Reliable prediction of regulator targets using 12 Drosophila genomes (original) (raw)
Abstract
Gene expression is regulated pre- and post-transcriptionally via _cis_-regulatory DNA and RNA motifs. Identification of individual functional instances of such motifs in genome sequences is a major goal for inferring regulatory networks yet has been hampered due to the motifs’ short lengths that lead to many chance matches and poor signal-to-noise ratios. In this paper, we develop a general methodology for the comparative identification of functional motif instances across many related species, using a phylogenetic framework that accounts for the evolutionary relationships between species, allows for motif movements, and is robust against missing data due to artifacts in sequencing, assembly, or alignment. We also provide a robust statistical framework for evaluating motif confidence, which enables us to translate evolutionary conservation into a confidence measure for each motif instance, correcting for varying motif length, composition, and background conservation of the target regions. We predict targets of fly transcription factors and miRNAs in alignments of 12 recently sequenced Drosophila species. When compared to extensive genome-wide experimental data, predicted targets are of high quality, matching and surpassing ChIP-chip microarrays and recovering miRNA targets with high sensitivity. The resulting regulatory network suggests significant redundancy between pre- and post-transcriptional regulation of gene expression.
Understanding gene expression and its regulation in response to developmental and environmental stimuli is one of the greatest challenges of modern biology. Regulatory control of gene expression occurs at many levels, both pre- and post-transcriptionally, generally based on short DNA and RNA signals known as regulatory motifs. These are recognized in a sequence-specific way by diverse protein and RNA regulators to direct transcription initiation, mRNA export, stability, and translation, ultimately leading to diverse gene-regulatory programs in organogenesis and development, and in response to environmental stimuli.
The sequence-based nature of regulatory control should in principle enable computational identification of regulator targets, by recognizing individual motif instances that constitute functional binding sites. However, due to their short lengths, motifs match very frequently to the genome or in fact any (random) nucleotide sequence by chance alone, and the majority of genome-wide motif occurrences do not lead to functional regulator binding, being either occluded by chromatin structure, separated from necessary cofactor motifs, or otherwise nonconsequential to transcriptional regulation (Wasserman and Sandelin 2004). To address the large signal-to-noise problem and predict functional regulatory elements, previous computational approaches have sought regions of motif clustering across several cooperating motifs, which are often associated with enhancer function (Berman et al. 2002; Markstein et al. 2004; Schroeder et al. 2004; Philippakis et al. 2006). Although these approaches have been successful in identifying novel enhancers, which are functional when tested in vivo, they only identify a small subset of all functional targets of each regulator and are only applicable when the specific combinations of factors are already known. In particular, they are unable to identify individual motif instances when these act in isolation or with diverse sets of cofactors.
Comparative genomics provides a general methodology for distinguishing functional regulatory motif instances, as biologically meaningful elements are typically under negative selection during evolution, with the type and extent of evolutionary conservation generally reflecting the specific requirements of the selected function (Ureta-Vidal et al. 2003; Miller et al. 2004). As closely related species often share substantial parts of their morphology and developmental programs, the expression of important genes, their regulatory connections, and the underlying regulatory elements are also likely conserved. In fact, some gene-regulatory network kernels involved in organogenesis, such as heart specification, are conserved in species as distant as flies and vertebrates (Davidson and Erwin 2006). Thus, although some processes are subject to more rapid divergence or positive selection (e.g., body color and pigmentation [Prud’homme et al. 2006]), this suggests that comparative genomics at a range of evolutionary distances should allow for the identification of many regulatory components of gene expression programs.
