Active genes dynamically colocalize to shared sites of ongoing transcription (original) (raw)
- Article
- Published: 07 September 2004
- Lyubomira Chakalova1,
- Karen E Brown2,
- David Carter1 nAff4,
- Alice Horton1,
- Emmanuel Debrand1,
- Beatriz Goyenechea1,
- Jennifer A Mitchell1,
- Susana Lopes3,4 nAff5,
- Wolf Reik3 &
- …
- Peter Fraser1
Nature Genetics volume 36, pages 1065–1071 (2004)Cite this article
- 10k Accesses
- 803 Citations
- 18 Altmetric
- Metrics details
Abstract
The intranuclear position of many genes has been correlated with their activity state, suggesting that migration to functional subcompartments may influence gene expression. Indeed, nascent RNA production and RNA polymerase II seem to be localized into discrete foci or 'transcription factories'. Current estimates from cultured cells indicate that multiple genes could occupy the same factory, although this has not yet been observed. Here we show that, during transcription in vivo, distal genes colocalize to the same transcription factory at high frequencies. Active genes are dynamically organized into shared nuclear subcompartments, and movement into or out of these factories results in activation or abatement of transcription. Thus, rather than recruiting and assembling transcription complexes, active genes migrate to preassembled transcription sites.
Similar content being viewed by others
Main
The activity state of a gene can often be correlated with its intranuclear localization to specific subregions or its position relative to constitutive heterochromatin1,2,3,4,5,6; however, it is unclear whether activation or repression of an individual gene is a cause or consequence of intranuclear position. Transcriptionally active genes have been proposed to associate with transcription factories, discrete nuclear sites of nascent RNA production and concentrated transcriptional components, such as RNA polymerase7,8,9,10,11,12. Calculations using the number of RNA polymerase II (RNAP II) transcription factories in HeLa cells (several thousand per nucleus) and estimates of the number of transcription units suggest that several active genes could occupy the same factory10. To investigate whether individual genes are grouped into discrete subcompartments or factories in a functional context, we used a combination of three-dimensional (3D) fluorescence in situ hybridization (FISH), immunofluorescence and chromosome conformation capture (3C) assays to assess the spatial organization of several genes in a 40-Mb region of distal mouse chromosome 7 (Fig. 1a).
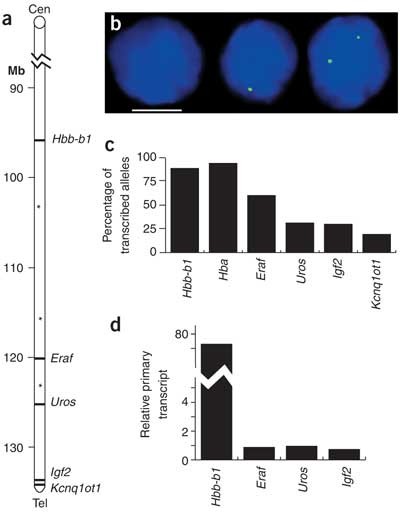
Figure 1: Transcription frequencies of genes on mouse chromosome 7.
(a) Schematic map of distal region of mouse chromosome 7 showing the positions of the Hbb gene cluster (96.0 Mb from centromere), Eraf (120.4 Mb), Uros (125.6 Mb), Igf2 (134.7 Mb) and Kcnq1ot1 (135.3 Mb). Asterisks indicate the locations of additional primers used in 3C analysis (Fig. 7). (b) RNA FISH on adult anemic spleen erythroid cells with intron probes for Uros. Note cell nuclei with zero, one or two signals (green). DAPI staining is blue. Scale bar, 5 μm. (c) The percentage of alleles with a gene transcription signal by RNA FISH for Hbb-b1, Hba, Eraf, Uros, Igf2 and Kcnq1ot1 in erythroid cells. (d) Relative primary transcript levels for Hbb-b1, Eraf, Uros and Igf2, measured by quantitative RT-PCR of intron sequences. Average values are shown with Eraf set to 1.
Results
Discontinuous transcription of 'active' genes
We first used RNA FISH to examine the transcriptional activity of active genes in single erythroid progenitors. For most genes, not every allele across the population had an RNA FISH signal (Fig. 1b). We counted the number of alleles with transcription foci and expressed this number as a percentage of all alleles for that gene in the population (Fig. 1c). We evaluated two erythroid-specific genes: the highly transcribed β-like globin Hbb-b1 (ref. 13) and Eraf (α-hemoglobin-stabilizing protein)14, located 96 Mb and 120.4 Mb, respectively, from the centromere of chromosome 7 (Fig. 1a). We also quantified transcription of the gene Uros (uroporphyrinogen III synthetase; 125.6 Mb), which is ubiquitously expressed but upregulated in erythroid cells. In addition, we examined two widely expressed, imprinted genes: Igf2 (insulin-like growth factor 2; 134.7 Mb) and Kcnq1ot1 (long QT intronic transcript; 135.3 Mb). Finally, we assessed the unlinked, erythroid-specific gene Hba (α-globin) located on chromosome 11. Nearly all Hba and Hbb-b1 alleles had signals in erythroid cells (Fig. 1c), confirming that there is near-continuous transcriptional activity at these gene loci13,15. The non-globin genes had transcription signals at substantially fewer alleles (ranging from 60% for Eraf to 19% for Kcnq1ot1). This category includes genes whose expression requires tight coordination with Hbb-b1 (Eraf and the essential heme biosynthetic enzyme gene Uros) and those that presumably do not require coordination (Igf2 and Kcnq1ot1). We considered the possibility that this pattern resulted from silencing of non-globin genes during terminal erythroid differentiation. To test this, we assayed transcription of Hbb-y, Uros and Eraf in embryonic day (E) 10.5 circulating embryonic erythroid cells, which do not enucleate and remain transcriptionally active in circulation until E12.5 (ref. 13). The percentage of transcription foci for each gene, relative to Hbb, was essentially unchanged (Supplementary Table 1 online), indicating that the differentiation state of the cell does not influence the relative frequencies of transcription. Another explanation for the observation that not all 'active' gene loci are associated with a detectable transcription spot is that genes alternate between 'on' and 'off' transcription states9,15,16,17,18,19. We tested this by comparing primary transcript steady-state levels for several of the genes by quantitative RT-PCR. The level of Hbb primary transcripts was ∼80 times higher than that of Eraf, Uros or Igf2 primary transcripts (Fig. 1d). It is unlikely that the 1.5-kb Hbb transcription unit could contain 80 times more elongating RNAP II complexes than the non-globin genes; this is far greater than could theoretically be contained in a single Hbb transcription unit and substantially more than biochemically determined for Hbb20 or observed for other highly transcribed genes21. This difference could be reconciled by near-constant transcription of all Hbb alleles, with only a proportion of the non-globin alleles being transcribed at any given time. We conclude, in agreement with others, that 'active' genes go through transcriptional on-off periods, and that many genes experience longer periods of transcriptional quiescence than activity17,19.
