Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs (original) (raw)
Abstract
MicroRNA (miRNA) genes give rise to small regulatory RNAs in a wide variety of organisms. We used computational methods to predict miRNAs conserved among Drosophila species and large-scale sequencing of small RNAs from Drosophila melanogaster to experimentally confirm and complement these predictions. In addition to validating 20 of our top 45 predictions for novel miRNA loci, the large-scale sequencing identified many miRNAs that had not been predicted. In total, 59 novel genes were identified, increasing our tally of confirmed fly miRNAs to 148. The large-scale sequencing also refined the identities of previously known miRNAs and provided insights into their biogenesis and expression. Many miRNAs were expressed in particular developmental contexts, with a large cohort of miRNAs expressed primarily in imaginal discs. Conserved miRNAs typically were expressed more broadly and robustly than were nonconserved miRNAs, and those conserved miRNAs with more restricted expression tended to have fewer predicted targets than those expressed more broadly. Predicted targets for the expanded set of microRNAs substantially increased and revised the miRNA-target relationships that appear conserved among the fly species. Insights were also provided into miRNA gene evolution, including evidence for emergent regulatory function deriving from the opposite arm of the miRNA hairpin, exemplified by mir-10, and even the opposite strand of the DNA, exemplified by mir-iab-4.
MicroRNAs (miRNAs) are ∼23-nt RNA species that direct the post-transcriptional repression of messenger RNAs (mRNAs) (Bartel 2004). They are generated from primary transcripts (pri-miRNAs) that can fold into characteristic hairpin secondary structures. In animals, those hairpins are typically first cleaved away from the rest of the primary transcript by the nuclear RNase III enzyme Drosha to generate miRNA precursors (pre-miRNA), and are then cleaved near their loops by the cytoplasmic RNase III enzyme Dicer to generate a heteroduplex of two ∼23-nt RNAs (Lee et al. 2003). The mature miRNA is preferentially packaged into the RNA-induced silencing complex (RISC), while the other species, known as the miRNA star (miRNA*), is discarded (Lau et al. 2001; Lim et al. 2003b). The decision as to which species is incorporated into the silencing complex is influenced by the difference in pairing stabilities between the two ends of the miRNA:miRNA* duplex, with preferential incorporation of the strand whose 5′ end is less stably paired (Khvorova et al. 2003; Schwarz et al. 2003).
Once incorporated into the silencing complex, metazoan miRNAs pair to the messages of their mRNA targets, primarily in 3′ untranslated regions (3′ UTRs). Complementarity between the message and a segment in the 5′ region of the miRNA known as the “seed” (miRNA nucleotides 2–7) appears to be the most crucial requirement of target recognition. Conserved pairing to the seed region is a feature of most genetically identified miRNA–target interactions (Lee et al. 1993; Wightman et al. 1993; Lai 2002). Indeed, the requirement of conserved pairing to the miRNA seed enables miRNA targets to be predicted in excess of the noise of false-positive predictions (Lewis et al. 2003; Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005). Short, 7- to 8-nt sites matching the seed region of the miRNA are not only important but sometimes can suffice for repression in reporter assays (Doench and Sharp 2004; Brennecke et al. 2005; Lai et al. 2005). Consistent with the in vivo sufficiency of 7-mer seed-matching sites in mediating repression, many messages preferentially coexpressed with a highly expressed miRNA are depleted in 7-mer sites matching that miRNA, presumably because of selective avoidance of miRNA-mediated repression during evolution (Farh et al. 2005; Stark et al. 2005). Moreover, miRNAs that share the same seed sequence but are diverse throughout the remainder of their sequences can be functionally redundant (Abbott et al. 2005; Lim et al. 2005), which justifies their grouping into members of the same miRNA “family” (Lewis et al. 2003). The seeds that define families are often conserved throughout diverse species even as the individual miRNA genes within the family vary (Ruby et al. 2006). The arms of the hairpin precursors are less conserved than the seeds, but are more conserved than either the surrounding genomic sequence or the intervening loop sequence (Lai et al. 2003; Lim et al. 2003b).
Most known miRNAs were discovered through the cloning and sequencing of small-RNA cDNAs (Griffiths-Jones 2004). However, this method can miss miRNAs expressed at low levels or in only specific cell types or conditions. One approach for identifying low-abundance miRNAs that has previously been applied in Drosophila is to identify candidate miRNA hairpins computationally and then validate their expressions using more directed, and thereby potentially more sensitive, experimental methods (Lai et al. 2003). Because identification of plausible candidates is aided by comparative genomics, this approach gains efficacy as the genome sequences of additional related species become available. A second approach for identifying rare miRNAs is simply to increase the scale of small-RNA sequencing well beyond the reach of prior efforts. This high-throughput sequencing approach has not been used previously in insects, but in other lineages it has revealed miRNAs and miRNA candidates that escaped earlier detection because they are rare or not well conserved in related genomes (Berezikov et al. 2006; Lu et al. 2006; Rajagopalan et al. 2006; Ruby et al. 2006; Fahlgren et al. 2007).
Here, we use the two complementary approaches described above—computational prediction and high-throughput sequencing—to identify nearly 60 additional fly miRNAs and to refine the descriptions of about half of those that had been previously annotated. These results provided insights into miRNA evolution, biogenesis, and expression in insects. When combined with improved target prediction, which used information from all 12 sequenced Drosophila genomes (Drosophila 12 Genomes Consortium 2007; Stark et al. 2007c) to increase prediction accuracy, these new and revised miRNAs substantially expanded and improved the view of miRNA-directed regulation in flies.
Results
Computational prediction of fly miRNAs
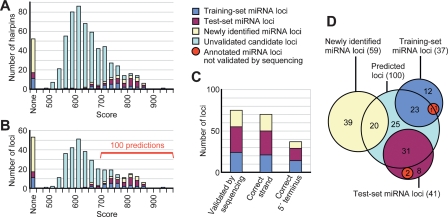
Novel miRNA genes were sought computationally as hairpins with secondary structure and conservation patterns resembling those of previously annotated miRNAs, using an approach with similarities to that described for MiRscan, which had previously been applied to nematodes and vertebrates (Lim et al. 2003a, b). For each of six Drosophila genomes (Drosophila melanogaster, Drosophila ananassae, Drosophila pseudoobscura, Drosophila mojavensis, Drosophila virilis, and Drosophila grimshawi) (Adams et al. 2000; Richards et al. 2005; Drosophila 12 Genomes Consortium 2007; Stark et al. 2007c), RNAfold (Hofacker et al. 1994) was used to identify candidate hairpins from across the entire genome. Candidate hairpins from each genome were first scored based on the relative frequencies of structural characteristics in the background candidate set versus a foreground training set of annotated miRNA hairpins. This training set comprised 37 miRNA hairpins from D. melanogaster that were selected randomly from the 78 previously annotated in miRBase v8.1 (Griffiths-Jones 2004); the remaining 41 miRNAs were reserved as a test set, the performance of which was not evaluated until after the completion of the prediction process. Candidates with scores far below that of the lowest foreground hairpin were removed from the background set, altering its aggregate properties. Unlike the previous method, candidates were then re-scored based on the properties of the minimized background, and the worst candidates were again eliminated. Following several rounds of eliminating candidates from each individual genome, candidates from different genomes (nodes) were paired as putative orthologs (edges) using BLAST (Altschul et al. 1990) and put through the same process of iterative scoring and elimination, now simultaneously evaluating conservation.
The surviving ortholog pairs included 565 hairpin candidates from D. melanogaster that could form complete networks between all six genomes, including all 15 possible edges. These successful candidates were ranked by the sum of the 15 pairwise scores from their first round of pairwise scoring and elimination (Supplemental Table S1). Of the 37 members of the training set, 26 survived and fell mostly within the higher scoring tail of the distribution (Fig. 1A). Examination of the test set revealed that 35 of 41 members of the test set survived and that these 35 hairpins had similar score distributions as the training set, indicating that our prediction method did not overfit to the training-set data (Fig. 1A).
Figure 1.
Performance of miRNA gene prediction. (A) The summed pairwise scores across all 15 two-species comparisons for each miRNA hairpin candidate. Those candidates overlapping the training, test, newly identified, and unvalidated sets of miRNA hairpins are colored as indicated in the key (right) and listed (Supplemental Table S1). (B) The candidate loci, following strand collapse and exon filtering, depicted as in A. The top 100 candidates, which had scores >698, were carried forward as the set of computational gene predictions (Supplemental Table S1). Of the remaining candidates, only a few were likely to be authentic miRNAs. (C) Specificity of the 100 predictions. Plotted are the number of predicted loci that were validated, the number that correctly identified the strand of the miRNA gene, and the number that correctly identified the miRNA 5′ end (Supplemental Table S1), colored as in A. (D) The overlap of the 100 predicted miRNA loci with the training set, test set, and newly identified miRNA loci. Two loci from the training set and two from the test set were not validated by sequencing (red).
