The control of phosphatidylinositol 3,4-bisphosphate concentrations by activation of the Src homology 2 domain containing inositol polyphosphate 5-phosphatase 2, SHIP2 (original) (raw)
Abstract
Activation of class Ia PI3K (phosphoinositide 3-kinase) produces PtdIns_P_3, a vital intracellular mediator whose degradation generates additional lipid signals. In the present study vanadate analogues that inhibit PTPs (protein tyrosine phosphatases) were used to probe the mechanisms which regulate the concentrations of these molecules allowing their independent or integrated function. In 1321N1 cells, which lack PtdIns_P_3 3-phosphatase activity, sodium vanadate or a cell permeable derivative, bpV(phen) [potassium bisperoxo(1,10-phenanthroline)oxovanadate (V)], increased the recruitment into anti-phosphotyrosine immunoprecipitates of PI3K activity and of the p85 and p110α subunits of class Ia PI3K and enhanced the recruitment of PI3K activity stimulated by PDGF (platelet-derived growth factor). However, neither inhibitor much increased cellular PtdIns_P_3 concentrations, but both diminished dramatically the accumulation of PtdIns_P_3 stimulated by PDGF or insulin and markedly increased the control and stimulated concentrations of PtdIns(3,4)_P_2. These actions were accounted for by the ability of PTP inhibitors to stimulate the activity of endogenous PtdIns_P_3 5-phosphatase(s), particularly SHIP2 (Src homology 2 domain containing inositol polyphosphate 5-phosphatase 2) and to inhibit types I and II PtdIns(3,4)_P_2 4-phosphatases. Thus bpV(phen) promoted the translocation of SHIP2 from the cytosol to a Triton X-100-insoluble fraction and induced a marked (5–10-fold) increase in SHIP2 specific activity mediated by enhanced tyrosine phosphorylation. The net effect of these inhibitors was, therefore, to switch the signal output of class I PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2. A key component controlling this shift in the balance of lipid signals is the activation of SHIP2 by increased tyrosine phosphorylation, an effect observed in HeLa cells in response to both PTP inhibitors and epidermal growth factor.
Keywords: lipid phosphatase, phosphoinositide 3-kinase (PI3K), protein phosphatase, reactive oxygen species
Abbreviations: EGF, epidermal growth factor; IGF-1, insulin-like growth factor-1; PDGF, platelet-derived growth factor; PI, phosphoinositide; PI3K, phosphoinositide 3-kinase; ROS, reactive oxygen species; bpV(phen), potassium bisperoxo(1,10-phenanthroline)oxovanadate (V); PTP, protein tyrosine phosphatase; PTEN, phosphatase and tensin homologue deleted on chromosome ten; SH2, Src homology 2; SHIP, SH2 domain containing inositol polyphosphate 5-phosphatase; siRNA, small interfering RNA; GFP, green fluorescent protein; SHP1, Src homology 2 domain tyrosine phosphatase 1; GST, glutathione transferase: PEI, polyethyleneimine
INTRODUCTION
PIs (phosphoinositides) play a central role in intracellular signalling, acting as substrates for phospholipase C or PI3K (PI 3-kinase), but also by recruiting or regulating the activity of modular proteins with appropriate PI-specific binding domains [1–4]. The receptor-regulated (class I) PI3Ks use PtdIns(4,5)P_2 as their substrate to synthesize PtdIns_P_3 [2], a vital lipid signal which regulates diverse cellular processes [3,4]. PtdIns_P_3 is degraded by the 3-phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome ten) [5] and by several 5-phosphatases including SHIP and SHIP2 [SH2 (Src homology 2) domain containing inositol polyphosphate 5-phosphatases 1 and 2] [6,7] but the two routes are not redundant, perhaps reflecting their distinct lipid products [8]. The removal of PtdIns_P_3 by 5-phosphatases produces PtdIns(3,4)P_2 which is then degraded, most probably to PtdIns3_P [9]. However, PtdIns(3,4)P_2 and PtdIns3_P also can be synthesized directly from PtdIns4_P and PtdIns by the non-receptor-regulated class II and III PI3Ks respectively [2]. The role of the PtdIns3_P_ produced by class III PI3K in the endocytic pathway is well-documented [10] but whether or not the formation of this lipid by PtdIns_P_3 metabolism gives rise to a functionally and/or spacially discrete pool of PtdIns3_P_ is uncertain. Similarly, the distinct functions of separate PtdIns(3,4)_P_2 pools remain unclear.
Nevertheless, several lines of evidence indicate that PtdIns(3,4)_P_2 is an important, independent lipid signal. First, mechanisms exist for its synthesis both independently of and co-ordinately with that of other 3-phosphorylated PIs. Secondly, two enzymes, the types I and II inositol polyphosphate 4-phosphatases, exhibit a strong preference for PtdIns(3,4)_P_2 over other possible PI or inositol phosphate substrates, at least in vitro [11–13]. Loss of the type I enzyme accounts for the genetic defect which underlies the neuronal loss characteristic of Weeble mice [14] and mouse embryo fibroblasts derived from these animals show elevated concentrations of PtdIns(3,4)_P_2 [15], implying an important role for this enzyme in PtdIns(3,4)_P_2 metabolism and a crucial function(s) for its substrate and/or product lipids. A previous study showing that the type I 4-phosphatase regulates cell growth downstream of the GATA-1 transcription factor [16] supports a similar conclusion. Finally, several proteins bind with high selectivity to PtdIns(3,4)_P_2 [17,18] whereas others show a similar preference for PtdIns(3,4)_P_2 and PtdIns_P_3 over other PIs [19], suggesting that this lipid has precise molecular targets through which its function(s) may be mediated.
The sequential production of PtdIns_P_3, PtdIns(3,4)P_2 and PtdIns3_P following recruitment of class I PI3Ks by activated growth-factor receptor tyrosine kinases and other cell-surface receptors suggests that these lipids occur at a common sub-cellular location and implies that their distinct functions may be integrated. To allow flexible signalling, however, mechanisms must exist beyond the activation of PI3K to allow the concentrations of each lipid to be modulated independently. The regulation of type I 4-phosphatase activity by Rab5 may reflect one such mechanism [15]. More pertinently, however, the regulation of appropriate enzyme activities by ROS (reactive oxygen species) may be crucial in this regard. ROS are generated endogenously in response to both physiological and pathological stimuli, including growth factors that activate class I PI3Ks and may play important roles in mediating the actions of these agents [20,21]. Key intracellular targets of ROS include members of the PTP (protein tyrosine phosphatase) family whose catalytic mechanism relies critically upon a redox-sensitive cysteine residue [21,22]. Enzymes in this category include the PTPs which reverse the tyrosine phosphorylation of growth factor receptors stimulated by ligand binding, but also may include the PtdIns_P_3 3-phosphatase, PTEN [23,24] and the 4-phosphatases that probably limit PtdIns(3,4)_P_2 concentrations [13]. Significantly, exogenous oxidative stress not only stimulates a PtdIns_P_3-dependent increase in the activity of Akt mediated by inhibition of PTEN [23] but also evokes selective accumulation of PtdIns(3,4)_P_2 [25]. Thus ROS may play an important physiological role as modulators of PI3K signalling [22,26].
In the present study we have used vanadate derivatives which are established inhibitors of PTP activity [27] to modify the balance of the 3-phosphorylated PIs produced following the stimulation of PI3K and to identify the vulnerable targets. These are likely to include endogenous tyrosine kinase/phosphatase cycles as well as other enzymes that are also common physiological targets of ROS. Our results in PTEN null, 1321N1 astrocytoma cells reveal endogenous mechanisms that allow independent control of the concentrations of the PtdIns_P_3 and PtdIns(3,4)_P_2 synthesized sequentially following stimulation of class I PI3K. This control, exerted unexpectedly by the concerted activation of PtdIns_P_3 5-phosphatase(s) and inhibition of cysteine-dependent PtdIns(3,4)_P_2 4-phosphatase(s), switches the output of class I PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2 and has important implications for the physiological regulation of PI3K signalling by growth factor receptors.
EXPERIMENTAL
Materials
Cells were from the European Tissue Culture Collection. Antibodies against the following were obtained from the sources indicated: p85 and anti-phosphotyrosine (clone 4G10) (Upstate Biotechnology); p110α (Cell Signalling Technology); type I inositol polyphosphate 4-phosphatase (Santa Cruz Biotechnology); anti-SHIP2 antibodies were raised in sheep against a peptide (DPAHKRLLLDTLQLSK) in the C-terminus of the human enzyme and purified by affinity chromatography on Sepharose columns coupled to the immunizing peptide by the Division of Signal Transduction Therapy, University of Dundee, Dundee, Scotland, U.K. SHP1 (Src homology 2 domain tyrosine phosphatase 1) was provided by Dr N. Leslie, Division of Molecular Physiology, University of Dundee, Dundee, Scotland, U.K. Potassium bpV(phen) [bisperoxo(1,10-phenanthroline)oxovanadate (V)] and TLC plates were from Merck. Sodium vanadate was treated prior to use as described previously [28]. EDTA-free protease inhibitor cocktail tablets were from Roche. Radiolabels were from PerkinElmer. Synthetic inositol phosphates and PIs (the dipalmitoyl form) were from Cell Signals. Other reagents were from sources described previously [29–31].
