The depletion attraction: an underappreciated force driving cellular organization (original) (raw)
Abstract
Cellular structures are shaped by hydrogen and ionic bonds, plus van der Waals and hydrophobic forces. In cells crowded with macromolecules, a little-known and distinct force—the “depletion attraction”—also acts. We review evidence that this force assists in the assembly of a wide range of cellular structures, ranging from the cytoskeleton to chromatin loops and whole chromosomes.
The depletion attraction
As biologists, we are all aware that ionic and hydrogen bonds, plus van der Waals and hydrophobic forces, act within and between macromolecules to shape the final structure. However, a distinct interaction, known as the “depletion attraction,” may also play a substantial role (Asakura and Oosawa, 1958; Yodh et al., 2001). This force is only seen in crowded environments like those found in cells, where 20–30% of the volume is occupied by soluble proteins and other macromolecules (Ellis, 2001; Minton, 2001, 2006). Crowding increases effective concentrations, which has important consequences (Box 1), but it also creates a force apparently out of nothing. We argue that this force drives the assembly of many large structures in cells.
Box 1. AO and related theories
The physics of an aqueous solution crowded with ions and macromolecules of different sizes is complicated, and various theories provide different perspectives on the underlying problems (Lebowitz et al., 1965; Ogston, 1970; Cotter, 1974; Mao et al., 1995; Minton, 1998; Parsegian et al., 2000; Kinjo and Takada, 2002; Spitzer and Poolman, 2005). The AO theory (Asakura and Oosawa, 1958) is one approximation, that shows that
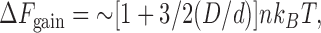
where Δ_F_ gain is the free energy gained when the two large spheres in Fig. 1 come into contact, D and d are the diameters of the large and small spheres, n the volume occupied by the small spheres, k B the Boltzmann constant, and T is the absolute temperature. This equation applies generally because particles of all sizes possess a hard core; it also applies to values of n up to ∼0.3, after which it becomes less accurate (Gotzelmann et al., 1998). In cells, n can be determined in various ways (i.e., by cell fractionation, electron microscopy, or gel filtration), and is (luckily) between 0.2–0.3 (Busch and Daskal, 1977; Zimmerman and Trach, 1991; Bohrmann et al., 1993). D thus determines the scale of the attraction (as d, n, and T are usually constant). Results obtained using “molecular tweezers” show the equation to be so accurate that it is being used to position particles within manmade nanostructures (Yodh et al., 2001).
We now consider how AO theory differs from two related theories. First, both the depletion attraction and hydrophobic effect (Chandler, 2002) tend to minimize the surface exposed to the macromolecular solute or water. They are also superficially similar in that one is purely, and the other mainly, driven by entropic effects. However, an increase in volume available to a macromolecular solute drives the depletion attraction, whereas an increase in hydrogen-bonding states available to water underlies the hydrophobic effect (Chandler, 2002). The second theory is known as “macromolecular crowding” in the biological literature. “Crowding” increases thermodynamic activities, and has been successfully used to compute effects on chemical reactions and equilibria (Ellis, 2001; Minton, 2001, 2006). Macromolecular crowding describes the same phenomenon as AO theory, but is based on scaled particle theory and so cannot be applied to the (concave) structures we consider (i.e., two touching large spheres; Minton, 1998). But if the large spheres are allowed to fuse to give one larger (convex) sphere, it then gives roughly equivalent results (unpublished data). Therefore, the hydrophobic effect differs in mechanism, and macromolecular crowding differs in technical treatment.
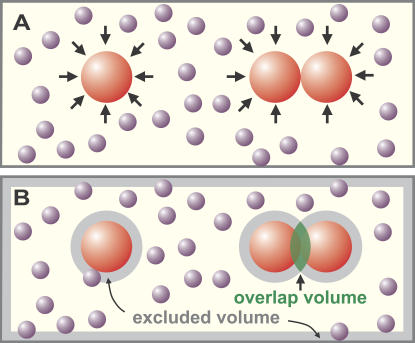
Consider Fig. 1 A, where many small and a few large spheres are contained in a box, representing the many small, crowding macromolecules and the fewer, larger complexes in a cell. In physicists' terminology, both types of sphere are “hard” and “noninteracting,” so that none of the forces familiar to biologists act between them. The small spheres bombard the large ones from all sides (arrows). When two large spheres approach one another, the small ones are excluded from the volume between the two. Therefore, the small ones exert an unopposed force equivalent to their osmotic pressure on opposite sides of the two large ones to keep them together. This osmotic effect depends on the volume that is inaccessible to the small spheres; if the small spheres could gain access to this (depleted) volume, they would force the two large ones apart. Fig. 1 B gives an alternative view. The centers of mass of the small spheres can access the yellow volume, but not the gray volumes, around each large sphere or abutting the wall. When one large sphere approaches another, these excluded volumes overlap; as a result, the small spheres can now access a greater volume. The resulting increase in entropy of the many small spheres generates a depletion attraction between the large spheres. At first glance, this seems like an oxymoron; entropy usually destroys the order that an attraction creates. But if we consider the whole system (not just the large spheres), the excluded volume is minimized and thus entropy is maximized (because there are so many small spheres).