Our concept of a candidate miRNA hairpin specified the genomic strand from which the hairpin was derived. However, the secondary structure and conservation properties of a genomic sequence frequently match those of its reverse complement. As a result, 174 of our 565 candidate hairpins were paired with a candidate locus from the opposing genomic strand and thus represented only 87 unique genomic loci. We therefore collapsed our predictions into 478 strand-independent genomic loci, which each included a prediction of which strand would give rise to the mature miRNA based on the higher score. Eliminating 151 candidates that overlapped the annotated exons of protein-coding genes (Supplemental Table S1) left 327 candidate loci (Fig. 1B). The top 100 candidate loci were carried forward as our predictions. These included 55 of the previously annotated genes (24 of the 26 surviving training-set genes), as well as 45 novel predictions.
Recent results from plants and worms demonstrate that for the validation of miRNA expression, large-scale sequence data sets are more reliable and sensitive compared to RNA blotting, and more reliable than and roughly equally sensitive as PCR assays (Rajagopalan et al. 2006; Ruby et al. 2006). We therefore used large-scale small RNA sequence data to evaluate the quality of our predictions.
High-throughput sequencing of small RNAs
To survey the miRNAs of flies, we performed high-throughput pyrosequencing (Margulies et al. 2005; Ruby et al. 2006) on libraries of small RNAs isolated from the following 10 D. melanogaster tissues or stages: very early embryo (0–1 h), early embryo (2–6 h), mid-embryo (6–10 h), late embryo (12–24 h), larvae (first and third instars), imaginal discs, pupae (0–4 d), adult heads, adult bodies, and tissue-culture cells (S2). Pyrosequencing yielded a total of 1.14 million small RNA reads (55,761–174,031 reads per library) that perfectly matched the D. melanogaster genome.
Refinement of prior miRNA annotations
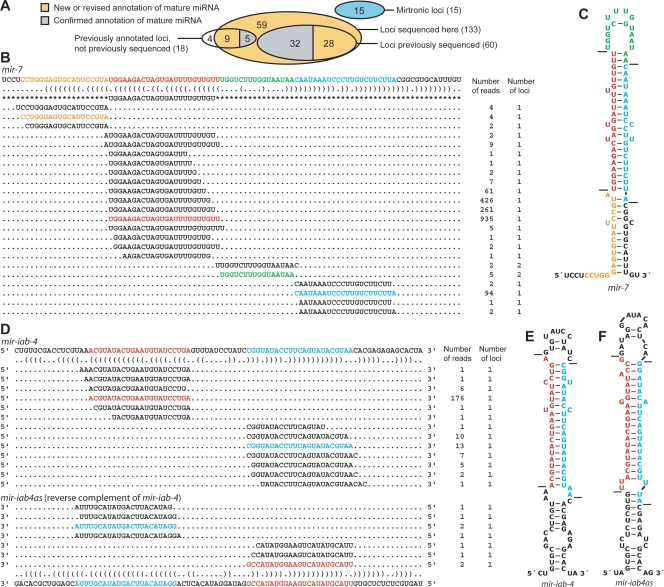
Of the 54 D. melanogaster miRNAs (corresponding to 60 hairpins) that had been previously cloned and sequenced (Lagos-Quintana et al. 2001; Aravin et al. 2003), all 54 were represented in our data set of 1.14 million small RNA reads, as exemplified by miR-7 and miR-iab-4 (Fig. 2), and detailed for all the miRNAs (Supplemental Table S2). For the 60 hairpins of these previously cloned miRNAs, read frequencies ranged from 60 (miR-303) to 20,049 (miR-14), with a median of 2415, and for each of these hairpins the miRNA* species was also recovered. Additional Drosophila miRNA genes are annotated in miRBase v.8.1 based on homology with other miRNAs or computational predictions supported by RNA blots (Aravin et al. 2003; Lai et al. 2003). Of these 18 genes for which small RNAs had not been previously cloned, 14 were represented in our data set (Fig. 2A). The four that were missing (mir-280, mir-287, mir-288, and mir-289) had been predicted computationally and experimentally supported by RNA blots of samples from early embryos, larvae and pupae, and adult males (Lai et al. 2003). Their absence in our libraries from these same developmental stages called their authenticity into question.
Figure 2.
Correspondence between previously annotated miRNA hairpins and sequenced miRNAs. (A) Overlap between previously annotated miRNA hairpins and the total set of 133 hairpins of canonical miRNAs supported by our high-throughput sequencing (Supplemental Table S2). Mirtronic loci are described elsewhere (Ruby et al. 2007). (B) Small RNAs derived from the mir-7 hairpin. A portion of the mir-7 transcript is shown above its bracket-notation secondary structure, mature miRNA annotation from miRBase v8.1 (Griffiths-Jones 2004) flanked by asterisks, and sequences from the present study. For each sequence, the number of reads giving rise to that sequence and the number of loci to which the sequence maps in the D. melanogaster genome are shown on the right. Highlighted are the most abundant sequences corresponding to the miRNA (red), miRNA* (blue), intervening loop (green), and fragment flanking the 5′ Drosha cleavage site (orange) (Supplemental Text). Analogous data for all previously annotated D. melanogaster miRNAs are provided (Supplemental Table S2). (C) The predicted hairpin structure of the mir-7 hairpin, colored as in B. Lines indicate inferred Drosha and Dicer cleavage sites. (D) Small RNAs derived from the mir-iab-4 and mir-iab4as hairpins, displayed as in B. (E) The predicted secondary structure of the sense mir-iab-4 hairpin precursor, formatted as in C. (F) The predicted secondary structure of the mir-iab-4 reverse complement, mir-iab4as, formatted as in C.
In half of the cases (37 of the 74 confirmed genes), the distribution of reads across the hairpin suggested that the mature miRNA differs from the one that had been previously annotated (Supplemental Table S2). Usually, the discrepancy was only at the 3′ terminus of the mature miRNA, as exemplified by miR-7 (Fig. 2B). Although proper 3′ annotation is needed for some miRNA expression profiling methods (Wang et al. 2007), reannotation of the miRNA 3′ terminus was of little consequence because 3′ heterogeneity is a hallmark of mature miRNAs (Lau et al. 2001; Basyuk et al. 2003; Lim et al. 2003b; Ruby et al. 2006). However, in 12 cases, there was discrepancy at the miRNA 5′ terminus (Supplemental Table S2). The reannotation of a miRNA 5′ terminus is far more consequential because of its role in defining the miRNA seed sequence, which, in turn, defines the set of targets. For example, shifting the 5′ terminus by a single nucleotide changes the identity of one or both of the two 7-mers used for target prediction (Lewis et al. 2005), thereby dramatically altering the set of predicted targets, and shifting it by two or more nucleotides would have an even greater effect. Seven of these 12 cases were corrections of annotations that have been based on computational or molecular evidence not expected to identify the 5′ termini with confidence (Supplemental Text). The other five cases were more interesting because they illustrated how a single miRNA hairpin or paralogous hairpins could spawn new miRNA function.
For miR-210, there were 917 reads with the originally annotated 5′ end and 1031 reads with an extra 5′ nucleotide, all of which mapped uniquely to the genome. Combined, the abundance and equivalence of reads indicated that miR-210 was a rare example of a single hairpin generating mature miRNAs with multiple abundant 5′ ends. As was done for miR-248 in Caenorhabditis elegans (Ruby et al. 2006), we annotated the species with an extended 5′ end as miR-210.1 and the species with the originally annotated 5′ end as miR-210.2, with the idea that both probably direct repression in the fly (Supplemental Table S2).
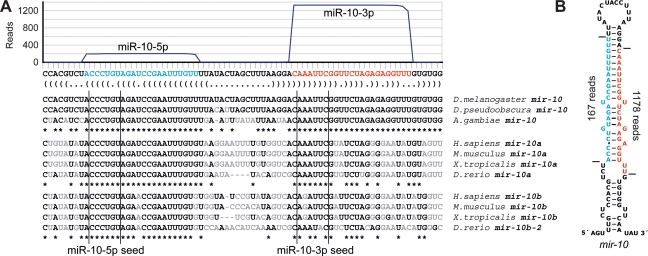
In the case of miR-10, the dominant read was from the arm of the hairpin precursor opposite the annotated miRNA and was sevenfold more abundant (Fig. 3), a result expanding on the observation that species from both arms are easily detected (Schwarz et al. 2003). Because conservation criteria supported the function of RNAs from both arms of the hairpin, and in conjunction with a parallel study (Stark 2007), we annotated the two major products of the mir-10 hairpin as miR-10-5p and miR-10-3p. The seed of the original miR-10 (miR-10-5p) was conserved throughout all annotated mir-10 genes, including those of vertebrates (Fig. 3A). The seed of the species more abundantly represented in our data set (miR-10-3p) was not conserved in all annotated mir-10 genes but was nonetheless conserved in at least one mir-10 gene of each species examined (typically the mir-10a paralogs of vertebrates; Fig. 3A).
