Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling (original) (raw)
Abstract
Amajor function of the endocytic system is the sorting of cargo to various organelles. Endocytic sorting of the yeast reductive iron transporter, which is composed of the Fet3 and Ftr1 proteins, is regulated by available iron. When iron is provided to iron-starved cells, Fet3p–Ftr1p is targeted to the lysosome-like vacuole and degraded. In contrast, when iron is not available, Fet3p–Ftr1p is maintained on the plasma membrane via an endocytic recycling pathway requiring the sorting nexin Grd19/Snx3p, the pentameric retromer complex, and the Ypt6p Golgi Rab GTPase module. A recycling signal in Ftr1p was identified and found to bind directly to Grd19/Snx3p. Retromer and Grd19/Snx3p partially colocalize to tubular endosomes, where they are physically associated. After export from the endosome, Fet3p–Ftr1p transits through the Golgi apparatus for resecretion. Thus, Grd19/Snx3p, functions as a cargo-specific adapter for the retromer complex, establishing a precedent for a mechanism by which sorting nexins expand the repertoire of retromer-dependent cargos.
Introduction
The endocytic system fulfills several vital roles for the cell, including internalization and degradation of macromolecules, down-regulation of activated signaling receptors, and spatial regulation of intracellular signaling. Another important function of the endocytic system is to regulate the protein composition of the plasma membrane through remodeling. For instance, the number of plasma membrane–localized transporters and receptors present at any one time arises from multiple sorting decisions made within the endosomal system (Lemmon and Traub, 2000). Indeed, given that constitutive or bulk endocytosis is constantly occurring, the steady-state localization of several plasma membrane proteins is maintained by endocytic recycling, a process in which internalized proteins are returned to the cell surface (Maxfield and McGraw, 2004). However, little is known regarding the cis-acting sorting signals and trans-acting protein machinery that mediate recycling of plasma membrane proteins.
After internalization, recycling integral plasma membrane proteins are sorted into transport carriers that bud from a tubular early endosomal compartment. These vesicles are then targeted either directly to the plasma membrane or to an intermediate destination (i.e., the endocytic recycling compartment or the Golgi apparatus) before cargo is delivered back to the plasma membrane. In yeast, the best-characterized plasma membrane recycling pathways involve transit of cargo through the Golgi for resecretion. For example, endocytic recycling of the exocytic v-SNARE Snc1p involves endosome-to-Golgi transport, followed by export from the Golgi to the plasma membrane (Lewis et al., 2000; Galan et al., 2001). Therefore, although the pathways that various cargo proteins take in returning back to the plasma membrane may be complex and involve multiple organelles, it is clear that sorting of cargo at endosomes is a common and key event in determining whether a particular protein will be recycled or targeted for degradation.
Considerable insight has been gained into the sorting, export, and retrieval of cargo proteins from endosomes. An evolutionarily conserved protein complex, known as retromer, has received much attention for its role in this process (Seaman, 2005). In yeast, retromer is composed of five subunits that form two subcomplexes: a complex thought to perform cargo selection, composed of Vps26p, Vps29p, and Vps35p, and a complex that mediates membrane association, composed of Vps5p and Vps17p (Seaman et al., 1998; Nothwehr et al., 1999, 2000; Seaman and Williams, 2002). Retrieval of the vacuolar hydrolase receptor Vps10p and the late Golgi proteins Kex2p and Ste13p from the prevacuolar compartment back to the Golgi has been shown to be dependent on a functional retromer complex (Horazdovsky et al., 1997; Nothwehr and Hindes, 1997). In mammals, retromer is composed of hVps26, hVps29, hVps35 (Haft et al., 2000), and the Bin/Amphiphysin/Rvs (BAR) domain–containing proteins Snx1 and Snx2 (Carlton et al., 2004). Retromer also functions in several trafficking pathways, including transcytosis of the polymeric immunoglobulin receptor (Verges et al., 2004) and retrieval of cargo, such as the cation-independent mannose-6-phosphate receptor from late endosomes back to the Golgi (Arighi et al., 2004; Seaman, 2004). Interestingly, recent studies in Caenorhabditis elegans have also implicated retromer function in establishing long-range Wnt signaling gradients, expanding the role of retromer to include developmental patterning (Coudreuse et al., 2006; Prasad and Clark, 2006). Thus, evidence indicates that retromer participates in endocytic sorting of multiple distinct cargos, but it is not yet known how such diverse cargo is selected by retromer.
The retromer components Vps5p and Vps17p (as well as the mammalian homologues Snx1 and Snx2) are members of the sorting nexin family of proteins, or SNXs, which has been broadly implicated in sorting within the endosomal system (Teasdale et al., 2001; Worby and Dixon, 2002; J. Carlton et al., 2005). This protein family is characterized by the presence of a SNX-PX (phox homology) domain that binds preferentially to phosphatidylinositol-3-phosphate (PtdIns[3]P), which is a lipid enriched on endosomal membranes (Seet and Hong, 2006). Currently, 28 mammalian and 10 yeast sorting nexin proteins have been identified (Worby and Dixon, 2002; Carlton and Cullen, 2005), and most of the data regarding the functions of these proteins in yeast points to their role in endocytic sorting. Snx4p, Snx41p, and Snx42p, for instance, form a complex that retrieves Snc1p from early endosomes (Hettema et al., 2003). Yeast cells that lack Grd19p (the yeast homologue of human Snx3) or cells that lack retromer mislocalize Ste13p and Kex2p to the vacuole (Nothwehr and Hindes, 1997; Voos and Stevens, 1998). In addition, Vps5p, Vps17p, and Mvp1p (another sorting nexin), have been shown to be essential for retrieval of Vps10p from late endosomes (Ekena and Stevens, 1995; Horazdovsky et al., 1997). Thus, there is evidence supporting the notion that these sorting factors cooperate to fulfill their functions in protein retrieval and recycling.
In S. cerevisiae, sorting decisions within the endosomal system have been shown to regulate the abundance, stability, and localization of various nutrient transporters. These sorting decisions are generally based on cues regarding the availability of particular nutrients. The general amino acid permease Gap1p, for instance, is highly expressed at the plasma membrane when cells are grown in media containing a poor nitrogen source, but is rapidly internalized when high concentrations of amino acids are present (Chen and Kaiser, 2002). Similarly, the uracil permease Fur4p is down-regulated when cells are exposed to toxic concentrations of uracil (Blondel et al., 2004). We have used the yeast high-affinity iron transporter (composed of the proteins Fet3p and Ftr1p) as a model to investigate endocytic sorting events in response to a limiting concentration of extracellular iron. We demonstrate that the plasma membrane localization of Fet3p–Ftr1p in the absence of iron is maintained by endocytic recycling and that the sorting nexin Grd19p mediates this recycling via interactions with both Ftr1p and the retromer complex.
Results
Retromer and Grd19/Snx3p are required to maintain Ftr1p-GFP at the plasma membrane via endocytic recycling
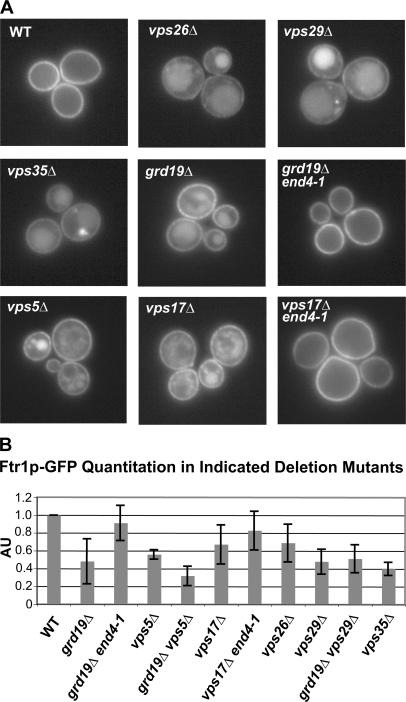
The high-affinity reductive iron transporter in S. cerevisiae is composed of two proteins, Fet3p and Ftr1p (Askwith et al., 1994; Stearman et al., 1996), whose biosynthesis and localization are regulated by the concentration of extracellular iron. When iron is limiting in the growth medium, the protein complex is highly expressed and is localized to the plasma membrane. However, when the extracellular concentration of iron is high, transcription ceases and the protein complex undergoes regulated endocytosis and delivery to the lysosome-like vacuole, where it is degraded (Felice et al., 2005). When cells are grown in media containing intermediate concentrations of iron (10–100 μM), transcription of FET3 and FTR1 is barely detectable, yet the abundance of the Fet3 and Ftr1 proteins remains high and the proteins are localized to the plasma membrane (Felice et al., 2005). As the abundance of several other nutrient transporters is regulated by protein sorting within the endosomal–lysosomal system (Blondel et al., 2004; Rubio-Texeira and Kaiser, 2006), we hypothesized that the localization and stability of Fet3p and Ftr1p might be due, at least in part, to recycling of endocytosed Fet3p–Ftr1p. According to this hypothesis, Fet3p–Ftr1p are internalized constitutively and are recycled back to the plasma membrane under low-iron conditions; if they were not recycled, they would be delivered to the vacuole and degraded. To test our hypothesis, a GFP tag was integrated upstream of the stop codon of the FTR1 locus in a collection of yeast deletion mutants with known defects in protein trafficking within the endocytic system (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200609161/DC1), and this collection was visually screened to identify mutants in which Ftr1p-GFP was partially localized to the vacuole when cells were grown in iron-deficient medium. This screen identified nine genes required to restrict Ftr1p-GFP localization to the plasma membrane under these conditions (Fig. 1 A). Five of these genes, VPS5, VPS17, VPS26, VPS29, and VPS35, encode subunits of retromer, which is a protein complex that has been implicated in the trafficking of cargo proteins from endosomes to the Golgi (Seaman et al., 1997, 1998). Another gene, GRD19/SNX3, encodes a member of the sorting nexin family of proteins, and it has also been implicated in trafficking from endosomes to the Golgi (Voos and Stevens, 1998). The other three genes, YPT6, RIC1, and RGP1, encode components of the Ypt6 Golgi Rab GTPase module that functions at the Golgi in endosome-to-Golgi trafficking pathways (Fig. S1 A; Siniossoglou et al., 2000; Bensen et al., 2001). In all of these mutants, Fet3p-GFP was also mislocalized to the vacuole (Fig. S1 B). We have focused here on investigating the roles of Grd19p and retromer in endocytic recycling.