Cell culture
1321N1 astrocytoma cells and HeLa cells were cultured in 6-well dishes as described previously [29,30].
Preparation of cell lysates
Cells were incubated with and without stimuli or inhibitors and detergent lysates prepared as described previously [31] or with the following modifications. Extracts for SHIP2 analysis were prepared using an alternative lysis buffer [lysis buffer A: 50 mM Tris (pH 7.5 with HCl), 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate, 1 mM sodium vanadate, 0.1% (v/v) 2-mercaptoethanol, 0.5% (w/v) Triton X-100, 0.1 mM benzamidine and 0.1 mM PMSF]. Cell lysates for analysis of inositol polyphosphate 4-phosphatase were prepared as follows. A 75 cm2 flask of cells (typically approx. 5–10 mg of protein) was rinsed twice with 10 ml of buffer containing 154 mM NaCl and 10 mM Hepes/NaOH at pH 7.4, then lysed in 2.5 ml of ice-cold buffer [lysis buffer B: 100 mM KCl, 20 mM NaCl, 25 mM Hepes/KOH (pH 7.4) and 0.25% (w/v) Triton X-100, freshly supplemented with 25 mM dithiothreitol, 0.25 mM PMSF, 0.25 mM benzamidine and 0.05 mM leupeptin]. The cell debris was collected and centrifuged at 20000 g for 10 min at 4 °C. The supernatant was then mixed with 0.25 ml of packed Chelex 100 beads (100–200 mesh, sodium form, pre-washed with lysis buffer B) on ice for 15 min to remove Mg2+ ions and thereby minimize in subsequent assays the activity of Ins(1,3,4)_P_3 1-phosphatase. The latter activity was eliminated more conveniently and routinely by using a final assay buffer containing an excess of EDTA over Mg2+ ions (5 mM and 2 mM respectively), but the ability of EDTA to chelate vanadate [32] precluded this approach in some experiments. Cell extracts were used immediately or stored at −80 °C.
Cell fractionation
The incubation buffer was aspirated and replaced with 1 ml of ice-cold hypotonic lysis buffer [20 mM Hepes (pH 7.5 with NaOH), 10 mM KCl, 1.5 mM MgCl2, 5 mM NaF, 5 mM sodium pyrophosphate, 5 mM sodium β-glycerophosphate and EDTA-free protease inhibitor cocktail] and the cells were allowed to swell on ice for 15 min. The cells were then scraped and the debris was passed five times through a 25 mm gauge needle before centrifugation at 20000 g for 10 min at 4 °C. The supernatants (designated as the cytosolic fraction) were adjusted to final concentrations of 1 mM EDTA, 1 mM EGTA and 0.5% (w/v) Triton X-100 and aliquots (30 μl) were analysed for SHIP2 by SDS/PAGE and immunoblotting. The pellets were washed once with 1 ml of ice-cold hypotonic lysis buffer then extracted on ice with 0.5 ml of ice-cold lysis buffer A for 15 min. After centrifugation at 20000 g for 10 min at 4 °C, the supernatants (reflecting the Triton-soluble membrane fraction) were collected and aliquots (30 μl) were analysed for SHIP2 as described above. The residual pellets (reflecting the Triton-insoluble membrane fraction) were washed once more with 1 ml of ice-cold lysis buffer A then solubilized in SDS/PAGE sample buffer (100 μl) and aliquots (30 μl) were also analysed for SHIP2.
Cellular [3H]PIs
Cellular [3H]PIs were analysed as described previously [29,30] except that for some experiments the final HPLC analysis used a modified gradient elution which afforded better resolution of the [3H]GroPIns_P_s derived from PtdIns3_P_, PtdIns4_P_ and PtdIns5_P_. This was achieved by elution with 1.0 M NH4H2PO4 adjusted to pH 3.0 with H3PO4 (B) at 1 ml/min and the following gradient: 0 min, 0% B; 5 min, 0% B; 25 min, 7.5% B; 55 min, 7.5% B; 130 min, 82.5% B; 131 min 0% B; 150 min 0% B.
Preparation of radiolabelled substrates
The preparation of [3-33P]PtdIns_P_3 and [3-32P] or [3-33P]Ins(1,3,4,5)_P_4 was as described previously [33,34] except that the latter product was purified by chromatography on Dowex anion exchange resin. This was achieved on a column (2.0 m×0.8 cm) of AG1X8 resin (200–400 mesh, formate form) eluted sequentially with water (10 ml), 0.35 M ammonium formate (pH 3.0 with formic acid; 40 ml, to remove residual ATP), 0.75 M ammonium formate/0.1 M formic acid (10 ml, to remove residual Ins_P_3) and 1.0 M ammonium formate/0.1 M formic acid (10 ml, to recover the Ins_P_4 product). The final fraction was spiked with 5 μmoles of NH4H2PO4 and dried under vacuum until all of the ammonium formate had sublimed. The [3-32P] or [3-33P]Ins(1,3,4,5)_P_4 recovered was dissolved in water and stored at −20 °C.
The degradation of [3-32P] or [3-33P]Ins(1,3,4,5)_P_4 to [3-32P] or [3-33P]Ins(1,3,4)_P_3 was achieved using a recombinant, catalytically active fragment of SHIP2 corresponding to amino acid residues 418–884 of the human protein. Radiolabelled Ins_P_4 was incubated at 37 °C for 3 h in 0.5 ml of buffer [100 mM KCl, 20 mM NaCl, 2 mM MgCl2, 50 mM Hepes/KOH (pH 7.4) and 1 mg/ml BSA] containing 25 μg/ml SHIP2. The radiolabelled Ins_P_3 produced was then extracted and purified as described previously [29], spiked with NH4H2PO4 and desalted as described above, then dissolved in water (0.25–0.5 ml) and stored at −20 °C. The radiochemical purity of the [3-32P] or [3-33P]Ins(1,3,4,5)_P_4 and of [3-32P] or [3-33P]Ins(1,3,4)_P_3 obtained was verified routinely by PEI (polyethyleneimine) TLC on plates developed in 2.5 M ammonium formate/1.0 M formic acid.
Immunoprecipitation and in vitro assays
PI3K was immunoprecipitated from cell lysates using 1 μg of anti-phosphotyrosine antibody and assayed using PtdIns as the substrate as described previously [31] except that after separation of the reaction products by TLC, the spots corresponding to [32P]PtdIns3_P_ were detected by phosphorimaging and quantified in arbitrary units using AIDA (Advanced Image Data Analysis) software. The identity of the [32P]PtdIns3_P_ formed was confirmed by analysis of its [32P]GroPIns3_P_ deacylation product in the presence of appropriate internal standards by HPLC performed as described previously [29,30] except that the separation of the GroPIns_P_ isomers was achieved by isocratic elution at a flow rate of 1 ml/min with 50 mM NH4H2PO4 adjusted to pH 3.0 with H3PO4. SHIP2 was immunoprecipitated with 2 μg of an anti-SHIP2 antibody from cell lysates pre-cleared by prior incubation with 5 μl of packed protein-G-coupled Sepharose beads for 45 min at 4 °C. The immunoprecipitated material was washed at 4 °C, once with 1 ml of lysis buffer A and twice with 1 ml of assay buffer (see below) lacking MgCl2. After aspirating the final wash, immunoprecipitates were mixed with 50 μl of assay buffer [50 mM Tris (pH 7.5 with HCl), 0.1 mM EGTA, 0.03% (v/v) Brij-35, 0.1% (v/v) 2-mercaptoethanol and 2.5 mM MgCl2] and [3-33P]Ins(1,3,4,5)_P_4 substrate (approx. 0.002 MBq/assay) and the samples were incubated with continuous shaking at 30 °C. At the times indicated aliquots (0.5 μl) of reaction mix were spotted on to PEI-TLC plates and these were developed as described above to separate the [3-33P]Ins(1,3,4)_P_3 released. The PEI-TLC plates were dried, phosphorimaged and radioactive spots quantified as described above. Alternatively, immunoprecipitated SHIP2 activity was measured using [3-33P]PtdIns_P_3 (1 μM, 0.002 MBq/assay) as substrate under identical conditions but with 1% (w/v) Triton X-100 in place of Brij-35. The reactions were stopped by the addition of 0.6 ml of methanol/chloroform/12 M HCl (80:40:1, by vol.), 0.2 ml of chloroform and 0.31 ml of 0.1 M HCl, the lower phases dried under vacuum and the [3-33P] lipid products separated by TLC on silica gel 60 plates pre-dipped in 50 mM potassium oxalate/5 mM EDTA at pH 8.5 in 50% (v/v) methanol and developed in chloroform/methanol/acetone/acetic acid/water (7:5:2:2:2, by vol.). The radioactive spots were quantified as described above.
