Global analysis of alternative splicing differences between humans and chimpanzees (original) (raw)
Abstract
Alternative splicing is a powerful mechanism affording extensive proteomic and regulatory diversity from a limited repertoire of genes. However, the extent to which alternative splicing has contributed to the evolution of primate species-specific characteristics has not been assessed previously. Using comparative genomics and quantitative microarray profiling, we performed the first global analysis of alternative splicing differences between humans and chimpanzees. Surprisingly, 6%–8% of profiled orthologous exons display pronounced splicing level differences in the corresponding tissues from the two species. Little overlap is observed between the genes associated with alternative splicing differences and the genes that display steady-state transcript level differences, indicating that these layers of regulation have evolved rapidly to affect distinct subsets of genes in humans and chimpanzees. The alternative splicing differences we detected are predicted to affect diverse functions including gene expression, signal transduction, cell death, immune defense, and susceptibility to diseases. Differences in expression at the protein level of the major splice variant of Glutathione _S_-transferase omega-2 (GSTO2), which functions in the protection against oxidative stress and is associated with human aging-related diseases, suggests that this enzyme is less active in human cells compared with chimpanzee cells. The results of this study thus support an important role for alternative splicing in establishing differences between humans and chimpanzees.
[Keywords: Alternative splicing, gene regulation, microarray profiling, primate evolution]
A major challenge of the post-genomic era is to identify the set of molecular characteristics that account for human-specific traits. Recent global comparisons of human and chimpanzee genomes and transcriptomes have begun to identify molecular differences that could underlie some of the unique attributes of these primate species (Preuss et al. 2004; Bustamante et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005; Khaitovich et al. 2006). These differences stem from sequence and regulatory diversity acting on both protein coding and noncoding transcripts (Pollard et al. 2006; Prabhakar et al. 2006), as well as from structural genomic alterations including segmental duplications, large-scale copy number variations, and deletions (Cheng et al. 2005; Newman et al. 2005; Perry et al. 2006). Despite the recent accumulation of data revealing molecular differences between humans and chimpanzees, additional possible sources of diversity, such as differential alternative splicing, have not yet been investigated.
Alternative splicing is the process by which splice sites in precursor mRNA transcripts are differentially selected to result in the production of different mRNA and protein isoforms (Cartegni et al. 2002; Matlin et al. 2005; Blencowe 2006). Two-thirds or more genes in human and mouse contain at least one alternative exon, and it is known that mammalian organs comprising many specialized cell types such as the brain are associated with relatively complex alternative splicing patterns (Xu et al. 2002; Johnson et al. 2003; Yeo et al. 2004a). Surprisingly, comparisons of human and mouse transcript sequences have revealed that <20% of alternative splicing events have been conserved during the ∼80- to 90-million-year interval separating these species (Modrek and Lee 2003; Nurtdinov et al. 2003; Sorek et al. 2004; Pan et al. 2005; Yeo et al. 2005). While these and related observations have suggested an important role for alternative splicing in the evolution of mammalian species (Ast 2004; Xing and Lee 2006), no experimental study has yet addressed the extent to which splicing patterns may differ between more closely related mammalian species such as humans and chimpanzees. The identification of orthologous human and chimpanzee transcripts with different splicing patterns represents a critical first step toward understanding the role of alternative splicing in the evolution of specific traits in these species.
In this study, we used comparative genomics and a quantitative alternative splicing microarray platform to compare alternative splicing patterns in human and chimpanzee tissues. Despite an overall sequence identity between human and chimpanzee genomic coding regions of 98%–99%, we observe that 6%–8% of surveyed alternative exons display pronounced splicing level differences. These differences affect a primarily nonoverlapping subset of genes compared with the subset of genes that display differences in steady-state transcript levels between humans and chimpanzees. Alternative splicing differences between humans and chimpanzees are consistently detected when comparing corresponding cell and tissue types from multiple individuals from each species, and the associated genes have diverse and important functional roles, including regulation of gene expression, signal transduction, apoptosis, immune defense, and disease. Detection of increased expression of the major, functional splice variant of Glutathione _S_-transferase omega-2 (GSTO2) at the protein level in chimpanzee cells suggests that this enzyme could play a more active role in protection against oxidative stress and diseases associated with loss of such protection in chimpanzees compared with humans. Our results thus provide evidence that alternative splicing changes have evolved rapidly and in parallel with changes in transcriptional regulation, affecting an additional set of genes and associated gene functions. The identification of alternative splicing differences between humans and chimpanzees provides a new basis for understanding the molecular mechanisms underlying the evolution of primate species-specific characteristics.
Results
Analysis of human and chimpanzee splicing patterns by comparative genomic and microarray profiling strategies
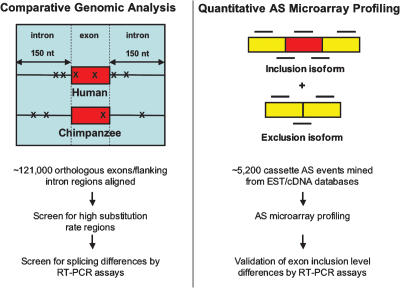
To compare global splicing patterns between humans and chimpanzees and identify individual alternative splicing events that differ between these species, we applied a comparative genomics strategy together with a previously described quantitative alternative splicing microarray profiling system (Fig. 1; Pan et al. 2004; Shai et al. 2006; refer to Materials and Methods). In these comparisons, we focused on splicing patterns involving orthologous exons. While it is possible that some splicing differences could involve exons that are unique to either genome, the lack of substantial transcript sequence data and of a polished chimpanzee genomic sequence makes such events difficult to detect in a reliable manner (Newman et al. 2005). Moreover, given the 98%–99% sequence identity between the coding regions of human and chimpanzee genomes (Chimpanzee Sequencing and Analysis Consortium 2005), few exons are expected to be unique to either genome. Accordingly, in the comparative genomics strategy, we aligned 120,951 orthologous human and chimpanzee exons and the 150 base pairs (bp) of intron sequence flanking these exons and screened these regions for high substitution rates. Previous studies have shown that short and degenerate motifs referred to as splicing enhancers and silencers, which specify splice site recognition and regulate alternative splicing, are enriched in exonic and intronic regions proximal to splice sites (Brudno et al. 2001; Zhang et al. 2003; Yeo et al. 2004b, 2007; for reviews, see Matlin et al. 2005; Blencowe 2006; Chasin 2007). Consequently, we expected that frequent nucleotide changes in exons and the flanking intron regions would be predictive of splicing differences between humans and chimpanzees.
Figure 1.
Strategies used to identify alternative splicing differences between humans and chimpanzees. (Left panel) In the comparative genomics strategy, regions including orthologous human and chimpanzee exons (red boxes) and the flanking 150 nucleotides of intron sequence were aligned. Nucleotide substitutions (indicated by Xs) in these regions were scored, and events displaying >5% substitution rates were analyzed for alternative splicing differences by RT–PCR assays using poly(A)+ RNA from human and chimpanzee tissues (see main text for details). (Right panel) In the quantitative alternative splicing microarray profiling strategy, labeled cDNA from the same poly(A)+ RNA samples were hybridized to an alternative splicing microarray designed to monitor inclusion levels of cassette-type alternative exons. Each alternative splicing event is monitored by a set of six oligonucleotide probes (black horizontal lines) (Pan et al. 2004). Predictions for alternative splicing differences between the corresponding human and chimpanzee tissues were validated by RT–PCR assays (see Table 1; Figs. 4, 5; Supplementary Fig. 1).