Figure 1.
Retromer and the sorting nexin Grd19p are required to maintain PM localization of Fet3p–Ftr1p under iron-limiting conditions. (A) Ftr1p-GFP is mislocalized to the lumen of the vacuole in cells that lack a functional retromer complex (_vps5_Δ, _vps17_Δ, _vps26_Δ, vps29_Δ, and vps35_Δ) or the sorting nexin Grd19p (_grd19_Δ). Cells expressing Ftr1p-GFP from the native FTR1 locus were grown in iron-deficient medium and analyzed by fluorescence microscopy. (B) Quantitative immunoblot analysis of steady-state levels of Ftr1p-GFP in wild-type (WT) cells and the indicated deletion mutants. Cells were grown in iron-deficient medium, and whole-cell extracts were prepared and immunoblotted with an anti-GFP antibody and an antibody to Pgk1p (3-phosphoglycerate kinase) which was used as a loading control. ECF was used to detect the antibodies, and the ECF signals were quantified using ImageQuant software and normalized to the loading control. AU, arbitrary units. The amount of Ftr1p-GFP detected from the wild-type strain was set to 1.0, and the means and SDs for each of the indicated strains were calculated from three independent experiments.
In principle, Grd19p and retromer could be functioning in one of two ways to localize Ftr1p to the plasma membrane under iron-limiting conditions. They could act in the biosynthetic pathway to deliver Fet3p–Ftr1p to the plasma membrane, or they could function after Fet3p–Ftr1p has been delivered to the plasma membrane to maintain them there. These models can be distinguished experimentally because in the latter model endocytosis would be required for delivery of Ftr1p-GFP to the vacuole in the mutants. To test this, double mutants were constructed in which the endocytosis-defective end4-1 allele was combined with _grd19_Δ or a retromer gene deletion, _vps17_Δ, and Ftr1p-GFP localization was determined. In both cases, no vacuolar GFP signal was observed with the double mutants (Fig. 1 A), indicating that the end4-1 mutation is epistatic to _grd19_Δ and _vps17_Δ. As an independent test of these observations, we used quantitative immunoblotting of cell extracts to determine steady-state levels of Ftr1p-GFP in all of these strains, in addition to the double mutant strains _grd19_Δ _vps5_Δ and _grd19_Δ _vps29_Δ. In all the retromer and grd19 mutant strains, the amount of Ftr1p-GFP was significantly reduced compared with a wild-type control extract, and the end4-1 mutation substantially restored levels of detected Ftr1p-GFP. These results indicate that, in these mutants, Ftr1p-GFP must first be delivered to the plasma membrane to be subsequently delivered to the vacuole and implicate retromer and Grd19p in a recycling pathway that maintains Fet3p–Ftr1p on the plasma membrane. These results further suggest that retromer and Grd19p are likely functioning together and do not act independently or in a sequential manner, as the Grd19p-retromer double mutants have reduced steady-state levels of Ftr1p-GFP that are essentially identical to the single mutants.
The C-terminal cytoplasmic tail of Ftr1p is required for endocytic recycling
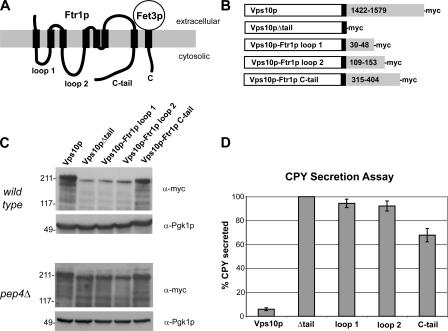
Because endocytic recycling signals are not well characterized, we sought to design a systematic strategy to identify those features of Fet3p and Ftr1p that are responsible for directing endocytic recycling. We designed a novel assay based on the trafficking itinerary of the vacuolar protein sorting receptor, Vps10p, which cycles between endosomes and the Golgi to mediate the sorting of cargo proteins to the vacuole (Marcusson et al., 1994; Cooper and Stevens, 1996). After delivery of Vps10p–cargo complexes to the late endosome, retrieval of Vps10p back to the Golgi requires retromer, which recognizes a retrieval signal in the C-terminal cytoplasmic tail of Vps10p (Nothwehr et al., 1999). Removal of the cytoplasmic tail results in vacuolar delivery and degradation of Vps10p, and, as a result, cargos normally destined for the vacuole, such as carboxypeptidase Y (CPY) are instead secreted (Cereghino et al., 1995; Cooper and Stevens, 1996). Because retromer is required for the stability of both Vps10p and Fet3p–Ftr1p, we reasoned that Fet3p–Ftr1p contains at least one endosome-to-Golgi sorting signal. This served as the basis of an assay aimed at identifying Fet3p–Ftr1p sequences, which can restore function to a Vps10p mutant lacking the cytoplasmic tail. We focused our analysis on Ftr1p because experiments, which are described in the following sections, indicated that Fet3p does not contain any post-Golgi sorting signals. Ftr1p contains four cytoplasmic portions; three of these are loops connecting membrane-spanning segments and the fourth is a C-terminal tail (Fig. 2 A). The third loop of Ftr1p, connecting the fifth and sixth membrane-spanning segments, was not investigated because it is predicted to consist of, at most, four amino acids. Fusion proteins in which the cytoplasmic tail of Vps10p was replaced with sequences encoding the remaining cytoplasmic regions of Ftr1p were constructed and integrated at the endogenous VPS10 locus. All constructs also contained 13 copies of the Myc epitope tag (13xMyc) at the C terminus. As controls, full-length Vps10p was tagged with a 13xMyc tag at its C terminus, and a deletion mutant that removed the entire cytoplasmic domain of Vps10p was constructed by integrating a 13xMyc epitope tag after Arg1421 (Fig. 2 B).
Figure 2.
The C-terminal cytoplasmic tail of Ftr1p contains an endosome-to-Golgi sorting/recycling signal. (A) The predicted topology of the S. cerevisiae high-affinity iron transporter. The iron oxidase Fet3p contains a single transmembrane domain, whereas Ftr1p, the iron permease, contains seven transmembrane domains connected by three cytoplasmic loops. Both proteins have cytoplasmic C-terminal tails. (B) Control proteins and chimeric Vps10p-Ftr1p proteins were generated to identify a potential recycling signal within Ftr1p. The indicated cytoplasmic regions of Ftr1p (numbers in gray rectangles) were fused to Vps10p just downstream of the single transmembrane domain (black rectangle). All constructs also contained a myc-epitope tag at the C terminus. (C) Steady-state immunoblot analysis of myc-tagged Vps10p-Ftr1p chimeras and control constructs. Whole-cell extracts were prepared and immunoblotted with an anti-myc antibody and an antibody to Pgk1p. An identical analysis of the abundance of the chimeric proteins was done with _pep4_Δ cells, which are deficient in vacuolar proteolysis. (D) Analysis of CPY secretion from Vps10p-Ftr1p strains by quantitative colony blot assay. Secretion of vacuolar CPY from strains expressing chimeric Vps10p-Ftr1p fusion proteins was quantified by colony immunoblotting with an anti-CPY monoclonal antibody. ECF was used to detect anti-CPY antibody, and the ECF signals were quantified using ImageQuant software. The amount of CPY secreted from the Vps10Δtail strain was set to 100%, and the means and SDs for each of the indicated strains were calculated from 10 independent measurements.