Inositol polyphosphate 4-phosphatase activity was measured in cell lysates. When required, these were immunodepleted of 4-phosphatase by two sequential treatments each with 5 μg of antibody coupled to protein-G-coupled Sepharose beads. The 4-phosphatase activity was determined by measuring the dephosphorylation of [3-32P] or [3-33P]Ins(1,3,4)_P_3 to [3-32P] or [3-33P]Ins(1,3)_P_2. Assays were performed at 37 °C in 25 μl of reaction buffer [100 mM KCl, 20 mM NaCl, 25 mM Hepes/KOH (pH 7.4), 10 mM dithiothreitol, 0.1 mM PMSF, 0.1 mM benzamidine, 0.02 mM leupeptin, 0.1% (w/v) Triton X-100 and, where appropriate, inhibitors as indicated or 5 mM EDTA and 2 mM MgCl2] containing 0.003–0.03 MBq of [3-32P] or [3-33P]Ins(1,3,4)_P_3 at the concentrations indicated. Assays were started by the addition of either substrate (5 μl) or cell extract (approx. 20–40 μg of protein) and aliquots (0.5 μl) of the reaction mix were collected immediately and at further intervals. The samples were spotted immediately on to 10 cm×10 cm PEI-TLC plates and these were developed in 1.25 M ammonium formate/0.5 M formic acid and the reaction products quantified as described above. The 4-phosphatase activity was calculated as the sum gain in radiolabelled Ins(1,3)_P_2 plus Pi above the same at zero time.
SDS/PAGE and immunoblotting
This was performed as described previously [31]. Primary antibodies were applied for either 2 h at room temperature (20 °C) at 0.1 μg/ml (SHIP2 and anti-phosphotyrosine) or overnight at 4 °C at 2 μg/ml (inositol polyphosphate 4-phosphatase). Proteins were visualized using appropriate horseradish peroxidase-coupled secondary antibodies and ECL® (enhanced chemiluminescence) reagent (Amersham).
SHIP2 dephosphorylation
SHIP2 was immunoprecipitated as described above. After washing twice with 1 ml of SHP1 phosphatase buffer [50 mM Tris (pH 7.5 with HCl), 0.1 mM EGTA, 0.03% (v/v) Brij-35 and 10 mM dithiothreitol] samples were shaken for 30 min at 30 °C with recombinant, full length GST (glutathione transferase)–SHP1 (0.4 mg/ml) or with their buffer controls. After transfer on to ice, washing twice with ice-cold lysis buffer A and twice with SHIP2 assay buffer, samples were analysed by immunoblotting or for activity as described above.
Fluorescence microscopy of 1321N1 cells transfected with GFP (green fluorescent protein)–SHIP2
1321N1 astrocytoma cells, grown to 70% confluence on glass coverslips, were transfected with full-length SHIP2 cloned into an EGFP (enhanced GFP) C1 vector (from Clontech), using FuGENE™ 6 according to the manufacturer's protocol. After 24 h, the medium was replaced with modified Krebs–Henseleit buffer as described previously [29] and cells were treated as indicated. After aspiration of the incubation buffer, cells were fixed with 3% (w/v) paraformaldehyde in PBS for 30 min at room temperature, washed 3 times with PBS and mounted in DABCO/Mowiol [13% (w/v) Mowiol 4-88, 33% (v/v) glycerol, 2.5% (w/v) DABCO and 13 mM Tris (pH 8.5 with HCl)]. Images were recorded using an Improvision microscope with a GFP filter set.
RESULTS
Inhibition of PTPs reveals a mechanism which switches the output of receptor-stimulated PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2
To study the effects of PTP inhibitors on the production of 3-phosphorylated PIs in response to growth factor stimulation of PI3K we used a 1321N1 astrocytoma cell line which lacks PTEN so that PtdIns(3,4,5)_P_3 catabolism is channelled exclusively via 5-phosphatase(s). Table 1 compares the effects on PI concentrations in these cells of two potent PTP inhibitors, sodium vanadate and bpV(phen), a cell permeant vanadate analogue [35]. The results show that these inhibitors evoked changes in the cellular concentrations of several PIs, inducing similar, though not identical effects, presumably reflecting some differences in both their cellular targets and accessibility. Most strikingly, however, sodium vanadate and bpV(phen) induced dramatic increases of >10–1000 fold in the concentration of PtdIns(3,4)_P_2, implying that inhibitor-sensitive targets are essential for the synthesis and/or degradation of this lipid in particular. The profound increase in the accumulation of PtdIns(3,4)_P_2 in response to bpV(phen), though not the similar smaller effect of sodium vanadate, was accompanied by a marked reduction in the concentration of PtdIns(4,5)_P_2, suggesting the conversion of this lipid into PtdIns(3,4)_P_2 via class I PI3K. Significantly, however, neither PTP inhibitor altered the concentration of PtdIns_P_3. Thus we next examined the effects of both inhibitors on the accumulation of PtdIns_P_3 and PtdIns(3,4)_P_2 stimulated by PDGF (platelet-derived growth factor) in these cells.
Table 1. The effects of vanadate analogues on cellular PI concentrations.
Cells pre-labelled with [3H]inositol were incubated for 30 min in the absence or presence of 1 mM sodium vanadate or 0.1 mM bpV(phen) before the extraction and analysis of the [3H]PIs. Values for control and sodium vanadate represent the means±S.E.M. of ten experiments performed in duplicate and for bpV(phen) of three experiments performed similarly. The lipid concentrations for control cells are expressed as percentages of the total cellular [3H]PI and for sodium vanadate and bpV(phen) treated cells as a proportion of the appropriate control. *Concentrations of PtdIns4_P_ and PtdIns5_P_ were measured as a combined total but in a subset of four experiments in which these lipids were determined separately the concentration of PtdIns5_P_ as a percentage of total PI was 0.085±0.018% and 0.247±0.017%** respectively in control and sodium vanadate treated cells. **Indicates a significant difference (_P_≤ 0.05) from the appropriate control determined by Student's t test.
| Stimulus | |||
|---|---|---|---|
| Lipid | Control (% total PI) | Sodium vanadate (fold control) | bpV(phen) (fold control) |
| PtdIns | 80.615±1.556 | 0.89±0.01 | 0.95±0.03 |
| PtdIns3_P_ | 0.425±0.039 | 1.73±0.01** | 3.95±1.86 |
| PtdIns4/5_P_* | 7.511±0.586 | 2.30±0.26** | 0.77±0.06 |
| PtdIns(3,5)_P_2 | 0.022±0.003 | 1.96±0.22** | 8.63±4.77 |
| PtdIns(3,4)_P_2 | 0.004±0.001 | 13.68±4.59** | 5164.00±2243** |
| PtdIns(4,5)_P_2 | 11.407±1.026 | 1.09±0.02 | 0.27±0.11** |
| PtdIns_P_3 | 0.011±0.001 | 1.15±0.015 | 2.17±1.25 |
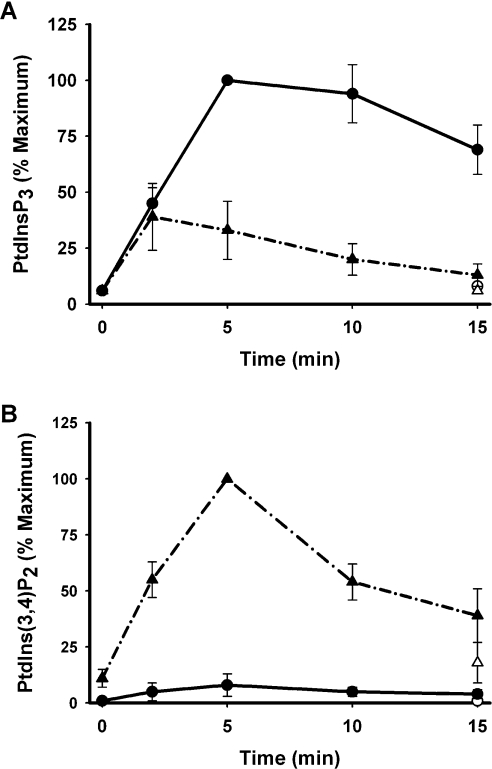
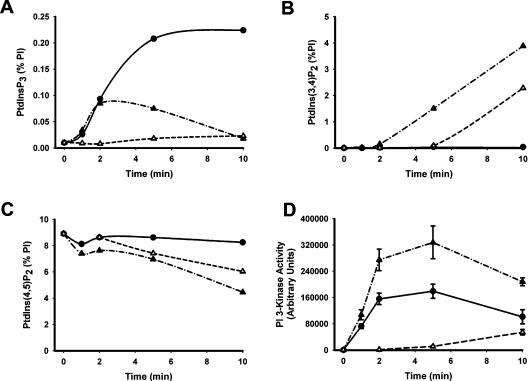
Figures 1(A) and 1(B) show that PDGF stimulated a ∼10–20-fold and a ∼5–10-fold increase in the cellular concentrations of PtdIns_P_3 and of PtdIns(3,4)_P_2 respectively, consistent with the established recruitment of class I PI3K by the PDGF receptor [36]. The increased concentrations of both lipids were persistent over at least 15 min and, importantly, the stimulated, steady-state concentration of PtdIns_P_3 was ∼10-fold greater than that of PtdIns(3,4)_P_2, as is also the case in response to activation of the IGF-1 (insulin-like growth factor 1) receptor in 1321N1 cells [30]. By comparison, sodium vanadate alone increased the concentration of PtdIns(3,4)_P_2 but not that of PtdIns_P_3. Unexpectedly however, sodium vanadate markedly altered the response to receptor stimulation, potentiating PDGF-stimulated accumulation of PtdIns(3,4)_P_2 but suppressing that of PtdIns_P_3. Figure 2 reveals similar effects of bpV(phen) but with some important distinctions. Figures 2(A)–2(C) show that bpV(phen) alone had little effect on the cellular concentration of PtdIns_P_3 but caused an impressive increase in the concentration of PtdIns(3,4)_P_2 which was manifest only after a delay of several minutes and was mirrored by an almost quantitatively identical reduction in the concentration of PtdIns(4,5)_P_2. When added together with PDGF, however, bpV(phen) attenuated rapidly and powerfully the accumulation of PtdIns_P_3 stimulated by receptor activation of PI3K but also induced a faster and more pronounced accumulation of PtdIns(3,4)_P_2 and loss of PtdIns(4,5)_P_2 than when present alone. Figure 2(D), however, also shows that, under the same conditions, bpV(phen) increased the recruitment of PI3K stimulated by PDGF by ∼2-fold. Sodium vanadate also increased the PDGF-stimulated PI3K activity measured in anti-phosphotyrosine or PDGF receptor immunoprecipitates (results not shown) and the IGF-1 receptor-mediated association of PI3K with insulin receptor substrate-1 [31]. Thus these results show that the net effect of these PTP inhibitors is to alter the balance of PtdIns_P_3 and PtdIns(3,4)_P_2 accumulated in response to the activation of class I PI3K by growth factor receptors and suggest that this is achieved by their concerted intervention at three key steps.