Indeed, previous comparative genomics studies have used the conservation of regulatory elements for the de novo discovery of regulatory motifs across related species (Cliften et al. 2003; Kellis et al. 2003; Chan et al. 2005; Ettwiller et al. 2005; Xie et al. 2005). These studies have relied on the average conservation of thousands of motif instances for each regulator, leading to a high genome-wide signal for motif discovery. However, it has remained unclear what fraction of conserved motif instances were functional and what fraction of functional instances were conserved, namely, whether in fact comparative genomics is applicable for high-specificity and high-sensitivity identification of individual motif instances. Moreover, the available genomes have been either too few for sufficient neutral divergence or too distantly related for motif instances to be conserved (e.g., Cooper et al. 2005; Ettwiller et al. 2005). Accurate motif instance identification would thus require many closely related species, which also present novel conceptual and methodological challenges, with respect to sequence coverage, alignment accuracy, and motif movement, gain, and loss (Boffelli et al. 2003; Margulies et al. 2003, 2007; Thomas et al. 2003; Cooper et al. 2005; Eddy 2005).
Methods such as phylogenetic footprinting, evolutionary rate profiling, and phylogenetic hidden Markov models (HMMs) have been successfully used to identify genomic regions under evolutionary selection (Wasserman et al. 2000; Margulies et al. 2003, 2007; Cooper et al. 2005; Siepel et al. 2005), but they cannot determine the regions’ functions that are selected for. Similar to more complex models of motif evolution (e.g., Moses et al. 2004; Zhou and Wong 2004), such methods are often restricted to regions that are well aligned and can be sensitive to motif movements or errors in sequencing, assembly, or alignment (Moses et al. 2004; Margulies et al. 2007). Further, methods to predict genomic regions with regulatory potential generally do not allow identification of regulatory targets for individual factors or miRNAs (Elnitski et al. 2003; Taylor et al. 2006). Lastly, the comparative prediction of miRNA binding sites in 3′ UTRs proved successful (for reviews, see Lai 2004; Rajewsky 2006) but has relied on site presence in defined sets of informant species, and a severe loss of sensitivity has been observed when the number of informant species was increased (Lewis et al. 2003; Grun et al. 2005; Stark et al. 2005).
In this paper, we develop a general methodology for identifying functional motif instances based on their evolutionary conservation across many related species and provide a robust statistical framework for evaluating motif confidence, enabling us to achieve both high sensitivity and high specificity. Our approach uses a phylogenetic framework, which allows for motif movements and local alignment inaccuracies and is robust against missing data due to artifacts in sequencing, assembly, or alignment. Our statistical framework enables us to translate evolutionary conservation into a confidence measure for each motif instance, correcting for varying motif length, composition, and background conservation of the target regions.
We apply our framework to whole-genome alignments of 12 recently sequenced Drosophila species (Drosophila 12 Genomes Consortium 2007; Stark et al. 2007) and predict targets of 83 transcription factors (TFs) and 78 miRNAs (57 distinct families), leading to 46,525 regulatory connections. We use genome-wide ChIP-chip experiments and direct tests of TF or miRNA targeting (independently published by us [Stark et al. 2005; Zeitlinger et al. 2007] and others [Abrams and Andrew 2005; Sandmann et al. 2006, 2007; Sethupathy et al. 2006]) to show that computationally predicted regulator targets are of very high quality, matching and surpassing ChIP-chip sensitivity and specificity, and can identify seemingly functional instances even when these are not bound in the conditions experimentally surveyed. Lastly, we study properties of the resulting network, which suggest significant redundancy between pre- and post-transcriptional regulation.
Assessing motif-instance conservation across many genomes
Unlike protein-coding and RNA genes, which are typically well aligned in the multiple sequence alignments of related species, many regulatory motifs are too short to guide alignment algorithms and thus may not appear at orthologous positions in multiple sequence alignments (Wray et al. 2003; Wasserman and Sandelin 2004). As motifs can act at a wide range of distances, individual motif instances may move, either by insertions and deletions, or by “birth” of new motifs and loss of old motifs via compensatory mutational changes (Ludwig et al. 2000). In addition, individual instances of regulatory motifs may actually diverge across different species, and may experience duplication, gain, and loss across the evolutionary tree (Ludwig et al. 2005; Prud’homme et al. 2006; McGregor et al. 2007). Lastly, comparison of many species introduces artifacts due to sequencing, assembly, and alignment, which may affect the alignment of equivalent regulatory motif instances (see Supplemental Fig. S1; Margulies et al. 2007).