Figure 1.
The depletion attraction and its role in cellular organization. (A) Many small spheres (purple) representing soluble macromolecules bombard three large spheres (red), representing cellular complexes, from all sides (arrows). When two large spheres come into contact (right), the small ones exert a force equivalent to their osmotic pressure on opposite sides of the two large ones to keep them together. (B) The shaded regions in this alternative view show regions inaccessible to the centers of mass of the small spheres. When one large sphere contacts another, their excluded volumes overlap to increase the volume available to the small spheres (increasing their entropy); then aggregation of the large spheres paradoxically increases the entropy of the system. An analogous effect is found when a large sphere contacts the wall.
The Asakura–Oosawa theory (“AO theory”; Asakura and Oosawa, 1958), allows us to estimate the scale of this depletion attraction (Box 1). In cells, the diameters of the large spheres are the major determinants, as the other variables in the equation in Box 1 are constant; larger spheres tend to cluster more than smaller ones (Fig. 2 A, compare i with ii). The attraction can easily be recognized in vitro; adding an inert crowding agent like a dextran or polyethylene glycol (PEG) promotes aggregation (by increasing the volume fraction, n, of the small spheres). However, the force has a maximum range of only ∼5 nm, which is the diameter of a typical crowding protein; it will be larger if the two large objects fit snugly together (or are “soft” enough to fuse into one with conservation of volume) and smaller if surface irregularities limit close contact (Marenduzzo et al., 2006).
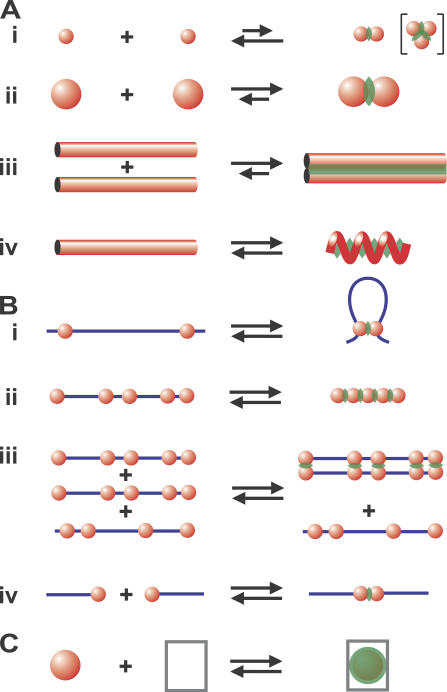
Figure 2.
Examples of AO theory. Overlap volumes are green; small spheres not depicted. (A) Interactions within and between proteins. (i and ii) The attraction increases as the overlap volume increases; larger spheres generate larger overlap volumes and so are more likely to aggregate. (brackets) Adding one large sphere to two large spheres cooperatively generates two (not one) extra overlap volumes. (iii) Aligning two rods (in the same or different proteins) generates a large overlap volume (and thus attraction). (iv) Folding a tube into a helix generates an overlap volume that stabilizes the helix. (B) Interactions involving chromatin. (i) When large spheres (polymerizing complexes and clusters of bound transcription factors) are threaded on a string (DNA or chromatin fiber) the attraction is countered by the entropic cost of looping. (ii) Beads (nucleosomes and heterochromatic clumps) on one string can collapse onto each other (to pack a chromatin fiber or mitotic chromosome). (iii). Similar strings of beads (factories and heterochromatic clumps) can align perfectly, whereas dissimilar ones cannot. (iv) Large beads (NORs and centromeric heterochromatin) on different strings can aggregate (into nucleoli, chromocenters). (C) Confined spaces. Enclosing a sphere in a confined space (a pore or a proteasome) generates a large overlap volume (and thus attraction).