Role of nucleotide substitutions in generating differences between human and chimpanzee splicing patterns
High substitution rates were rarely found in the aligned orthologous exons and flanking intron sequences: Of these regions, 0.3% displayed substitution rates of >5%, and substitution rates >10% were found in <0.1% of the data set (Supplementary Table 1). To assess whether regions with >5% substitution rates are predictive of splicing differences, semiquantitative RT–PCR assays were performed using primers specific for neighboring exon sequences and poly(A)+ RNA samples from human or chimpanzee frontal cortex and heart tissues. These tissue RNA samples were pooled, in each case, from several adult individuals (Supplementary Table 2). Of 31 regions with >5% substitution rates selected for analysis, 14 contained exons that displayed evidence of alternative splicing in humans from analysis of the corresponding EST/cDNA (complementary DNA) sequences. Five of the 31 regions displayed splicing level differences between humans and chimpanzees in at least one of the two tissues (see below). Interestingly, all five of these isoform ratio differences involved exons included among the 14 alternative exons identified in the test set. At this validation rate, <20 (∼0.02%) of the 120,951 orthologous exons associated with high substitution rates would be expected to display splicing level differences between humans and chimpanzees.
Since substitution rates of <5% could also result in splicing differences, especially if the substitutions target important _cis_-acting elements, a splicing element scoring system (Stadler et al. 2006) was also employed to predict alterations in splicing levels between humans and chimpanzees. However, RT–PCR analysis of 20 exons (including several alternative exons) associated with high differential scores did not yield detectable splicing differences (data not shown). This observation could reflect a current lack of knowledge of the full set of _cis_-elements and mechanisms responsible for splice site selection in endogenous transcripts. It is possible that relevant changes involving defined _cis_-regulatory elements could occur outside of the regions we analyzed. Another possibility is that selection pressure acting to reduce the frequency of substitutions proximal to most splice sites in humans and chimpanzees (Chimpanzee Sequencing and Analysis Consortium 2005; Gazave et al. 2007) results in a limited role for _cis_-acting differences in altering splicing patterns between the two species. Moreover, consistent with the results described above, high substitutions rates could have a greater impact on the splicing levels of alternative exons, since the regions surrounding these exons are, on average, more highly conserved than the regions surrounding constitutive exons (Sorek and Ast 2003; Yeo et al. 2005; Sugnet et al. 2006), and therefore potentially more susceptible to rare mutations that disrupt important splicing regulatory elements.
Detection of alternative splicing differences between humans and chimpanzees by microarray profiling
Splicing differences between humans and chimpanzees could also occur as a consequence of alterations in one or more _trans_-acting splicing regulators. Accordingly, to assess the extent of alternative splicing differences between human and chimpanzee tissues that are not the consequence of elevated rates of substitution in the alternative exons and flanking intron sequences, splicing patterns were next compared using quantitative alternative splicing microarray profiling (Fig. 1; refer to Materials and Methods). An alternative splicing microarray capable of profiling ∼5000 cassette-type alternative splicing events mined from human ESTs and cDNA sequences (Fig. 1) was hybridized with dye-labeled cDNAs synthesized from the frontal cortex and heart mRNA samples analyzed above. This microarray contains sets of exon body and splice junction probes that are capable of monitoring the levels of inclusion of both the EST/cDNA-mined human cassette alternative exons and the orthologous exons identified in alignments of the corresponding chimpanzee genome sequences. The processed microarray data was analyzed using the Generative model for Alternative Splicing Array Platform (GenASAP) algorithm (Pan et al. 2004; Shai et al. 2006), which generates for each profiled alternative splicing event a confidence-ranked estimate for the fraction of transcripts that include an alternative exon, represented below as percent inclusion (%in) values. After filtering steps (see Materials and Methods), the levels of inclusion of ∼1700 orthologous exons were available for comparison between the corresponding human and chimpanzee tissues. None of these profiled alternative splicing events were associated with a substitution rate of >5%.
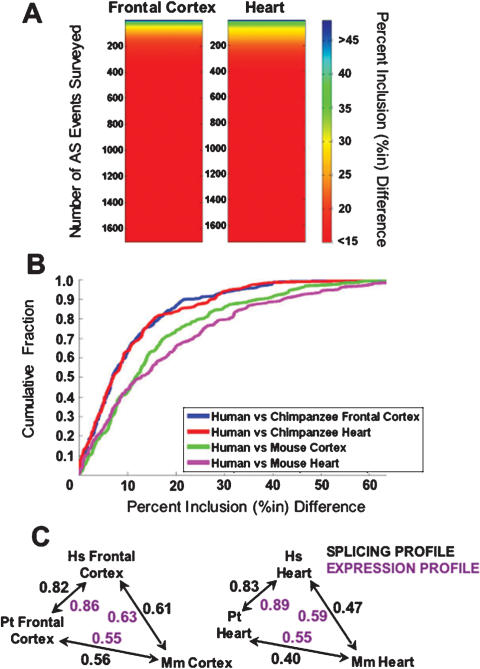
As expected, given the close evolutionary relationship between humans and chimpanzees, we found that the majority of the profiled orthologous exons in the two species have similar splicing levels. For example, when sorting genes based on the magnitude of the percent inclusion difference between human and chimpanzee frontal cortex and heart, ∼80% of the profiled exons display percent inclusion differences of <15%, which is close to the limit of sensitivity of our alternative splicing microarray profiling system (Fig. 2A; Pan et al. 2006). However, in addition to exons with similar splicing levels, a surprisingly large fraction of exons (6%–8%) show pronounced percent inclusion differences (i.e., >25%in). RT–PCR experiments were performed on 37 representative alternative splicing events displaying a range from no percent inclusion difference to a >25%in difference between the human and chimpanzee frontal cortex and/or heart. Of these events, 30 (81%) were validated as having the expected differential splicing patterns (Figs. 4, 5, below; Table 1; additional data not shown). Thus, despite the remarkable degree of conservation between the coding regions of the human and chimpanzee genomes, a substantial number of alternative exons that are not associated with high substitution rates in the alternative exons and flanking intron regions display pronounced splicing level differences between the two species.
Figure 2.
Quantitative alternative splicing microarray profiling reveals the extent of alternative splicing differences between human and chimpanzee orthologous exons, as well as the degree of divergence between splicing patterns over different evolutionary time periods. (A) Color spectrum plots indicating the number and magnitude of alternative splicing differences between human and chimpanzee frontal cortex and heart tissues. The _Y_-axes indicate the number of alternative splicing events profiled; these are sorted according to the magnitude of the absolute value of the percent exon inclusion level (%in) difference between the human and chimpanzee tissue being compared. The magnitude of the percentage inclusion difference is indicated by the color scale on the right. (B) Cumulative distribution plot displaying the distribution of percentage inclusion differences when comparing microarray data for 217 conserved alternative splicing events between the following pairs of tissues: human and chimpanzee frontal cortex (blue line), human and chimpanzee heart (red line), human frontal cortex and mouse cortex (green line), and human and mouse heart (purple line). (C) Spearman correlation coefficients are shown for pairwise comparisons between alternative splicing levels (black numbers) and transcript levels (purple numbers) for the set of 217 orthologous genes analyzed in human, chimpanzee, and mouse tissues in B. Double arrows indicate the pairs of species compared. (Hs) Homo sapiens; (Pt) Pan troglodytes; (Mm) Mus musculus.
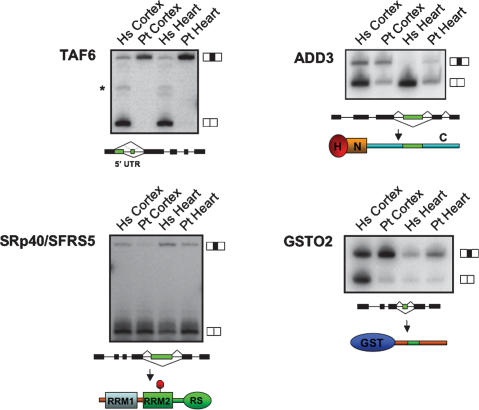
Figure 4.
