.
The N-terminal region of Rad18 binds to PCNA
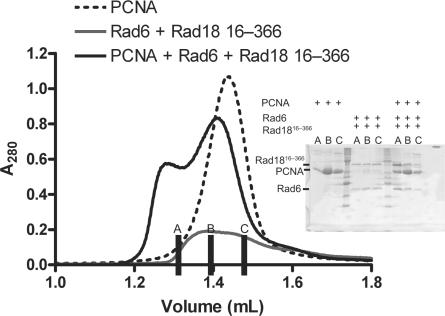
In an analytical gel filtration experiment, we could show that PCNA interacts with the Rad6/Rad18 complex: both peaks shift from their separate elution volumes to a higher apparent molecular weight.
The shorter construct of Rad1816–366 also could be shifted by interaction with PCNA in this manner (Figure 3) but the GST-Ring, GST-ZnF, GST-SAP or GST-ZnF-SAP could not increase the apparent molecular weight of the PCNA (data not shown). Therefore we conclude that the PCNA-binding motif is contained within the region 16–366, but cannot be localized to any of the domains in this region; the Ring, ZnF or SAP domains.
Figure 3.
Rad18 interaction with its target PCNA. S200 analytical gel filtration profile showing that mixtures of Rad6/Rad18 (16–366) and PCNA co-eluted at a higher molecular weight compared with the single proteins, as confirmed by SDS–PAGE (inset).
Rad18 interacts with ubiquitin covalently and non-covalently
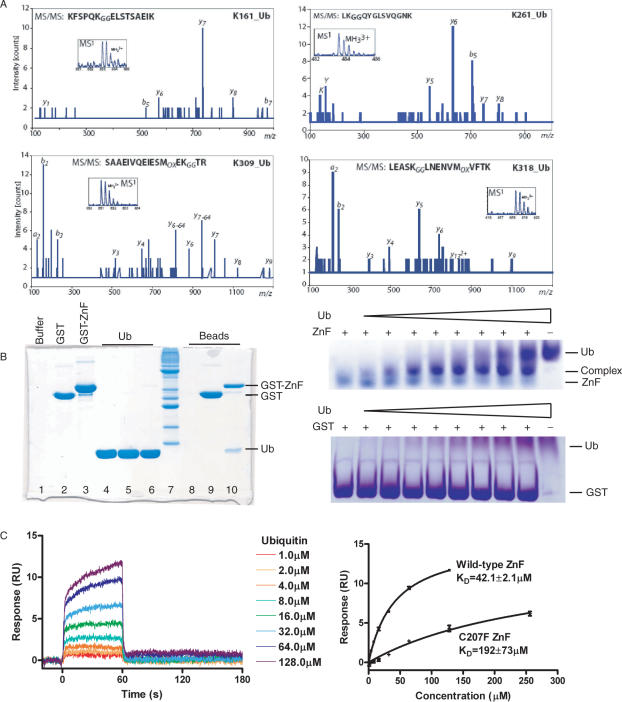
Rad18 has been shown to be monoubiquitinated in vivo (20) and in vitro (Figure 1C) in a Rad6-dependent manner. We have mapped the in vitro sites of monoubiquitination in murine Rad18, and found that ubiquitin can be attached to lysines 161, 261, 309 and 318 (Figure 4A). These residues are conserved in the human sequence.
Figure 4.