Colocalization of transcribed genes
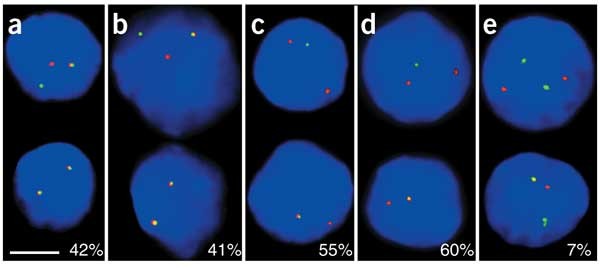
To determine whether widely separated genes colocalize when transcribed, we carried out double-label RNA FISH with a probe for each gene, together with a probe for Hbb-b1. The RNA FISH signals in cis were scored as colocalizing by epifluorescence if the red and green signals overlapped to create a visible yellow signal. The results showed a markedly high frequency of colocalization between transcription foci for Hbb-b1 and each gene, ranging from 42% to 60% yellow signals (Fig. 2a–d). This high frequency of colocalization was observed at all erythroid developmental stages (Supplementary Table 1 online). Even the transcription signals of the unlinked genes Hba and Hbb-b1 overlapped at a considerable, albeit much lower, frequency (Fig. 2e). These results showed that several transcriptionally active genes over a 40-Mb region colocalized with Hbb-b1 at high frequencies.
Figure 2: Colocalization of transcribed genes on mouse chromosome 7.
Double-label RNA FISH for Hbb-b1 gene transcripts and various other genes. Red signals are Hbb-b1 and green signals are Eraf (a), Uros (b), Igf2 (c), Kcnq1ot1 (d) and Hba (e). The percentage of signals that overlap with an Hbb-b1 signal is given at the bottom of each panel. DAPI staining is blue. Scale bar, 5 μm.
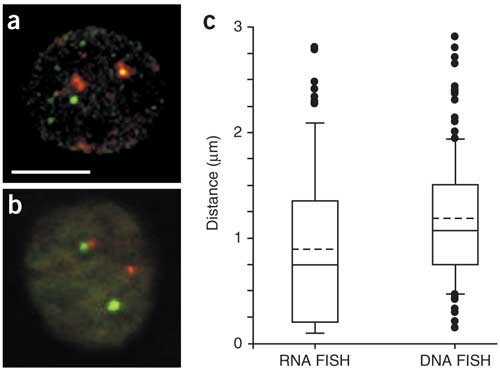
We questioned whether colocalization between genes was coupled to transcription, or whether temporarily nontranscribed alleles also colocalized with the Hbb locus. We carried out high-resolution, confocal microscopy and 3D measurements for Hbb-b1 and Eraf comparing RNA FISH (transcribed alleles) and DNA FISH (transcribed and temporarily nontranscribed alleles; Fig. 3). In these experiments, as well as those described above, we used techniques that preserve 3D nuclear structure4,11,22,23 and specific higher-order chromatin interactions24. We measured the distance from signal center to signal center to minimize the limitations of light microscopy, rather than scoring signals on the basis of overlaps between red and green signals (yellow) as above. Analysis of 79 randomly chosen loci in which Eraf and Hbb-b1 RNA FISH signals appear in cis showed that during transcription, the positions of Eraf were skewed strongly toward Hbb-b1, with a high proportion of signals that were <0.5 μm from the center of the Hbb-b1 signal (Fig. 3c). The separation between signal centers ranged from 0.1 μm (the minimum measurable distance) to 2.8 μm. The DNA FISH signals for Eraf and Hbb had a similar range, but the distribution was significantly different from that of the RNA FISH (Fisher's exact test, P < 2.7 × 10−8). On average, the Eraf signals were clearly shifted away from the Hbb locus, with substantially fewer signals <0.5 μm apart. Taken together, these data correlate transcriptional activity of Eraf with frequent juxtaposition to the transcribed Hbb locus; the transcriptionally silent Eraf alleles were, on average, further away from the site of Hbb transcription.
Figure 3: Colocalization of genes is transcription-dependent.