Two different methods were used to analyze the functionality of the Vps10p-Ftr1p chimeric proteins relative to the control constructs. Vps10p mutants that are not recycled, such as Vps10pΔtail, are delivered to the vacuole and degraded, resulting in low steady-state levels of the protein (Cereghino et al., 1995; Cooper and Stevens, 1996). Therefore, we compared the steady-state levels of the Vps10p-Ftr1p fusion proteins to full-length Vps10p and Vps10pΔtail in a wild-type strain and a _pep4_Δ strain that lacks the predominant vacuolar protease, and thus is severely compromised in vacuolar degradation (Fig. 2 C). In wild-type cells, the amount of the Vps10p-Ftr1p(C-tail) construct was nearly as high as native Vps10p, whereas the amounts of Vps10p-Ftr1p(loop1) and Vps10p-Ftr1p(loop2) proteins were similar to the level of Vps10pΔtail. The low amounts of these proteins were rescued in the _pep4_Δ strain, indicating that the low levels were caused by proteolysis in the vacuole.
As a second test, we quantified the secretion of CPY from these strains using a CPY colony blot assay (Fig. 2 D; Conibear and Stevens, 2002). The results show that CPY secretion by the Vps10p-Ftr1p(C-tail) strain was rescued by 33% compared with the Vps10pΔtail strain. No significant rescue of CPY sorting was observed in the Vps10p-Ftr1p(loop1) and Vps10p-Ftr1p(loop2) strains. It is unclear why the Vps10p-Ftr1p(C-tail) chimeric protein is not more effective at rescuing CPY sorting; however, it is likely that the Ftr1p sequence lacks TGN sorting signals that are present in Vps10p. Nevertheless, the corroborative results of these two assays suggest that the C-terminal tail of Ftr1p contains at least one signal that can partially substitute for the Vps10p endosome–Golgi retrieval signal.
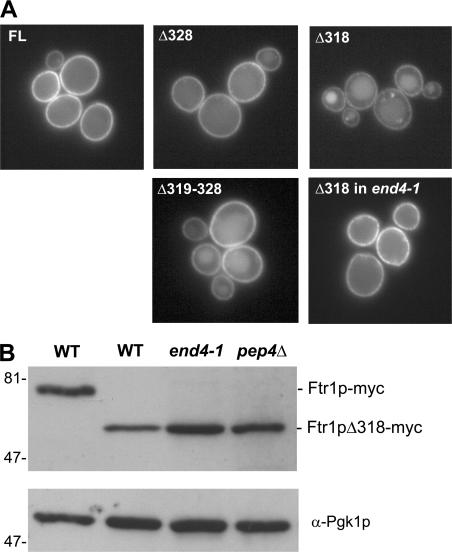
To further map the recycling signals within the cytoplasmic tail of Ftr1p, a series of truncation mutants was systematically constructed by integrating GFP at various points within the FTR1 locus and analyzing the localization of these truncated proteins under iron-limiting conditions. Truncation after residue 318 (Ftr1pΔ318) resulted in localization to the lumen of the vacuole in addition to localization at the plasma membrane. However, a mutant truncated after residue 328 (Ftr1pΔ328) was localized solely to the plasma membrane, implicating amino acids 319–328 as important for endocytic recycling. To confirm a requirement for this region, an internal deletion mutant lacking residues 319–328 (Ftr1pΔ319-328) was constructed and localized to the plasma membrane and vacuole lumen, although missorting of this internal deletion mutant was not as severe as the Ftr1pΔ318 mutant (Fig. 3 A), suggesting that other portions of the Ftr1p C-tail may also play a role in recycling. Importantly, none of the truncated proteins accumulated in the ER, indicating that their folding and association with Fet3p was not grossly affected by removal of these sequences.
Figure 3.
The Ftr1p recycling signal resides between residues 319–328. (A) Localization of GFP-tagged Ftr1p truncation mutants. Cells were grown in iron-deficient medium and imaged by fluorescence microscopy. Numbers indicate the last Ftr1p residue expressed (top). An internal deletion mutant lacking aa 319–328 was also tested (Δ319–328). (B) Analysis of steady-state levels of full-length Ftr1p-myc and Ftr1pΔ318-myc grown in iron-deficient medium. Whole-cell extracts were prepared from the indicated strains and were immunoblotted with an anti-myc antibody and an antibody to Pgk1p as a loading control.
To interpret these results from the perspective of endocytic sorting signals, it was important to distinguish if the Ftr1pΔ318-GFP–truncated protein was being targeted to the vacuole via endocytosis or the biosynthetic route. We again made use of the end4-1 strain to determine the localization of the Ftr1pΔ318-GFP construct under iron-starvation conditions. In the end4-1 strain, Ftr1pΔ318-GFP was localized exclusively to the plasma membrane (Fig. 3 A), and these results were confirmed by immunoblotting of cell extracts. Less Ftr1p was detected when the cytoplasmic tail of the protein was truncated, and levels of the truncated protein were stabilized in the end4-1 strain, as well as in a _pep4_Δ strain (Fig. 3 B). The results indicate that this Ftr1p truncation mutant is trafficked properly to the plasma membrane, but it is not maintained there. Together, these results demonstrate that aa 319–328 of Ftr1p are important for endocytic recycling.
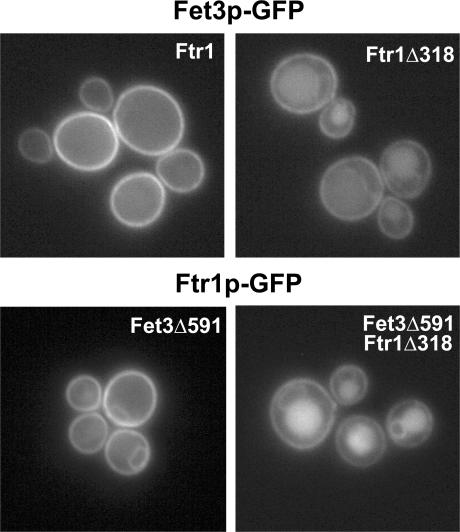
The delivery of newly synthesized Fet3p and Ftr1p to the plasma membrane depends on their coexpression, and iron-induced down-regulation of Fet3p parallels Ftr1p, indicating that these two proteins traffic together throughout the endomembrane system (Stearman et al., 1996; Sato et al., 2004; Felice et al., 2005). To determine if the aberrant trafficking of Ftr1p truncation mutants also impacts the trafficking of Fet3p, we examined the localization of Fet3p-GFP in the context of the Ftr1p-tail truncation. We integrated a GFP tag on Fet3p in strains expressing full-length Ftr1p or Ftr1pΔ318 and determined the localization of Fet3p-GFP under iron-limiting conditions by fluorescence microscopy. Fet3p-GFP was restricted to the plasma membrane when coexpressed with full-length Ftr1p, but it was also localized to the vacuole lumen (and a very minor fraction in the ER) when coexpressed with Ftr1pΔ318 (Fig. 4). In a strain with a deletion of most of the Fet3p cytoplasmic tail (deletion of the last 45 amino acids), Ftr1p-GFP was localized to the ER and the plasma membrane (Fig. 4), as expected from a previous study (Singh et al., 2006), suggesting that the Fet3p cytoplasmic tail is required for efficient ER export, but does not influence post-ER trafficking. However, in a strain with deletions of both the Fet3p and Ftr1p C-terminal tails (C-tails), both proteins were localized to the ER, plasma membrane, and the vacuole lumen (Fig. 4), indicating that once exported from the ER, the complex was sorted to the vacuole in the absence of the Ftr1p recycling signal. Collectively, the results indicate that a region of the C-terminal cytoplasmic tail of Ftr1p (aa 319–328; GHLPFTKNLQ) is necessary for endocytic recycling of both Fet3p and Ftr1p under iron-depleted conditions.
Figure 4.
The Ftr1p recycling signal is required for endocytic recycling of Fet3p. Yeast strains were constructed in which Fet3p-GFP was expressed in cells expressing full-length Ftr1p or Ftr1pΔ318 (top), or Ftr1p-GFP and Ftr1pΔ318-GFP in cells expressing a C-tail–truncated form of Fet3p, Fet3pΔ591 (bottom). The cells were grown in iron-deficient medium and imaged by fluorescence microscopy.