In what follows, free energy is expressed in k B T units; 1 k B T is ∼0.7 kcal/mol, which is roughly comparable to the energy associated with one hydrogen bond in a protein (Pace et al., 1996). Therefore, attractions of only a few k B T are within the range that biologists know can stabilize a structure.
A simple case: actin dimerization and bundling
It is widely believed that ATP hydrolysis provides most of the energy that drives actin dimerization. However, calculation shows the depletion attraction makes some contribution, ∼0.5 k B T (Fig. 2 A, i; Marenduzzo et al., 2006) compared with the experimentally determined free energy change of 1–2 k B T (Sept and McCammon, 2001; Dickinson et al., 2004). The attraction is nonspecific in the sense that it can bring two large spheres together, but it cannot orient them. Therefore, the addition of a third sphere would create the structure shown in Fig. 2 A (i, inset), and not a linear fiber. Long (F-actin) fibers will only form if specific forces augment the nonspecific attraction to orient monomers appropriately; then the overlap volume between two fibers (Fig. 2 A, iii) becomes large enough (i.e., many tens of k B T per micrometer) that adding a crowding agent causes fiber “bundling” (Hosek and Tang, 2004). Similar aggregation is seen with other spheres (e.g., bovine pancreatic trypsin inhibitor; Snoussi and Halle, 2005) and rods (e.g., tobacco mosaic virus; Adams and Fraden, 1998; Adams et al., 1998).
Secondary structures, tertiary structures, and helices
Within a protein, the scale of the attraction is small relative to hydrogen bonding. For example, forming a linear tube into a helix generates an overlap volume (Fig. 2 C, iv) so the attraction can stabilize a helix (Maritan et al., 2000; Snir and Kamien, 2005). But in the case of an α helix (with four hydrogen bonds per helical turn), it contributes only ∼0.07 k B T per turn (calculated using a helix with a 0.25-nm radius and 0.54-nm pitch, and assuming d = 5 nm and n = 0.2; unpublished data). The attraction created by folding a tube into a β-sheet (to produce two cylinders lying side-by-side, as in Fig. 2 A, iii), where each amino acid makes two hydrogen bonds and strands are 0.35 nm apart, is similarly small (i.e., <0.02 k B T per amino acid; not depicted). This is consistent with experimental observations and calculations showing that crowding agents increase the rates of refolding of lysozyme and the β-sheet WW domain by two- to fivefold (van den Berg et al., 2000; Cheung et al., 2005). The attraction also contributes ∼0.8 k B T per 14-nm turn in a coiled coil (calculated using two 0.5-nm cylinders; unpublished data), and <1 k B T per 10 bp of DNA (not depicted). Again, this is consistent with crowding agents slightly increasing the melting temperature of DNA (Woolley and Wills, 1985; Goobes et al., 2003).
Abnormal interactions: sickle cell hemoglobin and amyloid fibrils
In larger structures, the attraction becomes more prominant. For example, sickle cell hemoglobin results from the substitution of valine for glutamic acid at the β6 site of hemoglobin; this drives end-to-end polymerization of deoxygenated hemoglobin into fibers, followed by side-by-side “zippering” into bundles. As a result, red blood cells become more rigid and so pass less rapidly through capillaries, reducing oxygen exchange and causing sickle cell anemia. As with actin, the attraction contributes slightly to dimerization (Fig. 1 C, i), but contributes many tens of k B T per micrometer of fiber length to bundling (Fig. 1 C, iii; Jones et al., 2003). It may similarly drive aggregation in many other pathologies (e.g., into amyloid fibrils in Alzheimer's, type 2 diabetes, and the transmissible spongiform encephalopathies; Hatters et al., 2002; Ellis and Minton, 2006). As tissue hydration falls slightly on ageing (Barber et al., 1995), this may increase the volume fraction, n, and promote aggregation, which is consistent with the increased incidence seen with age.