Confocal microscopy and 3D measurement of separation distances between Hbb-b1 and Eraf. (a) RNA FISH 3D reconstructed image from confocal stack on anemic spleen erythroid cell. Hbb-b1 transcription foci (red) and Eraf (green). Scale bar, 5 μm. (b) DNA FISH 3D reconstructed image from confocal stack on anemic spleen erythroid cell. Hbb (red) and Eraf (green). (c) Box and whiskers plot of the distributions of 3D measurements of the separation distance between Hbb-b1 and Eraf using RNA FISH (n = 79) and DNA FISH (n = 130). Lower and upper whiskers denote the 10th and 90th percentiles, respectively, of the distribution. The lower and upper limits of the boxes denote the 25th and 75th percentiles, respectively. Solid and dashed lines in the boxes indicate the median and mean, respectively. Outliers (i.e., values above and below the 10th and 90th percentiles) are shown as filled circles.
In an attempt to understand the relevance of this difference, we carried out additional DNA FISH experiments, measuring the distance between various combinations of active and inactive gene loci in erythroid and nonerythroid cells (Supplementary Fig. 1 online). All the DNA FISH measurements had few signals that were <0.5 μm apart, whereas many of the Eraf and Hbb RNA FISH signals were <0.5 μm apart. This difference was statistically significant (Fisher's exact test, P < 0.0006). Pairwise comparisons among the DNA FISH distributions showed very little difference in the range from 0 μm to 0.5 μm.
Distal transcribed genes can share RNAP II factories
We considered two possibilities that could account for the differences between the RNA FISH and DNA FISH measurements: first, comparing DNA FISH and RNA FISH directly may have inherent difficulties owing to differences in the techniques; and second, RNA FISH (which detects only the transcribed subpopulation of alleles) may highlight a genuine transcription-coupled juxtaposition between a subset of Eraf and Hbb alleles, whereas DNA FISH assesses all alleles (transcribed and temporarily nontranscribed), potentially diluting the colocalizing subset of transcribed alleles. To differentiate between these possibilities, we devised an immuno-FISH system to compare directly transcribed and temporarily nontranscribed alleles in the same cell by virtue of their relationship to RNAP II foci.
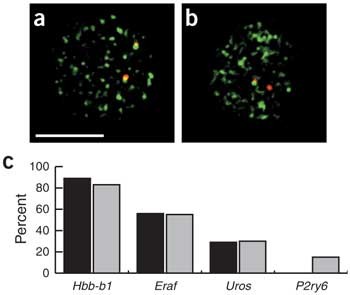
We first established that there is a tight correlation between transcribed alleles and association with RNAP II foci. Using RNA immuno-FISH with antibodies against RNAP II, we observed that ∼90% of transcribed alleles were associated with distinct RNAP II foci (Fig. 4a), indicating that most gene transcription occurs in RNAP II foci. We then carried out DNA immuno-FISH and observed that the percentage of genes associated with RNAP II foci matched exactly the percentage of transcribed alleles in the whole population (Fig. 4b,c). These data clearly show that actively transcribed alleles are associated with RNAP II foci and that temporarily inactive alleles are not.
Figure 4: Actively transcribed genes associate with RNAP II foci.
(a) RNA immuno-FISH of Hbb-b1 transcription (red) with RNAP II staining (green) in anemic spleen erythroid cells. Scale bar, 5 μm. (b) DNA immuno-FISH of Eraf (red) with RNAP II staining (green). (c) Comparison of the percentage of alleles exhibiting a gene transcription signal by RNA FISH (black), with the percentage of loci that overlap with an RNAP II focus by DNA FISH (gray) for Hbb-b1 (n = 83), Eraf (n = 59), Uros (n = 47) and P2ry6 (n = 79).
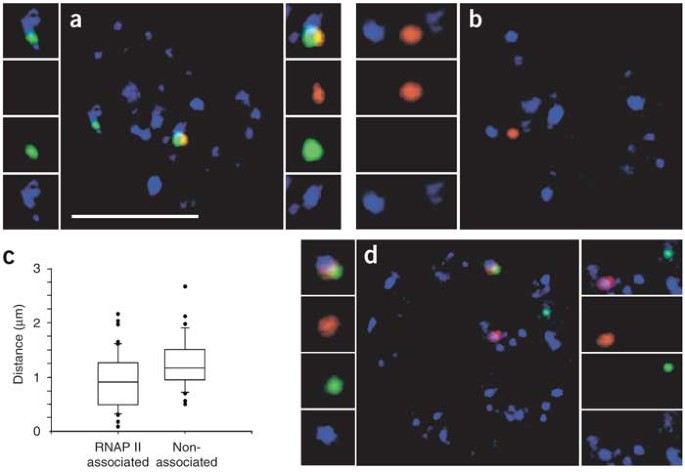
We then carried out triple-label DNA immuno-FISH to visualize Hbb and Eraf along with RNAP II (Fig. 5a,b). We discerned the transcribed alleles from the temporarily inactive alleles by virtue of their association with RNAP II foci. We collected 3D image stacks of randomly selected image fields and measured the distances between _cis_-linked Hbb and Eraf signals before visualizing the RNAP II immunofluorescence. We categorized the measurements into two subpopulations: RNAP II–associated (transcribed alleles), in which each gene was associated with RNAP II, and nonassociated (temporarily inactive alleles), in which one or both genes were not associated with RNAP II. In the RNAP II–associated subpopulation, >25% of the gene pairs were <0.5 μm apart, whereas in the nonassociated subpopulation, none of the genes were within 0.5 μm (Fisher's exact test, P < 0.0016; Fig. 5c). Again, we observed a distinct subset of transcribed alleles that were close together. The temporarily inactive alleles, however, were not juxtaposed. This finding discounts the possibility that the differences were the result of technical problems in comparing RNA FISH and DNA FISH and supports our previous observations of transcription-coupled juxtaposition of alleles (Fig. 3).