Figure 1. Sodium vanadate attenuates receptor-mediated accumulation of PtdIns_P_3 but potentiates that of PtdIns(3,4)_P_2.
Cells pre-labelled with [3H]inositol were treated for 30 min in the absence (circles) or presence (triangles) of 1 mM sodium vanadate then further incubated as indicated in the absence (open symbols) or presence (closed symbols) of PDGF (50 ng/ml) and the [3H]PIs then determined. The results are expressed as a percentage of the maximum accumulation of either PtdIns_P_3 (A) or PtdIns(3,4)_P_2 (B) and represent the means±S.E.M. of three experiments performed in duplicate.
Figure 2. bpV(phen) enhances PDGF-stimulated PI3K activity but switches its cellular product from PtdIns_P_3 to PtdIns(3,4)_P_2.
(A–C) Cellular PI concentrations. Cells labelled with [3H]inositol were incubated as indicated with 50 ng/ml PDGF (●), 0.1 mM bpV(phen) (△) or both reagents (▲) before the extraction and measurement of the [3H]PIs shown. The concentration of each lipid is expressed as a percentage of the total cellular PI and the results reflect the mean values of duplicate measurements in a single experiment representative of two that gave similar results. (D) PI3K activity. Cells were incubated as indicated with 50 ng/ml PDGF (●), 0.1 mM bpV(phen) (△) or both reagents (▲) and the PI3K activity present in anti-phosphotyrosine immunoprecipitates was then measured. The results represent the means±S.E.M. of three experiments performed in duplicate.
PTP inhibitors promote selective PtdIns(3,4)_P_2 accumulation by activation of class I PI3K, inhibition of 4-phosphatases and stimulation of PtdIns_P_3 5-phosphatase(s)
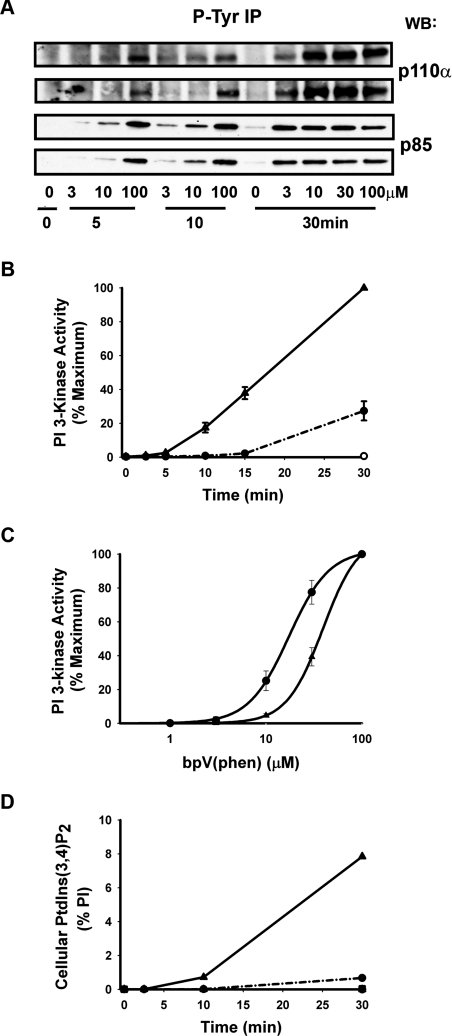
Figure 3(A) shows that bpV(phen) stimulated the recruitment of the p85 and p110α regulatory and catalytic subunits respectively, of class Ia PI3K into complexes immunoprecipitated by anti-phosphotyrosine antibodies. This occurred in a time- and concentration-dependent manner which correlated closely with the recruitment of PI3K activity into similar immunoprecipitates as shown in Figures 3(B) and (C). Furthermore, the product of this activity when determined using PtdIns as a substrate was confirmed as PtdIns3_P_ (results not shown), excluding the possibility that this reflects activity other than that of 3-kinases and consistent with the in vitro capability of class I PI3K(s) to 3-phosphorylate each of the three major PIs [2]. Most importantly, however, Figure 3(D) demonstrates that the cellular accumulation of PtdIns(3,4)_P_2 stimulated by bpV(phen) also occurs with a very similar time- and concentration-dependence suggesting that the synthesis of this lipid in response to bpV(phen) is mediated predominantly by class Ia PI3K. In support of this, Figure 3(D) additionally shows that the bpV(phen)-stimulated accumulation of PtdIns(3,4)_P_2 was prevented almost completely by pre-treatment of cells with wortmannin, a broad spectrum inhibitor of PI3K(s). However, the dramatic, linear accumulation of this lipid shown in Figure 3(D) cannot result from activation of PI3K alone and must also reflect the ability of bpV(phen) to inhibit PtdIns(3,4)_P_2 hydrolysis.
Figure 3. bpV(phen) stimulates a time- and concentration-dependent recruitment of class Ia PI3K that accounts for the cellular accumulation of PtdIns(3,4)_P_2.
(A) The recruitment of class Ia PI3K. Cells were incubated for the times and with the concentrations of bpV(phen) indicated and the recruitment of the p85 and p110α subunits of class Ia PI3K into anti-phosphotyrosine immunoprecipitates was then determined. The results show duplicate determinations from one experiment representative of three which gave similar data. (B) Time-course of PI3K activity. Cells were incubated with bpV(phen) at 0.01mM (●) or 0.1 mM (▲) for the times indicated and the PI3K activity associated with anti-phosphotyrosine immunoprecipitates was then measured. The results are the means±S.E.M. of three experiments performed in duplicate and are expressed relative to the maximum activity achieved. (C) Concentration-dependence of PI3K activity. Cells were incubated for 10 min (▲) or 30 min (●) with the concentrations of bpV(phen) indicated before measurement of the PI3K activity as described above. The results are expressed as a percentage of the maximum activity achieved at the separate times and represent the means±S.E.M. of five experiments performed in duplicate (▲) or the mean and range of two experiments performed in duplicate (●). (D) The cellular accumulation of PtdIns(3,4)_P_2. Cells labelled with [3H]inositol were treated for 30 min without (circles and triangles) or with (squares) wortmannin (100 nM), then further incubated with 0.01mM (●) or 0.1 mM (▲ and ■) bpV(phen) for the times indicated before the measurement of PtdIns(3,4)_P_2. Values are the means of duplicate determinations in a single experiment representative of two which gave similar results and show PtdIns(3,4)_P_2 concentrations as a percentage of total cellular PI.
To address directly the susceptibility of PtdIns(3,4)_P_2 degradation to PTP inhibitors, we developed an assay for the inositol polyphosphate 4-phosphatases thought to regulate the cellular concentrations of this lipid. This assay exploits the ability of these 4-phosphatases to hydrolyse Ins(1,3,4)_P_3 in vitro [11–13] providing a specific index of 4-phosphatase activity in crude cell extracts. Immunodepletion experiments using antibodies specific for the type I 4-phosphatase revealed that the type I enzyme accounted for approx. 60% of the activity in cell lysates with the remainder presumably reflecting the presence of the type II enzyme. Our results suggest that both enzymes are potently and non-competitively inhibited by sodium vanadate and bpV(phen) with indistinguishable IC50 values of ∼0.1 μM (results not shown). These results establish the vulnerability of both 4-phosphatases to PTP inhibitors and support the view that blockade of these enzymes is likely to contribute to the cellular accumulation of PtdIns(3,4)_P_2 induced by these agents.