Large nuclear bodies and membrane-bound structures
Nucleoli and promyelocytic leukemia bodies disassemble when nuclei from human hematopoietic cells are immersed in a low concentration of monovalent cations; both reassemble (and nucleolar transcription recovers) when a crowding agent like PEG is added (Rosania and Swanson, 1995; Hancock, 2004). This points to a role for crowding, perhaps acting through cooperative effects and the depletion attraction (Fig. 1 C, i). If so, the attraction could also shape other large nuclear structures, such as splicing speckles and Cajal bodies (Spector, 2003). PEG is also used routinely to induce cell fusion during hybridoma production, and the attraction drives the first step, which is cell aggregation (Kuhl et al., 1996; Chu et al., 2005); it also induces thylakoid membranes to stack (Kim et al., 2005). Thus, thermodynamics could give direction to vesicular traffic—toward clustering (through the attraction) and membrane fusion (by minimizing surface curvature).
Genome looping
There are entropic costs associated with forming DNA or chromatin into a loop, but these can be overcome if large enough complexes are bound to the template (Fig. 2 B, i; Marenduzzo et al., 2006). Consider two transcription complexes; each might contain a multisubunit polymerase, the transcript and its neutralizing proteins, plus associated ribosomes (in bacteria) or spliceosome (in eukaryotes). When they come into contact, the resulting attraction will keep them together, thus looping the intervening DNA. A cost/benefit analysis of the energies involved enabled correct prediction of various types of organization. First, looping should depend on ongoing transcription (as only then is the complex associated with the template); it does. For example, loops are present in all transcriptionally active cells examined (from bacteria to man), but not in inactive ones like chicken erythrocytes and human sperm (Jackson et al., 1984; Cook, 2002). And as chicken erythroblasts mature into erythrocytes, transcription falls progressively as loops are lost, until no activity or loops remain (Cook and Brazell, 1976). Recent evidence also shows that loops detected using “chromosome conformation capture” are tied through active polymerizing complexes (Cook, 2003). Thus, the Hbb-b1 (β-globin) gene lies tens of kilobase pairs away, on chromosome 7, from its locus control region, and ∼25 Mbp away from a gene (Eraf) encoding the α-globin–stabilizing protein; it contacts the locus control region and Eraf in erythroid nuclei (where all three are transcribed), but not in brain nuclei (where all are inactive; Osborne et al., 2004). Second, active polymerases cluster, as predicted. Thus, in higher eukaryotes, ∼8 active polymerase II units cluster into nucleoplasmic “factories” (Cook, 1999; Faro-Trindade and Cook, 2006), and bacterial ribosomal DNA operons aggregate similarly (Cabrera and Jin, 2003). Active DNA-polymerizing complexes in both pro- and eukaryotes also cluster into analogous replication factories (Cook, 1999), and the bacterial ones separate (Bates and Kleckner, 2005) just when the looping cost exceeds the attraction. In all cases, the scale of the attraction relative to the looping cost correlates with the clustering seen.
Conclusions
We have argued that an osmotic depletion attraction drives the organization of many cellular structures. Unlike other noncovalent interactions (i.e., ionic and hydrogen bonds, van der Waals and hydrophobic forces), this one only becomes significant in crowded environments like those in cells. It is nonspecific in the sense that it can bring spheres together without orienting them. It also depends on size and shape; the larger the overlap volume, the larger the attraction. Just as the entropy of the solvent (water) mainly underlies the hydrophobic effect, that of the solute (the crowding macromolecules) creates the attraction. These generalizations come with caveats because the underlying physics is complicated, and AO theory involves several simplifications (e.g., it becomes less accurate when n is >0.3, and it takes no account of kinetics). Nevertheless, the concept of a hydrophobic force is useful to biologists despite the underlying complexity, and we believe the concept discussed in this work will be similarly useful, especially because its scale can be calculated so simply.