Rad18 interacts with ubiquitin covalently and non-covalently. (A) Identification of ubiquitinated lysines in Rad18 by tandem mass spectrometry. The fragmentation pattern of the triply charged tryptic peptides derived from murine Rad18 contain the ubiquitinated K161, K261, K309 and K318, respectively. The most abundant C-terminus containing fragments (y ion series) are indicated in the figure as well as the b ion series (N-terminus containing fragments). The inset in each tandem mass spectra show the peptide precursor ion that was selected for sequencing. (B) Left. Glutathione beads were loaded with buffer, GST or GST-ZnF until saturated. The flow through is shown on lanes 1–3 of the 15% SDS–PAGE gel, respectively. The beads were then saturated with ubiquitin. The flow through is shown on lanes 4–6, respectively. The beads were washed with buffer and loaded onto lanes 8–10. Ubiquitin bound to GST-ZnF (lane 10) but not GST (lane 9) or glutathione beads alone (lane 8). Right. ZnF traveled to a lesser extent on a native gel as the concentration of ubiquitin was increased, indicative of a ZnF–Ub interaction, while no interaction of GST with Ub could be observed in a control experiment. (C) The interaction of GST–ZnF was studied further by surface plasmon resonance. Wild type or C207F GST-ZnF was immobilized to a CM5 chip and ubiquitin was flowed over the chip at 1, 2, 4, 8, 16, 32, 64, 128 and 256 μM (C207F). Right. Langmuir binding curves showing the variation of mean equilibrium binding responses with ubiquitin concentration. The _K_D of the interaction was estimated to 42.1 μM for WT-ZnF or 192 μM for C207F-ZnF by non-linear curve fitting. Error bars are SD from three repetitions of the experiment.
E3 enzymes of the Ring type generally interact with the ubiquitin E2 enzyme and with their target, but several E3 enzymes also have ubiquitin interaction motifs. It has been proposed that monoubiquitination of ubiquitin-binding proteins may result in an autoubiquitin-induced conformational switch to regulate the functions of the target (29). Since the Zn-finger domain (UbZ) in Pol-eta was shown to be involved in DNA binding (30), we decided to study ubiquitin binding to the Rad18 ZnF domain.
In a pull-down experiment using GST-ZnF we could show that GST-ZnF, but not GST, will pull down ubiquitin (Figure 4B). To study this interaction in more detail we loaded the ZnF domain with increasing amounts of ubiquitin on a native PAGE gel. At higher concentrations of ubiquitin, we could see a clear shift of the Rad18 band. To quantify this interaction between ubiquitin and Rad18 ZnF domain we used surface plasmon resonance (Figure 4C). We used GST-ZnF or GST-C207F ZnF covalently coupled to a CM5 chip with ubiquitin bound at a range of concentrations. The _K_D in this flow-cell system was 42.1 μM for the WT-ZnF with fast-on/fast-off kinetics. This mutant had a 5-fold lower _K_D of 192 μM, indicating that the ZnF is indeed important for ubiquitin binding (Figure 4C). Other Zinc-fingers have been shown to interact with ubiquitin, such as the UbZ domain in pol-eta (30), with affinities in a similar range. Thus, the binding of Rad18 to ubiquitin resembles that of other ubiquitin-binding domains (31).
The C207F mutant was shown in cells to be important for the monoubiquitination of Rad18 itself (20), whereas PCNA is still ubiquitinated normally by this mutant following UV damage. In mutant cells, Rad18 altered its subcellular localization, from the cytoplasm to the nucleus and the ubiquitination of Rad18 affected its stability in a proteasome-dependent manner. We tested the C207F Rad18 complex with Rad6 in our in vitro ubiquitin conjugation assay (Supplementary Figure 1). In these assays we could not see a difference in ubiquitin conjugation of either PCNA or in the levels of autoubiquitinated Rad18. Thus there seems to be a clear discrepancy between the in vitro and in vivo results. It remains possible that the ZnF–ubiquitin interaction is indeed important for in vivo autoubiquitination to regulate the functions of Rad18 (20), due to differences in components or other regulatory events.
After submission of this manuscript, Bish and Myers (32) also showed interaction of the Rad18 Znf with ubiquitin, and reported a specificity for polyubiquitin chains, although they were not able to detect an interaction with monoubiquitin. The interaction of C2HC zinc fingers with ubiquitin appears to be general; Werner helicase interacting protein 1 interacts with ubiquitin via its C2HC zinc finger (32). The in vivo relevance of this interaction requires further study.