Figure 5: Actively transcribed genes colocalize to shared transcription factories.
(a) Single optical section of a triple-label DNA immuno-FISH on erythroid cell, showing Hbb (green), Eraf (red) and RNAP II foci (blue). The merged and separate channels of the signals are shown in the side panels. On the left of the main panel, an Hbb signal alone associates with an RNAP II focus. On the right, two colocalizing signals associate with the same RNAP II focus. Scale bar, 5 μm. (b) A separate optical section of the same cell showing the second Eraf allele, which does not associate with an RNAP II focus. (c) Box and whiskers plot of the distributions of 3D measurements of the separation distance between Hbb and Eraf loci (n = 84), divided into RNAP II–associated versus nonassociated. (d) Triple-label RNA immuno-FISH on erythroid cell showing Hbb-b1 (red), Eraf (green) and RNAP II (blue). Left panels, colocalized transcription signals associating with the same RNAP II focus. Right panels, separate transcription signals associating with distant RNAP II foci.
Notably, the RNAP II–associated signals included a subset of alleles for which the two gene loci were close together, occupying the same RNAP II focus. Triple-label RNA immuno-FISH also showed that colocalizing transcribed alleles associated with the same RNAP II focus (Fig. 5d). These results indicate that two transcribed genes can, and often do, occupy the same factory. Taken together, these data show that actively transcribed genes are associated with transcription factories and associate with the same factory at high frequencies, whereas identical inactive alleles in the same cells are repositioned away from transcription factories.
Extra-territorial position correlates with potential
Several studies suggest that positioning of genes relative to their chromosome territories is a contributing factor in determining gene expression. An expressed gene is preferentially located outside of its chromosome territory, whereas a closely linked, inactive gene is located inside25. The Hbb locus is more often looped outside its chromosome territory in mouse erythroleukemia cells than is an inactive gene26. We questioned whether the on-off transcription and in-out position of active genes, relative to factories, was related to differential positioning relative to the chromosome territory. To test this, we assessed the position of the infrequently transcribed gene Uros relative to the chromosome 7 territory (Supplementary Fig. 2 online). Although Uros is actively transcribed only 29% of the time, it was found outside its chromosome territory in 79% of cases. In contrast, the inactive gene Fgfr2 was outside the chromosome territory in only 19% of cases (Supplementary Fig. 2 online). These results confirm that expressed genes are often located outside chromosome territories and inactive genes are more often inside chromosome territories. But these data do not show a correlation between positioning relative to the chromosome territory and the on-off transcriptional behavior of active genes. Instead, our data suggest that genes with transcriptional potential are preferentially located outside chromosome territories, but this alone is not sufficient for transcription.
RNAP II factories are limiting in vivo
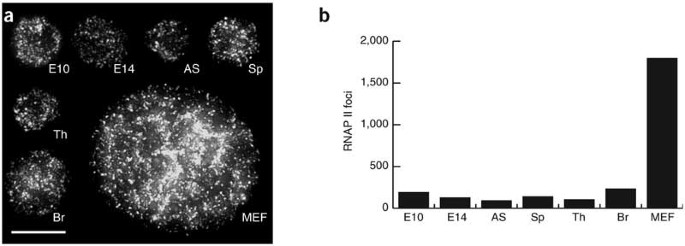
We noticed that the number of RNAP II foci in erythroid cells was markedly lower than that reported for fibroblast-like cell lines. Figure 6 shows deconvoluted, projected images derived from 3D image stacks showing all the RNAP II transcription factories in single cell nuclei from various tissues. We found that erythroid cells had, on average, only 100–300 RNAP II foci per nucleus. Many other tissue types have equivalent numbers of RNAP II foci, suggesting that erythroid cells do not have abnormally low numbers of RNAP II foci. In contrast, limited-passage mouse embryonic fibroblasts (MEFs) have a much greater number and higher density of RNAP II foci, similar to previous reports for HeLa and fibroblast cell lines. We conclude that the number of transcription factories in tissues is far more restricted than indicated by previous estimates from cultured cells. It is, perhaps, not surprising that colocalization of transcribed genes was not observed in a recent study using cultured fibroblast-like cells27. Our data indicate that erythroid and other differentiated or committed tissue types have a limited number of available transcription sites. Coupled with estimates from expressed-sequence tag databases, which show that erythroid cells express at least 4,000 genes (data not shown), we conclude that many genes are obliged to seek out and share the same factory.
Figure 6: Comparison of RNAP II foci in several tissue types and MEFs.
(a) Deconvoluted maximum-intensity projections of image stacks of nuclei immunostained for RNAP II. E10, embryonic blood; E14, fetal liver erythroid; AS, adult anemic spleen erythroid; Sp, normal adult spleen; Th, adult thymus; Br, fetal brain. Scale bar, 10 μm. (b) Numbers of RNAP II foci counted for each nucleus shown in a.
3C analysis
Finally, we corroborated the colocalization of transcribed alleles by a completely independent method. 3C generates a population-average measurement of interaction frequency between any two genomic loci, thereby providing information on their relative proximity in the nucleus28. We compared erythroid and brain tissues using the interaction between Hbb-b1 and the upstream β-globin locus control region (LCR) as an erythroid-specific positive control, and ligation between two fragments of the ubiquitously expressed gene Calr as a control for both tissues24,29. With short fixation times, the LCR–Hbb-b1 interaction was readily detectable in erythroid, but not brain, tissues (Fig. 7a). We also detected an erythroid-specific ligation product between Eraf and Hbb-b1 but did not detect ligation between Hbb-b1 and three inactive regions (Fig. 1a). These results indicate that Eraf is frequently in proximity to Hbb in expressing cells, whereas inactive, intervening regions are not. An erythroid-specific ligation product between Hbb-b1 and Uros was detectable with longer fixation times (Fig. 7b), consistent with the lower transcription frequency of Uros. A ligation product was produced between the LCR and Hbb-b1 in brain with extended fixation, although the product was still six times more abundant in erythroid cells, as determined by quantitative PCR. We did not detect a product between Hbb-b1 and Igf2, even though they colocalized by RNA FISH. These biochemical analyses are consistent with our cytological observations and indicate that distal active genes are frequently proximal to Hbb in erythroid cells.