Many questions remain. On the theoretical side, what happens when n increases above 0.3, and the AO equation becomes less precise and the theory much more complicated (Gotzelmann et al., 1998)? What are the relative advantages and disadvantages of the different theories of crowded solutions (Box 1)? On the experimental side, what exactly is the volume fraction within a cell, and how closely can typical proteins approach each other? Could the attraction help nucleosomes strung along DNA pack into the chromatin fiber (Fig. 2 B, ii). Can clumps of heterochromatin be treated as spheres that are subject to the attraction? If so, the attraction could underpin the condensation of an (interphase) string of such clumps into the mitotic chromosome (Fig. 2 B, ii; Manders et al., 1999). Could it also underpin the pairing of chromosomes seen during meiosis and polytenization, where a string bearing a unique array of factories and heterochromatic clumps aligns in perfect register with a homologue, but not with others carrying different arrays (Fig. 2 B, iii; Cook, 1997)? Could it drive end-to-end pairing of chromosomes? For example, diploid human lymphocytes contain 10 chromosomes encoding nucleolar organizing regions (NORs), but only ∼6 NORs are transcribed, and only these aggregate to form nucleoli (Wachtler et al., 1986). Does the attraction act through the thousands of active polymerizing complexes associated with each active NOR to drive nucleolar assembly (Fig. 2 B, iv)? Could it similarly drive the clustering of heterochromatic centromeres into chromocenters (Fig. 2 B, iv)? We have also seen how the attraction contributes to protein folding, but what of the special case where a protein is so confined that the overlap volume resulting from contact with the surrounding wall becomes significant (Fig. 2 C)? Do pores, and the barrels formed by chaperonins, proteasomes, and exosomes (Lorentzen and Conti, 2006), all exploit the attraction to promote ingress of their target proteins (Martin, 2004; Cheung et al., 2005; see Ellis, 2006, for a review of how crowding affects protein folding in confined spaces)? Clearly, we need to extend the experimental studies on the simple model systems reviewed in this study to complex subcellular assemblies, much as Hancock (2004) describes.
As soon as cellular structures become larger than ∼75 nm, the overlap volume can generate an attraction of ∼5 k B T; this is probably sufficient to promote irreversible aggregation when cooperative effects are included (Fig. 2 A, i, inset; Marenduzzo et al., 2006). This begs the obvious question: why don't all large structures in the cell end up in one aggregate (just as overexpressed bacterial proteins form inclusion bodies)? We suggest they will tend to do so unless energy is spent to stop aggregation and/or inert mechanisms prevent it. For example, anchorage to a larger structure (e.g., the cytoskeleton), surface irregularities (Jones et al., 2003), or charge interactions could all prevent close contact, and thus reduce the attraction. All seem to operate; for example, >70% of proteins in Escherichia coli and Bacillus subtilis (and >90% of the most abundant ones) are anionic at cellular pH, and thus would be expected to repel each other (Eymann et al., 2004; Weiller et al., 2004). We also note that structures like the cytoskeleton and membrane-bound vesicles are not rigid and permanent; rather, they continually turn over, to reduce their effective size and ensure that a large structure does not persist long enough to aggregate (Misteli, 2001; Altan-Bonnet et al., 2004). Nature, although constrained by the second law of thermodynamics, finds ways around it.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, Cancer Research UK, the Medical Research Council, and the Wellcome Trust for support.
K. Finan is supported by the E.P. Abraham Trust, a Clarendon Fund award from the University of Oxford, and an Overseas Research Student award from the UK government.
D. Marenduzzo and K. Finan contributed equally to this paper.
Abbreviations used in this paper: AO, Asakura–Oosawa; NOR, nucleolar organizing region; PEG, polyethyleneglycol.
References
- Adams, M., and S. Fraden. 1998. Phase behavior of mixtures of rods (tobacco mosaic virus) and spheres (polyethylene oxide, bovine serum albumin). Biophys. J. 74:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M., Z. Dogic, S.L. Keller, and S. Fraden. 1998. Entropically driven microphase transitions in mixtures of colloidal rods and spheres. Nature. 393:349–352. [Google Scholar]
- Altan-Bonnet, N., R. Sougrat, and J. Lippincott-Schwartz. 2004. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16:364–372. [DOI] [PubMed] [Google Scholar]