Examples of alternative splicing differences between humans and chimpanzees confirmed by semiquantitative RT–PCR and sequencing. RT–PCR assays were performed using primers specific for sequences in constitutive exons flanking the alternative exons predicted to be differentially spliced by the comparative genomic or alternative splicing microarray profiling analyses. Corresponding tissues from human (Hs) and chimpanzee (Pt) are indicated. Major splice isoforms that include and skip alternative exons (black boxes) are indicated on the right of each panel. Diagrams below each gel illustrate the predicted consequence of alternative splicing changes at the protein and/or transcript levels for TAF6, ADD3, the SR-repeat family protein splicing factor of 40 kDa (SFRS5/SRp40) and GSTO2 (refer to Table 1 and main text for details). Protein domains are labeled as follows: (H) head domain; (N) neck domain; (C) C-terminal tail region known to interact with spectrin; (RRM1) RNA recognition motif 1; (RRM2) RNA recognition motif 2; (RS) arginine/serine-rich domain; (GST) glutathione _S_-transferase domain. The stop sign indicates the insertion of a PTC. The asterisk indicates an additional TAF6 splice isoform detected in human but not chimpanzee tissue, as confirmed by sequencing.
Figure 5.
Comparisons of AS levels in RNA samples from macaque, human, and chimpanzee individuals. (A) RT–PCR assays were performed using rhesus macaque total RNA from frontal cortex and heart samples from a number of different individuals. RT–PCR primers were designed to anneal to the exons neighboring each alternative exon, resulting in the amplification of two products (the isoforms including and skipping the alternative exon, as indicated in each of the panels). Asterisks denote novel isoforms observed primarily in one species but not the other. The transcripts shown correspond to the ADD3 (top panel), GSTO2 (second panel), HVEM/TNFRS14 (third panel), and ARL3 (bottom panel) genes. The macaque is used as an outgroup to define the ancestral splicing pattern for each gene. Examples of chimpanzee lineage-specific splicing differences (ADD3 and ARL3 transcripts) and human lineage-specific splicing differences (GSTO2 and TNFRSF14) are shown (see also Supplementary Table 3 for additional examples). (B) RT–PCR experiments were performed using total RNA from each of the individual samples that were pooled for analysis in the comparative genomic and microarray experiments. The same alternative splicing events displayed for macaque experiments in A are also shown in B for comparison. The labels for each gel lane represent the macaque, human, and chimpanzee individuals listed in Supplementary Table 2.
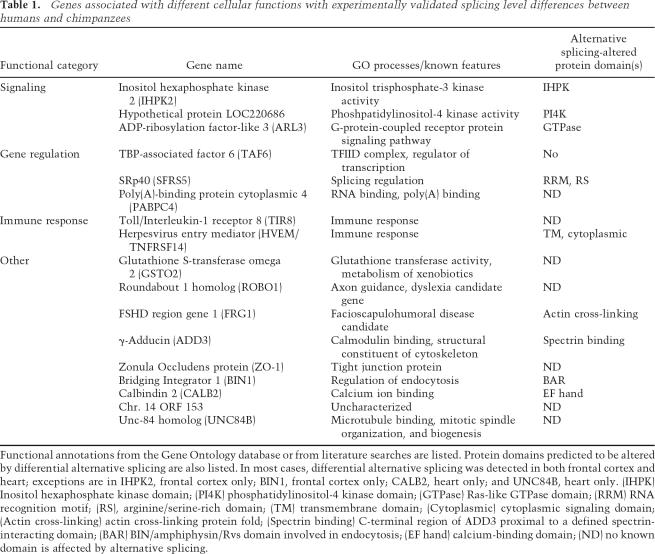
Table 1.
Genes associated with different cellular functions with experimentally validated splicing level differences between humans and chimpanzees
Evidence for stabilizing selection pressure acting to preserve the majority of splicing levels of orthologous human, chimpanzee, and mouse alternative exons
The similarity between the splicing levels for the majority of the profiled orthologous exons could reflect stabilizing selection pressure acting to conserve the inclusion levels of most human and chimpanzee orthologous exons. However, it is also possible that insufficient evolutionary time has accumulated to result in a higher incidence of pronounced inclusion level differences than observed above. To investigate these possibilities, we compared the extent of divergence between alternative splicing profiles representing a longer evolutionary time span, namely the 80- to 90-million-year period separating the common ancestor of human or chimpanzee and mouse. Percent inclusion values for 217 alternative splicing events conserved between humans and the mouse were obtained from RT–PCR-validated, quantitative alternative splicing microarray profiling data from mouse heart and brain cortex tissues (Fagnani et al. 2007) and compared with percent inclusion values from the human and chimpanzee datasets described above (Fig. 2B,C; see the Supplemental Material).
Notably, although the splicing levels of the mouse exons are also very similar to the orthologous exons in the corresponding human and chimpanzee tissues, they have diverged to a significantly greater extent in both tissues (P < 1 × 10−5, Mann-Whitney _U_-test) (Fig. 2B, cf. blue and green lines, and red and purple lines). Importantly, these increased differences in the alternative splicing profiles were not due to any inherent percent inclusion level biases in the datasets, since “control” profiles generated by randomly pairing exons from the same datasets were not significantly different (data not shown). Moreover, transcript level profiles obtained from the same datasets (see the Supplemental Material) also correlate to a higher degree between human and chimpanzee than between either primate species and mouse (Fig. 2C).
The analysis described above reveals that a significant proportion of alternative splicing patterns involving orthologous human, chimpanzee, and mouse alternative exons are conserved in the corresponding tissues, despite sharing an 80- to 90-million-year period of divergence. These results therefore support the conclusion that stabilizing selection pressure has acted to preserve the inclusion levels of the majority of orthologous exons between mammals. Nevertheless, 6%–8% of orthologous human and chimpanzee exons display pronounced deviations from this overall pattern of conservation. Such exons, in addition to those identified in the comparative genomic analysis, most likely represent alternative splicing events that could underlie important functional and phenotypic differences between the two species.
Primarily nonoverlapping subsets of genes have evolved splicing and transcript level differences between humans and chimpanzees
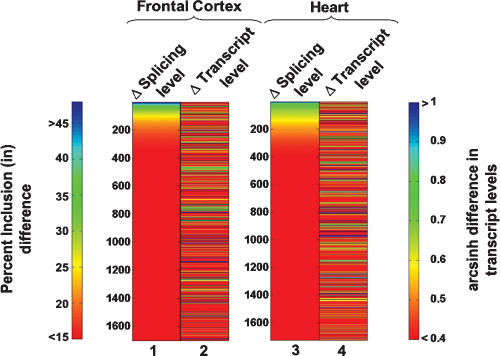
The 6%–8% of exons with pronounced splicing level differences detected in the above analysis involve ∼4% of the microarray profiled genes. A comparable proportion of orthologous genes has been found to display at least twofold transcript level differences between corresponding human and chimpanzee tissues (Preuss et al. 2004; Khaitovich et al. 2005). Remarkably, however, despite a parallel divergence in alternative splicing and transcriptional patterns during the evolution of humans and chimpanzees, primarily nonoverlapping subsets of genes are associated with these changes (Fig. 3). By sorting genes according to the magnitude of the microarray-detected percent inclusion alternative splicing level differences (Fig. 3, columns 1 and 3), it is evident that relatively few genes display coincident changes in transcriptional levels (Fig. 3 in cf. columns 1 and 2, and columns 3 and 4). Among the 160–240 splicing events with predicted percent inclusion differences >20% in frontal cortex and/or heart tissues, only 25% are predicted to also have twofold or greater steady-state transcript level changes. This observation extends previous results indicating that predominantly different subsets of genes are regulated in a cell-specific/tissue-specific or activity-dependent manner at the transcriptional and alternative splicing levels (Le et al. 2004; Pan et al. 2004; Li et al. 2006; Fagnani et al. 2007; Ip et al. 2007). In particular, the present results show that these two layers of regulation have evolved rapidly to affect different subsets of genes, and indicate that alternative splicing has served as an additional mechanism for diversifying gene regulation during the 5–7 million years of evolution separating humans and chimpanzees.