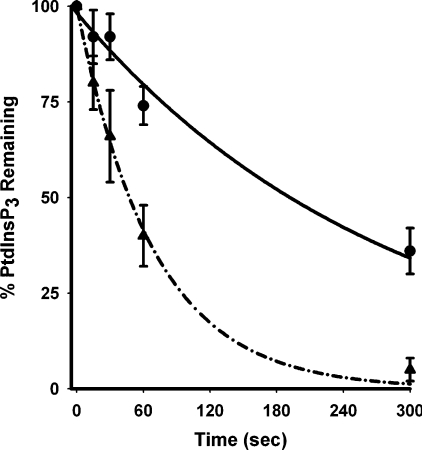
Our results showing the ability of PTP inhibitors to attenuate agonist-stimulated PtdIns_P_3 accumulation suggest that they also activate PtdIns_P_3 5-phosphatases. As 1321N1 cells lack PTEN, this issue could be addressed directly by determining the rate of PtdIns_P_3 hydrolysis in these cells as illustrated in Figure 4. This shows the results of experiments in which cells pre-treated in the absence or presence of sodium vanadate were then stimulated briefly with PDGF to promote accumulation of PtdIns_P_3 before the addition of a high concentration of wortmannin to rapidly prevent further PI3K activity and allow measurement of the rate at which existing PtdIns_P_3 was then removed by 5-phosphatases. Figure 4 shows that the PtdIns_P_3 accumulated in response to PDGF alone was metabolized with a half-time of ∼3 min. By contrast, the PtdIns_P_3 accumulated in the co-presence of PDGF and sodium vanadate was removed with a half-time of ∼30–60 s demonstrating a several-fold increase in the rate of 5-phosphatase activity that is consistent with the lower accumulation of PtdIns_P_3 under these conditions (see Figure 1A). These results clearly show that PTP inhibitors can increase PtdIns_P_3 5-phosphatase activity, but it should be emphasized that they probably underestimate the maximum extent to which this can occur. Thus although for practical purposes we elected to use sodium vanadate for these experiments, it is apparent that bpV(phen) must act more powerfully, since despite increasing PDGF-stimulated PI3K activity ∼2-fold, it abolished concomitantly, the 10–20-fold PDGF-stimulated rise in PtdIns_P_3 concentrations (compare with Figures 2A and 2D), suggesting an increase in PtdIns_P_3 5-phosphatase activity by 1–2 orders of magnitude.
Figure 4. Sodium vanadate increases the rate of cellular PtdIns_P_3 5-phosphatase activity.
Cells labelled with [3H]inositol were treated for 30 min in the absence (●) or presence (▲) of 1 mM sodium vanadate then incubated for 10 min in the presence of PDGF (50 ng/ml) before the addition of wortmannin (10 μM) and the [3H]PtdIns_P_3 remaining at the intervals shown was then measured. The results are expressed as percentages of the PtdIns_P_3 concentrations achieved in the absence and presence of sodium vanadate respectively, immediately prior to addition of wortmannin and represent the means±S.E.M. of five experiments performed in duplicate.
PTP inhibitors stimulate the translocation of SHIP2 and a tyrosine phosphorylation-mediated increase in SHIP2 specific activity
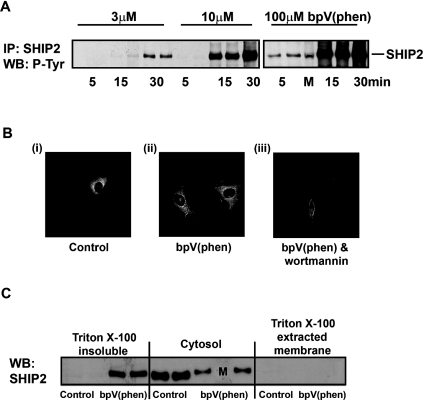
SHIP2 is a widely expressed PtdIns_P_3 5-phosphatase [6,7] which undergoes reversible tyrosine phosphorylation [37,38] and can be localized through its interaction with a variety of tyrosine phosphorylated binding partners [6,7]. Thus we speculated that this enzyme might be responsible for the increased 5-phosphatase activity induced by PTP inhibitors. Figure 5(A) shows that SHIP2 is expressed in 1321N1 cells and that its tyrosine phosphorylation is markedly increased above negligible resting levels in response to bpV(phen). Similar results were also obtained using sodium vanadate (results not shown). Both the time- and concentration-dependent tyrosine phosphorylation of SHIP2 evoked by bpV(phen) correlated with that for the recruitment of class I PI3K and the accumulation of PtdIns(3,4)_P_2 induced by this agent, consistent with a role for SHIP2 in mediating the increase in 5-phosphatase activity which minimizes the contemporary accumulation of PtdIns_P_3. The results presented in Table 2, however, suggest that only limited phosphorylation of SHIP2 may be required, showing that as little as 3 μM bpV(phen) dramatically reduced the cellular accumulation of PtdIns_P_3 stimulated by PDGF or insulin. Figures 5(B) and 5(C) demonstrate further that bpV(phen) treatment also induced a profound re-localization of over-expressed and endogenous SHIP2 respectively in 1321N1 cells. Thus in resting cells GFP-tagged SHIP2 was predominantly cytosolic but translocated to the cell periphery in response to bpV(phen). Interestingly, however, this re-localization appeared to be independent of PI3K activity as it was not blocked by concentrations of wortmannin that prevented the bpV(phen)-stimulated increase in cellular PtdIns(3,4)_P_2 concentrations by ≥95% and which markedly altered the resting cell morphology. Figure 5(C) shows that a significant proportion (≥50%) of endogenous SHIP2 also translocated from the cytosol of 1321N1 cells following bpV(phen) treatment and was recovered predominantly in the Triton X-100-insoluble fraction, consistent with the known ability of SHIP2 to interact with cytoskeletal proteins [6,7], a feature which may be associated intimately with its tyrosine phosphorylation.
Figure 5. bpV(phen) stimulates the tyrosine phosphorylation and translocation of SHIP2.
(A) Tyrosine phosphorylation of endogenous SHIP2. Cells were incubated for the times and with the concentrations of bpV(phen) indicated, then lysed and the SHIP2 protein immunoprecipitated with an anti-SHIP2 antibody and its tyrosine phosphorylation determined by SDS/PAGE and immunoblotting. The results show duplicate determinations from a single experiment representative of three that gave similar results. The lane labelled M contained markers only. (B) Translocation of GFP–SHIP2. Cells expressing GFP–SHIP2 were treated for 30 min in the absence [(i) and (ii)] or presence (iii) of wortmannin (100 nM) then further incubated for 20 min without (i) or with [(ii) and (iii)] bpV(phen) (20 μM) before the cells were fixed and examined by fluorescence microscopy. The images shown are representative of two experiments. (C) Translocation of endogenous SHIP2. Cells were incubated for 30 min with and without bpV(phen) (20 μM) and the cellular fractions indicated then prepared and analysed for SHIP2 by SDS/PAGE and immunoblotting. The results are from one experiment of three which gave similar results. The lane labelled M contained markers only.
Table 2. Receptor-mediated accumulation of PtdInsP3 is suppressed by concentrations of bpV(phen) that induce limited SHIP2 tyrosine phosphorylation.
Cells pre-labelled with [3H]inositol were treated for 30 min in the absence or presence of 3 μM bpV(phen) then incubated for 10 min in the absence or presence of PDGF (50 ng/ml) or insulin (1 μM) before the extraction and measurement of [3H]PIs. PtdIns_P_3 concentrations are expressed as a percentage of the total cellular PI and represent the mean value and range of two experiments performed in duplicate. **Indicates a significant difference (_P_≤ 0.05) from the appropriate control determined by Student's t test.
| Cellular PtdIns_P_3 (% total PI) | ||
|---|---|---|
| Stimulus | −bpV(phen) | +bpV(phen) |
| None | 0.015±0.003 | 0.012±0.006 |
| PDGF | 0.223±0.03 | 0.025±0.010** |
| Insulin | 0.097±0.032 | 0.025±0.001** |
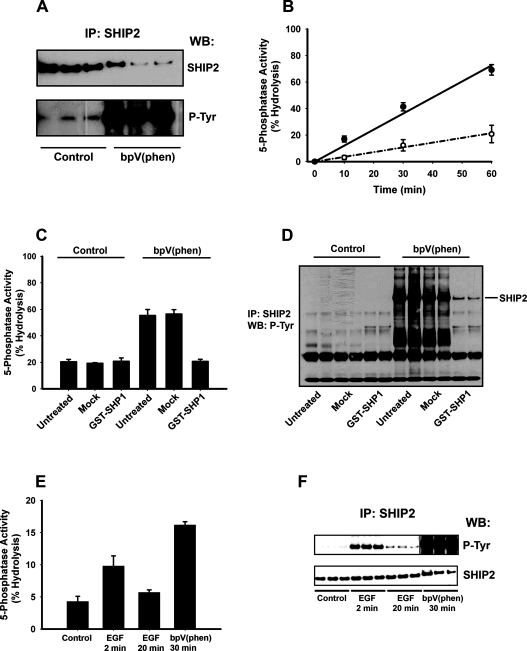
Earlier studies, however, have not shown that increased tyrosine phosphorylation of SHIP2 causes an enhanced capacity of this enzyme to degrade PtdIns_P_3 [39–41]. In contrast, our results imply that this is probable and the results presented in Figure 6 confirm this view. Figures 6(A) and 6(B) compare directly SHIP2 tyrosine phosphorylation and activity measured in immunoprecipitates of this protein from Triton X-100 lysates of control and bpV(phen)-treated 1321N1 cells. Figure 6(A) indicates the relative amount of SHIP2 immunoprecipitated from control and stimulated cells (top panel) and its tyrosine phosphorylation (bottom panel) and confirms that bpV(phen) stimulates both a reduction of ≥2-fold in the amount of SHIP2 recovered in the Triton X-100-soluble fraction from these cells and a marked increase in its phosphorylation. Figure 6(B) shows the SHIP2 activity, measured using Ins(1,3,4,5)_P_4, the water-soluble headgroup of PtdIns_P_3 against which SHIP2 is also active [42], as a substrate. These results show that although a reduced amount of SHIP2 protein is present in immunoprecipitates from stimulated cells, this nevertheless hydrolysed Ins(1,3,4,5)_P_4 ∼3–4-fold faster than that from control cells, revealing that bpV(phen) stimulates an increased tyrosine phosphorylation of SHIP2 and a coincident increase of ∼5–10-fold in its specific activity. A very similar increase in the SHIP2 activity stimulated by bpV(phen) was also measured with PtdIns_P_3 as substrate (results not shown). By contrast, however, bpV(phen) did not stimulate SHIP2 activity from control or stimulated cells when added directly to in vitro assays but rather inhibited activity modestly (∼25%) at high concentrations (100 μM), consistent with an indirect cellular action of this reagent.