Figure 3.
Expression and conservation of mir-10. (A) The sequence and bracket-notation secondary structure of the mir-10 hairpin, highlighting the mature miR-10-5p (blue) and the mature miR-10-3p (red), with read abundance along the length of the sequence plotted above and orthologous hairpins aligned below. Nucleotides differing from the D. melanogaster identities are in gray. Vertical lines indicate the edges of the 6-nt seed of each mature RNA. (B) The mir-10 hairpin predicted secondary structure, colored as in A. Horizontal lines indicate the inferred Drosha and Dicer cleavage sites.
The mir-281 and mir-2 paralogs illustrated how highly related miRNA genes could have divergent functions. For both sets of paralogs, miRNAs deriving from the miRNA arms of the hairpins could be mapped to multiple related hairpins. Nonetheless, the miRNA* species, which mapped uniquely to their hairpins, revealed the likely processing of each hairpin and indicated that one of the two mir-281 hairpins and two of the five mir-2 hairpins gave rise to miRNA species that differed from those previously annotated (Supplemental Text).
Novel miRNAs
Having revised many of the previous miRNA annotations of Drosophila, we next examined the overlap between our predicted miRNA loci and the small RNA reads. Of the 100 predictions, 45 had not been previously annotated as miRNA genes. Of those, 20 were supported by the reads. In all 20 cases, our prediction method identified the correct strand of the miRNA gene (as well as 21 of 24 cases from the training set and 29 of 31 cases from the test set). Given correct identification of the miRNA strand, the miRNA 5′ terminus was correctly predicted in eight of 20 cases (plus 14 of 21 cases from the training set and 15 of 29 cases from the test set; Fig. 1C). Some of the remaining 25 predictions that lacked experimental confirmation also might be authentic miRNAs. However, because most might be false positives and because their 5′ termini were not predicted with high confidence, we did not consider any of these 25 suitable for annotation or target-prediction studies.
Looking more broadly at the small RNA reads to identify the miRNAs that were more difficult to recognize computationally increased our count of newly identified miRNA loci from 20 to 59. To confidently identify these as miRNA genes, we considered the following criteria: (1) the pairing characteristics of the hairpin; (2) the miRNA expression, as measured by the abundance of sequence reads sharing the same 5′ terminus; (3) evolutionary conservation, as evaluated by the apparent conservation of the hairpin in other fly species and grouping of the miRNA candidate into a family based on its seed sequence; (4) the absence of annotation suggesting non-miRNA biogenesis; and (5) the presence of reads corresponding to the predicted miRNA* species.
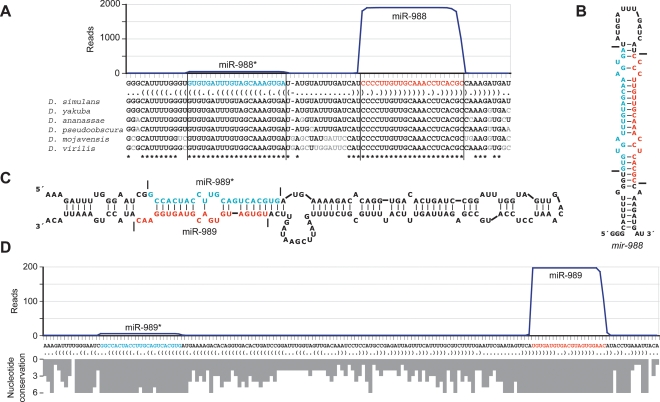
The observation of both a candidate miRNA and a candidate miRNA* in a set of reads provides particularly compelling evidence for Dicer-like processing from an RNA hairpin (Rajagopalan et al. 2006; Ruby et al. 2006; Fahlgren et al. 2007). As illustrated for mir-988 (Fig. 4A), 40 newly identified genes satisfied all five of our criteria, and 19 others satisfied a subset of the criteria deemed sufficient for confident annotation as miRNAs (Table 1). Nine additional candidates fell within predicted miRNA-like hairpins and were sequenced more than once (Supplemental Table S2). However, they were considered unlikely to be miRNAs because they did not satisfy the other criteria sufficiently and they mostly mapped to either protein-coding transcripts (candidates 1–5) or heterochromatic DNA (candidates 6–8). Ten of the newly identified miRNAs derived from loci that were among the top 200 predicted to form miRNA precursor hairpins in a previous effort (Lai et al. 2003). Nine of those predictions correctly identified the genomic strand from which the miRNA was derived, but prediction of the mature miRNA had not been attempted.
Figure 4.
Newly identified miRNAs. (A) The sequence and bracket-notation secondary structure of the mir-988 hairpin, highlighting the miRNA (red) and the miRNA* (blue), with read abundance along the length of the sequence plotted above and orthologous hairpins aligned below; nonconserved nucleotides in gray (Drosophila Comparative Genome Sequencing and Analysis Consortium 2007a, b). Vertical lines indicate the inferred Drosha and Dicer cleavage sites. Analogous data for all newly identified D. melanogaster miRNAs are provided (Supplemental Table S2). (B) The predicted secondary structure of the mir-988 hairpin, colored as in A. Horizontal lines indicate the inferred Drosha and Dicer cleavage sites. (C) The unusually large hairpin of mir-989, colored as in A. (D) The sequence and bracket-notation secondary structure of the mir-989 hairpin, with coloring and read-abundance display as in A. Conservation across the length of the hairpin is shown below as a histogram, with bar depth indicating for each nucleotide the number of orthologs from the organisms shown in A with that nucleotide conserved.
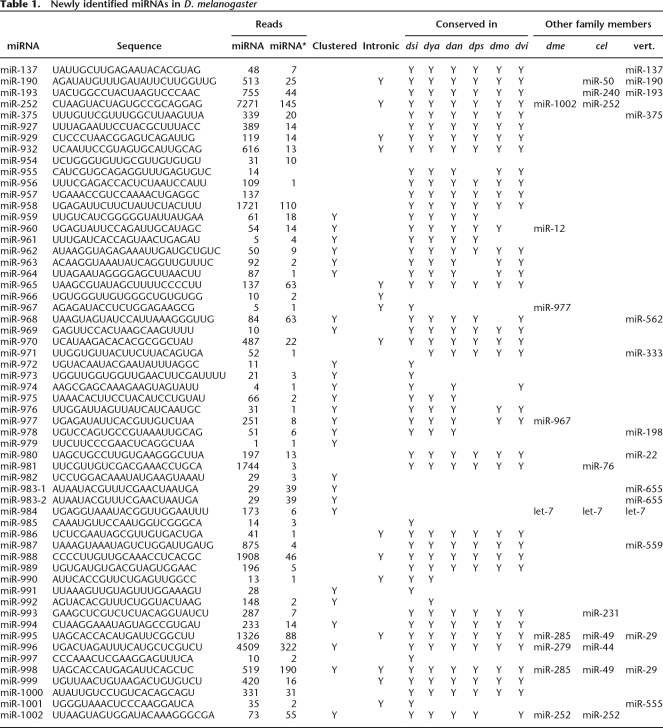
Table 1.
Newly identified miRNAs in D. melanogaster
Two-thirds of the novel miRNAs appeared to be broadly conserved in the Drosophila genus (Table 1). Orthologs were sought in six species spanning both the Sophophora subgenus (Drosophila simulans, Drosophila yakuba, D. ananassae, and D. pseudoobscura) and the Drosophila subgenus (D. mojavensis and D. virilis). Putative orthologs were found in all six species for 28 of the miRNAs and in five of six species for another nine. In 12 of the remaining cases, orthologs were found in two or fewer of the Drosophila species.
One newly identified locus, mir-996, resided 1.5 kb downstream from a related miRNA (mir-279) and within the transcript of CG31044, which is annotated as encoding a 140-amino-acid protein (Crosby et al. 2007). We suggest that miR-996, not the putative protein, is the functional product of this gene. Consistent with this proposal, the observed miRNA was perfectly conserved across a wide scope of fly species, whereas in the open reading frame (ORF), sequence polymorphisms in the closely related species D. simulans, D. yakuba, D. ananassae, and D. pseudoobscura introduced nonsense mutations at codons 73, 56, 13, and 40, respectively.