Figure 3.
Transcript and AS level differences between humans and chimpanzees involve largely distinct subsets of genes. The color spectrum plots compare splicing level differences and transcript level differences for the same set of genes expressed in the frontal cortex and heart. For each tissue comparison, the left column shows splicing differences (measured as the magnitude of percent inclusion difference, columns 1 and 3) and the right column measures differences in gene transcript level (measured as the magnitude of the hyperbolic arcsine [arcsinh] difference, columns 2 and 4). An hyperbolic arcsine difference of ∼0.4 corresponds to a 1.5-fold change in expression level, and a difference of ∼0.7 corresponds to a twofold difference in expression level.
Alternative splicing differences between humans and chimpanzees affect transcripts from genes associated with diverse cellular functions
Genes with alternative splicing differences between humans and chimpanzees detected using the methods described above, and confirmed by the RT–PCR assays, are associated with diverse biological processes including gene expression, signaling pathways, immune defense, and disease (Table 1). In most cases the alternative splicing differences were observed in both frontal cortex and heart tissues (Figs. 4 5) as well as in cell lines from similar origins from both species (Supplementary Fig. 1; see below). An example of an alternative splicing difference detected using the comparative genomics approach involves transcripts encoding the TATA-box-binding protein-associated factor 6 (TAF6) (Fig. 4; Supplementary Fig. 1; see Table 1 and below for additional examples and information). Examples of alternative splicing differences detected in the microarray data affect transcripts encoding the herpesvirus entry mediator receptor (HVEM/TNFRSF14), the spectrin-binding protein γ-Adducin (ADD3), ADP-ribosylation factor-like 3 (ARL3), serine/arginine (SR)-repeat family protein splicing factor of 40 kDa (SRp40/SFRS5), and GSTO2 (Figs. 4, 5; see Table 1 and below for additional examples and information). To assess whether these observed differences are specifically associated with the human or chimpanzee lineage, RT–PCR assays were also performed using RNA extracted from frontal cortex and heart tissue from rhesus macaques, an Old World monkey, as an outgroup comparison. The results of this analysis are summarized below and in Supplementary Table 3, and representative examples are shown in Figure 5A.
Alternative splicing differences are consistent between human and chimpanzee individuals
Since the analyses described above employed samples of poly(A)+ mRNA pooled, in each case, from several human individuals and from several chimpanzee individuals (Supplementary Table 2), it was important to assess the extent to which the alternative splicing differences might be explained by variations between individuals from each species. Individual samples comprising the pools of tissue mRNAs were therefore analyzed separately for splicing level differences. In each case examined, similar alternative splicing differences were observed between the several human and chimpanzee individuals (Fig. 5B; data not shown). These data indicate that it is highly unlikely that the overall differences in splicing levels measured using the pooled samples are attributed to differences associated with individual variation within each species. This conclusion was further supported by an analysis of alternative splicing level differences in primary fibroblasts and lymphoblastoid cells, also from multiple additional individuals from each species (Supplementary Fig. 1). Again, in all cases examined, similar species-associated splicing level differences were detected between the different individuals as detected in the tissue samples (Supplementary Fig. 1; see below). The results from this last experiment further indicate that the majority of the splicing level differences we observed are unlikely related to possible contributions from environmental or physiological differences, such as diet or stress, since the corresponding cell lines from both species were cultured in parallel for several weeks under identical growth conditions, prior to harvesting.
Examples of validated human and chimpanzee alternative splicing differences
The alternative splicing difference detected in TAF6 transcripts is human lineage specific and is pronounced in both the frontal cortex and heart (Fig. 4). TAF6 is a subunit of the general transcription factor TFIID, which is involved in gene activation (Sauer et al. 1995). It has been reported that TAF6 isoforms are associated with the control of apoptosis and cell cycle arrest (Wang et al. 1997, 2004; Bell et al. 2001). The alternative splicing difference found between humans and chimpanzees lies in the 5′ untranslated region (UTR), and could therefore affect TAF6 regulation at the post-transcriptional or translational levels. Consistent with this proposal are previous observations of alternative splicing events in 5′ UTRs that affect translational efficiency (Wang et al. 1999; Singh et al. 2005). Such a difference impacting TAF6 expression could, in turn, account for some of the differences in transcriptional profiles that have been observed between humans and chimpanzees (Preuss et al. 2004; Khaitovich et al. 2006).
The variation in alternative splicing of SRp40, which is also human lineage specific, is intriguing in light of the known roles of SR family members in both constitutive and regulated splicing (Graveley 2000; Sanford et al. 2005; Lin and Fu 2007). These proteins generally function in splicing by binding to exonic enhancer sequences via their RNA recognition motifs (RRMs) (Fig. 4). The alternative splicing difference in SRp40 transcripts results in the increased inclusion of a highly conserved premature termination codon (PTC)-containing exon (located between exons 4 and 5) in human transcripts, in both frontal cortex and heart tissue, and correspondingly reduced levels of the shorter transcript encoding the full-length protein, particularly in the heart. Several RNA-binding proteins including SR family members have been shown previously to regulate their own expression levels by activating the splicing of PTC-containing isoforms that are subsequently targeted by the process of nonsense-mediated mRNA decay (Lareau et al. 2007; Ni et al. 2007 and references within). Individual SR family members are essential and tightly regulated proteins (Graveley 2000; Sanford et al. 2005; Lin and Fu 2007). It is therefore possible that the differential expression of the two SRp40 isoforms described above could be related to some of the splicing differences we observed between humans and chimpanzees, including those that are not associated with nucleotide substitutions.
The alternative splicing difference affecting GSTO2 transcripts results in pronounced skipping of exon 4 of this gene, specifically in human frontal cortex tissue (Fig. 3; Supplementary Table 3). GSTO2 and its closely related paralog GSTO1 are important enzymes in the biotransformation of endogenous and exogenous compounds and in protection against oxidative stress (Schmuck et al. 2005), which plays an important role in defense against aging-associated diseases including cancers and neurodegeneration. In this regard, it is interesting to note that in some studies single nucleotide polymorphisms in GSTO2 and GSTO1 have been tentatively linked to certain human cancers and the age at onset of familial Alzheimer’s and Parkinson’s disease (Li et al. 2003; Whitbread et al. 2005; Pongstaporn et al. 2006; see Discussion).
An important question raised by the present and previous studies on alternative splicing is the extent to which differences in splice variant levels detected at the transcript level correspond to actual differences at the protein level. It should be borne in mind that comparisons of datasets from large-scale protein mass spectrometry analyses and from microarray profiling of mRNA from the same mammalian tissues have revealed good overall correlations between steady-state protein and transcript abundance (Kislinger et al. 2006). To investigate this question in the context of splice variants that differ between humans and chimpanzees, we next asked whether the alternative splicing level difference involving skipping of the frame-preserving exon 4 of GSTO2 transcripts could result in species-specific differences in the expression of GSTO2 variants at the protein level (Fig. 6).