Figure 6. The specific activity of SHIP2 is increased by bpV(phen)- or EGF-stimulated tyrosine phosphorylation.
(A) and (B) 1321N1 cells were incubated for 30 min in the absence or presence of bpV(phen) (20 μM) as indicated and then lysed and SHIP2 immunoprecipitates analysed either for (A) SHIP2 protein and SHIP2 tyrosine phosphorylation or for (B) the corresponding SHIP2 5-phosphatase activity from control (○) and stimulated (●) cells. The results show (A) triplicate determinations and (B) the means±S.D. of the corresponding 5-phosphatase activity from the same samples in a single experiment representative of three which gave similar results. (C) and (D) Immunoprecipitates of SHIP2 from 1321N1 cells incubated as controls or treated for 30 min with bpV(phen) (20 μM) were analysed either for (C) 5-phosphatase activity or (D) SHIP2 tyrosine phosphorylation either directly (untreated) or after incubation for 30 min at 30 °C with GST–SHP1 (0.4 mg/ml) (GST–SHP1) or with buffer alone (Mock). The results in (C) show the mean and range of duplicate determinations from a single experiment and in (D) the corresponding duplicate anti-phosphotyrosine immunoblots and are representative of three experiments which gave similar results. (E) and (F) HeLa cells were incubated in the absence or presence of EGF (50 ng/ml) or bpV(phen) (100 μM) for the times indicated then lysed and SHIP2 immunoprecipitates analysed either for (E) SHIP2 5-phosphatase activity or for (F) SHIP2 protein and SHIP2 tyrosine phosphorylation. The results show the means±S.D. activity measured in a 30 min assay or the corresponding triplicate immunoblots for the same samples and are representative of five experiments which gave similar results. The vertical line dividing the upper panel in (F) indicates where images from the same immunoblot have been merged to eliminate a molecular mass marker lane and allow alignment with the lower panel.
To confirm the causal relationship between the tyrosine phosphorylation and the increased specific activity of SHIP2, we prepared immunoprecipitates from control and bpV(phen)-treated cells and incubated these in the absence or presence of the phosphotyrosine specific phosphatase SHP1 prior to measurement of SHIP2 activity and tyrosine phosphorylation as shown in Figures 6(C) and 6(D) respectively. The results show that SHP1 had no effect on the activity of SHIP2 from control cells, consistent with the minimal tyrosine phosphorylation of this protein under resting conditions, and that mock incubations did not alter the activity or tyrosine phosphorylation of SHIP2 from either control or stimulated cells. By contrast, however, SHP1 completely reversed both the increase in SHIP2 activity and tyrosine phosphorylation stimulated by bpV(phen). This demonstrates unequivocally that enhanced tyrosine phosphorylation of SHIP2 can increase directly the specific activity of this PtdIns_P_3 5-phosphatase and provides strong support for the role of SHIP2 in the mechanism by which PTP inhibitors switch the output of PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2. To date our attempts to confirm this using SHIP2-directed siRNAs (small interfering RNAs) have been confounded by insufficiently reduced protein expression, making adequate assessment of the effects on cellular PtdIns_P_3 concentrations impractical. However, the limited reduction in SHIP2 expression achieved with siRNAs was sufficient to show a corresponding reduction in the phosphatase activity and tyrosine phosphorylation measured in SHIP2 immunoprecipitates (results not shown), suggesting that both reflect properties of SHIP2 itself rather than of other contaminating proteins.
EGF (epidermal growth factor) receptor-stimulated tyrosine phosphorylation of SHIP2 correlates with increased SHIP2 5-phosphatase activity
To determine how phosphorylation modulates the activity of SHIP2, we have recently begun to map the SHIP2 residues phosphorylated in response to bpV(phen) stimulation of 1321N1 and HeLa cells. The preliminary results of these studies indicate a number of sites at which phosphorylation is induced by this reagent, including the NPXY motif C-terminal to the active site and tyrosine residue 1162 which is also located toward the C-terminus. As the phosphorylation of these residues in response to EGF has been reported previously [43,44], we compared the ability of EGF and bpV(phen) to stimulate SHIP2 activity and tyrosine phosphorylation. In 1321N1 cells EGF had little effect on either parameter (results not shown) but in HeLa cells the growth factor induced a transient increase in both SHIP2 activity and tyrosine phosphorylation as shown in Figures 6(E) and 6(F) respectively. Thus within 2 min EGF increased the specific activity of SHIP2 ∼2-fold and induced a marked increase in tyrosine phosphorylation whereas by 20 min after EGF addition both the activity and phosphorylation returned close to control values. By comparison, stimulation with bpV(phen) resulted in a more profound increase in SHIP2 tyrosine phosphorylation and increased 5-phosphatase specific activity ∼3-fold. In contrast with earlier studies [39–41], these results show that receptor-stimulated tyrosine phosphorylation of SHIP2 can modify the specific activity of this 5-phosphatase. Importantly, however, our results also imply that PTP inhibitors and receptor stimuli that activate PI3K signalling can induce tyrosine phosphorylation of SHIP2 at co-incident sites, including at least some of those through which increases in SHIP2 phosphatase activity is mediated. Thus the phosphorylation-mediated regulation of SHIP2 activity that we have demonstrated in response to PTP inhibitors may also serve to regulate the balance of lipid signals produced in response to physiological stimulation of PI3K.
DISCUSSION
In the present study we have examined the effects of vanadate analogues which are powerful inhibitors of PTPs to determine how the concentrations of the separate 3-phosphorylated PI signals generated by the activation of class I PI3K can be regulated independently. Our results show how the action of these agents at key points can switch the balance of lipid signals generated in response to the activation of class I PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2. Thus, although PTP inhibitors increase the productivity of PI3K, they also stimulate the activity of PtdIns_P_3 5-phosphatase(s) and inhibit the action of PtdIns(3,4)_P_2 4-phosphatases. Our results therefore explain means by which endogenous mediators might exert independent control over the concentrations of the separate 3-phosphorylated PIs produced in response to the activation of a single class of PI3K. The established sensitivity of PTEN to inhibition by ROS [23,24] may confer further flexible control, whereas the ability of hydrogen peroxide to induce a marked accumulation of PtdIns(3,4)_P_2 but not of PtdIns_P_3 [25] suggests that exogenously applied ROS evoke effects qualitatively similar to PTP inhibitors. The regulatory mechanisms revealed by PTP inhibitors are important not only in the context of endogenous ROS however, but also have wider physiological and therapeutic relevance. First, because the stimulation of SHIP2 activity by PTP inhibitors also occurs in response to ligands for endogenous receptors that activate PI3K. Secondly, because although vanadate analogues are powerful insulin mimetics [27] and inhibit PTEN [45], the present results show that their net effect on PI3K output can be to reduce rather than to augment cellular PtdIns_P_3 concentrations.
PtdIns(3,4)_P_2 can, in principle, arise from the degradation of PtdIns_P_3 or can be synthesized directly by class II PI3Ks. Several lines of evidence presented support the former as the predominant route leading to PtdIns(3,4)_P_2 accumulation in response to PTP inhibitors. First, PTP inhibitors acting alone cause recruitment of both the regulatory (p85) and catalytic (p110α) components of class Ia PI3K into anti-phosphotyrosine immunoprecipitates which also display appropriate PI3K activity. Secondly, this occurs with time- and concentration-dependences that correlate closely with that for the wortmannin-sensitive accumulation of PtdIns(3,4)_P_2 induced by PTP inhibitors. Thirdly, sodium vanadate and bpV(phen) accelerate the accumulation of PtdIns(3,4)_P_2 produced in response to the stimulation of growth factor receptors known to recruit class Ia PI3K. Finally, the dramatic accumulation of PtdIns(3,4)_P_2 evoked by bpV(phen) occurs at the expense of PtdIns(4,5)_P_2, the physiological substrate for class I PI3Ks [2]. Interestingly, this implies that PTP inhibitors stimulate the phosphorylation of PtdIns(4,5)_P_2 by PI3K but also impede the mechanisms that normally replenish this lipid. Hence, the fraction of cellular PtdIns(4,5)_P_2 accessible to class I PI3K can be estimated at ≥70%, a proportion similar to that also available to receptor-activated phospholipase C in 1321N1 cells [30,46], suggesting that these two signalling enzymes share a substantially overlapping pool of PtdIns(4,5)_P_2 substrate. However, comparison with our previous data [29,30] suggests that the maximal rate of PtdIns(4,5)_P_2 consumption following activation of endogenous PI3K in these cells is ∼100-fold slower than by phospholipase C.