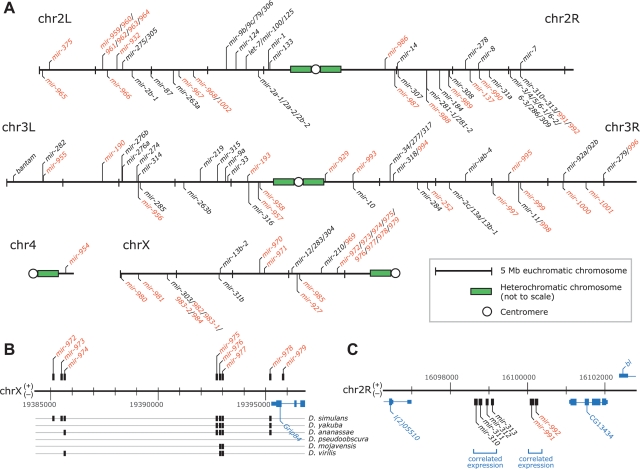
Like many known miRNA genes (Griffiths-Jones 2004), 26 of the 59 newly identified loci were clustered with other miRNA loci (Table 1; Fig. 5A), and 13 fell within annotated introns (and from the same genomic strand as the intron; Table 1). Thus, more than half (69 of 133) of the canonical Drosophila miRNAs were clustered (Fig. 5A), and more than a quarter (36 of 133) were intronic (Supplemental Table S1).
Figure 5.
Genomic landscape of miRNA genes. (A) The distribution of miRNA genes and clusters across the D. melanogaster genome, with newly identified miRNAs indicated (red). Euchromatic portions of the genome are drawn to scale, with (+) strand annotations marked above each chromosome and (−) strand annotations marked below. MicroRNA gene clusters, listed together (with gene numbers separated by slashes), were each defined as series of miRNA loci on the same strand of a given chromosome with no intervening gaps >10 kb. (B) Genomic arrangement and conservation of members of the mir-972∼979 cluster. Detection of an ortholog in the specified species is indicated (black box). (C) Genomic arrangement of the mir-310 cluster. Expression profiles among the constituent miRNAs of each labeled subcluster indicate that the two subclusters were expressed independently (Fig. 6E).
Although most of the novel miRNA genes closely resembled those previously annotated, three of the hairpin precursors were much larger than those observed previously in animals. For the vast majority of previously annotated fly genes, fewer than 30 nucleotides separated the miRNA and miRNA*, and all had fewer than 60 intervening nucleotides. The distribution of intervening sequence lengths was generally similar among the newly identified miRNAs. However, mir-956, mir-989, and mir-997 had abnormally large intervening sequences of 82 nt, 99 nt, and 112 nt, respectively (Fig. 4C,D; Supplemental Table S2). Each of these hairpins gave rise to miRNA* reads, and in each case, the dominant ends of the miRNA versus the miRNA* exhibited 1- or 2-nt 3′ overhangs. In no case was there EST evidence of an intron helping to bridge the distance between the miRNA and miRNA* loci (Karolchik et al. 2003; Crosby et al. 2007). The lack of constraint on the length of the intervening sequence was illustrated by mir-989, whose mature miRNA sequence was perfectly conserved across all seven of the Drosophila species examined but whose intervening sequence length varied widely, dipping as low as 52 nt in D. pseudoobscura (Fig. 4D).
MicroRNA biogenesis in flies
As in nematodes (Ruby et al. 2006), examining the multitude of reads arising from the previously annotated miRNA hairpins provided insights into the specificity and precision of Drosha and Dicer processing (Supplemental Table S2). These RNase III enzymes preferentially leave 2-nt 3′ overhangs when cleaving perfect RNA duplexes (Basyuk et al. 2003; Lee et al. 2003), and this configuration between the ends of miRNA and miRNA* species was observed in our data set. The 5′ ends of both the miRNA and miRNA* species were more homogeneous than the 3′ ends, and the miRNA 5′ ends were more consistent than the miRNA* 5′ ends, and the miRNA* 3′ ends were more consistent than the miRNA 3′ ends, regardless of which was generated by Drosha and which by Dicer (Supplemental Table S2; Supplemental Text). The heightened precision of either enzyme when it defined the miRNA seed implied that Dicer does not simply measure from the site of the Drosha cleavage and suggested that additional determinants must be used when needed to more accurately define Dicer cleavage. Similar conclusions arise from the sequencing data in nematodes (Ruby et al. 2006).
As reported in other species (Li et al. 2005; Ruby et al. 2006), untemplated nucleotide addition also contributed to a minor fraction of 3′ heterogeneity. In Drosophila, we observed evidence for processivity of the terminal-transferase activity, with a preference for adding adenosines (Supplemental Text).
Reads that were antisense to either the miRNA or miRNA* appeared for four previously annotated hairpins (mir-iab-4, mir-124, mir-305, mir-307). The mir-iab-4 hairpin gave rise to nine antisense reads (Fig. 2D); each of the remaining hairpins gave rise to one read. Antisense transcription from mir-iab-4 has been noted previously (Cumberledge et al. 1990). Our reads from either side of the antisense hairpin paired to each other with 2-nt 3′ overhangs, indicating that the antisense transcript was processed by the miRNA biogenesis machinery and likely produced a miRNA (miR-iab4as) that enters the silencing complex.
MicroRNA expression patterns
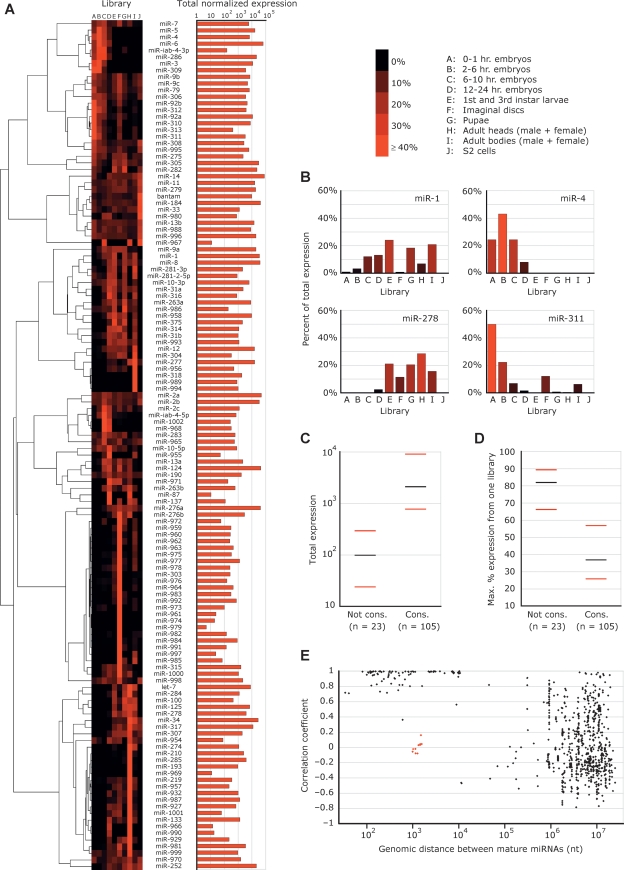
The collection of reads from a variety of developmental stages and anatomical contexts permitted analyses of miRNA expression profiles and overall miRNA expression (Fig. 6A). MicroRNAs were clustered based on their relative expression across all 10 libraries, with the expression values for a particular miRNA in a particular library set to the number of reads corresponding to that miRNA in the given library divided by the total number of reads matching miRNA hairpins in that library. For previously annotated miRNAs, this normalization scheme generated expression profiles similar to those observed previously using 2S rRNA-normalized Northern blots (Fig. 6B) (Aravin et al. 2003), whereas other schemes, such as normalizing to the total reads from each library or to the number of sequenced ribosomal RNA fragments, did not generate profiles matching the published Northern results (data not shown).
Figure 6.