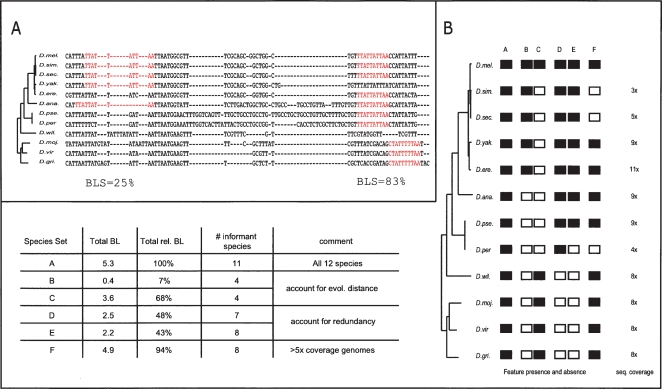
To account for these unique evolutionary and alignment properties of regulatory motifs, we developed a phylogenetic framework for motif instance identification which tolerates motif movement and loss, while recognizing their clear selective pressure across the phylogenetic tree. Briefly, we search for motif instances in each of the aligned genomes and, given the set of species that contain motif instances within tolerable distances of the D. melanogaster instance, we evaluate the total evolutionary branch length over which the motif appears conserved. The overall score of a motif instance becomes this total branch length of the phylogenetic tree over which the motif is conserved, which we call the Branch Length Score, or BLS (Fig. 1). We thus implicitly assume that all motif instances in D. melanogaster are potentially ancestral and count instances in the informant species as evidence when they are conserved. We do not interpret presence/absence patterns of motif instances as evolutionary gain- and loss events, as they could arise from artifacts in sequencing or alignment. The BLS value of a given motif instance ranges from BLS = 0.0 (nonconserved) to BLS = 1.0 (fully conserved), representing the fraction of the total phylogenetic tree covered by the species containing the motif.
Figure 1.
BLS measure (Branch Length Score) for assessing motif conservation in many genomes. (A) Conservation level and corresponding BLS scores for two Mef-2 motif instances. The BLS measure scores the total branch length of the subtree connecting the species with motif instances, as a fraction of the total branch length of all twelve species. As shown in these examples (Mef-2 motif: YTAWWWWTAR), BLS accounts for local alignment inaccuracies, gaps, motif movement, and motif loss. Species abbreviations as follows: Drosophila melanogaster (D. mel.), D. simulans (D. sim.), D. sechellia (D. sec.), D. yakuba (D. yak.), D. erecta (D. ere.), D. ananassae (D. ana.), D. pseudoobscura (D. pse.), D. persimilis (D. per.), D. willistonii (D. will.), D. mojavensis (D. moj.), D. virilis (D. vir.), and D. grimshawii (D. gri.). (B) BLS scores for different instance conservation scenarios. Given the pattern of presence (black) and absence (white) within a phylogenetic tree, BLS evaluates the total branch length of the subtree connecting the species that contain the motif: When all species are present, BLS is 100% (column A); different sets of species lead to different BLS scores based on their evolutionary distances: distantly related species lead to higher scores as they span larger evolutionary distances (columns B,C); species that are very closely related to each other lead to only small incremental contributions, due to their phylogenetic redundancy (columns D,E); sequencing, assembly, and alignment artifacts are not penalized, such as those stemming from lower-coverage genomes, as redundancy of branches between close species complements BLS (column F). Information about sequence coverage is from Drosophila 12 Genomes Consortium (2007) and Richards et al. (2005).