Grd19p binds to the Ftr1p recycling signal
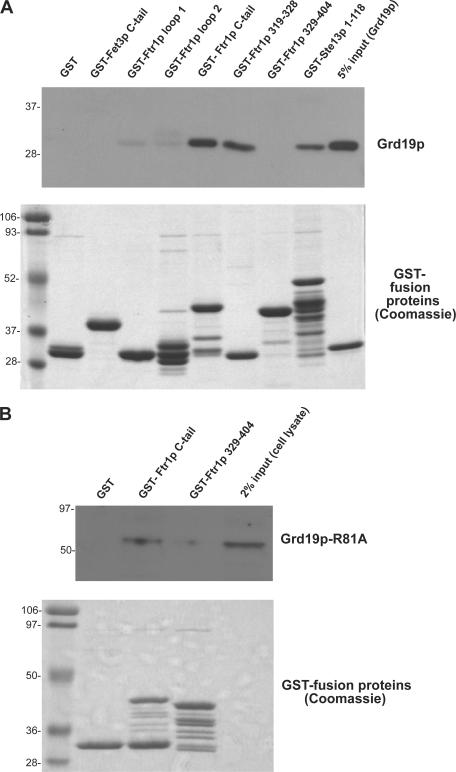
We next sought to address whether any of the endosomal sorting factors identified in the initial screen recognize the Ftr1p recycling signal. Previous work has shown that epitope-tagged Grd19p present in cell extract, but not the retromer subunit Vps5p, can be captured by a GST-Ste13p fusion protein containing the entire Ste13p cytoplasmic domain (Voos and Stevens, 1998). We first tested binding of purified recombinant Grd19p to a panel of GST fusion proteins containing each of the cytoplasmic regions of Fet3p and Ftr1p, as well as several smaller portions of the Ftr1p C-tail (Fig. 5 A). Grd19p bound the Ftr1p C-tail, but not any of the other cytoplasmic portions of Ftr1p or Fet3p. Remarkably, Grd19p also bound to a GST fusion protein consisting of only aa 319–328 of Ftr1p, but did not bind to the Ftr1p C-tail deletion mutant lacking these residues, narrowing the site of Grd19p interaction to aa 319–328 of Ftr1p. A recent study demonstrated that the C-terminal tails of Fet3p and Ftr1p physically associate via a region that includes the Grd19p binding site of Ftr1p (Singh et al., 2006), and we therefore tested if binding of Grd19p to the Ftr1p cytoplasmic tail in vitro is influenced by coincubation with the Fet3p cytoplasmic tail, revealing that it was not (unpublished data). Finally, we also tested if the retromer complex binds to the recycling signal in the Fet3p–Ftr1p complex using cell lysate prepared from a strain expressing Vps29p- myc; however, no Vps29p was captured by any of the GST fusion proteins (unpublished data). Although this does not rule out that retromer does not bind Ftr1p, all of the binding experiments indicate that Grd19p specifically recognizes the Fet3p–Ftr1p endocytic recycling signal.
Figure 5.
Grd19p binds directly to the Ftr1p recycling signal. (A) The following GST-fusion proteins were expressed and purified from bacteria and immobilized on glutathione–Sepharose beads: GST-Fet3p cytoplasmic tail (residues 581–636), GST-Ftr1p cytoplasmic loop 1 (residues 30–48), GST-Ftr1p cytoplasmic loop 2 (residues 109–153), GST-Ftr1p cytoplasmic tail (residues 315–404), GST-Ftr1p(319–328), which is the putative recycling signal, and GST-Ftr1p(329–404), which contains the entire C-terminal domain except for the recycling signal. Purified fusion proteins were incubated with purified recombinant His6-Grd19p, which contains a T7 epitope tag on the N terminus. The beads were washed, and bound proteins were eluted in SDS sample buffer. After SDS-PAGE, Grd19p was detected using an anti-T7 epitope antibody (top) and the GST fusion proteins were visualized by Coomassie blue staining (bottom). (B) Yeast strains expressing Grd19p-R81A-myc and the indicated GST fusion proteins were grown in synthetic media containing 4% galactose. Cells were harvested, converted to spheroplasts, and the bifunctional chemical cross-linking reagent DSP was added to a final concentration of 4 mM. The cells were then lysed, and the GST fusion proteins were captured on glutathione–Sepharose beads. The cross-links were disrupted by reduction in SDS sample buffer, and the samples were probed by immunoblotting with anti-myc antibodies (top). GST fusion proteins were visualized by Coomassie blue staining (bottom).
To confirm an interaction between Ftr1p and Grd19p in vivo, we attempted to coimmunoprecipitate endogenous epitope-tagged Ftr1p and Grd19p from cell extract; however, we were unable to detect a binding interaction using this approach, almost certainly because of the observation that, at steady-state, only a very small proportion of Ftr1p-GFP resides within endosomal compartments (Fig. 1 A). To circumvent this issue, we overexpressed the indicated GST-Ftr1p C-tail fusion proteins or GST alone in a yeast strain expressing an endogenous epitope-tagged mutant form of Grd19p, Grd19p-R81A-myc (Fig. 5 B). Arginine81 lies within the highly conserved PtdIns(3)P-binding pocket in the PX domain of Grd19p, and mutation of this residue to alanine results in mislocalization of the protein to the cytosol (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200609161/DC1). Cells grown in synthetic media were converted to spheroplasts, the bifunctional membrane-permeable chemical cross-linker dithiobis succinimidyl proprionate (DSP) was added before lysis, and the GST fusion proteins were captured on glutathione–Sepharose beads. The purified material was then probed with antibodies to Grd19p-R81A-myc. The results demonstrate that Grd19p-R81A was only coprecipitated from cells expressing the GST-Ftr1p C-tail (315–404) fusion protein, but not from cells expressing the GST-Ftr1p C-tail (329–404) construct, which lacks the putative endocytic recycling signal. These results indicate that Grd19p can bind the Ftr1p C-tail in cells and further confirms that this interaction requires aa 319–328 of Ftr1p.
Retromer and Grd19p colocalize on tubular endosomes
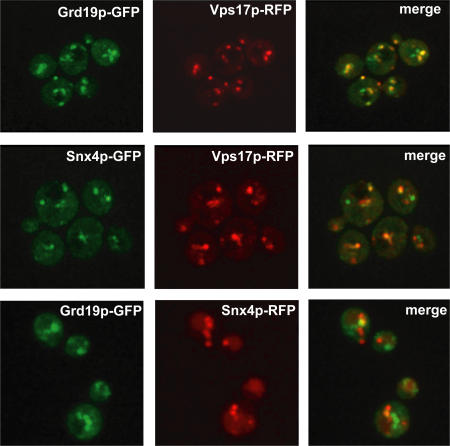
The most straightforward model to explain the dual requirement for Grd19p and retromer in Fet3p–Ftr1p sorting is that they function together at a common sorting step, and this predicts that some portion of Grd19p and retromer colocalize on endosomes. To test this, we constructed strains that expressed functional, C-terminally tagged Grd19p-GFP and Vps17p-RFP, and then imaged the entire volume of these cells by spinning disc confocal microscopy. Three-dimensional reconstructions and two-dimensional maximum projections were generated from the image z stacks (Fig. 6 and Video 1). The majority of labeled organelles contained both Grd19p-GFP and Vps17p-RFP, although some organelles were labeled by only one of the tagged proteins. The three-dimensional representations allowed us to characterize the morphology of the endosomes labeled by Grd19p and Vps17p, many of which had a distinct tubular shape. Retromer proteins have been localized to the tubular domains of early sorting endosomes in cultured mammalian cells (Zhong et al., 2002; Arighi et al., 2004; Carlton et al., 2004; Seaman, 2004; J.G. Carlton et al., 2005), and this is thought to reflect their roles in a geometric sorting mechanism that segregates integral membrane proteins from lumenal content (Maxfield and McGraw, 2004). Furthermore, human Snx3 is also enriched on the tubular domains of endosomes (Xu et al., 2001). Our results clearly indicate that a substantial proportion of Grd19p and the retromer subunit Vps17p colocalize on tubular endosomes, suggesting that they may function together, although the exclusive presence of each protein on some endosomes indicates that they function autonomously as well.
Figure 6.
Retromer and Grd19p partially colocalize on tubular endosomal membranes. Live cells coexpressing endogenous C-terminally tagged Grd19p-GFP and Vps17p-RFP, Snx4p-GFP and Vps17p-RFP, and Grd19p-GFP and Snx4p-RFP were imaged by spinning disc confocal microscopy. Images acquired using the individual red and green channels were then merged to determine extent of colocalization. Shown are two-dimensional maximum projections of the entire cell volumes.