Figure 7: 3C analysis.
PCR detection of unique ligation products between Hbb-b1 and various restriction fragments in E18.5 fetal liver (E) and fetal brain cells (B). The nuclei were fixed for either 5 min (a) or 10 min (b). The restriction fragments assayed were the Hbb LCR, three intervening genomic regions (104, 116, 124; shown in Fig. 1) and Eraf, Uros and Igf2. Detection of ligated products between two restriction fragments of Calr was used as a positive control for fetal liver and brain, as described29. M, DNA size marker.
Discussion
We showed that widely separated genes colocalize in shared RNAP II factories in a transcription-dependent manner. Most 'active' genes undergo transcription on-off cycles. Our data link the 'on' state with transcription factory occupancy and the 'off' state with repositioning away from factories. This is important in the context of studies showing that gene induction can occur through a rapid and transient increase in the number of transcribed alleles30. Combined with recent studies showing that chromatin is highly mobile31,32,33, our data provide a crucial link substantiating the concept that genes are dynamically recruited to transcription factories. We conclude that most genes move in and out of transcription factories, resulting in activation and abatement of transcription (Fig. 8). As inactive genes can also come into proximity with factories (Fig. 4c), we suggest that genes must be potentiated to engage in a functional association. The classical effectors of gene expression, including _trans_-acting factors, enhancers, chromatin modifications and higher-order structure, presumably affect the affinity of individual genes for such a site or the residence time within that site. Thus, mechanisms regulating recruitment of genes into factories would be expected to have a fundamental role in gene expression.
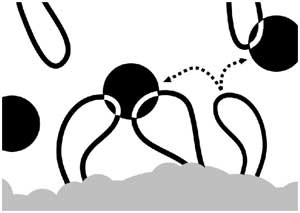
Figure 8: Model of dynamic associations of genes with transcription factories.
Schematic representation of chromatin loops (black) extruding from a chromosome territory (gray). Transcribed genes (white) in RNAP II factories (black circles). Potentiated genes (free loops) that are not associated with RNAP II factories are temporarily not transcribed. Potentiated genes can migrate to a limited number of preassembled RNAP II factories to be transcribed (dotted arrows). We propose that both cis and trans associations are possible.
Our findings stress the importance of assessing the transcribed subpopulation of alleles to observe the colocalization phenomenon. DNA FISH is not sufficient to tease out differences between transcribed and temporarily nontranscribed alleles in a population. With RNA FISH, the colocalizing subset of transcribed alleles can be observed in the distribution, even though it is partially diluted by those alleles transcribed far from the Hbb locus. With DNA FISH, the colocalizing subset is further obscured by the inclusion of temporarily nontranscribed alleles. We observed a high proportion of juxtaposed transcribed alleles using both RNA FISH and DNA immuno-FISH. But direct comparison of the distribution of distances between transcribed alleles by RNA FISH (Fig. 3c) and DNA immuno-FISH (RNAP II–associated; Fig. 5c) shows that the techniques differ slightly. A higher number of signals are close together in the RNA FISH experiments. We believe that this is the result of differences in probe sizes and the targets they detect. RNA FISH probes recognize primary transcripts clustered around relatively small transcription units, whereas 100- to 300-kb DNA FISH probes detect large genomic regions that may represent entire chromatin loops (Fig. 8). Measuring the distance between signals of two colocalizing genes from center to center would logically result in DNA FISH signals being, on average, slightly farther apart than corresponding RNA FISH signals.
Our results suggest that genes with transcriptional potential often reposition to existing sites of ongoing transcription when activated, rather than assembling a new transcription site. If each gene assembled its own transcription site de novo upon activation, we predict that transcription signals would rarely colocalize. Although we cannot rule out a de novo assembly pathway, or the possibility that genes initially engage RNAP II outside foci, the observed high frequency of colocalization with Hbb-b1 and the limited number of factories provide strong evidence that genes migrate to preassembled factories for transcription. This concept questions the traditional view that active genes or promoters recruit RNAP II and instead posits that active genes are recruited to RNAP II compartments.
We propose that two linked genes are more likely to occupy a factory than are two genes in trans. Chromosomal regions can extend far beyond the frontiers of their territories23,34,35, however, suggesting that there is a certain degree of intermingling between chromosomes. The observation that active genes dynamically operate outside the chromosome territory suggests that genes from neighboring chromosomes may occupy a factory together. We estimated the expected frequency of random colocalization between two genes in trans to be <1%, based on the volume of the nucleus and individual gene signals. Notably, we detected 7% colocalization between the unlinked genes Hba and Hbb-b1 (Fig. 2e). These may represent genuine trans associations of genes on different chromosomes with the same RNAP II factory, the frequency of which may reflect the spatial orientation of one chromosome relative to its neighbor and the frequency and heritability of such positioning36,37,38,39,40,41. Trans associations have obvious implications in chromosomal translocations39, whereas cis associations between distal transcribed regions may function in V(D)J recombination42.