Figure 6.
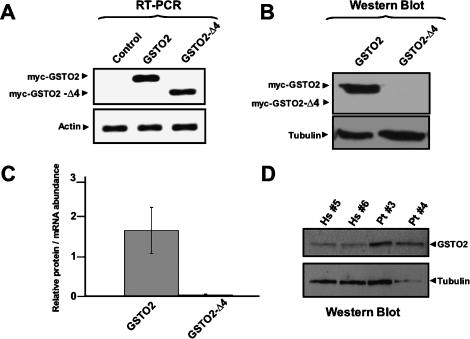
Increased skipping of exon 4 in GSTO2 transcripts reduces overall levels of functional GSTO2 protein in humans compared with chimpanzees. (A) RT–PCR experiments were performed to measure transcript abundance of individual myc-tagged GSTO2 isoforms transfected into HeLa cells. Primers were designed to anneal to exons neighboring the alternative exon in order to amplify isoforms including and skipping exon 4. β-Actin transcript levels were measured to normalize for input of total RNA between samples. (B) Western blotting experiments on the same transfected samples from A. Anti-c-myc antibodies were used to detect protein expression of individual GSTO2 splice variants. α-Tubulin protein levels were measured to control for loading input. (C) Measure of the relative protein/mRNA abundance for each of the transfected samples in A and B. Measurements represent the average of three independent transfection experiments, and standard deviations are shown. (D) Western blotting using samples from human and chimpanzee lymphoblastoid cells and antibodies to detect endogenous GSTO2. α-Tubulin protein levels were measured to control for loading input.
Expression of the functional splice variant of GSTO2 differs between humans and chimpanzees at the protein level
To initially assess the stability of the GSTO2 protein variants, expression vectors containing c-myc epitope-tagged GSTO2 cDNAs, with and without exon 4, were transiently expressed in HeLa cells. RT–PCR experiments using primers specific to constitutive exon sequences 5′ and 3′ to exon 4 indicated that comparable levels of the two splice variant transcripts are produced (Fig. 6A). Surprisingly, however, Western blotting of the protein lysates from the transfected cells with an anti-myc epitope antibody revealed that only the GSTO2 variant containing exon 4 results in significant protein expression (Fig. 6B,C); minor levels of the GSTO2 variant lacking exon 4 could be detected, but only after prolonged exposure of the blot (Fig. 6C; data not shown). These results indicate that GSTO2 splice variants lacking exon 4 are unstable at the protein level, and that levels of active GSTO2 are therefore likely determined by the relative expression of the splice variant containing exon 4. Also in support of this proposal are recent findings indicating that residues overlapping exon 4 of GSTO2 are important for both the stability and activity of this class of enzymes in vitro (Schmuck et al. 2005). From comparisons of the levels of the GSTO2 variants between different tissues and cell types it is apparent that increased skipping of exon 4 can result in reduced steady-state levels of exon 4-containing transcripts in humans compared with chimpanzees and other nonhuman primates, although in some cell and tissue sources differences in overall GSTO2 transcript levels may also account in part for increased levels of this variant in chimpanzees (Figs. 4, 5; Supplementary Fig. 1). Western blotting of lysates from lymphoblastoid cells from two human and two chimpanzee individuals confirms that the exon 4-containing splice variant is expressed at higher levels in the chimpanzee than human cells (Fig. 6D). This result therefore suggests that GSTO2 is likely more active in chimpanzee cells compared with the corresponding human cells.
Discussion
This study represents the first large-scale comparative analysis of alternative splicing patterns in humans and chimpanzees, and indeed between any pair of species separated by a relatively short period of evolutionary history. The results indicate that at least 4% of genes in humans and chimpanzees have one or more cassette alternative exons that display pronounced splicing level differences between the two species (refer to the Supplemental Material). This is comparable with the proportions (2%–8%) of orthologous human and chimpanzee protein coding genes reported to have significant transcript level differences in corresponding tissues (Preuss et al. 2004; Khaitovich et al. 2005). However, the majority of orthologous human and chimpanzee genes with splicing level differences do not overlap genes that display transcript level differences. These results indicate that alternative splicing has evolved rapidly to substantially increase the number of protein coding genes with altered patterns of regulation between humans and chimpanzees. Furthermore, the alternative splicing differences identified between humans and chimpanzees are predicted to affect a diverse range of critical functions, including the ability of cells to protect against oxidative damage.
Our result revealing a difference in the level of expression of the active splice variant of GSTO2 between humans and chimpanzees has interesting implications for how some alternative splicing differences could impact the evolution of important physiological and phenotypic differences between two species. Although GSTO2 transcripts that include and skip exon 4 have similar stabilities, only minor levels of protein expression were detected from the exon 4-skipped splice variant. This indicates that skipping of this exon leads to the expression of an unstable protein. Thus, even though it is unlikely that the exon 4-skipped splice variant of GSTO2 has a significant functional role at the protein level, differential inclusion of exon 4 appears to impact, at least in part, the relative expression levels of GSTO2 enzyme between humans and chimpanzees. This finding raises an important question: What is the extent to which alternative splicing events result in the expression of stable and functionally distinct protein products? Recent structural modeling of several splice variants from genes in the 1% of the genome surveyed by the ENCODE Consortium has predicted that many alternative splicing events are unlikely to result in correct protein folding, since the alternatively spliced regions are expected to often disrupt core structural domains (Tress et al. 2007). Based on this observation, it was concluded that a large proportion of splice variants detected in sequenced transcripts may not have functional roles. An alternative possibility, which is supported by the results in the present study, is that some alternative splicing events that do not lead to expression of functional or active protein could nevertheless impact overall levels of functional protein, either as a consequence of regulated splicing decisions within a species, or as a consequence of the evolution of differences in splicing levels between species.
In regard to the above, what are the possible functional consequences of altered splicing levels of GSTO2 exon 4? As mentioned earlier, GSTO2 functions in the biotransformation of toxic compounds and in the protection against oxidative stress, and SNPs in the GSTO2 gene, as well as in the paralogous GSTO1 gene, have been tentatively linked to human aging-associated diseases including cancer (Pongstaporn et al. 2006), and Alzheimer’s and Parkinson’s diseases (Li et al. 2003). GSTO2 appears to be more highly expressed in frontal cortex compared with heart, and it is also detected in other cells and tissues (Wang et al. 2005; Whitbread et al. 2005). Moreover, the differences we detected between human and chimpanzee GSTO2 transcripts appear to be quite specific, since we did not detect differences between the splicing or expression patterns of GSTO1 (data not shown), which nevertheless is closely related to GSTO2, although it has distinct substrate specificities (Schmuck et al. 2005; Wang et al. 2005). Reduced expression levels of GSTO2 in human cells would therefore be expected to specifically impact GSTO2-dependent functions. Intriguing in this regard are apparent major differences between humans and chimpanzees in relation to aging-associated neurodegeneration. Normal brain aging in humans is associated with the aberrant phosphorylation of the microtubule protein Tau, resulting in intraneuronal accumulations and formation of paired helical filaments and neurofibrillary tangles, a process that is greatly accelerated in Alzheimer’s disease (Hof and Morrison 2004). Comparable normal and pathological changes have not been identified in studies of aged chimpanzees or other nonhuman primates (Walker and Cork 1999). It is therefore possible that differences in gene regulation that impact the levels of proteins that function in the protection against oxidative damage, including GSTO2, could be linked to aging-associated normal and disease-related differences between humans and chimpanzees.