Rad18 interacts with DNA through its SAP domain
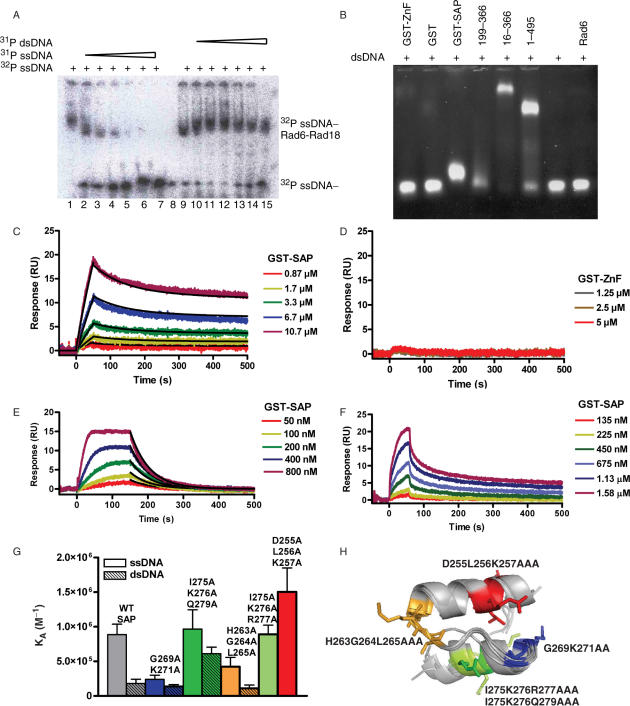
Rad18 has been reported to bind to DNA, with a preference for single-stranded DNA compared with double-stranded DNA (15). To confirm this specificity, we performed a competition experiment. Unlabeled single-stranded DNA competed efficiently for the radiolabeled single-stranded DNA whereas double-stranded DNA would not, at concentrations of up to 10 μM (Figure 5A).
Figure 5.
Rad18 interacts with DNA through its SAP domain (A) Competition experiments of ssDNA (lanes 2–7) and dsDNA (lanes 10–15) for 32P-ssDNA on a 4% native gel. Lanes 1 and 9 contain no competing DNA while lane 8 was left empty. (B) Electrophoresis mobility shift assays of dsDNA on agarose gels. The dsDNA (lane 7) was shifted by GST-SAP, Rad6/Rad18 199–366, Rad6/Rad18 16–366 and Rad6/Rad18 1–495 (lanes 3–6, respectively) but not GST-ZnF (lane 1), GST (lane 2) or Rad6 (lane 8). (C) The interaction of GST-SAP with single-stranded DNA was studied by surface plasmon resonance. 40T-biotin was immobilized to a SA biacore chip and GST-SAP was flowed over the chip at 10.7, 6.7, 3.3, 1.7 and 0.87 μM. Clear components to the off rate can be seen as a result of a bivalent interaction from dimerization of the SAP domain via the GST tag. The kinetics of the interaction was estimated by fitting the binding profile by a bivalent interaction model. (D) GST-ZnF was flowed over the chip at concentrations of 5, 2.5 or 1.25 μM. Binding to the 40T was not observed. (E–F) Equilibrium responses could be estimated for dsDNA and ssDNA at long contact times and low immobilization densities of DNA. (G) Affinities derived from Langmuir-binding isotherms of SPR data for double and triple point mutants to alanine binding to ss- and ds-DNA. Values are not stated for the interaction of I275K276R277AAA or D255L256K257AAA with dsDNA as an insufficient portion of the binding curve was sampled to accurately estimate the affinity. (H) Superposition of the SAP domains from structures 1ZBH, 1ZRJ, 2DO1, 1H1J and 1KCF. The groups of residues mutated to alanine are shown as stick representations on 1ZBH and colored blue (G269K271AA), orange (H263G264L265AAA), red (D255L256K257AAA) or lime/green (I275K276R277/Q279AAA).