Although the activation of class I PI3K is clearly essential, the near linear accumulation of PtdIns(3,4)_P_2 in response to bpV(phen) also strongly suggests that PTP inhibitors must impair the catabolism of this lipid. The types Ia and IIa inositol polyphosphate 4-phosphatases are candidate PtdIns(3,4)_P_2 phosphatases [11–13]. Previous studies [15,47] in cells under- or over-expressing the type I enzyme have shown significant effects on the cellular concentrations of PtdIns(3,4)_P_2. In the present study we have shown that both enzymes are present in 1321N1 cells and are potently inhibited by PTP inhibitors as expected of these PTP-like, cysteine-dependent phosphatases [13]. The comprehensive inhibition of these enzymes in vitro by sodium vanadate and bpV(phen) suggests that a similar action in cells contributes to the dramatic accumulation of PtdIns(3,4)_P_2 which these agents induce. Conversely, the powerful cellular action of these inhibitors reflected by increases of up to 1000-fold in the concentrations of PtdIns(3,4)_P_2 provides compelling support for the limited available evidence that either or both 4-phosphatase(s) function as essential regulators of 3-phosphorylated PI signals and reinforces the notion that these enzymes may be important targets of endogenously generated ROS. Indeed, as the PTPs which dephosphorylate growth factor receptors and PTEN are also ROS-sensitive, our results make apparent the extent to which even limited similar susceptibility of the 4-phosphatases would drive the output of PI3K towards PtdIns(3,4)_P_2. In addition, they suggest that PTP inhibitors may be useful pharmacological tools with which to probe the functions of these enzymes and of the lipids whose concentrations they control.
The most exciting and unexpected aspect of our study is the observation that PTP inhibitors modulate cellular PtdIns_P_3 concentrations by activating appropriate 5-phosphatases. This is demonstrated both by the action of these agents to attenuate receptor-mediated increases in the accumulation of PtdIns_P_3 and by their capacity to accelerate the rate of removal of PtdIns_P_3 in PTEN-null 1321N1 cells. Our results provide strong correlative evidence that SHIP2 is an important component of this process and suggest that the novel regulation of SHIP2 activity uncovered by PTP inhibition occurs by a mechanism that is shared by physiological stimuli of PI3K.
A family of 5-phosphatases which use PIs or inositol polyphosphates as substrates has now been identified [6,7] and evidence suggests that several function physiologically as PtdIns_P_3 5-phosphatases. These include SHIP1 and 2 [6,7], SKIP (skeletal muscle inositol polyphosphate phosphatase) [48] and PIPP (proline-rich inositol polyphosphate phosphatase) [49]. SHIP1 expression is restricted primarily to cells of haematopoietic origin and was not detectable in 1321N1 cells by immunoblotting (results not shown). SHIP2, however, is widely expressed and interacts with a diverse group of intracellular proteins through its N-terminal SH2 domain and its C-terminal proline-rich domain, but also undergoes reversible tyrosine phosphorylation at an NPXY motif C-terminal to its active site [6,7]. Both the tyrosine phosphorylation of SHIP2 and its translocation, mediated via its interaction with appropriate binding partners, are thought to contribute to the regulation of this enzyme, but the details of the relationship between these events are not yet clear and a direct correlation between SHIP2 tyrosine phosphorylation and the specific activity of this enzyme has not been established [39–41]. Similarly, the functional consequence(s) of agonist-stimulated SHIP2 phosphorylation at other sites [43,44] remains obscure. Our results demonstrate that PTP inhibitors induce a pronounced re-localization of SHIP2 to a Triton X-100-insoluble fraction at the cell periphery, consistent with the interaction of SHIP2 with components of the cytoskeleton [6,7], and a marked increase in the tyrosine phosphorylation of this enzyme. The tyrosine phosphorylation of SHIP2 evoked by PTP inhibitors occurs with a time- and concentration-dependence that correlates closely with that for the stimulated accumulation of PtdIns(3,4)_P_2 and causes a 5–10-fold increase in SHIP2 5-phosphatase activity. This is the first demonstration that phosphorylation can modulate the activity of this enzyme directly and alone can account for a significant fraction of the increase in the cellular capacity to degrade PtdIns_P_3 induced by PTP inhibitors. Furthermore, as the translocation of SHIP2 also is likely to influence the cellular efficiency of this enzyme, our results are consistent with SHIP2 being the predominant 5-phosphatase contributing to the cellular action of these agents. Importantly, neither the tyrosine phosphorylation nor the re-localization of SHIP2 stimulated by PTP inhibitors was sensitive to the inhibition of PI3K by wortmannin, consistent with our proposal that the ratio of the PtdIns_P_3/PtdIns(3,4)_P_2 produced by PI3K can be controlled independently through the regulation of 5-phosphatase activity. Our results also illustrate the wide dynamic range displayed by cellular PtdIns_P_3 5-phosphatase activity from resting to stimulated conditions and emphasize the capacity of SHIP2 for activation. This suggests that unlike the 3-phosphatase PTEN, which regulates basal PtdIns_P_3 concentrations, 5-phosphatases like SHIP2 operate at only a fraction of their maximal capacity in resting cells and may be geared specifically to control stimulated increases in PtdIns_P_3 concentrations.
In contrast with earlier studies [39–41], our results clearly show that the specific activity of SHIP2 is increased by tyrosine phosphorylation. This may reflect methodological differences and, for the increased activity stimulated by PTP inhibitors, the intense tyrosine phosphorylation of SHIP2 induced, arguably including phosphorylation at non-physiological sites. The latter is unlikely because EGF acts similarly and because PTP inhibition would be expected primarily to amplify the steady-state phosphorylation level of proteins responsive to endogenous kinase/phosphatase cycles rather than to drive other phosphorylation events. This view is supported by our analysis of the SHIP2 sites phosphorylated in response to bpV(phen) as these show phosphorylation of tyrosine residues known to be phosphorylated in response to EGF. Further studies will be required to fully elucidate the role of tyrosine phosphorylation in the regulation of SHIP2. By revealing the existence of hitherto unrecognized mechanisms which allow the powerful activation of cellular 5-phosphatases such as SHIP2, our results highlight novel targets for potential therapeutic intervention, the significance of which is implied by knockout studies [50,51] which suggest that SHIP2 plays a particularly important role in insulin signalling.
In conclusion, in the present study we have used PTP inhibitors to uncover novel, powerful mechanisms capable of switching the predominant output of class I PI3K from PtdIns_P_3 to PtdIns(3,4)_P_2. We propose that growth factor receptor stimuli which activate PI3K act, perhaps indirectly, through the generation of endogenous ROS to modulate the same key targets to provide the flexible, independent control that is required for the PtdIns_P_3, PtdIns(3,4)P_2 and PtdIns3_P produced following the stimulation of class I PI3K to function as separate but integrated lipid signals. Thus elevated ROS concentrations would act to amplify PtdIns_P_3 formation via their inhibition of PTEN [23,24] and receptor tyrosine phosphatase activities [20,21] whereas the contemporary inhibition of 4-phosphatases by ROS and activation/translocation of SHIP2 mediated by receptor stimulation would act concertedly and adaptably to control the balance of other PtdIns_P_3-derived signals and ultimately to modify the outcome of PI3K activation.
Acknowledgments
We are grateful to Dr N. Morrice (Medical Research Council Protein Phosphorylation Unit, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.) for help and advice in performing the phosphorylation site mapping studies. Financial support from Medical Research Council, U.K. (Programme Grant No.G9403619) is gratefully acknowledged.