Analysis of the human transcriptome map showed that genes are nonrandomly arranged, clustered in gene-dense chromosomal regions called regions of increased gene expression (RIDGEs), suggestive of functional organization of the genome43,44. The human homologs of Hbb, Igf2 and Kcnq1ot1 are located within 3 Mb of each other on a RIDGE at chromosome 11p15.5. In the mouse, nonsyntenic regions containing Eraf and Uros intervene between Hbb and the Igf2_–_Kcnq1ot1 cluster, increasing the distance between them to ∼40 Mb (Supplementary Fig. 3 online). Our data show that associations between genes on different RIDGEs are possible, but this does not necessarily mean that RIDGEs have no functional importance. We cannot overlook the possibility that the Hbb genes and LCR contribute to the formation of an unusually stable transcription site that attracts or holds distal genes. Previous studies showed that the Hbb LCR affects the probability and stability of transcription of the Hbb genes18,45, as well as their nuclear location3, suggesting that it may tether the locus to a factory or be involved in nucleation or stabilization of a factory. Other areas of the genome that are not similarly endowed may benefit from clustering of genes that dynamically cooperate in the self-organization46,47 of a local factory. Characterization of additional regions and generation of an erythroid transcriptome map will undoubtedly help to clarify these issues.
Methods
Tissue collection and culture of MEFs.
We prepared adult erythroid cells from spleens of anemic mice 5 d after treating them with phenylhydrazine48. We measured the erythroid cell population of the anemic spleen to be 93%, based on the percentage of cells that contain an Hba RNA FISH signal, an Hbb-b1 RNA FISH signal or both. E18.5 fetal livers were 87% erythroid. We disrupted fresh brain, thymus and spleen tissues into a single-cell suspension in ice-cold phosphate-buffered saline and fixed them immediately, as described previously49. To prepare MEFs, we washed embryos, from which we removed the livers, three times in phosphate-buffered saline and then disrupted them in trypsin–EDTA for 30 min at room temperature. We cultured the cell suspension in Dulbecco's modified Eagle medium plus 10% fetal bovine serum for 24–48 h, or until confluency was reached, and then froze them. We then thawed aliquots and grew them for another 24 h before use.
Quantitative RT-PCR.
We quantified steady-state primary transcript levels by real-time RT-PCR. We isolated total RNA from E14.5 fetal liver cells using RNA-Bee reagent (AMS Biotechnology) and reverse-transcribed it with Superscript II (Invitrogen). We assessed the abundance of individual intron sequences in the cDNA by SYBR Green real-time PCR, carried out on an ABI Prism 7,000 Sequence Detection System (Applied Biosystems; primer sequences are available on request). We determined relative primary transcript levels against genomic DNA standards. We tested amplicons in intron 1 and intron 2 of Hbb-b1, which gave similar results in comparison with the non-globin genes.
RNA FISH.
We carried out RNA FISH as described previously49. We visualized Hbb-b1 and Hbb-y transcription with antisense, dinitrophenol-labeled, oligonucleotide intron probes and Texas Red detection. We prepared digoxigenin-labeled, single-stranded DNA probes to detect Eraf, Uros, Igf2 and Kcnq1ot1 as described50 (primers sequences used to make probes are available on request).
DNA FISH.
We carried out DNA FISH as previously described4. We obtained BAC clones from BACPAC Resources, labeled them by nick translation and used them as DNA FISH probes. We used the following BAC clones: 195-kb RP24-344M21 for Hbb; 170-kb RPCI-23-353J20 for Eraf; 221-kb RP24-132K17 for Uros; 176-kb RP23-452J21 for P2ry2; 212-kb RP24-562C11 for Fgfr2; and 130-kb RP24-127G8 for Gprc5b. For double-label experiments, we labeled one of the DNA FISH probes directly with Alexa Fluor 594, labeled the other probe with digoxigenin and detected it with fluorescein-conjugated antibodies. We visualized mouse chromosome 7 with cyanine 3–labeled StarFISH chromosome paint obtained from Cambio. We labeled the gene probes used in conjunction with the chromosome paint with digoxigenin and detected them with fluorescein.
Immunofluorescence and immuno-FISH.
We detected RNAP II using a mouse antibody to RNAP II IgG (clone CTD4H8; Upstate) followed by a goat antibody to mouse conjugated with Alexa Fluor 350 or fluorescein isothiocyanate (Molecular Probes). Immuno-detection of RNAP II was combined with DNA FISH as described1. We carried out RNAP II detection, in combination with RNA FISH, by adding the antibody to RNAP II combination after RNA detection. For triple-label experiments, we labeled one of the DNA FISH probes directly with Alexa Fluor 594, labeled the other probe with biotin and detected it with Alexa Fluor 350–conjugated streptavidin. We visualized RNAP II with fluorescein isothiocyanate.
Microscopy and image analysis.
We examined RNA FISH signals on an Olympus BX41 epifluorescence microscope. We assessed at least 100 loci. We captured images of representative nuclei with a CCD camera. To measure distances between genes in cis by RNA and DNA FISH, we collected image stacks using Leica TCS 4D and SP2 confocal microscopes. For RNA FISH, we chose chromosomes with both Eraf and Hbb-b1 signals in cis at random by directly observing and selecting cells under the microscope using the green channel, which permits detection of the Eraf signal only. We then captured confocal stacks for these cells using green and red channels to detect both Eraf and Hbb-b1. For DNA FISH, we collected confocal stacks of randomly chosen fields and included all cells in the analysis. We measured the distances between signals on 3D-reconstructed image stacks using Volocity image analysis software. In all cases, we made measurements from center to center of the two gene signals. To assess the association of FISH signals and RNAP II foci, we captured image stacks of nuclei using an epifluorescence Olympus BX41 microscope fitted with a motorized stage.
3C assay.