The ZnF domain has been proposed as a potential DNA interaction region of Rad18 (18). In order to map the site of the DNA interaction in the human protein, we performed an electrophoresis mobility shift assay using agarose gels. We did not, however, find any DNA interaction for GST-ZnF, using an agarose gel-shift assay. In contrast we saw clear DNA binding for the adjacent SAP domain. In addition, we also found that any protein construct that contains this SAP domain results in a shift of both double-stranded (Figure 5B) and single-stranded DNA (data not shown).
We used plasmon resonance analysis in a Biacore setup to characterize the interaction of Rad18 with DNA. Single-stranded 40-T was immobilized to an SA chip and GST-ZnF or GST-SAP flowed over the chip at different concentrations. A clear binding curve was observed for the GST-SAP domain but the GST-ZnF did not interact (Figure 5C–D).
The binding curve of the SAP domain for single-stranded DNA could be fit using a bivalent analyte model, probably due to dimerization of the GST tag. The kinetics (Table 1) are consistent with an apparent affinity from the divalent interaction of the SAP domain dimers in the 1 μM range. At longer contact times and very low immobilization densities, the binding approached equilibrium and the slow component of the dissociation was no longer apparent. The dissociation could be fitted by a single exponential decay (Figure 5E), presumably because binding of a single SAP domain to the DNA was preferred at equilibrium. Fitting the subsequently derived Langmuir-binding isotherm gave an apparent affinity of 1.1 μM, which is highly consistent with the fits of the kinetic parameters (Table 1). For double-stranded DNA, analysis by SPR indicated a slightly lower affinity (5.6 μM), predominantly due to a faster off-rate (Figure 5F).
Table 1.
Kinetic parameters derived from fitting the interaction of GST-SAP with single-stranded DNA, using a bivalent interaction model
| _k_on | _k_off | |
|---|---|---|
| 1st component | 96.4 M−1 s−1 | 4.25 × 10−3 s−1 |
| 2nd component | 3.40 × 10−5 RU−1 s−1 | 2.06 × 10−4 s−1 |
In other proteins such as Ku and Pias, the SAP region is known to interact with DNA. The estimated affinity for the Pias1 N-terminal domain is in the low micromolar range (33). We tested the relevance of the Rad18 SAP domain for DNA binding by mutational analysis. Based on structural analysis of known SAP domain structures, we made three sets of mutations on the predicted surface of the SAP domain (Figure 2H) and measured their affinity for DNA (Figure 5G). An ∼4-fold reduction in affinity of G269K271AA for ssDNA versus wild type was observed while the affinity of H263G264L265AAA also seemed to be modestly reduced. Both groups of mutants lie on a loop region between the two helices of the SAP domain (Figure 2H). These regions were also implicated in binding to DNA in Pias1 by NMR-based chemical shift perturbation studies, while the regions probed by the other sets of mutants were not significantly perturbed in the Pias1 study. This implies that the mode of binding may be conserved between Rad18 and Pias1 (33). The affinity of a D255L256K257AAA appeared to be slightly higher than the wild-type interaction, although the difference was not significant, and the I275K276Q279AAA and I275K276R277AAA mutations showed no significant change in affinity.
The interaction of the Rad18 SAP domain with DNA is in good agreement with the data of Nakajima et al. (21) who showed that any Rad18 construct that contains the SAP domain can localize to DNA damage, with the single exception of the C207F mutant, located in the ZnF. Their interpretation is that it is the ZnF that is important for the DNA localization, while the SAP domain is required for pol-eta containing focus formation. Since their constructs either have both ZnF and the SAP domains or neither, their observed localization of Rad18 at sites of DNA damage may be due to the SAP domain in vivo.