Expression of D. melanogaster miRNAs. (A) The expression profiles of the D. melanogaster miRNAs across the 10 libraries (left) and total level of expression (right). For each library, miRNA reads are normalized to the total reads deriving from miRNA hairpins in that library. Increasing red color intensity indicates an increasing percentage of normalized reads deriving from that library. Read counts and normalized counts for each miRNA in each library are provided (Supplemental Tables S3 and S4). The summed normalized expressions across all 10 libraries are shown on the right; units are the number of miRNA reads per 100,000 total miRNA hairpin reads per library. The tree and image on the left were generated using the publicly available software packages Cluster (Eisen et al. 1998) and MapleTree (L. Simirenko, UC Berkeley). (B) The expression profiles following normalization of four miRNAs whose profiles can be compared to those determined by stage-specific Northern blot (Aravin et al. 2003). (C) The relationship between miRNA conservation and magnitude of total expression. MicroRNAs were separated into two groups based on whether they were conserved (Cons.) or not conserved (Not cons.) beyond the subgenus Sophophora. (Black bars) The median expression for each category; (red bars) the 25th and 75th percentiles. Total expression is defined as in A. (D) The relationship between conservation and breadth of expression, portrayed as in C. The _Y_-axis indicates the maximum percentage of expression for a given miRNA derived from a single library. (E) The relationship between the genomic distances separating miRNAs and the correlation of their expressions. Each point represents a pair of miRNAs from A, including all pairs from the same strand of the same chromosome, but excluding those that can be attributed to multiple genomic loci. The _X_-axis indicates the distance between the mature miRNAs in nucleotides. The _Y_-axis indicates the Pearson correlation coefficient between the normalized expression patterns of the two miRNAs, as displayed in A. The red dots represent miR-991 or miR-992 paired with members of the miR-310∼313 cluster, and miR-283 paired with miR-12/304. Despite their proximity, these subclusters appeared to be expressed independently.
Most miRNAs were observed across several libraries. However, several large sets of miRNAs exhibited strong preference for expression in a single context. The 33 miRNAs that exhibited the narrowest ranges of expression (>70% of their library-normalized reads deriving from a single library) were prevalent in the imaginal discs, adult heads, and to a lesser extent, adult bodies and late embryos (61%, 24%, 12%, and 3% of narrowly expressed miRNAs, respectively), and most were first sequenced in this study (88% of narrowly expressed miRNAs; Fig. 6A).
The normalization of read counts across libraries also permitted an approximate but informative assessment of relative overall expression (Fig. 6A). As reported in vertebrates, worms, and plants (Bartel 2004; Rajagopalan et al. 2006; Ruby et al. 2006), miRNA abundance correlated strongly with the extent of conservation, with those miRNAs found only within the subgenus Sophophora expressed significantly less than those conserved beyond that clade (Fig. 6C, Wilcoxon rank-sum test, P < 2.7 × 10−9). Notably, the more highly conserved miRNAs were also observed more evenly across the 10 libraries examined (Wilcoxon rank-sum test, P < 8.5 × 10−7; Fig. 6D).
As observed previously in worms and mammals (Lau et al. 2001; Sempere et al. 2004; Baskerville and Bartel 2005), miRNAs that were clustered in the Drosophila genome usually had similar expression profiles (Fig. 6E). The correlation of miRNA expression patterns diminished as the distance separating miRNAs surpassed 10,000 nt. Proximally located miRNAs are thought to derive generally from common primary transcripts (Lagos-Quintana et al. 2001; Lau et al. 2001). The mir-991/992 and mir-310∼313 clusters, separated from each other by only 1.0 kb, provided a counterexample (Fig. 5C). Although these two clusters each exhibited internally consistent expression patterns, there was little correlation of expression between the two clusters (Fig. 6E), implying that the mir-991/992 and mir-310∼313 clusters derived from independent transcripts. A more intriguing example was provided by mir-283, mir-12, and mir-304, all three of which map within a single intron. The expression patterns of mir-12 and mir-304 correlated very closely with each other (Pearson correlation coefficient = 0.94), but neither correlated well with that of mir-283 (correlation coefficients of 0.16 and −0.05, respectively), which resided only 1.0 kb upstream of mir-304 and 1.5 kb upstream of mir-12.
MicroRNA targets
In order to gain insight into the functional consequences of the known D. melanogaster miRNAs, including those whose annotations were established or modified here, we predicted their targets using comparative genomics of the sequenced genomes of the Drosophila genus. As done previously, sites were identified in annotated D. melanogaster 3′ UTRs that matched the seed region of each miRNA. Two types of 7-mer sites were sought: the perfect Watson-Crick match to miRNA nucleotides 2–8 (Lewis et al. 2003; Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005) and the perfect Watson-Crick match to miRNA nucleotides 2–7, supplemented with an adenosine opposite miRNA position 1 (Lewis et al. 2005).
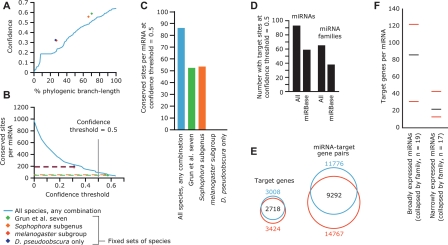
Conservation of 7-mer sites was assessed using a multi-genome alignment of 12 Drosophila species (Adams et al. 2000; Richards et al. 2005; Drosophila 12 Genomes Consortium 2007; Stark et al. 2007c). The phylogenetic distribution of each seed-match motif was used to calculate the total branch length score (BLS), a measure of evolutionary distance across which the motif was conserved (Kheradpour et al. 2007). Requiring perfect conservation across all of the available species maximized confidence in predicted targets, defined as the fraction of sites that were conserved above chance expectation, but also substantially reduced sensitivity (Fig. 7A–C). This trend extended to arbitrary subsets of the currently available species, including subsets that have been used for other prediction efforts in flies (Enright et al. 2003; Stark et al. 2003; Brennecke et al. 2005; Grun et al. 2005). Such loss of sensitivity is partly attributed to artifacts in sequencing coverage, assembly, or alignment, whose impacts on predictions increase with the number of genomes considered (Grun et al. 2005). Discarding the traditional requirement for perfect conservation within a species set and replacing it with a BLS cutoff enabled confident predictions to be reported in spite of the absence of the motif in particular genomes (Kheradpour et al. 2007). The confidence of miRNA target predictions increased with the total branch length and approached a maximum, averaged over all conserved miRNAs, of 0.64 (Fig. 7A), corresponding to a signal-to-noise ratio of 2.7:1. These improved predictions for the expanded and revised set of miRNAs are available at http://targetscan.org.
Figure 7.
MicroRNA target predictions. (A) Confidence of miRNA target prediction versus phylogenic branch length over which sites were conserved in the Drosophila genus. Confidence increased with branch length within 12 Drosophila species (blue line). Confidence versus branch length values for the following fixed sets of species, strictly requiring conservation in every species, are shown as dots of the indicated colors. (Green) Seven species used by Grun et al. (2005) (D. melanogaster, D. erecta, D. yakuba, D. ananassae, D. pseudoobscura, D. mojavensis, D. virilis); (orange) members of the Sophophora subgenus (D. melanogaster, D. sechellia, D. simulans, D. erecta, D. yakuba, D. ananassae, D. persimilis, D. pseudoobscura, D. willistoni); (red) members of the melanogaster subgroup (D. melanogaster, D. sechellia, D. simulans, D. erecta, D. yakuba, D. ananassae); (purple) D. melanogaster and D. pseudoobscura only (Enright et al. 2003; Stark et al. 2003). (B) Sensitivity of target prediction, shown as the average number of sites per conserved miRNA, versus confidence threshold; colored as in A. Note that strict conservation requirements cannot accommodate reduced confidence thresholds, as illustrated by dashed lines. (C) Average number of retained target sites per miRNA for each analysis depicted in A and B at a confidence threshold of 0.5, colored as in A. (D) The number of miRNAs and miRNA families with targets above a confidence threshold of 0.5. Numbers for miRNAs from miRBase v8.1 (Griffiths-Jones 2004) are compared to those for our expanded/corrected set of miRNA annotations. (E) Change to the scope of the predicted miRNA–target network (left) and set of genes predicted to be targeted by miRNAs (right) as a result of miRNA annotation additions and changes. Target-miRNA pairs and target genes identified based on miRBase v8.1 annotations (Griffiths-Jones 2004) are in blue; those based on the expanded/corrected set of miRNA annotations provided by the present study are in red. (F) Specifically expressed miRNAs had fewer predicted targets than did broadly expressed miRNAs. Sets of the most broadly and narrowly expressed miRNAs were collapsed into families based on 6-nt seeds, including only miRNAs conserved beyond the Sophophora subgenus. The number of predicted targets for each family was set to the maximum number of predicted targets of any family member. The median (black bars) and 25th and 75th percentiles (red bars) of the number of targets per miRNA family are indicated for each set.