We also tested the idea that retromer might be required for association of Grd19p with endosomal membranes, as many phosphoinositide-binding modules require other interactions for stable association with intracellular membranes (Lemmon, 2003). Grd19p consists of a PtdIns(3)P-binding PX domain with a short N-terminal extension (Zhou et al., 2003). PtdIns(3)P-binding is critical for endosomal targeting and function, as the R81A point mutant resulted in missorting of Ftr1p-GFP to the vacuole (Fig. S2 B). We examined localization of Grd19p-GFP in _vps29_Δ cells and found that it still localized to endosomes, although the compartments decorated by Grd19p appeared smaller and more numerous (Fig. S2 C). We conclude that retromer does not influence recruitment of Grd19p to endosomes from the cytosol, but it may influence the cellular distribution of Grd19p-positive endosomes, and these results further indicate that Grd19p and retromer are targeted to endosomes independently.
In the course of these experiments, we also determined the relative localization of Vps17p to another sorting nexin, Snx4p, which functions in a retromer-independent early endosome-to-Golgi pathway (Hettema et al., 2003) and is not required for Fet3p–Ftr1p recycling. Although yeast retromer is considered to function solely in late endosome-to-Golgi trafficking pathways, a substantial degree of colocalization between Snx4p-GFP and Vps17p-RFP was observed (Fig. 6). These observations suggest that the localization of retromer in the yeast endocytic system is more widespread than currently appreciated. In addition, we visualized fluorescently labeled Grd19p and Snx4p within the same cell (Fig. 6). Grd19p-GFP– and Snx4p-RFP-labeled organelles exhibited little to no colocalization, indicating that Grd19p and Snx4p are localized almost exclusively to distinct subsets of endosomes. This is consistent with published studies reporting that these two sorting nexins function in retrieval pathways from different populations of endosomes (Hettema et al., 2003).
Co-purification of retromer and Grd19p from cell extracts
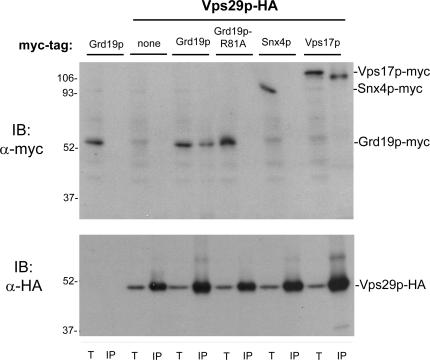
Based on the results so far, we hypothesized that Grd19p and retromer may physically cooperate, with Grd19p linking cargo recognition to retromer-dependent export from endosomes. To test this, we purified epitope-tagged Vps29p retromer subunit from a cell lysate and determined if Grd19p copurified with it. Strains were constructed that expressed 3xHA epitope-tagged Vps29p and Grd19p tagged with a 13xMyc epitope, and as a control, Vps29p-3xHA and another retromer subunit, Vps17p, tagged with a 13xMyc epitope. Cells were converted to spheroplasts and lysed, and Vps29p was immunopurified under native conditions, and the precipitates were probed with antibodies to Grd19p-myc. Under these conditions, no Grd19p-myc copurified with Vps29p-HA, but Vps17p-myc did (unpublished data). However, when DSP was added to intact spheroplasts before lysing the cells, both Grd19p and Vps17p copurified with Vps29p (Fig. 7). As a control for this experiment, we used a strain that simultaneously expressed Vps29p-HA and Snx4p-myc and found that Snx4p did not copurify with retromer (Fig. 7), despite the fact that substantial amounts of Snx4p and retromer colocalized on endosomal membranes. Furthermore, cross-linking depended on colocalization of Grd19p and retromer on endosomes as the Grd19p-R81A mutant, which is localized to the cytosol (Fig. S2 A), did not cross-link to retromer (Fig. 7). These results indicate that Grd19p and retromer interact in vivo on PtdIns(3)P-containing endosomes.
Figure 7.
Grd19p copurifies with retromer after chemical cross-linking. Strains expressing the indicated epitope-tagged proteins were grown in synthetic medium and converted to spheroplasts, and DSP was added to a final concentration of 5 mM. The cells were lysed, and Vps29-HA was captured on anti-HA beads. The cross-links were disrupted by reduction in SDS sample buffer, and the samples were probed by immunoblotting with anti-myc (top) or anti-HA antibodies (bottom). Note that the 9E10 (anti-myc) antibody detects a faint 55-kD background band that migrates just above Grd19p-myc, and that Vps17p-myc recovered by immunoprecipitation runs ∼10 kD below Vps17p-myc detected in whole-cell extract. T, 2% total (whole-cell extract); IP, immunoprecipitates.
Discussion
Our results suggest that the sorting nexin Grd19/Snx3p functions as a cargo-specific accessory component of the retromer complex, which is required for endocytic recycling of the Fet3p–Ftr1p iron transporter. In support of this model, we report that in null mutants of grd19 and each of the five retromer subunits, Fet3p–Ftr1p is missorted to the vacuole when cells are grown under conditions that favor recycling, that Grd19p directly binds a sequence in Ftr1p required for endocytic recycling, that Grd19p-GFP and Vps17p-RFP colocalize substantially on tubular endosomes, and that Grd19p can be chemically cross-linked to the retromer complex in vivo. Although the full range of functions and the specific mechanisms by which retromer operates in membrane trafficking have not been elucidated, it is clear that it is a general endosomal sorting factor required for the proper sorting and export of a diverse set of cargo molecules from endosomes. The work presented in this study is relevant for understanding how cargo is identified by retromer, and the results suggest that that the repertoire of retromer-dependent cargos is extended by its interaction with Grd19p. We speculate that Grd19p functions with retromer in a manner analogous to vesicle coat protein adapters that link cargo selection to coat protein recruitment.
Recycling of Fet3p–Ftr1p is likely to be initiated through recognition of Ftr1p by Grd19p because the PX domain of Grd19p binds PtdIns(3)P with the highest affinity of all the PX domains encoded in the yeast genome (Yu and Lemmon, 2001). In contrast, the PX domains of the retromer subunits Vps5p and Vps17p bind PtdIns(3)P with at least 100-fold lower affinity than Grd19p (Yu and Lemmon, 2001). Preliminary analysis of the binding interaction between Grd19p and the Ftr1p recycling signal by surface plasmon resonance indicates that the affinity is relatively weak (K d between 10 and 100 μM; unpublished data), which is consistent with PtdIns(3)P binding providing the driving force for recruitment of Grd19p to endosomes. Because the abundances of Grd19p and Vps17p appear to be similar (Fig. 7), Grd19p is expected to preferentially accumulate on early endosomes containing relatively low amounts of PtdIns(3)P. Retromer will subsequently load onto endosomes as they accrue higher levels of PtdIns(3)P during maturation, facilitated by interactions with other factors, such as Grd19p and cargo molecules. In this manner, Grd19p could serve as both a coincidence sensor that detects the presence of Fet3p–Ftr1p on PtdIns(3)P-containing endosomes and as an adaptor that recruits retromer to cargo to initiate export from the endosome. Grd19p and retromer appear to be sufficient for export from the endosome because we have not observed any recycling defects in other known cargo-sorting factors, including the clathrin adaptor AP-1 complex, other sorting nexins, or the recently identified GSE–EGO complex involved in endosome-to-plasma membrane sorting of the general amino acid permease Gap1p (Gao and Kaiser, 2006; Table S1). Once exported from the endosome, Fet3p–Ftr1p is probably delivered to the Golgi for resecretion because deletion of the Golgi Rab GTPase Ypt6p and its regulators also results in defective recycling, and because the Ftr1p C-terminal tail can direct Vps10p from the endosome back to the Golgi.
In yeast, five proteins have been identified that are sorted via the Grd19p–retromer pathway. These include native proteins that cycle between the Golgi and endosomes, Ste13p, Kex2p, and Pep12p (Nothwehr et al., 2000; Hettema et al., 2003), and Fet3p–Ftr1p, which uses the Grd19p–retromer pathway to be sorted back to the plasma membrane. The Golgi retrieval signals in Ste13p and Kex2p contain key aromatic residues, although their relevance to Grd19p-mediated sorting is not clear because Grd19p (supplied in a cell extract) still bound well to the cytoplasmic tail of Ste13p, even when the aromatic residue-based retrieval signal had been deleted (Voos and Stevens, 1998). Consistent with this, mutation of the single aromatic residue within the Ftr1p recycling signal (Phenylalanine323 to Alanine) did not affect recycling (unpublished data), so a more systematic analysis of this signal is required to identify its key features. It is also interesting that vacuolar targeting of the Ftr1p mutant lacking the Grd19p binding site (Ftr1pΔ319-328) was not so robust, perhaps implying that other sorting determinants are present within the C-terminal tail of Ftr1p. Inasmuch as the available data suggest that the sequences of the signals which confer Grd19p- and retromer-dependent trafficking are diverse, Grd19p probably recognizes structural features of cargo proteins rather than a strict linear amino acid sequence.