- Asakura, S., and F. Oosawa. 1958. Interactions between particles suspended in solutions of macromolecules. J. Polym. Sci. [B]. 33:183–192. [Google Scholar]
- Barber, B.J., R.A. Babbitt, S. Parameswaran, and S. Dutta. 1995. Age- related changes in rat interstitial matrix hydration and serum proteins. J. Gerontol. A. Biol. Sci. Med. Sci. 50:B282–B287. [DOI] [PubMed] [Google Scholar]
- Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 121:899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann, B., M. Haider, and E. Kellenberger. 1993. Concentration evaluation of chromatin in unstained resin-embedded sections by means of low-dose ratio-contrast imaging in STEM. Ultramicroscopy. 49:235–251. [DOI] [PubMed] [Google Scholar]
- Busch, H., and Y. Daskal. 1977. Methods for isolation of nuclei and nucleoli. Methods Cell Biol. 16:1–43. [DOI] [PubMed] [Google Scholar]
- Cabrera, J.E., and D.J. Jin. 2003. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 50:1493–1505. [DOI] [PubMed] [Google Scholar]
- Chandler, D. 2002. Hydrophobicity: two faces of water. Nature. 417:491. [DOI] [PubMed] [Google Scholar]
- Cheung, M.S., D. Klimov, and D. Thirumalai. 2005. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. USA. 102:4753–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y.S., S. Dufour, J.P. Thiery, E. Perez, and F. Pincet. 2005. Johnson-Kendall-Roberts theory applied to living cells. Phys. Rev. Lett. 94:028102. [DOI] [PubMed] [Google Scholar]
- Cook, P.R. 1997. The transcriptional basis of chromosome pairing. J. Cell Sci. 110:1033–1040. [DOI] [PubMed] [Google Scholar]
- Cook, P.R. 1999. The organization of replication and transcription. Science. 284:1790–1795. [DOI] [PubMed] [Google Scholar]
- Cook, P.R. 2002. Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32:347–352. [DOI] [PubMed] [Google Scholar]
- Cook, P.R. 2003. Nongenic transcription, gene regulation and action at a distance. J. Cell Sci. 116:4483–4491. [DOI] [PubMed] [Google Scholar]
- Cook, P.R., and I.A. Brazell. 1976. Conformational constraints in nuclear DNA. J. Cell Sci. 22:287–302. [DOI] [PubMed] [Google Scholar]
- Cotter, M.A. 1974. Hard-rod fluid: scaled particle theory revisited. Phys. Rev. A. 10:625–636. [Google Scholar]
- Dickinson, R.B., L. Caro, and D.L. Purich. 2004. Force generation by cytoskeletal end-tracking proteins. Biophys. J. 87:2838–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R.J. 2001. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26:597–604. [DOI] [PubMed] [Google Scholar]
- Ellis, R.J. 2006. Protein folding: inside the cage. Nature. 442:360–362. [DOI] [PubMed] [Google Scholar]
- Ellis, R.J., and A.P. Minton. 2006. Protein aggregation in crowded environments. Biol. Chem. 387:485–497. [DOI] [PubMed] [Google Scholar]
- Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam le, K. Buttner, G. Buurman, C. Scharf, S. Venz, U. Volker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics. 4:2849–2876. [DOI] [PubMed] [Google Scholar]
- Faro-Trindade, I., and P.R. Cook. 2006. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value. Mol. Biol. Cell. 17:2910–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goobes, R., N. Kahana, O. Cohen, and A. Minsky. 2003. Metabolic buffering exerted by macromolecular crowding on DNA-DNA interactions: origin and physiological significance. Biochemistry. 42:2431–2440. [DOI] [PubMed] [Google Scholar]
- Gotzelmann, B., R. Evans, and S. Dietrich. 1998. Depletion forces in fluids. Phys. Rev. E. 57:6785–6800. [Google Scholar]
- Hancock, R. 2004. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 146:281–290. [DOI] [PubMed] [Google Scholar]
- Hatters, D.M., A.P. Minton, and G.J. Howlett. 2002. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C–II. J. Biol. Chem. 277:7824–7830. [DOI] [PubMed] [Google Scholar]
- Hosek, M., and J.X. Tang. 2004. Polymer-induced bundling of F actin and the depletion force. Phys. Rev. E. Nonlin. Soft Matter Phys. 69:051907. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., S.J. McCready, and P.R. Cook. 1984. Replication and transcription depend on attachment of DNA to the nuclear cage. J. Cell Sci. Suppl. 1:59–79. [DOI] [PubMed] [Google Scholar]
- Jones, C.W., J.C. Wang, F.A. Ferrone, R.W. Briehl, and M.S. Turner. 2003. Interactions between sickle hemoglobin fibers. Faraday Discuss. 123:221–236. [DOI] [PubMed] [Google Scholar]
- Kim, E.H., W.S. Chow, P. Horton, and J.M. Anderson. 2005. Entropy-assisted stacking of thylakoid membranes. Biochim. Biophys. Acta. 1708:187–195. [DOI] [PubMed] [Google Scholar]