For comparison, we used the same procedure to predict targets for the D. melanogaster miRNAs as annotated in miRBase v8.1 (Griffiths-Jones 2004). By both increasing the number of annotated conserved miRNAs and correcting the identities of previously annotated miRNAs, our study increased the numbers of both miRNAs and miRNA families with significantly conserved targets (confidence ≥ 0.5) by 1.6-fold (Fig. 7D). While 9292 miRNA–target gene pairs were unaffected by the miRNA annotation additions and changes, 2484 were removed and 5475 were added, thereby changing the predicted network of miRNAs and targets in D. melanogaster by 68% [(2484 + 5475)/(9292 + 2484)]. Of the 3424 unique genes predicted to be conserved targets of miRNAs, 706 had conserved sites for only novel miRNAs. Conversely, 290 genes were erroneously predicted to be conserved targets due to miRNA annotations that were adjusted based on our sequencing data (Fig. 7E).
The scope of miRNA targeting varied between those miRNAs broadly expressed across many libraries compared to those expressed more narrowly, independent of the relationship between breadth of expression and conservation discussed above. Of those miRNAs conserved beyond the scope of the Sophophora subgenus, the narrowly expressed miRNAs tended to have fewer predicted target genes (Fig. 7F, Wilcoxon rank-sum test, P < 0.0015).
Discussion
Hairpin characteristics
The sets of miRNAs initially identified in nematodes, flies, and mammals derive from short hairpins, whereas many of those identified in plants derive from longer precursors (Bartel 2004). Three somewhat longer exceptions have been noted (Drosophila mir-31b [Aravin et al. 2003], C. elegans mir-229 [Ambros et al. 2003; Lim et al. 2003b], and Caenorhabditis briggsae mir-72 [Ohler et al. 2004]), but the prevalence of short hairpin precursors has seemed to justify limiting the length of the sequenced folded during the initial steps of many prediction protocols (Grad et al. 2003; Lai et al. 2003; Lim et al. 2003b), including the approach described here. Several protocols even explicitly evaluate the distance between the predicted miRNA and miRNA* as a characteristic feature of miRNA hairpins (Bentwich et al. 2005), again including the approach described here. Although imposing these constraints likely boosts the specificity of miRNA prediction, our sequencing results indicated that this comes at the cost of missing some miRNAs with unusually long hairpins, particularly in Drosophila, where we found three hairpins (mir-956, mir-989, and mir-997) with at least 80 nt separating the miRNA and miRNA* strands (Fig. 4C,D; Supplemental Table S2).
Our observation that metazoan miRNA precursors can be much longer than previously recognized confirmed that minimal sequence or structural requirements are imposed on the loops of miRNA hairpins (Lai et al. 2003; Berezikov et al. 2005; Han et al. 2006) but raised the question of why long miRNA hairpins are not more frequent in animals. A large, open loop can lead to Drosha processing on the incorrect end of the hairpin by mimicking the single-stranded RNA normally present at the base (Han et al. 2006). This opportunity for dead-end side reactions implies selective pressure for the maintenance of a tight loop. Consistent with this idea, mir-956, mir-989, and mir-997 each exhibited extensive secondary structure in the segment connecting the miRNA and miRNA* (Supplemental Table S2). Deletions can tighten a loop even if they disrupt secondary structure, making them more tolerable than insertions, which must be accompanied by compensatory changes in order to maintain the tightness of the loop. Thus, miRNA hairpins might be expected to shorten rather than lengthen over evolutionary time. Another possibility is that shorter pre-miRNAs might be more suitable substrates for downstream events such as nuclear export, and longer pre-miRNAs might only rarely bypass these constraints.
The evolutionary origins of novel miRNA genes
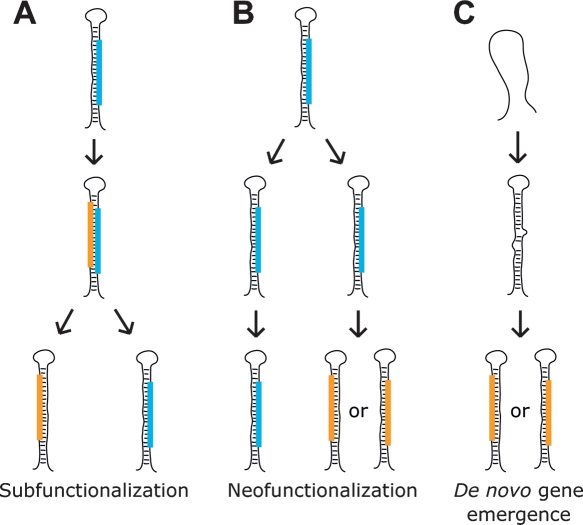
High-throughput sequencing of miRNAs in D. melanogaster provided insight into the origins of novel miRNA genes and how their origins might differ from those of protein-coding genes. Generally, the first step in the emergence of a new gene is the duplication of all or part of an ancestral gene (Ohno 1970). A redundant copy of a gene eventually faces one of three fates: the accumulation of mutations that render the copy functionless (nonfunctionalization), the accumulation of mutations that confer a novel and independently selectable function (neofunctionalization), or, in cases in which the ancestral gene had multiple functions, the accumulation of complementary degenerative mutations in both gene copies that specialize each to perform one of the parental functions (subfunctionalization) (Force et al. 1999). Protein-coding genes provide some examples consistent with subfunctionalization and others consistent with neofunctionalization. We observed examples of miRNA genes that were consistent with each of these models and also examples that appeared to be the products of de novo emergence.
The process of subfunctionalization first requires that an ancestral gene acquire multiple functions. Mechanisms that could impart multiple functions on a miRNA locus include imprecise processing that generates alternative miRNA 5′ termini, like that observed for mir-210, and transcription from both orientations with subsequent processing of both pri-miRNAs, as observed for mir-iab-4. But perhaps the most available mechanism for acquiring new functions is bringing the miRNA* into service. MicroRNA* species are initially generated at an obligate 1:1 stoichiometric ratio compared to mature miRNAs and to varying degrees are incorporated into RISC just like their complementary counterparts, albeit at a generally lower frequency. They thereby represent an easily accessible substrate for the evolution of novel functionality (Lai et al. 2004) (Fig. 8A). Examples of genes in which conservation data, read abundance, or experimental data suggested that both strands could be functional included mir-10, mir-iab-4, and mir-313. The miR-313* seed was only conserved within the melanogaster complex, which diverged from the yakuba and erecta complexes of the melanogaster subgroup 6–15 million yr ago (Lachaise et al. 1988). Although the melanogaster complex species were insufficiently diverged to conclude selective maintenance of the seed, the high abundance of the miRNA* implied the capacity to affect the expressions of target messages.
Figure 8.
Three models for the genesis of miRNA genes. (Blue bars) Ancestral miRNAs; (orange bars) novel miRNAs. (A) An example of subfunctionalization: a miRNA* acquires function; following gene duplication, one daughter copy maintains the function of the original miRNA, while the other maintains the function of the former miRNA*. Another example of subfunctionalization begins with heterologous 5′ processing. (B) Neofunctionalization: a miRNA gene duplicates; one daughter copy maintains the function of the original miRNA, while the other accumulates mutations that confer novel functionality to either the former miRNA or miRNA*. (C) De novo gene emergence: an unselected portion of a pre-existing transcript, such as an intron or part of a pri-miRNA, acquires the capacity to fold into a hairpin that can be processed into a mature miRNA. That product is selectively maintained because of the fortuitous benefit of gene silencing guided by its seed.
If a locus with multiple miRNA products, such as one of those listed above, were to duplicate, selective pressure would diminish for each daughter copy to continue producing all miRNA species, and eventually each daughter copy might retain the ability to produce just one of the functional miRNA products. This process might be in progress for the vertebrate mir-10 paralogs; miR-10-3p is maintained in all of the mir-10a, but not the mir-10b, hairpins (Fig. 3).
Neofunctionalization requires that gene copies find novel selectable functions after duplication and prior to loss of expressional competence. Because 5′ heterogeneity was very rare among the known miRNAs, the divergent processing of both the mir-2 paralogs and the tandemly duplicated mir-281 paralogs likely emerged after duplication and thus represent attractive candidates for neofunctional origins. In the case of mir-281, divergent processing not only shifted the seed of the ancestral miRNA, thereby potentially altering its target specificity, but also changed the miRNA:miRNA* pairing asymmetry, which significantly enhanced expression of the presumptive ancestral miRNA* species (Supplemental Text). We speculate that future drift of the two loci, with one increasingly specialized to produce a mature miRNA from the former star strand, would result in two genes with common ancestry yet no recognizable sequence identity.