Importantly, our results showing that Grd19p directly recognizes Ftr1p and Ste13p and links them to retromer establish for the first time how a sorting nexin (other than Snx1) and retromer cooperate to recognize cargo. Although direct interactions between cargo and any subunit of retromer have not yet been confirmed using purified proteins, the current view posits that cargo is recognized by Vps35p, although other sorting nexins, including human Snx1, also have the capacity to bind cytoplasmic regions of some endocytic cargo proteins (Wang et al., 2002). Moreover, the recent discovery that Vps26 has an arrestin-like structure raises the possibility that Vps26p, like arrestins, may also interact with cargo (Shi et al., 2006). Another possible function of retromer is suggested by the crystal structure of Vps29, which has a protein fold resembling that of phosphoesterases (Collins et al., 2005; Wang et al., 2005), and recent studies have shown that Vps29p exhibits protein phosphatase activity (Damen et al., 2006). Regardless of the specific functions of the individual retromer subunits, it is clear that multiple cargo recognition mechanisms must contribute to the general function of retromer in endosomal sorting because Grd19p is not required for all retromer-dependent trafficking.
In mammalian cells, the early endosomal system is comprised of vacuolar domains connected to an extensive network of tubules, which are enriched in integral membrane cargo proteins that are subsequently sorted to a variety of different organelles (Bonifacino and Rojas, 2006). On the basis of the large surface area/volume ratio of tubes compared with spherical structures, it has been proposed that the packaging of integral membrane proteins into tubes is a highly efficient geometry-based mechanism for segregating membrane and lumenal components (Helenius et al., 1983; Marsh et al., 1986; Geuze et al., 1987). Human retromer appears to be involved in this sorting mechanism through its cargo-binding activities and the ability of the BAR domain–containing Snx1 subunit to sense regions of high membrane curvature (Carlton et al., 2004). The role of this geometric sorting mechanism in yeast is not known due, in large part, to the difficulty in visualizing cargo within domains of yeast endosomes by light microscopy. However, visualization of yeast endosomes by electron microscopy has provided clear evidence that a subset of them is indeed tubular in shape (Prescianotto-Baschong and Riezman, 1998; Quenneville et al., 2006). These results support the notion that retromer-mediated sorting in yeast and mammalian cells involves the same mechanisms, and they open the door to evaluating the roles of various retromer proteins and auxiliary factors, such as Grd19p in the biogenesis of these organelles. We expect that the interaction between Grd19p and retromer in yeast holds true for the human orthologues, as this interaction could explain the observation that overexpression of human Snx3 in cultured cells leads to a huge expansion of tubular early endosomal compartments through enhanced recruitment of the BAR domain–containing Snx1 component of retromer (Xu et al., 2001).
The results presented in this work demonstrate that recognition of recycling protein cargo by retromer can be initiated by a sorting nexin (Grd19p) that functions as an adaptor to link cargo to the cellular recycling machinery. With the identification of the endocytic recycling machinery and insight into how it mediates recycling of Fet3p–Ftr1p, the opportunity now exists to explore how, in response to changes in extracellular iron concentration, the iron transporter is channeled into either recycling or degradative pathways.
Materials and methods
Yeast strains, media, and growth conditions
Unless otherwise indicated, all yeast strains were constructed by integration using recombination of gene-targeted, PCR-generated DNAs using the method of Longtine et al. (1998) to ensure expression from the native loci. The strain background expressing the end4-1 allele is SEY6210 (_MAT_a ura3-52, his3-200, trp1-901, lys2-801, suc2-9, and leu2-3) and the strain background for yeast strains expressing Ftr1p-Vps10p chimeric proteins is BHY10 (SEY6210 _leu2-3_∷pBHY11 [CPY-Inv _LEU2_]). All other yeast strains were constructed in the BY4742 background (_MAT_α his3-1, leu2-0, met15-0, and ura3-0).
For the initial screen, a GFP tag was integrated in the FTR1 ORF immediately preceding the stop codon in yeast deletion strains from the EUROSCARF KAN MX deletion collection. To induce iron starvation, cells were grown overnight to OD600 ≈ 1.0 in synthetic media containing 50 μM of the iron chelator bathophenanthrolinedisulfonic acid. For all other experiments, cells were grown in either yeast extract/peptone/dextrose, or yeast nitrogen base supplemented with the appropriate nutrients as necessary.
Construction of strains coexpressing GFP- and RFP-tagged proteins was accomplished by generating PCR DNA to integrate a RFP tag at the VPS17 or SNX4 locus using pKT359 (provided by Kurt Thorn, University of California, San Francisco, San Francisco, CA) as a template (pFA6a-link-tdTomato-HIS3MX6; Sheff and Thorn, 2004) into strains already expressing integrated copies of Grd19p-GFP or Snx4p-GFP (integrated with the KANMX cassette using the method of Longtine et al. [1998]).
Strains expressing Vps10p-Ftr1p chimeric proteins were constructed using recombination of PCR-generated DNAs. All chimeric genes were expressed from the endogenous VPS10 promoter by integrating FTR1 sequences after codon 1,421 (immediately downstream of the membrane-spanning segment) of the VPS10 ORF. The FTR1 segments were chosen based on a published topology analysis (Severance et al., 2004). To construct the VPS10-FTR1(C-tail) fusion gene, the region encoding Ftr1p aa 315–404, followed by a 13xMyc epitope tag and the HIS3MX6 selectable marker, was amplified from genomic DNA of strain TSY30 (FTR1∷13XMyc∷HIS3MX6), and the PCR DNA was used to transform a wild-type strain (BHY10). The VPS10-FTR1 chimeras containing portions of FTR1 that encode internal loops were constructed by cotransformation with two PCR DNAs; one encoding the FTR1 sequence and a second containing a 13xMyc epitope tag followed by the HIS3MX6 selectable marker. The DNAs encoding the VPS10-FTR1 fusion site were amplified such that the FTR1 segment was flanked by 40 bases of the VPS10 locus, and on the other end it was flanked by 40 bases of the Myc epitope tag. The second DNA encoded a 13xMyc epitope tag, followed by the HIS3MX6 selectable marker, followed by 51 bases of sequence that matched immediately downstream of the VPS10 stop codon. Recombination between the two PCR DNAs within the Myc epitope tag sequences generated _FTR1_-13xMyc-His3MX6 DNAs flanked by VPS10 sequences, and recombination of these at the VPS10 locus generated the chimeric VPS10-FTR1 genes. The internal FTR1 portions encoded aa 30–48 (loop1) and 109–153 (loop 2). For all constructs, His+ transformants were screened by immunoblotting using the 9E10 antibody to identify those which expressed myc-tagged proteins of the correct size. PCR of genomic DNA from these strains using primers upstream of the VPS10 integration site and within the Myc epitope tag were then used to amplify the VPS10-FTR1 junctions and all of the FTR1 sequences. These PCR DNAs were then sequenced to confirm the constructs. Control strains to compare the functionality of the Vps10-Ftr1p fusion proteins included a Vps10p truncation mutant in which a 13xMyc-epitope tag was integrated after codon 1,421 of the VPS10 ORF, and full-length VPS10 tagged on its C terminus with the 13xMyc epitope. Both control strains were constructed by transformation of PCR-generated DNAs using pFA6a-13Myc-HIS3MX6 as the template (Longtine et al., 1998).
Strains expressing mutant and tagged integrated copies of Grd19p were constructed as follows. The GRD19 ORF including promoter and terminator regions was amplified by PCR as a BamHI–SalI fragment and cloned into vector pRS416. Site-directed mutagenesis (QuikChange Site-Directed Mutagenesis kit; Stratagene) was used to generate the R81A mutation in this plasmid, and this mutation was confirmed by sequencing. A GRD19_Δ_∷KANMX strain was then cotransformed with the BamHI–SalI fragment released from this plasmid along with GFP∷HIS3MX or 13Xmyc∷HIS3MX PCR DNA, with flanking sequences targeted to the GRD19 ORF (Longtine et al., 1998). Transformants were screened for growth on plates that lacked histidine, for G418 sensitivity, and for the presence of the GFP tag by microscopy or the 13Xmyc tag by immunoblotting using the 9E10 antibody.
Antibodies and reagents
Enzymes used in DNA manipulations were purchased from New England Biolabs or Promega. Standard molecular biological and microbiological techniques were used throughout. Primary mouse monoclonal antibodies used in these studies included: 9E10 anti-myc (1:10,000; University of Pennsylvania Cell Center), anti-HA (1:1,000; Covance), anti-PGK (1:10,000; Invitrogen), anti-T7 (1:10,000; Novagen), anti-CPY (1:1,000; Invitrogen), and anti-GFP (1:2,000; Covance). Secondary sheep anti–mouse HRP-conjugated antibodies (GE Healthcare) were used at 1:5,000.