- Kinjo, A.R., and S. Takada. 2002. Effects of macromolecular crowding on protein folding and aggregation studied by density functional theory: statics. Phys. Rev. E. Nonlin. Soft Matter Phys. 66:031911. [DOI] [PubMed] [Google Scholar]
- Kuhl, T., Y. Guo, J. Aldefer, A. Berman, D. Leckband, J. Israelachvili, and S. Hui. 1996. Direct measurement of PEG induced depletion attraction between bilayers. Langmuir. 12:3003–3014. [Google Scholar]
- Lebowitz, J.L., E. Helfand, and E. Praestgaard. 1965. Scaled particle theory of fluid mixtures. J. Chem. Phys. 42:774–779. [Google Scholar]
- Lorentzen, E., and E. Conti. 2006. The exosome and the proteasome: nano-compartments for degradation. Cell. 125:651–654. [DOI] [PubMed] [Google Scholar]
- Manders, E.M.M., H. Kimura, and P.R. Cook. 1999. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 144:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y., M.E. Cates, and N.H.W. Lekkerkerker. 1995. Depletion stabilization by semidilute rods. Phys. Rev. Lett. 75:4548–4551. [DOI] [PubMed] [Google Scholar]
- Marenduzzo, D., C. Micheletti, and P.R. Cook. 2006. Entropy-driven genome organization. Biophys. J. 90:3712–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritan, A., C. Micheletti, A. Trovato, and J.R. Banavar. 2000. Optimal shapes of compact strings. Nature. 406:287–290. [DOI] [PubMed] [Google Scholar]
- Martin, J. 2004. Chaperonin function – effects of crowding and confinement. J. Mol. Recognit. 17:465–472. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 1998. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 295:127–149. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 2001. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 256:10577–10580. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 2006. Macromolecular crowding. Curr. Biol. 16:R269–R271. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogston, A.G. 1970. On the interaction of solute molecules with porous networks. J. Phys. Chem. 74:668–669. [Google Scholar]
- Osborne, C.S., C. Chakalova, K.E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J.A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically co-localize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., B.A. Shirley, M. McNutt, and K. Gajiwala. 1996. Forces contributing to the conformational stability of proteins. FASEB J. 10:75–83. [DOI] [PubMed] [Google Scholar]
- Parsegian, V.A., R.P. Rand, and D.C. Rau. 2000. Osmotic stress, crowding, preferential hydration, and binding: a comparison of perspectives. Proc. Natl. Acad. Sci. USA. 97:3987–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosania, G.R., and J.A. Swanson. 1995. Effects of macromolecular crowding on nuclear size. Exp. Cell Res. 218:114–122. [DOI] [PubMed] [Google Scholar]
- Sept, D., and J.A. McCammon. 2001. Thermodynamics and kinetics of actin filament nucleation. Biophys. J. 81:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snir, Y., and R.D. Kamien. 2005. Entropically driven helix formation. Science. 307:1067. [DOI] [PubMed] [Google Scholar]
- Snoussi, K., and B. Halle. 2005. Protein self-association induced by macromolecular crowding: a quantitative analysis by magnetic relaxation dispersion. Biophys. J. 88:2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573–608. [DOI] [PubMed] [Google Scholar]
- Spitzer, J.J., and B. Poolman. 2005. Electrochemical structure of the crowded cytoplasm. Trends Biochem. Sci. 30:536–541. [DOI] [PubMed] [Google Scholar]
- van den Berg, B., R. Wain, C.M. Dobson, and R.J. Ellis. 2000. Macromolecular crowding perturbs protein refolding kinetics: implications for folding inside the cell. EMBO J. 19:3870–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler, F., A.H. Hopman, J. Wiegant, and H.G. Schwarzacher. 1986. On the position of nucleolus organizer regions (NORs) in interphase nuclei. Exp. Cell Res. 167:227–240. [DOI] [PubMed] [Google Scholar]
- Weiller, G.F., G. Caraux, and N. Sylvester. 2004. The modal distribution of protein isoelectric points reflects amino acid properties rather than sequence evolution. Proteomics. 4:943–949. [DOI] [PubMed] [Google Scholar]
- Woolley, P., and P.R. Wills. 1985. Excluded-volume effect of inert macromolecules on the melting of nucleic acids. Biophys. Chem. 22:89–94. [DOI] [PubMed] [Google Scholar]
- Yodh, A.G., K.H. Lin, J.C. Crocker, A.D. Dinsmore, R. Verma, and P.D. Kaplan. 2001. Entropically driven self-assembly and interaction in suspension. Philos. Trans. R. Soc. Lond. A. 359:921–937. [Google Scholar]
- Zimmerman, S.B., and S.O. Trach. 1991. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of _Escherichia coli._J. Mol. Biol. 222:599–620. [DOI] [PubMed] [Google Scholar]