Many pairs of apparently unrelated modern miRNA hairpins might have arisen from common ancestors through the processes of subfunctionalization and neofunctionalization. However, common descent is not always easy to identify, and the two mechanisms can be difficult to distinguish from each other even when descent from a common ancestor appears evident. For example, the mir-4, the three mir-9, and the mir-79 loci appeared to have all derived from a common ancestor, as did the mir-5 and the three mir-6 loci (Lai et al. 2004). However, in each of these cases, the structure of the gene family tree was ambiguous.
Novel protein-coding genes derive from duplication and divergence of an ancestral gene, and the active sites of their products generally evolve within the context of the ancestral tertiary structures. For protein-coding genes, the requirements of transcription, translation, protein folding, and protein function impose a myriad of informational constraints, making the completely de novo evolution of novel protein-coding genes highly improbable and therefore exceedingly rare. Canonical miRNAs, in contrast, have much more limited requirements. They must be transcribed, and the subsequent transcript must be capable of folding into a secondary structure that is competent for Drosha/Dicer processing (Han et al. 2006). The secondary structure requirements imposed on miRNA hairpin precursors are not excessively stringent, with a wide variety of bulge distributions, hairpin lengths, and loop sizes tolerated among the miRNAs of any given organism. The minimal informational requirements for miRNA-target interactions make it likely that any expressed miRNA will have a physiological consequence, enabling a young miRNA to find a selectively advantageous physiological role. Perhaps the most difficult obstacle for the emergence of new functional miRNA genes would be the plethora of coexpressed messages with fortuitous sites in their 3′ UTRs whose expression would be dampened as the expression of the emergent miRNA became consequential.
The genomic contexts of many miRNAs, including many of the youngest (most narrowly conserved) miRNAs described here, suggested that the pliancy of the miRNA processing machinery facilitates the emergence of new miRNA genes. Most derived from introns or miRNA clusters. In either of those contexts, a miRNA gene can emerge from otherwise unconstrained portions of pre-existing transcripts with little or no effect on the other products of those transcripts, thereby circumventing the otherwise required de novo acquisition of transcriptional competence. The varying extents of conservation observed within the mir-972∼979 cluster, which was preferentially expressed in the imaginal discs, reflected a variety of ages for the miRNA genes of that transcript (Fig. 5B). The oldest hairpins, mir-974/976/977, spanned the Drosophila genus, indicating that they are >30 million yr old (Beverley and Wilson 1984). In contrast, the other hairpins of the same cluster appeared to have emerged after the D. melanogaster/simulans split, indicating that they are <2.5 million yr old (Lachaise et al. 1988). The presence of hairpins with intermediate scopes of conservation, limited to the melanogaster species group (mir-975/978) or complex (mir-972/973), implied a model of functional miRNA genes emerging and presumably disappearing with some temporal regularity from the context of this transcript.
Two other miRNA genes that appeared to have emerged after the D. melanogaster/simulans split deserved special mention. The first, mir-984, expressed a miRNA whose 6-nt seed matched that of the let-7 miRNA and thus could repress many of the same target mRNAs (Lewis et al. 2005). Despite their seed identity, mir-984 and let-7 shared little sequence identity and had clearly distinct expression profiles (Fig. 6A; Supplemental Table S2), suggesting that mir-984 emerged de novo rather than as a paralog of let-7. The second gene was mir-954, notable for being the first miRNA gene to be identified on the dot chromosome of D. melanogaster, Chromosome 4 (Fig. 5A). Portions of the euchromatic chromosome 4 exhibit some heterochromatin-like properties such as variegated expression of inserted reporter constructs, and two such sites of variegated expression flank the mir-954 locus (Riddle and Elgin 2006).
The scope of miRNA genes and targets in flies
Our current tally of confidently identified miRNA genes in D. melanogaster stands at 148. These include 74 of the previously annotated genes, 59 novel genes reported in this study, and another 15 novel genes (mir-1003∼1017) whose transcripts bypass Drosha processing (Ruby et al. 2007). Forty-five of our top 47 computational predictions and 75 of our 100 predictions were either previously known or newly validated miRNAs (Fig. 1; Supplemental Table S1). Independent predictions from a parallel effort had even greater specificity (Stark 2007), which might be attributed to the use of different training sets; the set used here was smaller and included miRNAs annotated in miRBase but whose authenticity is now in doubt (miR-280, and miR-289), as well as other miRNAs whose 5′ termini appear to have been incorrectly annotated (miR-2a-2, miR-33, miR-274, miR-284, and miR-303). The high specificity of both approaches implied that very few highly conserved miRNAs remain to be discovered in flies. However, most of the miRNAs identified by our sequencing were missed by both prediction methods because these miRNAs were insufficiently conserved. Because the less broadly conserved fly miRNAs tended to be expressed at lower levels, it was impossible to use the cloning results to estimate a lower limit on the overall specificity of the computational gene predictions and thereby derive a meaningful upper limit on the number of miRNAs remaining to be identified in flies. Reliable upper limits on miRNA gene numbers face similar constraints in mammals, worms, and plants (Bartel 2004; Rajagopalan et al. 2006; Ruby et al. 2006).
The implication that there are many more miRNAs to be discovered in flies but almost none of them will be widely conserved, relates to the observed correlation between miRNA conservation and breadth of expression (Fig. 6D), which was likely understated here. All of the libraries from which small RNAs were sampled, with the exception of the S2 library, comprised a conglomerate of cell types, and many of the librariessurveyed thick slices of developmental time. The stronger direct correlation between conservation and total magnitude of expression that was observed here and in other systems may imply that the scarce miRNAs were actually expressed in even narrower contexts that contributed only a small fraction to their encompassing libraries. Thus, most of the remaining undiscovered miRNAs will inhabit niches of increasingly restricted physiological and evolutionary scopes.
Following that conclusion, another observation becomes relevant: given a consistent scope of conservation, the number of predicted targets decreased with more narrow breadth of miRNA expression (Fig. 7F). The regulatory reach of miRNAs, as indicated by the abundance of genes with conserved miRNA target sites, is likely quite vast. However, the as-yet-undiscovered miRNAs appear to have remained hidden thanks to the narrow scopes of their expression. Consequentially, the set of consequential miRNA targets will likely grow at a diminishing rate relative to the catalog of fly miRNAs, and our overall picture of the biological reach of miRNAs will likely not change substantially. This being said, the biology of some of the as-yet-undiscovered miRNAs is still likely to be quite interesting. As illustrated by lsy-6 in C. elegans (Johnston and Hobert 2003), a single miRNA expressed in only a few cells and acting on a limited set of targets can make quite a difference to the animal.
Methods
MicroRNA gene prediction
MicroRNA gene prediction is described in the Supplemental Text.
Library construction and sequencing
Total RNA was extracted from Canton S D. melanogaster and from S2 cells using TRIzol. Embryos were collected using a population cage whose food had been changed regularly to minimize egg withholding. Staged collections of 0–1 h, 2–6 h, 6–10 h, and 12–24 h embryos were obtained by culturing at 25°C. First-instar larvae were obtained by aging a 0–12-h embryo collection on a plate for 24 h. Wandering third-instar larvae were collected from vial cultures and rinsed several times in PBS to remove excess food. Total imaginal discs, brains, and salivary glands were isolated from wandering instar larvae to make a pooled “disc” preparation. Separate collections of 0–1 d, 1–2 d, and 2–4 d pupae were prepared and pooled to make a pupal library. Equal numbers of 1- to 5-d-old adult female and male flies were frozen at −80°C, vortexed, and sieved onto dry ice blocks to obtain adult head and body fractions. S2 cells were grown in Schneider’s medium and rinsed several times in PBS prior to extraction. A cDNA library was generated from each RNA sample as described (Lau et al. 2001) and was prepared for high-throughput pyrophosphate sequencing (Margulies et al. 2005) as described for run 4 of Ruby et al. (2006). Each library underwent a single sequencing run except for the 2–6-h embryo library, which underwent two sequencing runs. A total of 2,514,465 reads were generated. The processing of sequencing data is described in the Supplemental Text.
Expression analysis
Expression analysis is described in the Supplemental Text.
Target prediction and analysis
For each miRNA, we defined two 7-mer motifs that corresponded to the two types of 7-mers matching the seed region (the Watson-Crick match to miRNA nucleotides 2–8 and the Watson-Crick match to miRNA nucleotides 2–7 followed by an A). All occurrences of the two motifs were identified within annotated D. melanogaster 3′ UTRs from FlyBase Release 4.3 (Crosby et al. 2007), and the conservation of each of these sites was assessed using whole-genome alignments of D. melanogaster and 11 additional Drosophila species (Schwartz and Pachter 2007). To allow for alignment errors or gaps, sites were scored as conserved if they fell within 50 nt of the aligned site in each informant species. For each site, evolutionary conservation was evaluated as the total branch length corresponding to its species distribution (BLS) as described (Kheradpour et al. 2007). A site was considered conserved if its BLS met the specified cutoff. To prevent double-counting of 8-mer sites that contained both of the two 7-mers, target-prediction results reported nonoverlapping sites, obtained by first removing sites that did not meet the specified conservation cutoff and then removing overlapping sites, such that the maximum possible number of nonoverlapping sites was retained.