CPY secretion assay
This assay is based on the method of Conibear and Stevens (2002), with the following exceptions. Filters were incubated with AP-conjugated goat anti–mouse secondary antibodies (Jackson ImmunoResearch Laboratories) for 2 h at room temperature before development by enhanced chemifluorescence (ECF) and analysis on a Molecular Dynamics Storm 860 PhosphorImager (GE Healthcare). The use of ECF instead of ECL was essential here for obtaining linear signals. The amount of secreted CPY was quantified using ImageQuant software version 5.2 (GE Healthcare) and plotted in graphical form using Excel (Microsoft).
Protein expression and purification
GST fusion proteins were constructed by amplifying the indicated sequences by PCR as BamHI–SalI fragments using wild-type genomic DNA as a template. The amplified products were cloned in-frame into the corresponding restriction sites of vector pGEX-KG-KAN (Novagen) for expression in bacteria or vector pEG(KG) (Mitchell et al., 1993) for expression in yeast. All constructs were sequenced to ensure that no mutations were present.
For bacterial expression, plasmids were transformed into E. coli BL21 (DE3; Novagen). Expression was induced by the addition of 1 mM IPTG at 37°C for 3 h. Cell pellets were resuspended in lysis buffer (PBS [1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4] + 1× Complete Mini protease inhibitor cocktail [Roche] + 1 mM AEBSF [Calbiochem]). Cell pellets were lysed either by sonication or by using a French Press pressure cell, followed by centrifugation (20,000 g for 20 min) to produce clarified lysates. For expression in yeast, plasmids were transformed into strain TSY103 (GRD19-R81A∷13XMyc∷HIS3MX6). Expression was induced by overnight growth in synthetic media lacking uracil and containing 4% galactose as the sole carbon source according to a published protocol (Mitchell et al., 1993).
For the production of His6-Grd19p, the GRD19 ORF was amplified by PCR as a BamHI–SalI fragment using wild-type genomic DNA as template. The amplified product was cloned in-frame into the corresponding sites of vector pET28a (Novagen), and the resulting plasmid was sequenced. The plasmid was transformed into E. coli BL21 (DE3; Novagen), and expression was induced by the addition of 1 mM IPTG at 37°C for 4 h. The cell pellet was resuspended in binding buffer ([0.5 M NaCl and 20 mM sodium phosphate, pH 7.4] + 1× Complete Mini protease inhibitor cocktail) and lysed using a French Press pressure cell. His6-Grd19p was then purified according to a published protocol (Setty et al., 2003).
In vitro binding assays
GST fusion proteins were affinity purified on glutathione–Sepharose 4B (GE Healthcare) by rocking for 1 h at room temperature in 0.5 ml PBS. Beads were washed three times with 1 ml PBS, and then resuspended in 300 μl of binding buffer (1X PBS, 1 mM DTT, 5 mM MgCl2, and 0.1% Triton X-100). 10 μg of purified His6-Grd19p or 250 μg (total protein) of yeast extract expressing Vps29p-myc was added to the beads. Binding reactions were incubated for 2 h at 4°C with rocking. Beads were then gently spun down and washed three times with 300 μl of binding buffer. Proteins bound to the beads were eluted by the addition of 2× sample buffer and were separated by 10% SDS-PAGE followed by Western blotting with anti-T7 or anti-myc (9E10) antibodies.
Microscopy
Fluorescence microscopy of Ftr1p-GFP and Fet3p-GFP constructs was done on live cells using a microscope (Eclipse E800; Nikon) fitted with a cooled, high-resolution charge-coupled device camera (model C4742-95; Hamamatsu). Images were acquired using Phase 3 Imaging software (Phase 3 Imaging Systems) and were subsequently manipulated using PhotoShop Elements version 2.0 (Adobe).
For colocalization studies and for imaging of Grd19p-GFP, microscopy was performed on live cells using a spinning disk confocal scanner (Perkin Elmer) combined with an inverted microscope (TE2000E; Nikon), equipped with 100×/1.45 NA Plan Apo objective (Nikon), electronically controlled filter wheels (Sutter Instruments), and a camera (ORCAII-ERG; Hamamatsu). In confocal mode, the 488- and 568-nm laser lines of an Argon/Krypton laser (Melles Griot) were used for sequential excitation of GFP and mRFP (1-ms exposures captured at optical sections of 0.3 μm) in combination with a triple bandpass dichroic mirror. Microscope control, image acquisition, and image analysis and manipulation were done using MetaMorph software version 7.0 (Universal Imaging). All imaging was done at room temperature (∼24°C).
Chemical cross-linking and immunoprecipitation
The in vivo cross-linking procedure is based on a published method (Hettema et al., 2003). In brief, 50 OD600 of the indicated strains were grown to log phase (OD600 ≈ 1.0) in synthetic media, harvested, washed once with water, and converted to spheroplasts. Spheroplasts were resuspended in 1 ml cross-linking buffer (25 mM potassium phosphate, 0.2 M sorbitol, and 1× EDTA-free protease inhibitor cocktail, pH 7.4), and 2–5 mM dithiobis succinimidyl proprionate (DSP; Pierce Chemical Co.) prepared freshly in DMSO was added. Cross-linking reactions were incubated for 30 min at 4°C. 25 mM Tris, pH 7.5, was added for 15 min to quench the reaction, and then 1% Triton X-100 was added to solubilize the material. Tubes were spun at 13,000 g for 10 min to remove insoluble debris, and the cross-linked cell lysate was incubated with 25 μl packed volume glutathione–Sepharose beads (GE Healthcare) or anti-HA affinity matrix (Roche) and allowed to rock overnight at 4°C to capture Vps29p-HA or for 2 h to capture the GST fusion proteins. Beads were pelleted and washed five times with 1 ml wash buffer (1× PBS and 1% Triton X-100). Proteins bound to the beads were eluted, cross-links were cleaved with 2× sample buffer containing 20 mM DTT, and eluted proteins were separated by 10% SDS-PAGE, followed by Western blotting with anti-HA and/or anti-myc (9E10) antibodies.
Online supplemental material
Table S1 displays the yeast deletion mutants screened for mislocalization of Ftr1p-GFP when cells were grown in iron-deficient medium. Fig. S1 shows the localization of Fet3p-GFP in retromer and _grd19_-null mutants, as well as the localization of Ftr1p-GFP and Fet3p-GFP in cells deleted for components of the Ypt6p GTPase module. Fig. S2 shows the cytosolic localization of Grd19p-R81A-GFP, the localization of Ftr1p-GFP in cells expressing wild-type Grd19p vs. Grd19p-R81A, and the localization of Grd19p-GFP in _vps29_Δ cells. The video displays a three-dimensional reconstruction of live cells coexpressing Grd19p-GFP and Vps17p-RFP. The online version of this article is available at http://www.jcb.org/cgi/content/full/jcb.200609161/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We thank Diane McVey Ward, and Andy Dancis in particular, for encouraging our work and for experimental advice. We also thank Phong Tran, Subba Rao Gangi Setty, Kate Ferguson, and Briana Schmiedekamp for assistance with experiments, and Mark Lemmon, Mickey Marks, Erfei Bi, Beverly Wendland, and Margaret Chou for stimulating discussions.
This work was supported by a grant from the National Institutes of Health (GM61221) to C.G. Burd and by a predoctoral fellowship from the American Heart Association (0615411U) to T.I. Strochlic.
Abbreviations used in this paper: BAR, Bin/Amphiphysin/Rvs; CPY, carboxypeptidase Y; DSP, dithiobis succinimidyl proprionate; ECF, enhanced chemifluorescence; PtdIns(3)P, phosphatidylinositol-3-phosphate; PX, phox homology.