A major challenge for the future will be to definitively establish which differences in the expression levels of splice variants are associated with functional and phenotypic attributes of species, including humans and chimpanzees. This challenge equally applies to the growing list of differences that have been detected recently at the levels of gene structure and composition (Bustamante et al. 2005; Cheng et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005; Newman et al. 2005; Perry et al. 2006), as well as at other levels of gene regulation (Pollard et al. 2006; Prabhakar et al. 2006). Another important question in relation to all of these studies is the degree to which evolutionary changes in gene structure or expression are required to establish major differences in morphological and phenotypic characteristics. While the accumulation of small-effect changes at multiple loci probably underlie many species-specific differences, differences in the expression of individual genes that are integrally involved in developmental processes can also result in the evolution of major morphological changes (McGregor et al. 2007). Likewise, the differences we detected in the levels of splicing involving 6%–8% of orthologous cassette alternative exons, which in general are highly consistent between individuals from each species, could have a significant impact on morphological and other phenotypic differences between humans and chimpanzees. The results of the present study provide the basis for future investigations directed at elucidating the functional and phenotypic consequences of alternative splicing differences between humans and chimpanzees.
Materials and methods
Comparative genomic analysis of human and chimpanzee exons/flanking introns
Human mRNA and EST sequences were aligned to the human genome sequence as described previously (Kim et al. 2007) using human UniGene EST data (January 2006) (ftp://ftp.ncbi.nih.gov/repository/UniGene) and human genome assembly version hg17 (http://hgdownload.cse.ucsc.edu/goldenPath/hg17). Internal exons were detected as genomic regions flanked by two consensus splice sites. Orthologous chimpanzee sequences were identified using the University of California at Santa Cruz human versus chimpanzee pairwise genome alignments (http://hgdownload.cse.ucsc.edu/goldenPath/hg17/vsPanTro2). The percent nucleotide divergence rate (number of substituted nucleotides within each region divided by the total number of nucleotides within the region) was determined for regions including each orthologous exon and the upstream and downstream 150 bp of flanking intronic sequence. Regions associated with elevated substitution rates were manually inspected for sequencing errors, and to ensure correct alignment to the orthologous genes and not to more distantly related paralogs.
Tissue samples, cell lines, and cell culture
Tissue samples were obtained post-mortem from nine humans (Homo sapiens), six common chimpanzees (Pan troglodytes), and six rhesus macaques (Macaca mulatta). Information on the sex, age, and origin of the samples from each individual is provided in Supplementary Table 2. Tissue samples were immediately frozen in liquid nitrogen after dissection and kept at −80°C. Brain cortical tissue samples were taken from the frontal pole (FP) of the left hemisphere of each species. Mouse brain cortex and heart tissue samples were obtained from approximately five mice, dissected immediately after sacrificing and flash-frozen in liquid nitrogen. Primary fibroblast and lymphoblast cell lines from human and chimpanzee individuals were a kind gift from Stephen Scherer (The Hospital for Sick Children, Toronto, Ontario, Canada). The lymphoblast cell lines (human and chimpanzee) were grown in RPMI medium supplemented with 15% FCS, sodium pyruvate, L-glutamine, and antibiotics. Fibroblasts were grown in α minimal essential medium supplemented with 10% FCS and antibiotics. HeLa cells (American Type Culture Collection) were used for all experiments involving the GSTO2 overexpression constructs and were grown in DMEM supplemented with 10% FBS and antibiotics. All cells were incubated at 37°C and 5% CO2.
Microarray hybridization, data extraction, and analysis
Total RNA was extracted from 1–2 g of each tissue sample using the Trizol reagent (Invitrogen) as per the manufacturer’s recommendations. Portions of the total RNA samples from the same tissue from each of the individuals were pooled and poly(A)+ mRNA was purified from these samples using oligo-dT cellulose resin (New England Biolabs), as described previously (Pan et al. 2004). cDNA synthesized from the pooled poly(A)+ RNA samples was separately labeled with cyanine 3 and cyanine 5 fluorescent dyes (Amersham Pharmacia), and hybridized in dye swap experiments to a custom human oligonucleotide microarray manufactured by Agilent Technologies, Inc., as described previously (Pan et al. 2004). Microarrays were washed and scanned with a Genepix 4000A scanner (Axon Instruments), and images were processed and normalized as described previously (Pan et al. 2004). Processed intensity values from the microarray scans were input into the GenASAP algorithm (Pan et al. 2004; Shai et al. 2006) to obtain confidence-ranked percent inclusion level predictions for 5183 unique cassette alternative splicing events. Additional details of data analysis procedures are given in the Supplemental Material.
RT–PCR assays and quantification
Pooled poly(A)+ RNA samples were normalized for RNA concentration by comparing the amplified band intensities to a portion of the coding region of the human β-actin transcript. In addition, small aliquots of total RNA from each of the human, chimpanzee, and macaque individuals were analyzed by RT–PCR assays (Fig. 5; Supplementary Fig. 1). In each RT–PCR reaction, 0.2 ng of poly(A)+ RNA or 20 ng of total RNA were used as input and cDNA synthesis and amplification was performed using the One-Step RT–PCR kit (Qiagen) as per the manufacturer’s recommendations, with the following changes: Reactions were performed in a 10-μL volume, and 0.3 μCi of α-32P-dCTP was added to the reaction. The number of amplification cycles was 22 for β-actin and 30 for all other transcripts analyzed. All reaction products were resolved by using 6% denaturing polyacrylamide gels. The gels were subsequently dried and analyzed using a Typhoon Trio PhosphorImager and software (Amersham). Percent inclusion levels from RT–PCR reactions were calculated as the percent of the isoform including an alternative exon over the total abundance of the isoforms including and excluding the alternative exon. In the case of candidates from the comparative genomic sequence analysis, the presence of novel bands on gels were also considered and validated by sequencing. All primer sequences used in this study are available on request.
Plasmids
The full-length GSTO2 ORF was amplified by PCR from a plasmid (a kind gift from Dr. Richard Weinshilboum, Mayo Clinic College of Medicine, Rochester, MN) using the following primers: 5′-CGGAAGCTTATGTCTGGGGATGCGACCAGG-3′ and 5′-CGGGGATCCTCAGCACAGCCCAAAGTCAAAG-3′. The insert was subcloned into pCMV-Myc (Rosonina et al. 2005) using HindIII and BamHI sites to generate pCMV-Myc-GSTO2. The construct expressing the cDNA corresponding to the GSTO2 isoform lacking exon 4 was generated by PCR amplification using pCMV-Myc-GSTO2 and the following primers: 5′-ATTCTTGAGTATCAGAACACCACCTTCTT-3′ and 5′-CTTACAAAATAGCTCCAATAACATCTTTTGG-3′. The amplified product was then ligated with T4 DNA ligase (Fermentas) to generate pCMV-Myc-GSTO2-Δ4.
Transfection and Western blotting
HeLa cells were transfected with 24 μg of either pCMV-Myc-GSTO2 or pCMV-Myc-GSTO2-Δ4 with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Cells were harvested 48 h post-transfection. Total RNA was prepared as described above from both the transfected HeLa cells and the human and chimpanzee lymphoblast and primary fibroblast lines. Protein lysates were prepared by the addition of 100–200 μL of RIPA buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.5, 500 μM EDTA, 100 μM EGTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) to cell pellets, followed by lysis. Thirty micrograms of protein lysate from each transfected sample or 100 μg of human and chimpanzee lymphoblast cell lysates were run on 12% SDS–polyacrylamide gels. Immunoblotting was performed using anti-c-myc (Sigma), anti-GSTO2 (a kind gift of Dr. Richard Weinshilboum), or anti-α-tubulin (Sigma) antibodies and chemiluminescence reagents and secondary antibodies at dilutions recommended by the manufacturers.