We adapted the 3C assay28,29 as follows. We fixed E18.5 liver and brain cells in 2% formaldehyde for 5 or 10 min at room temperature. We digested nuclei (1 × 106) overnight with 600 U of _Bgl_II. We ligated digested chromatin (2 μg) with 2,000 U of T4 DNA ligase in a final volume of 2 ml. We then used sample DNA (300 ng) in two rounds of PCR with nested primers (except for Calr). We amplified the _Hbb-b1_–LCR product for 25 cycles in the first round, followed by 35 cycles with nested primers, and all other products for two rounds of 35 cycles. We cloned ligation products and verified them by DNA sequencing. We prepared control ligation products by mixing equimolar amounts of multiple PCR products containing the _Bgl_II sites in their genomic orientation. We then digested this mixture with _Bgl_II and ligated it to generate a library of randomly ligated control fragments. We tested the specificity of the 3C primers by PCR using 2 ng of the randomly ligated control fragment mixture with 200 ng of genomic DNA for 35 cycles. All produced equivalent amounts of product. 3C primer sequences are available on request.
Note: Supplementary information is available on the Nature Genetics website.
References
- Brown, K.E. et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91, 845–854 (1997).
Article CAS Google Scholar - Brown, K.E., Baxter, J., Graf, D., Merkenschlager, M. & Fisher, A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3, 207–217 (1999).
Article CAS Google Scholar - Francastel, C., Walters, M.C., Groudine, M. & Martin, D.I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell 99, 259–269 (1999).
Article CAS Google Scholar - Brown, K.E. et al. Expression of α- and β-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3, 602–606 (2001).
Article CAS Google Scholar - Schubeler, D. et al. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev. 14, 940–950 (2000).
CAS PubMed PubMed Central Google Scholar - Wang, J. et al. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164, 515–526 (2004).
Article CAS Google Scholar - Jackson, D.A., Hassan, A.B., Errington, R.J. & Cook, P.R. Visualization of focal sites of transcription within human nuclei. EMBO J. 12, 1059–1065 (1993).
Article CAS Google Scholar - Iborra, F.J., Pombo, A., Jackson, D.A. & Cook, P.R. Active RNA polymerases are localized within discrete transcription 'factories' in human nuclei. J. Cell Sci. 109, 1427–1436 (1996).
CAS PubMed Google Scholar - Grande, M.A., van der Kraan, I., de Jong, L. & van Driel, R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 110, 1781–1791 (1997).
CAS PubMed Google Scholar - Jackson, D.A., Iborra, F.J., Manders, E.M. & Cook, P.R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 (1998).
Article CAS Google Scholar - Verschure, P.J., van Der Kraan, I., Manders, E.M. & van Driel, R. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147, 13–24 (1999).
Article CAS Google Scholar - Zaidi, S.K. et al. Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc. Natl. Acad. Sci. USA 99, 8048–8053 (2002).
Article CAS Google Scholar - Trimborn, T., Gribnau, J., Grosveld, F. & Fraser, P. Mechanisms of developmental control of transcription in the murine α- and β-globin loci. Genes Dev. 13, 112–124 (1999).
Article CAS Google Scholar - Kihm, A.J. et al. An abundant erythroid protein that stabilizes free α-haemoglobin. Nature 417, 758–763 (2002).
Article CAS Google Scholar - Wijgerde, M., Grosveld, F. & Fraser, P. Transcription complex stability and chromatin dynamics in vivo. Nature 377, 209–213 (1995).
Article CAS Google Scholar - Ross, I.L., Browne, C.M. & Hume, D.A. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol. 72, 177–185 (1994).
Article CAS Google Scholar - Kimura, H., Sugaya, K. & Cook, P.R. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159, 777–782 (2002).
Article CAS Google Scholar - Milot, E. et al. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87, 105–114 (1996).
Article CAS Google Scholar - Levsky, J.M., Shenoy, S.M., Pezo, R.C. & Singer, R.H. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Article CAS Google Scholar - Hunt, J.A. Rate of synthesis and half-life of globin messenger ribonucleic acid. Rate of synthesis of globin messenger ribonucleic acid calculated from data of cell haemoglobin content. Biochem. J. 138, 499–510 (1974).
Article CAS Google Scholar - Osheim, Y.N., Miller, O.L., Jr. & Beyer, A.L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell 43, 143–151 (1985).
Article CAS Google Scholar - Solovei, I. et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 276, 10–23 (2002).
Article CAS Google Scholar - Mahy, N.L., Perry, P.E. & Bickmore, W.A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159, 753–763 (2002).
Article CAS Google Scholar - Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F. & Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32, 623–626 (2002).
Article CAS Google Scholar - Chambeyron, S. & Bickmore, W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004).
Article CAS Google Scholar - Ragoczy, T., Telling, A., Sawado, T., Groudine, M. & Kosak, S.T. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11, 513–525 (2003).
Article CAS Google Scholar - Shopland, L.S., Johnson, C.V., Byron, M., McNeil, J. & Lawrence, J.B. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162, 981–990 (2003).
Article CAS Google Scholar - Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Article CAS Google Scholar - Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002).
Article CAS Google Scholar - Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Article CAS Google Scholar - Vazquez, J., Belmont, A.S. & Sedat, J.W. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11, 1227–1239 (2001).
Article CAS Google Scholar - Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S.M. Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186 (2001).
Article CAS Google Scholar - Chubb, J.R., Boyle, S., Perry, P. & Bickmore, W.A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Article CAS Google Scholar - Volpi, E.V. et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113, 1565–1576 (2000).
CAS PubMed Google Scholar - Mahy, N.L., Perry, P.E., Gilchrist, S., Baldock, R.A. & Bickmore, W.A. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157, 579–589 (2002).