CONCLUSIONS
Using biochemical and biophysical analysis we were able to unravel the protein recognition roles of various Rad18 domains and define their function. We have confirmed the previously published Rad6 interaction domains and shown that the functional unit of Rad18 activity is a dimer of Rad6-Rad18 heterodimers. We have shown that the N-terminal half of the Rad18 protein binds to PCNA by size-exclusion chromatography. Our mapping shows that the DNA recognition region in mammalian Rad18 is not localized to the ZnF domain, but to the SAP domain (Figure 6). Only fragments containing the SAP domain were able to bind DNA, as demonstrated in an electrophoresis mobility shift assay and the ZnF alone fails to do so. The SAP domain binds to DNA as shown by surface plasmon resonance. The affinity is ∼1 μM, as previously reported, and mutagenesis studies revealed that the mode of DNA binding may be conserved with other SAP family members (15). This role is not unusual for SAP domains, as these are found in other DNA-interacting proteins such as KU and PARP, the histone mRNA 3′ exonuclease, Saf-A and Pias. The importance of this domain for DNA targeting fits with the data of Nakajima et al. who found that the SAP domain of Rad18 is important for the formation of pol-eta containing foci (21).
Figure 6.
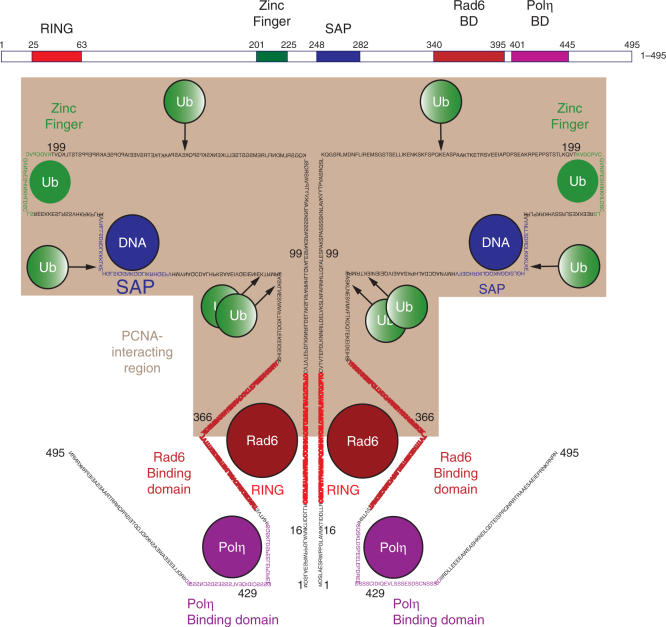
Schematic representation of ligand-interacting regions of human Rad18. The ZnF binds to ubiquitin, while the SAP domain binds to DNA. Rad6 interacts with the Ring domain and between residues 340 and 395 towards the C-terminus. Polη binds between residues 401 and 445 and the PCNA-interacting region is contained within residues 16–366 (shaded). Four sites of autoubiquitination of Rad18 were detected at K161, K261, K309 and K318, which are conserved between murine and human Rad18.
Rather than DNA binding, we clearly demonstrate that the Rad18 ZnF domain plays a critical role in binding ubiquitin. This capacity is in line with other zinc fingers that form ubiquitin-binding motifs. The affinity that we measure is in the range determined for UBMs. The precise role of such ubiquitin-binding motifs is under much debate. Apparently this region is not critical for PCNA modification (20), thus implying a role at a different step in the Rad18 function. The effect of the C207F mutation on the dimerization and autoubiquitination (20) could not be reproduced in vitro. Nevertheless, it is possible that in vivo ubiquitin binding is a step required for autoubiquitination, which subsequently regulates Rad18 localization and degradation.