Acknowledgments
We thank Arneet Saltzman, Ofer Shai, Matthew Fagnani, and Christine Misquitta for help with data processing and analysis. We also thank Chris Burge and James Thomas for support, Steve Scherer and The Centre for Applied Genomics (Hospital for Sick Children, Toronto) for technical assistance, and the staff of the Yerkes National Primate Research Center of Emory University and the Brain and Tissue Bank for Developmental Disorders at the University of Maryland for their assistance in obtaining tissue samples. Chris Burge, Jim Ingles, Quaid Morris, Mathieu Gabut, Arneet Saltzman, Matthew Fagnani, and Sidrah Ahmad kindly provided critical comments and helpful suggestions on the manuscript. Research in B.J.B’s laboratory was supported by grants from the Canadian Institutes of Health Research and National Cancer Institute of Canada, and in part by a grant from Genome Canada funded through the Ontario Genomics Institute. T.M.P. is supported by a James S. McDonnell Foundation grant (JSMF 21002093), the Center for Behavioral Neuroscience under the Science and Technology Center program of the National Science Foundation (IBN-9876754), and the Yerkes National Primate Research Center under National Center for Research Resources grant RR00165. C.L. and Y.X. were supported by NIH grant U54-RR021813 and a Dreyfus Foundation Teacher-Scholar Award. J.A.C. was supported by a scholarship from the Natural Science and Engineering Research Council of Canada, and M.C. was supported in part by the Ramón y Cajal Program (Ministerio de Educación y Ciencia, Spain)
Footnotes
References
- Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Bell B., Scheer E., Tora L., Scheer E., Tora L., Tora L. Identification of hTAF(II)80 δ links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell. 2001;8:591–600. doi: 10.1016/s1097-2765(01)00325-2. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Brudno M., Gelfand M.S., Spengler S., Zorn M., Dubchak I., Conboy J.G., Gelfand M.S., Spengler S., Zorn M., Dubchak I., Conboy J.G., Spengler S., Zorn M., Dubchak I., Conboy J.G., Zorn M., Dubchak I., Conboy J.G., Dubchak I., Conboy J.G., Conboy J.G. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., White T.J., Sninsky J.J., Hernandez R.D., Sninsky J.J., Hernandez R.D., Hernandez R.D., et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Chew S.L., Krainer A.R., Chew S.L., Krainer A.R., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chasin L.A. Searching for splicing motifs. In: Blencowe B.J., Graveley B.R., Graveley B.R., editors. Alternative splicing in the postgenomic era. Landes Bioscience; Georgetown, TX: 2007. pp. 86–107. [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Church D., DeJong P., Wilson R.K., Paabo S., DeJong P., Wilson R.K., Paabo S., Wilson R.K., Paabo S., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Fagnani M., Barash Y., Ip J., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Barash Y., Ip J., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Ip J., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., Shai O., Lee L., Rozenhek A., Mohammad N., Lee L., Rozenhek A., Mohammad N., Rozenhek A., Mohammad N., Mohammad N., et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E., Marques-Bonet T., Fernando O., Charlesworth B., Navarro A., Marques-Bonet T., Fernando O., Charlesworth B., Navarro A., Fernando O., Charlesworth B., Navarro A., Charlesworth B., Navarro A., Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P.R., Morrison J.H., Morrison J.H. The aging brain: Morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. doi: 10.1016/j.tins.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Ip J.Y., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W., Topp J.D., Blencowe B.J., Lynch K.W., Blencowe B.J., Lynch K.W., Lynch K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Schadt E.E., Stoughton R., Shoemaker D.D., Stoughton R., Shoemaker D.D., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S., Franz H., Weiss G., Lachmann M., Paabo S., Weiss G., Lachmann M., Paabo S., Lachmann M., Paabo S., Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P., Enard W., Lachmann M., Paabo S., Enard W., Lachmann M., Paabo S., Lachmann M., Paabo S., Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Kim N., Alekseyenko A.V., Roy M., Lee C., Alekseyenko A.V., Roy M., Lee C., Roy M., Lee C., Lee C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–D98. doi: 10.1093/nar/gk1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Gramolini A.O., Morris Q., Hallett M.T., Morris Q., Hallett M.T., Hallett M.T., et al. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Green R.E., Wengrod J.C., Brenner S.E., Wengrod J.C., Brenner S.E., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Le K., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Wang Q., Xu Q., Nelson S.F., Lee C., Xu Q., Nelson S.F., Lee C., Nelson S.F., Lee C., Lee C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.J., Oliveira S.A., Xu P., Martin E.R., Stenger J.E., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Oliveira S.A., Xu P., Martin E.R., Stenger J.E., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Xu P., Martin E.R., Stenger J.E., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Martin E.R., Stenger J.E., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Stenger J.E., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Scherzer C.R., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Hauser M.A., Scott W.K., Small G.W., Nance M.A., Scott W.K., Small G.W., Nance M.A., Small G.W., Nance M.A., Nance M.A., et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum. Mol. Genet. 2003;12:3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- Li H.R., Wang-Rodriguez J., Nair T.M., Yeakley J.M., Kwon Y.S., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Wang-Rodriguez J., Nair T.M., Yeakley J.M., Kwon Y.S., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Nair T.M., Yeakley J.M., Kwon Y.S., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Yeakley J.M., Kwon Y.S., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Kwon Y.S., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Bibikova M., Zheng C., Zhou L., Zhang K., Downs T., Zheng C., Zhou L., Zhang K., Downs T., Zhou L., Zhang K., Downs T., Zhang K., Downs T., Downs T., et al. Two-dimensional transcriptome profiling: Identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- Lin S., Fu X.-D., Fu X.-D. SR proteins and related factors in alternative splicing. In: Blencowe B.J., Graveley B.R., Graveley B.R., editors. Alternative splicing in the postgenomic era. Landes Bioscience; Georgetown, TX: 2007. pp. 108–123. [Google Scholar]
- Matlin A.J., Clark F., Smith C.W., Clark F., Smith C.W., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- McGregor A.P., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Srinivasan D.G., Payre F., Stern D.L., Payre F., Stern D.L., Stern D.L. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Modrek B., Lee C.J., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Newman T.L., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., McGrath S.D., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Shiue L., Clark T.A., Blume J.E., Ares M., Clark T.A., Blume J.E., Ares M., Blume J.E., Ares M., Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtdinov R.N., Artamonova I.I., Mironov A.A., Gelfand M.S., Artamonova I.I., Mironov A.A., Gelfand M.S., Mironov A.A., Gelfand M.S., Gelfand M.S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., Babak T., Siu H., Hughes T.R., Morris Q.D., Siu H., Hughes T.R., Morris Q.D., Hughes T.R., Morris Q.D., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Frey B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Blencowe B.J., Blencowe B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Maquat L.E., Frey B.J., Blencowe B.J., Frey B.J., Blencowe B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., Tyler-Smith C., Scherer S.W., Eichler E.E., Scherer S.W., Eichler E.E., Eichler E.E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Katzman S., King B., Onodera C., Siepel A., King B., Onodera C., Siepel A., Onodera C., Siepel A., Siepel A., et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Pongstaporn W., Rochanawutanon M., Wilailak S., Linasamita V., Weerakiat S., Petmitr S., Rochanawutanon M., Wilailak S., Linasamita V., Weerakiat S., Petmitr S., Wilailak S., Linasamita V., Weerakiat S., Petmitr S., Linasamita V., Weerakiat S., Petmitr S., Weerakiat S., Petmitr S., Petmitr S. Genetic alterations in chromosome 10q24.3 and glutathione S-transferase omega 2 gene polymorphism in ovarian cancer. J. Exp. Clin. Cancer Res. 2006;25:107–114. [PubMed] [Google Scholar]