References
- Arighi, C.N., L.M. Hartnell, R.C. Aguilar, C.R. Haft, and J.S. Bonifacino. 2004. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith, C., D. Eide, A. Van Ho, P.S. Bernard, L. Li, S. Davis-Kaplan, D.M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 76:403–410. [DOI] [PubMed] [Google Scholar]
- Bensen, E.S., B.G. Yeung, and G.S. Payne. 2001. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell. 12:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, M.O., J. Morvan, S. Dupre, D. Urban-Grimal, R. Haguenauer-Tsapis, and C. Volland. 2004. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell. 15:883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and R. Rojas. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7:568–579. [DOI] [PubMed] [Google Scholar]
- Carlton, J., M. Bujny, B.J. Peter, V.M. Oorschot, A. Rutherford, H. Mellor, J. Klumperman, H.T. McMahon, and P.J. Cullen. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14:1791–1800. [DOI] [PubMed] [Google Scholar]
- Carlton, J., M. Bujny, A. Rutherford, and P. Cullen. 2005. Sorting nexins–unifying trends and new perspectives. Traffic. 6:75–82. [DOI] [PubMed] [Google Scholar]
- Carlton, J.G., and P.J. Cullen. 2005. Sorting nexins. Curr. Biol. 15:R819–R820. [DOI] [PubMed] [Google Scholar]
- Carlton, J.G., M.V. Bujny, B.J. Peter, V.M. Oorschot, A. Rutherford, R.S. Arkell, J. Klumperman, H.T. McMahon, and P.J. Cullen. 2005. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J. Cell Sci. 118:4527–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino, J.L., E.G. Marcusson, and S.D. Emr. 1995. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell. 6:1089–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.J., and C.A. Kaiser. 2002. Amino acids regulate the intracellular trafficking of the general amino acid permease of _Saccharomyces cerevisiae._Proc. Natl. Acad. Sci. USA. 99:14837–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B.M., C.F. Skinner, P.J. Watson, M.N. Seaman, and D.J. Owen. 2005. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol. 12:594–602. [DOI] [PubMed] [Google Scholar]
- Conibear, E., and T.H. Stevens. 2002. Studying yeast vacuoles. Methods Enzymol. 351:408–432. [DOI] [PubMed] [Google Scholar]
- Cooper, A.A., and T.H. Stevens. 1996. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 133:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse, D.Y., G. Roel, M.C. Betist, O. Destree, and H.C. Korswagen. 2006. Wnt gradient formation requires retromer function in Wnt-producing cells. Science. 312:921–924. [DOI] [PubMed] [Google Scholar]
- Damen, E., E. Krieger, J.E. Nielsen, J. Eygensteyn, and J.E. van Leeuwen. 2006. The human Vps29 retromer component is a metallo-phosphoesterase for a cation-independent mannose 6-phosphate receptor substrate peptide. Biochem. J. 398:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena, K., and T.H. Stevens. 1995. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol. Cell. Biol. 15:1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice, M.R., I. De Domenico, L. Li, D.M. Ward, B. Bartok, G. Musci, and J. Kaplan. 2005. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 280:22181–22190. [DOI] [PubMed] [Google Scholar]
- Galan, J.M., A. Wiederkehr, J.H. Seol, R. Haguenauer-Tsapis, R.J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., and C.A. Kaiser. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8:657–667. [DOI] [PubMed] [Google Scholar]
- Geuze, H.J., J.W. Slot, and A.L. Schwartz. 1987. Membranes of sorting organelles display lateral heterogeneity in receptor distribution. J. Cell Biol. 104:1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft, C.R., M. de la Luz Sierra, R. Bafford, M.A. Lesniak, V.A. Barr, and S.I. Taylor. 2000. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol. Biol. Cell. 11:4105–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A., I. Mellman, D. Wall, and A. Hubbard. 1983. Endosomes. Trends Biochem. Sci. 78:245–250. [Google Scholar]
- Hettema, E.H., M.J. Lewis, M.W. Black, and H.R. Pelham. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky, B.F., B.A. Davies, M.N. Seaman, S.A. McLaughlin, S. Yoon, and S.D. Emr. 1997. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell. 8:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M.A. 2003. Phosphoinositide recognition domains. Traffic. 4:201–213. [DOI] [PubMed] [Google Scholar]
- Lemmon, S.K., and L.M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457–466. [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., B.J. Nichols, C. Prescianotto-Baschong, H. Riezman, and H.R. Pelham. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 11:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in _Saccharomyces cerevisiae._Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Marcusson, E.G., B.F. Horazdovsky, J.L. Cereghino, E. Gharakhanian, and S.D. Emr. 1994. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 77:579–586. [DOI] [PubMed] [Google Scholar]
- Marsh, M., G. Griffiths, G.E. Dean, I. Mellman, and A. Helenius. 1986. Three-dimensional structure of endosomes in BHK-21 cells. Proc. Natl. Acad. Sci. USA. 83:2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield, F.R., and T.E. McGraw. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.A., T.K. Marshall, and R.J. Deschenes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 9:715–722. [DOI] [PubMed] [Google Scholar]
- Nothwehr, S.F., and A.E. Hindes. 1997. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J. Cell Sci. 110:1063–1072. [DOI] [PubMed] [Google Scholar]
- Nothwehr, S.F., P. Bruinsma, and L.A. Strawn. 1999. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol. Biol. Cell. 10:875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr, S.F., S.A. Ha, and P. Bruinsma. 2000. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 151:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, B.C., and S.G. Clark. 2006. Wnt signaling establishes anteroposterior neuronal polarity and requires retromer in C. elegans. Development. 133:1757–1766. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong, C., and H. Riezman. 1998. Morphology of the yeast endocytic pathway. Mol. Biol. Cell. 9:173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville, N.R., T.Y. Chao, J.M. McCaffery, and E. Conibear. 2006. Domains within the GARP subunit Vps54 confer separate functions in complex assembly and early endosome recognition. Mol. Biol. Cell. 17:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Texeira, M., and C.A. Kaiser. 2006. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol. Biol. Cell. 17:3031–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., K. Sato, and A. Nakano. 2004. Endoplasmic reticulum quality control of unassembled iron transporter depends on Rer1p-mediated retrieval from the Golgi. Mol. Biol. Cell. 15:1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N. 2005. Recycle your receptors with retromer. Trends Cell Biol. 15:68–75. [DOI] [PubMed] [Google Scholar]
- Seaman, M.N., and H.P. Williams. 2002. Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell. 13:2826–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., E.G. Marcusson, J.L. Cereghino, and S.D. Emr. 1997. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., J.M. McCaffery, and S.D. Emr. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet, L.F., and W. Hong. 2006. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta. 1761:878–896. [DOI] [PubMed] [Google Scholar]
- Setty, S.R., M.E. Shin, A. Yoshino, M.S. Marks, and C.G. Burd. 2003. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 13:401–404. [DOI] [PubMed] [Google Scholar]
- Severance, S., S. Chakraborty, and D.J. Kosman. 2004. The Ftr1p iron permease in the yeast plasma membrane: orientation, topology and structure-function relationships. Biochem. J. 380:487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, M.A., and K.S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. [DOI] [PubMed] [Google Scholar]
- Shi, H., R. Rojas, J.S. Bonifacino, and J.H. Hurley. 2006. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat. Struct. Mol. Biol. 13:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A., S. Severance, N. Kaur, W. Wiltsie, and D.J. Kosman. 2006. Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in _Saccharomyces cerevisiae._J. Biol. Chem. 281:13355–13364. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., S.Y. Peak-Chew, and H.R. Pelham. 2000. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19:4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearman, R., D.S. Yuan, Y. Yamaguchi-Iwai, R.D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 271:1552–1557. [DOI] [PubMed] [Google Scholar]
- Teasdale, R.D., D. Loci, F. Houghton, L. Karlsson, and P.A. Gleeson. 2001. A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 358:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges, M., F. Luton, C. Gruber, F. Tiemann, L.G. Reinders, L. Huang, A.L. Burlingame, C.R. Haft, and K.E. Mostov. 2004. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nat. Cell Biol. 6:763–769. [DOI] [PubMed] [Google Scholar]
- Voos, W., and T.H. Stevens. 1998. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J. Cell Biol. 140:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., M. Guo, Z. Liang, J. Fan, Z. Zhu, J. Zang, Z. Zhu, X. Li, M. Teng, L. Niu, et al. 2005. Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. J. Biol. Chem. 280:22962–22967. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Y. Zhou, K. Szabo, C.R. Haft, and J. Trejo. 2002. Down-regulation of protease-activated receptor-1 is regulated by sorting nexin 1. Mol. Biol. Cell. 13:1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby, C.A., and J.E. Dixon. 2002. Sorting out the cellular functions of sorting nexins. Nat. Rev. Mol. Cell Biol. 3:919–931. [DOI] [PubMed] [Google Scholar]
- Xu, Y., H. Hortsman, L. Seet, S.H. Wong, and W. Hong. 2001. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3:658–666. [DOI] [PubMed] [Google Scholar]
- Yu, J.W., and M.A. Lemmon. 2001. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276:44179–44184. [DOI] [PubMed] [Google Scholar]
- Zhong, Q., C.S. Lazar, H. Tronchere, T. Sato, T. Meerloo, M. Yeo, Z. Songyang, S.D. Emr, and G.N. Gill. 2002. Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. USA. 99:6767–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C.Z., I.L. de La Sierra-Gallay, S. Quevillon-Cheruel, B. Collinet, P. Minard, K. Blondeau, G. Henckes, R. Aufrere, N. Leulliot, M. Graille, et al. 2003. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278:50371–50376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]