The identification of the domains responsible for the component interactions of Rad18 with DNA, ubiquitin, Rad6 and PCNA will be useful for designing mutants of Rad18 with altered ligand-binding properties to study the role of Rad18 further. Meanwhile these domains should not be envisaged as beads on a string. The presence of multiple domains that interact with Rad6 and PCNA make this obvious. The role of the dimer of Rad6/Rad18 is also of interest. Various different modes of interaction can be envisaged in which the multiple Ub and DNA-binding domains interact in different ways, and it will be interesting to see how these communicate with each other in time and space to execute the Rad18 function in translesion DNA synthesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Material]
ACKNOWLEDGEMENTS
Heinz Jacobs and Mark Luna-Vargas for critical reading of the manuscript. Jaap-Willem Back for cross-linking Mass-spec, Gert Vriend for bioinformatics analysis of Ring domains, Alexander Fish for static light-scattering, Joyce Lebbink for PCNA constructs, Puck Knipscheer for assistance with ubiquitin assays, Roald van der Laan, Henk Roest and Jan Hoeijmakers for Rad6/Rad18 cDNA, Alan Lehman for pol-eta and human Rad18 cDNA, Rene Bernards for PCNA construct, Arie Geerlof for pETM30 vector, NWO-CW for Pionier grant to T.K.S., Lymphoma and Leukemia for grant to V.N., EU-SPINE and RUBICON for enabling collaboration with M.M. Funding to pay the Open Access publication charges for this article was provided by NWO-CW.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 2.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 3.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 4.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulrich HD, Jentsch S. Two Ring finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Laan R, Uringa EJ, Wassenaar E, Hoogerbrugge JW, Sleddens E, Odijk H, Roest HP, de Boer P, Hoeijmakers JH, et al. Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J. Cell Sci. 2004;117:5023–5033. doi: 10.1242/jcs.01368. [DOI] [PubMed] [Google Scholar]
- 9.Masuyama S, Tateishi S, Yomogida K, Nishimune Y, Suzuki K, Sakuraba Y, Inoue H, Ogawa M, Yamaizumi M. Regulated expression and dynamic changes in subnuclear localization of mammalian Rad18 under normal and genotoxic conditions. Genes Cells. 2005;10:753–762. doi: 10.1111/j.1365-2443.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- 10.van der Laan R, Roest HP, Hoogerbrugge JW, Smit EM, Slater R, Baarends WM, Hoeijmakers JH, Grootegoed JA. Characterization of mRAD18Sc, a mouse homolog of the yeast postreplication repair gene RAD18. Genomics. 2000;69:86–94. doi: 10.1006/geno.2000.6220. [DOI] [PubMed] [Google Scholar]
- 11.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly V, Prakash S, Prakash L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 16.Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal. Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 18.Jones JS, Weber S, Prakash L. The Saccharomyces cerevisiae RAD18 gene encodes a protein that contains potential zinc finger domains for nucleic acid binding and a putative nucleotide binding sequence. Nucleic Acids Res. 1988;16:7119–7131. doi: 10.1093/nar/16.14.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 20.Miyase S, Tateishi S, Watanabe K, Tomita K, Suzuki K, Inoue H, Yamaizumi M. Differential regulation of Rad18 through Rad6-dependent mono- and polyubiquitination. J. Biol. Chem. 2005;280:515–524. doi: 10.1074/jbc.M409219200. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima S, Lan L, Kanno S, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 23.Romier C, Ben Jelloul M, Albeck S, Buchwald G, Busso D, Celie PH, Christodoulou E, De Marco V, van Gerwen S, et al. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- 24.Perry J, Kleckner N, Borner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl Acad. Sci. USA. 2005;102:17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: Ring domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 26.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric Ring-Ring complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 27.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell. Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 30.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bish RA, Myers MP. Werner helicase interacting protein 1 (WRNIP1) binds polyubiquitin via its zinc finger domain. J. Biol. Chem. 2007;282:23184–23193. doi: 10.1074/jbc.M701042200. [DOI] [PubMed] [Google Scholar]
- 33.Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, et al. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]