- Prabhakar S., Noonan J.P., Paabo S., Rubin E.M., Noonan J.P., Paabo S., Rubin E.M., Paabo S., Rubin E.M., Rubin E.M. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Preuss T.M., Caceres M., Oldham M.C., Geschwind D.H., Caceres M., Oldham M.C., Geschwind D.H., Oldham M.C., Geschwind D.H., Geschwind D.H. Human brain evolution: Insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Ip J.Y., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Ip J.Y., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., McCracken S., Tucker P., Ingles C.J., Blencowe B.J., Tucker P., Ingles C.J., Blencowe B.J., Ingles C.J., Blencowe B.J., Blencowe B.J. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol. Cell. Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Ellis J., Caceres J.F., Ellis J., Caceres J.F., Caceres J.F. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- Sauer F., Hansen S.K., Tjian R., Hansen S.K., Tjian R., Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- Schmuck E.M., Board P.G., Whitbread A.K., Tetlow N., Cavanaugh J.A., Blackburn A.C., Masoumi A., Board P.G., Whitbread A.K., Tetlow N., Cavanaugh J.A., Blackburn A.C., Masoumi A., Whitbread A.K., Tetlow N., Cavanaugh J.A., Blackburn A.C., Masoumi A., Tetlow N., Cavanaugh J.A., Blackburn A.C., Masoumi A., Cavanaugh J.A., Blackburn A.C., Masoumi A., Blackburn A.C., Masoumi A., Masoumi A. Characterization of the monomethylarsonate reductase and dehydroascorbate reductase activities of omega class glutathione transferase variants: Implications for arsenic metabolism and the age-at-onset of Alzheimer’s and Parkinson’s diseases. Pharmacogenet. Genomics. 2005;15:493–501. doi: 10.1097/01.fpc.0000165725.81559.e3. [DOI] [PubMed] [Google Scholar]
- Shai O., Morris Q.D., Blencowe B.J., Frey B.J., Morris Q.D., Blencowe B.J., Frey B.J., Blencowe B.J., Frey B.J., Frey B.J. Inferring global levels of alternative splicing isoforms using a generative model of microarray data. Bioinformatics. 2006;22:606–613. doi: 10.1093/bioinformatics/btk028. [DOI] [PubMed] [Google Scholar]
- Singh S., Bevan S.C., Patil K., Newton D.C., Marsden P.A., Bevan S.C., Patil K., Newton D.C., Marsden P.A., Patil K., Newton D.C., Marsden P.A., Newton D.C., Marsden P.A., Marsden P.A. Extensive variation in the 5′-UTR of Dicer mRNAs influences translational efficiency. Biochem. Biophys. Res. Commun. 2005;335:643–650. doi: 10.1016/j.bbrc.2005.07.138. [DOI] [PubMed] [Google Scholar]
- Sorek R., Ast G., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Shamir R., Ast G., Shamir R., Ast G., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Stadler M.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B., Yeo G.W., Schneider A., Xiao X., Burge C.B., Schneider A., Xiao X., Burge C.B., Xiao X., Burge C.B., Burge C.B. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1372/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet C.W., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Williams A., Kulp D., Blume J.E., Haussler D., Kulp D., Blume J.E., Haussler D., Blume J.E., Haussler D., Haussler D., et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress M.L., Martelli P.L., Frankish A., Reeves G.A., Wesselink J.J., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Martelli P.L., Frankish A., Reeves G.A., Wesselink J.J., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Frankish A., Reeves G.A., Wesselink J.J., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Reeves G.A., Wesselink J.J., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Wesselink J.J., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Yeats C., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Olason P.L., Albrecht M., Hegyi H., Giorgetti A., Albrecht M., Hegyi H., Giorgetti A., Hegyi H., Giorgetti A., Giorgetti A., et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl. Acad. Sci. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.C., Cork L.C., Cork L.C. The neurobiology of aging in nonhuman primates. Lippincott Williams & Wilkins; Philadelphia: 1999. [Google Scholar]
- Wang S., Dibenedetto A.J., Pittman R.N., Dibenedetto A.J., Pittman R.N., Pittman R.N. Genes induced in programmed cell death of neuronal PC12 cells and developing sympathetic neurons in vivo. Dev. Biol. 1997;188:322–336. doi: 10.1006/dbio.1997.8655. [DOI] [PubMed] [Google Scholar]
- Wang Y., Newton D.C., Robb G.B., Kau C.L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Newton D.C., Robb G.B., Kau C.L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Robb G.B., Kau C.L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Kau C.L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A., VanDamme S., Wilcox J.N., Marsden P.A., Wilcox J.N., Marsden P.A., Marsden P.A. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Nahta R., Huper G., Marks J.R., Nahta R., Huper G., Marks J.R., Huper G., Marks J.R., Marks J.R. TAFII70 isoform-specific growth suppression correlates with its ability to complex with the GADD45a protein. Mol. Cancer Res. 2004;2:442–452. [PubMed] [Google Scholar]
- Wang L., Xu J., Ji C., Gu S., Lv Y., Li S., Xu Y., Xie Y., Mao Y., Xu J., Ji C., Gu S., Lv Y., Li S., Xu Y., Xie Y., Mao Y., Ji C., Gu S., Lv Y., Li S., Xu Y., Xie Y., Mao Y., Gu S., Lv Y., Li S., Xu Y., Xie Y., Mao Y., Lv Y., Li S., Xu Y., Xie Y., Mao Y., Li S., Xu Y., Xie Y., Mao Y., Xu Y., Xie Y., Mao Y., Xie Y., Mao Y., Mao Y. Cloning, expression and characterization of human glutathione S-transferase omega 2. Int. J. Mol. Med. 2005;16:19–27. [PubMed] [Google Scholar]
- Whitbread A.K., Masoumi A., Tetlow N., Schmuck E., Coggan M., Board P.G., Masoumi A., Tetlow N., Schmuck E., Coggan M., Board P.G., Tetlow N., Schmuck E., Coggan M., Board P.G., Schmuck E., Coggan M., Board P.G., Coggan M., Board P.G., Board P.G. Characterization of the omega class of glutathione transferases. Methods Enzymol. 2005;401:78–99. doi: 10.1016/S0076-6879(05)01005-0. [DOI] [PubMed] [Google Scholar]
- Xing Y., Lee C., Lee C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Xu Q., Modrek B., Lee C., Modrek B., Lee C., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Holste D., Kreiman G., Burge C.B., Holste D., Kreiman G., Burge C.B., Kreiman G., Burge C.B., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004a;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Hoon S., Venkatesh B., Burge C.B., Hoon S., Venkatesh B., Burge C.B., Venkatesh B., Burge C.B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl. Acad. Sci. 2004b;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B., Van Nostrand E., Holste D., Poggio T., Burge C.B., Holste D., Poggio T., Burge C.B., Poggio T., Burge C.B., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Nostrand E.L., Liang T.Y., Nostrand E.L., Liang T.Y., Liang T.Y. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Heller K.A., Hefter I., Leslie C.S., Chasin L.A., Heller K.A., Hefter I., Leslie C.S., Chasin L.A., Hefter I., Leslie C.S., Chasin L.A., Leslie C.S., Chasin L.A., Chasin L.A. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 2003;13:2637–2650. doi: 10.1101/gr.1679003. [DOI] [PMC free article] [PubMed] [Google Scholar]