This BLS conservation measure has many attractive properties, which enable us to define the conservation level of motif instances across a complete genome, to select conservation thresholds for defining all genome-wide instances of a regulatory motif, and to assign confidence values to the observed conservation, as we describe below. Moreover, because missing instances in the aligned species are not interpreted as evolutionary loss events and are not explicitly penalized, the BLS measure is robust against missing sequence due to low-coverage sequencing, assembly errors, or alignment artifacts. Lastly, BLS provides a direct estimate of the expected neutral divergence of the species compared (Felsenstein 2004), accounting for different divergence times between species and correcting for redundant contributions of individual species in a complex tree and their different rates of divergence (Fig. 1).
Establishing confidence levels for BLS conservation scores
To translate this BLS conservation score to a robust statistic that can be used across different motifs and different types of genomic regions (e.g., promoters, introns, 5′ or 3′ UTRs, etc.), we mapped each BLS score to a confidence value between 0% and 100%, representing the probability that a given motif instance is functional. This probability reflects the increased conservation of motif instances compared to overall sequence similarity and is estimated using control motifs, similar to the signal-to-noise ratio for miRNA target predictions (Lewis et al. 2003). Evaluated in a motif- and region-specific way, it corrects for differences in motif length and composition and for different average conservation levels and nucleotide composition of different genomic regions. Intuitively, longer and highly specific motifs are very unlikely to be conserved by chance and thus result in high confidence levels, even for modest BLS thresholds. Further, regions of overall high conservation (such as protein-coding exons) are likely to contain many conserved motif instances by chance alone and thus require more stringent BLS thresholds to achieve a desired confidence level. Lastly, AT-rich motifs are likely to have many conserved occurrences in AT-rich regions due to chance alone (and GC-rich motifs in GC-rich regions), and thus require higher BLS thresholds if the corresponding control motifs show similarly high conservation.
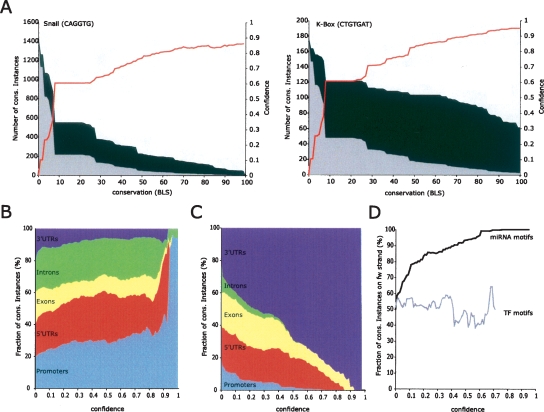
We found that the number of random motif instances generally decreased rapidly for increasing BLS values, while the number of instances for known motifs remained high (Fig. 2A). For example, at BLS ≥ 0.50, the motif for Snail (CAGGTG), has 229 occurrences in promoter regions, compared to 54 motif instances on average for a pool of 10 control motifs. Therefore, we would expect that of these 229 Snail instances, 54 are likely due to chance while 175 of them (76%) are nonrandom, leading to a confidence of _C_0.5 = 76% for each of these motif instances, at BLS ≥ 0.5. At a more stringent conservation threshold of BLS ≥ 0.70, Snail shows 152 instances while the control motifs show 24 instances on average, leading to a confidence of _C_0.7 = 128/152 = 84%. Similarly, the miRNA K-Box motif (Lai et al. 1998) (CTGTGAT; 5′ seed motif of Drosophila miRNAs 2, 6, 11, 13, and 308) reaches confidence values >75% at BLS = 0.4 and >90% at BLS = 0.76 (Fig. 2A). We note that the confidence measure is conservative by nature: 76% confidence for example means that 76% of conserved instances are conserved above background and are thus likely functional. The remaining 24% of conserved instances might contain functional instances that cannot be discerned from noise, suggesting a maximum false-positive rate of up to 24%.
Figure 2.