Article CAS Google Scholar - Lukasova, E. et al. Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia. Hum. Genet. 100, 525–535 (1997).
Article CAS Google Scholar - Neves, H., Ramos, C., da Silva, M.G., Parreira, A. & Parreira, L. The nuclear topography of ABL, BCR, PML, and _RAR_α genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood 93, 1197–1207 (1999).
CAS PubMed Google Scholar - Parada, L.A., McQueen, P.G., Munson, P.J. & Misteli, T. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 12, 1692–1697 (2002).
Article CAS Google Scholar - Roix, J.J., McQueen, P.G., Munson, P.J., Parada, L.A. & Misteli, T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 34, 287–291 (2003).
Article CAS Google Scholar - Walter, J., Schermelleh, L., Cremer, M., Tashiro, S. & Cremer, T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 160, 685–697 (2003).
Article CAS Google Scholar - Gerlich, D. et al. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112, 751–764 (2003).
Article CAS Google Scholar - Bolland, D.J. et al. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 5, 630–637 (2004).
Article CAS Google Scholar - Versteeg, R. et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13, 1998–2004 (2003).
Article CAS Google Scholar - Caron, H. et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291, 1289–1292 (2001).
Article CAS Google Scholar - Sutherland, H.G., Martin, D.I. & Whitelaw, E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol. Cell. Biol. 17, 1607–1614 (1997).
Article CAS Google Scholar - Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Article CAS Google Scholar - Cook, P.R. Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32, 347–352 (2002).
Article CAS Google Scholar - Dickerman, H.W., Cheng, T.C., Kazazian, H.H. Jr. & Spivak, J.L. The erythropoietic mouse spleen-a model system of development. Arch. Biochem. Biophys. 177, 1–9 (1976).
Article CAS Google Scholar - Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5, 377–386 (2000).
Article CAS Google Scholar - Chakalova, L., Carter, D. & Fraser, P. RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol. 375, 479–493 (2004).
Article CAS Google Scholar
Acknowledgements
We thank A. Corcoran and G. Kelsey for critical review of the manuscript; M. Weiss and Y. Kong for an Eraf genomic clone; and S. Andrews, C. Hennessy, E. Walters, S. Amoils and E. Astoul for their assistance. D.C. was supported by Biotechnology and Biological Sciences Research Council Studentships; A.H. is supported by a Medical Research Council Studentship; J.A.M. is supported by a Canadian Institute of Health Research Postdoctoral Fellowship; K.E.B. is supported by a Wellcome Trust Research Career Development Fellowship; and P.F. is a Senior Fellow of the Medical Research Council. This work was supported in part by the Medical Research Council and Biotechnology and Biological Sciences Research Council, UK.
Author information
Author notes
- David Carter
Present address: Sir William Dunn School of Pathology, Oxford University, South Parks Road, Oxford, OX1 3RE, UK - Susana Lopes
Present address: The Wellcome Trust/Cancer Research UK Institute, Tennis Court Road, Cambridge, CB2 1QR, UK
Authors and Affiliations
- Laboratory of Chromatin and Gene Expression, The Babraham Institute, Babraham Research Campus, Cambridge, CB2 4AT, UK
Cameron S Osborne, Lyubomira Chakalova, David Carter, Alice Horton, Emmanuel Debrand, Beatriz Goyenechea, Jennifer A Mitchell & Peter Fraser - Chromosome Biology Group, Imperial College Faculty of Medicine, Hammersmith Hospital Campus, Du Cane Road, London, W12 ONN, UK
Karen E Brown - Laboratory of Developmental Genetics and Imprinting, The Babraham Institute, Babraham Research Campus, Cambridge, CB2 4AT, UK
Susana Lopes & Wolf Reik - Sir William Dunn School of Pathology, Oxford University, South Parks Road, Oxford, OX1 3RE, UK
Susana Lopes
Authors
- Cameron S Osborne
You can also search for this author inPubMed Google Scholar - Lyubomira Chakalova
You can also search for this author inPubMed Google Scholar - Karen E Brown
You can also search for this author inPubMed Google Scholar - David Carter
You can also search for this author inPubMed Google Scholar - Alice Horton
You can also search for this author inPubMed Google Scholar - Emmanuel Debrand
You can also search for this author inPubMed Google Scholar - Beatriz Goyenechea
You can also search for this author inPubMed Google Scholar - Jennifer A Mitchell
You can also search for this author inPubMed Google Scholar - Susana Lopes
You can also search for this author inPubMed Google Scholar - Wolf Reik
You can also search for this author inPubMed Google Scholar - Peter Fraser
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toPeter Fraser.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Distributions of 3D measurements of the separation distances between genes. (PDF 39 kb)
Supplementary Fig. 2
Positioning of active and inactive genes relative to the chromosome 7 territory. (PDF 84 kb)
Supplementary Fig. 3
Positions of Hbb, Eraf, Uros, Igf2 and Kcnq1ot1 gene loci in relation to gene dense regions of mouse chromosome 7. (PDF 85 kb)
Supplementary Table 1
Quantitation of Eraf and Uros transcription foci in erythroid cells and frequency of co-localization with Hbb foci. (PDF 2 kb)
Rights and permissions
About this article
Cite this article
Osborne, C., Chakalova, L., Brown, K. et al. Active genes dynamically colocalize to shared sites of ongoing transcription.Nat Genet 36, 1065–1071 (2004). https://doi.org/10.1038/ng1423
- Received: 22 June 2004
- Accepted: 14 August 2004
- Published: 07 September 2004
- Issue Date: 01 October 2004
- DOI: https://doi.org/10.1038/ng1423