References
- 1.Berridge M. J. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M. A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 4.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 5.Leslie N. R., Downes C. P. PTEN: the down side of PI 3-kinase signalling. Cell. Signalling. 2002;14:285–295. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 6.Backers K., Blero D., Paternotte N., Zhang J., Erneux C. The termination of PI3K signalling by SHIP1 and SHIP2 inositol 5-phosphatases. Adv. Enzyme Regul. 2003;43:15–28. doi: 10.1016/s0065-2571(02)00043-2. [DOI] [PubMed] [Google Scholar]
- 7.Dyson J. M., Kong A. M., Wiradjaja F., Astle M. V., Gurung R., Mitchell C. A. The SH2 domain containing inositol polyphosphate 5-phosphatase-2: SHIP2. Int. J. Biochem. Cell Biol. 2005;37:2260–2265. doi: 10.1016/j.biocel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Lazar D. F., Saltiel A. R. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat. Rev. Drug Discov. 2006;5:333–342. doi: 10.1038/nrd2007. [DOI] [PubMed] [Google Scholar]
- 9.Majerus P. W., Kisseleva M. V., Norris F. A. The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 1999;274:10669–10672. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- 10.Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 11.Norris F. A., Majerus P. W. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J. Biol. Chem. 1994;269:8716–8720. [PubMed] [Google Scholar]
- 12.Norris F. A., Auethavekiat V., Majerus P. W. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J. Biol. Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 13.Norris F. A., Atkins R. C., Majerus P. W. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splicing in the 4-phosphatase family. J. Biol. Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- 14.Nystuen A., Legare M. E., Shultz L. D., Frankel W. N. A null mutation in inositol polyphosphate 4-phosphatase type I causes selective neuronal loss in weeble mutant mice. Neuron. 2001;32:203–212. doi: 10.1016/s0896-6273(01)00468-8. [DOI] [PubMed] [Google Scholar]
- 15.Shin H. W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M. R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vyas P., Norris F. A., Joseph R., Majerus P. W., Orkin S. H. Inositol polyphosphate 4-phosphatase type I regulates cell growth downstream of transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13696–13701. doi: 10.1073/pnas.250476397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M., et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Dowler S., Currie R. A., Downes C. P., Alessi D. R. DAPP1: a dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem. J. 1999;342:7–12. [PMC free article] [PubMed] [Google Scholar]
- 20.Finkel T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 21.Rhee S. G. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 22.Leslie N. R., Lindsay Y., Ross S. H., Downes C. P. Redox regulation of phosphatase function. Biochem. Soc. Trans. 2004;32:1018–1020. doi: 10.1042/BST0321018. [DOI] [PubMed] [Google Scholar]
- 23.Leslie N. R., Bennett D., Lindsay Y. E., Stewart H., Gray A., Downes C. P. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S. R., Yang K. S., Kwon J., Lee C., Jeong W., Rhee S. G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 25.Van der Kaay J., Beck M., Gray A., Downes C. P. Distinct phosphatidylinositol 3-kinase lipid products accumulate upon oxidative and osmotic stress and lead to different cellular responses. J. Biol. Chem. 1999;274:35963–35968. doi: 10.1074/jbc.274.50.35963. [DOI] [PubMed] [Google Scholar]
- 26.Leslie N. R. The redox regulation of PI 3-kinase-dependent signaling. Antioxid. Redox Signal. 2006;8:1765–1774. doi: 10.1089/ars.2006.8.1765. [DOI] [PubMed] [Google Scholar]
- 27.Morinville A., Maysinger D., Shaver A. From Vanadis to Atropos: vanadium compounds as pharmacological tools in cell death signalling. Trends Pharmacol. Sci. 1998;19:452–460. doi: 10.1016/s0165-6147(98)01257-7. [DOI] [PubMed] [Google Scholar]
- 28.Gordon J. A. Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. Methods Enzymol. 1991;201:477–482. doi: 10.1016/0076-6879(91)01043-2. [DOI] [PubMed] [Google Scholar]
- 29.Batty I. H., Downes C. P. The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary, selective, consequences of inositol depletion in 1321N1 cells. Biochem. J. 1994;297:529–537. doi: 10.1042/bj2970529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batty I. H., Downes C. P. Thrombin receptors modulate insulin-stimulated phosphatidylinositol 3,4,5-trisphosphate accumulation in 1321N1 astrocytoma cells. Biochem. J. 1996;317:347–351. doi: 10.1042/bj3170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batty I. H., Fleming I. N., Downes C. P. Muscarinic-receptor-mediated inhibition of insulin-like growth factor-1 receptor-stimulated phosphoinositide 3-kinase signalling in 1321N1 astrocytoma cells. Biochem. J. 2004;379:641–651. doi: 10.1042/BJ20031700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huyer G., Liu S., Kelly J., Moffat J., Payette P., Kennedy B., Tsaprailis G., Gresser M. J., Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem. 1997;272:843–851. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
- 33.McConnachie G., Pass I., Walker S. M., Downes C. P. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 2003;371:947–955. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Kaay J., Batty I. H., Cross D. A., Watt P. W., Downes C. P. A novel, rapid, and highly sensitive mass assay for phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] and its application to measure insulin-stimulated PtdIns(3,4,5)P3 production in rat skeletal muscle in vivo. J. Biol. Chem. 1997;272:5477–5481. doi: 10.1074/jbc.272.9.5477. [DOI] [PubMed] [Google Scholar]
- 35.Posner B. I., Faure R., Burgess J. W., Bevan A. P., Lachance D., Zhang-Sun G., Fantus I. G., Ng J. B., Hall D. A., Lum B. S., et al. Peroxovanadium compounds. A new class of potent phosphotyrosine phosphatase inhibitors which are insulin mimetics. J. Biol. Chem. 1994;269:4596–4604. [PubMed] [Google Scholar]
- 36.Heldin C. H., Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 37.Habib T., Hejna J. A., Moses R. E., Decker S. J. Growth factors and insulin stimulate tyrosine phosphorylation of the 51C/SHIP2 protein. J. Biol. Chem. 1998;273:18605–18609. doi: 10.1074/jbc.273.29.18605. [DOI] [PubMed] [Google Scholar]
- 38.Prasad N., Topping R. S., Decker S. J. Src family tyrosine kinases regulate adhesion-dependent tyrosine phosphorylation of 5′-inositol phosphatase SHIP2 during cell attachment and spreading on collagen I. J. Cell Sci. 2002;115:3807–3815. doi: 10.1242/jcs.00070. [DOI] [PubMed] [Google Scholar]
- 39.Blero D., De Smedt F., Pesesse X., Paternotte N., Moreau C., Payrastre B., Erneux C. The SH2 domain containing inositol 5-phosphatase SHIP2 controls phosphatidylinositol 3,4,5-trisphosphate levels in CHO-IR cells stimulated by insulin. Biochem. Biophys. Res. Commun. 2001;282:839–843. doi: 10.1006/bbrc.2001.4639. [DOI] [PubMed] [Google Scholar]
- 40.Giuriato S., Blero D., Robaye B., Bruyns C., Payrastre B., Erneux C. SHIP2 overexpression strongly reduces the proliferation rate of K562 erythroleukemia cell line. Biochem. Biophys. Res. Commun. 2002;296:106–110. doi: 10.1016/s0006-291x(02)00787-8. [DOI] [PubMed] [Google Scholar]
- 41.Taylor V., Wong M., Brandts C., Reilly L., Dean N. M., Cowsert L. M., Moodie S., Stokoe D. 5′ phospholipid phosphatase SHIP-2 causes protein kinase B inactivation and cell cycle arrest in glioblastoma cells. Mol. Cell. Biol. 2000;20:6860–6871. doi: 10.1128/mcb.20.18.6860-6871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesesse X., Moreau C., Drayer A. L., Woscholski R., Parker P., Erneux C. The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity. FEBS Lett. 1998;437:301–303. doi: 10.1016/s0014-5793(98)01255-1. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Steen H., Kuster B., Fernandez M., Pandey A., Mann M. Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem. 2002;277:1031–1039. doi: 10.1074/jbc.M109992200. [DOI] [PubMed] [Google Scholar]
- 45.Schmid A. C., Byrne R. D., Vilar R., Woscholski R. Bisperoxovanadium compounds are potent PTEN inhibitors. FEBS Lett. 2004;566:35–38. doi: 10.1016/j.febslet.2004.03.102. [DOI] [PubMed] [Google Scholar]
- 46.Batty I. H., Downes C. P. The mechanism of muscarinic receptor-stimulated phosphatidylinositol resynthesis in 1321N1 astrocytoma cells and its inhibition by Li+ J. Neurochem. 1995;65:2279–2289. doi: 10.1046/j.1471-4159.1995.65052279.x. [DOI] [PubMed] [Google Scholar]
- 47.Ivetac I., Munday A. D., Kisseleva M. V., Zhang X. M., Luff S., Tiganis T., Whisstock J. C., Rowe T., Majerus P. W., Mitchell C. A. The type Iα inositol polyphosphate 4-phosphatase generates and terminates phosphoinositide 3-kinase signals on endosomes and the plasma membrane. Mol. Biol. Cell. 2005;16:2218–2233. doi: 10.1091/mbc.E04-09-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ijuin T., Mochizuki Y., Fukami K., Funaki M., Asano T., Takenawa T. Identification and characterization of a novel inositol polyphosphate 5-phosphatase. J. Biol. Chem. 2000;275:10870–10875. doi: 10.1074/jbc.275.15.10870. [DOI] [PubMed] [Google Scholar]
- 49.Mochizuki Y., Takenawa T. Novel inositol polyphosphate 5-phosphatase localizes at membrane ruffles. J. Biol. Chem. 1999;274:36790–36795. doi: 10.1074/jbc.274.51.36790. [DOI] [PubMed] [Google Scholar]
- 50.Clement S., Krause U., Desmedt F., Tanti J. F., Behrends J., Pesesse X., Sasaki T., Penninger J., Doherty M., Malaisse W., et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 51.Sleeman M. W., Wortley K. E., Lai K. M., Gowen L. C., Kintner J., Kline W. O., Garcia K., Stitt T. N., Yancopoulos G. D., Wiegand S. J., Glass D. J. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]