To estimate the conservation expected by chance, we repeated the target-prediction analyses using control motifs and compared the conservation frequencies of their sites with the conservation frequencies of sites obtained for the authentic miRNA. For each miRNA, nine controls were generated for each of the two motifs. For the motif matching miRNA nucleotides 2–8, each control had equal nucleotide composition and a similar number of matches in D. melanogaster 3′ UTRs (deviation <15%) as the authentic motif. The last six nucleotides of these controls were each extended by an A to obtain the nine controls for the other motif. Signal-to-noise ratios were calculated for each individual miRNA by dividing the frequency of conservation for the authentic sites by the average frequency of the control sites. Signal-to-noise ratios for all miRNAs combined were calculated in the same manner, aggregating all sites for all miRNAs under consideration and their controls. In each case, the number of conserved sites expected by chance was determined by multiplying the total number of sites by the control conservation frequency. Confidence was defined as the number of conserved sites above those expected by chance (i.e., above noise) divided by the total number of conserved sites. Confidence reflected the likelihood of a single conserved site being under selection.
For analysis of expression breadth versus number of predicted targets, miRNAs whose conservation did not extend beyond the Sophophora subgenus were not considered. A set of narrowly expressed miRNAs was defined as those with >70% of their library-normalized reads deriving from a single library, and a set of broadly expressed miRNAs as those with no more than 25% of their library-normalized reads deriving from a single library. Each set was collapsed into families based on their 6-nt seeds, resulting in 17 narrowly expressed families and 19 broadly expressed families. The number of predicted targets was determined for each family in each set by requiring targets to be conserved across 70% of the available branch length. In cases in which the number of predicted target genes differed among family members because of differences at microRNA nucleotide 8 (which changes one of the two 7-mer sites), the largest number of predicted target genes for the family was used.
Acknowledgments
We thank Ann Hammonds, Michael Axtell, and Gerald Rubin for assistance and support during library construction; Joseph Rodriguez, Robin Ge, Katherine Gurdziel, George Bell, and Fran Lewitter for constructing the TargetScanFly database of predicted targets (http://targetscan.org); and Ramya Rajagopalan for comments on the manuscript. This work was supported by a grant from the NIH (to D.P.B.). D.P.B is an HHMI Investigator. E.C.L. was supported by grants from the Burroughs Wellcome Foundation, the Leukemia and Lymphoma Society, and the V Foundation for Cancer Research. A.S. was supported in part by the Schering AG/Ernst Schering Foundation and in part by the Human Frontier Science Program Organization (HFSPO).
Footnotes
References
- Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V., Bartel D.P., Horvitz H.R., Ambros V., Horvitz H.R., Ambros V., Ambros V. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D., Lee R.C., Lavanway A., Williams P.T., Jewell D., Lavanway A., Williams P.T., Jewell D., Williams P.T., Jewell D., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Snyder B., Gaasterland T., Meyer J., Tuschl T., Gaasterland T., Meyer J., Tuschl T., Meyer J., Tuschl T., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuk E., Suavet F., Doglio A., Bordonne R., Bertrand E., Suavet F., Doglio A., Bordonne R., Bertrand E., Doglio A., Bordonne R., Bertrand E., Bordonne R., Bertrand E., Bertrand E. Human let-7 stem–loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–6597. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev V., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., Guryev V., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., Wienholds E., Plasterk R.H., Cuppen E., Plasterk R.H., Cuppen E., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Bontrop R., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Beverley S.M., Wilson A.C., Wilson A.C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J. Mol. Evol. 1984;21:1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Principles of microRNA–target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby M.A., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Strelets V.B., Zhang P., Gelbart W.M., Zhang P., Gelbart W.M., Gelbart W.M. FlyBase: Genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian A., Sakonju S., Zaratzian A., Sakonju S., Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 1990;87:3259–3263. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007 doi: 10.1038/nature06341. (in press) [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D., Spellman P.T., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S., John B., Gaul U., Tuschl T., Sander C., Marks D.S., Gaul U., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Law T.F., Grant S.R., Dangl J.L., Grant S.R., Dangl J.L., Dangl J.L., et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of miRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Amores A., Yan Y.L., Postlethwait J., Yan Y.L., Postlethwait J., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y., Aach J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Aach J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Church G.M., Ruvkun G., Kim J., Ruvkun G., Kim J., Kim J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Langenberger D., Gunsalus K.C., Rajewsky N., Gunsalus K.C., Rajewsky N., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N., Cho Y., Zhang B.T., Kim V.N., Zhang B.T., Kim V.N., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P., Bonhoeffer L.S., Tacker M., Schuster P., Tacker M., Schuster P., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Johnston R.J., Hobert O., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P., Stark A., Roy S., Kellis M., Stark A., Roy S., Kellis M., Roy S., Kellis M., Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007 doi: 10.1101/gr.7090407. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D., Reynolds A., Jayasena S.D., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., da Piedade I., Gunsalus K.C., Stoffel M., Gunsalus K.C., Stoffel M., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lachaise D., Cariou M.L., David J.R., Lemeunier F., Tsacas L., Ashburner M., Cariou M.L., David J.R., Lemeunier F., Tsacas L., Ashburner M., David J.R., Lemeunier F., Tsacas L., Ashburner M., Lemeunier F., Tsacas L., Ashburner M., Tsacas L., Ashburner M., Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 1988;22:159–225. [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Rauhut R., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tomancak P., Williams R.W., Rubin G.M., Tomancak P., Williams R.W., Rubin G.M., Williams R.W., Rubin G.M., Rubin G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C., Wiel C., Rubin G.M., Wiel C., Rubin G.M., Rubin G.M. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C., Tam B., Rubin G.M., Tam B., Rubin G.M., Rubin G.M. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes & Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V., Feinbaum R.L., Ambros V., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Yim J., Lee J., Provost P., Radmark O., Kim S., Lee J., Provost P., Radmark O., Kim S., Provost P., Radmark O., Kim S., Radmark O., Kim S., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X., Yang Z., Yu B., Liu J., Chen X., Yu B., Liu J., Chen X., Liu J., Chen X., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Yekta S., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Vertebrate microRNA genes. Science. 2003a;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Rhoades M.W., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003b;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., Muthu Valliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Kulkarni K., Souret F.F., Muthu Valliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Souret F.F., Muthu Valliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Muthu Valliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Jacobsen S.E., Wang W., Green P.J., Wang W., Green P.J., Green P.J., et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Yekta S., Lim L.P., Bartel D.P., Burge C.B., Yekta S., Lim L.P., Bartel D.P., Burge C.B., Lim L.P., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; Berlin, New York: 1970. [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P., Vaucheret H., Trejo J., Bartel D.P., Trejo J., Bartel D.P., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N.C., Elgin S.C., Elgin S.C. The dot chromosome of Drosophila: Insights into chromatin states and their change over evolutionary time. Chromosome Res. 2006;14:405–416. doi: 10.1007/s10577-006-1061-6. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Lee W., Nusbaum C., Ge H., Bartel D.P., Nusbaum C., Ge H., Bartel D.P., Ge H., Bartel D.P., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., Bartel D.P., Jan C.H., Bartel D.P., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A.S., Pachter L., Pachter L. Multiple alignment by sequence annealing. Bioinformatics. 2007;23:e24–e29. doi: 10.1093/bioinformatics/btl311. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Du T., Xu Z., Aronin N., Zamore P.D., Xu Z., Aronin N., Zamore P.D., Aronin N., Zamore P.D., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Moss E., Dmitrovsky E., Ambros V., Dmitrovsky E., Ambros V., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. http://genomebiology.com/2004/5/3/R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M., Brennecke J., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Brennecke J., Hodges E., Hannon G.J., Kellis M., Hodges E., Hannon G.J., Kellis M., Hannon G.J., Kellis M., Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007 doi: 10.1101/gr.6593807. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Rasmussen M.D., Roy S., Deoras A.N., Roy S., Deoras A.N., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007c doi: 10.1038/nature06340. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ach R.A., Curry B., Ach R.A., Curry B., Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G., Ha I., Ruvkun G., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]