High-confidence recovery of individual motif instances. (A) Mapping BLS scores to confidence values. Recovery of conserved motif instances the transcriptional repressor Snail (CAGGTG) in promoter regions (2-kb regions upstream of transcription start sites), and the K-box miRNA (CTGTGAT) in 3′ UTRs, at different BLS cutoffs (X-axis). Instances of shuffled control motifs (gray area) decrease much more rapidly than instances of real motifs (height of black curve), leading to a large fraction of motif instances conserved above background (black area). The motif-confidence score (red line) is calculated as the fraction of conserved instances above background. Random motifs are selected to have equal frequency as real motifs at BLS = 0. (B,C) Increasing confidence values select functional motif instances. With increasing confidence cutoffs (_X_-axis), transcription factor (TF) motif instances fall increasingly in promoter regions (light blue), 5′ UTRs (red), and introns (green), at the exclusion of 3′ UTRs (dark blue) and coding regions (yellow). In contrast, miRNA motif instances fall increasingly into 3′ UTRs to the exclusion of promoters and other regions. Relative size of regions is normalized at BLS = 0. (D) miRNA motif instances at increasing confidence cutoffs are increasingly on the transcribed strand of 3′ UTRs (black curve), while no such trend is seen for TF motifs (gray). Curves are truncated when <10 instances reach the respective confidence.
We found that with increasing confidence levels motifs were predominantly found in regions in which they are known to function. For example, with increasing confidence, the normalized fraction of TF motif instances within promoter regions rises from 20% to 90%, and that of miRNA motif instances within 3′ UTRs from 20% to 100% (Fig. 2B,C). In addition, the percentage of miRNA motif instances on the transcribed strand of 3′ UTRs rises from essentially random (uniform 50%) to exclusively on the transcribed strand (100%), while promoter motifs do not show any strand preference (Fig. 2D). These results illustrate the effectiveness of region-specific confidence values (which require more stringent BLS thresholds for more highly conserved regions), as high-confidence motif instances were not simply biased toward regions with overall high conservation but specifically selected in regions they are known to act.
Effect of allowing motif movements on instance identification
Using confidence cutoffs also allowed us to assess the influence of tolerating motif movements on the recovery of functional motif instances. Allowing for motif movement permits capturing functionally equivalent instances across genomes, independent of their relative positions in the alignment. However, while this approach will always increase the number of conserved instances recovered for real motifs, it also increases the number of spurious motif instances that appear conserved due to increased background conservation for large tolerated movements.
The number of motif instances recovered at a given confidence value presents a robust measure of overall discovery power, as it evaluates sensitivity at a fixed specificity. If the window of tolerated motif movement is too small, many true motif instances will be missed. Conversely, if the window of tolerated motif movement is too large, we would expect both real and control motifs to show increased conservation, thus reducing the confidence and leading to fewer confidently identified instances. Between these two extremes, we would expect the number of high-confidence motif instances to peak for an optimal window of tolerated motif movement, and decrease for lower or higher values.
Indeed, we found that allowing for motif movements of 10–500 nucleotides relative to the D. melanogaster instance often increased the number of confident motif instances, while allowing for large movements generally decreased this effect (Fig. 3A). Different window sizes were optimal for different motifs: Longer TF motifs with higher information content peaked for longer windows (Pearson correlation 0.40 for information content, 0.33 for length), while motifs with many matches in D. melanogaster showed shorter optimal windows (correlation −0.27 for TF, −0.26 for miRNA motifs). We also found a correlation with GC content for miRNA motifs (0.28), as expected since 3′UTRs are AT-rich, but there was little correlation for TF motifs (−0.15). These results illustrate the more rapidly increasing noise levels for motifs with low information content, while motifs with higher information content are less likely to appear by chance within the length of the tolerated window.
Figure 3.
Discovery power for motif instance prediction. (A) Effect of tolerated motif movement. Number of recovered motif instances at 60% confidence for TF and miRNA motifs. (Left panel) For both TF motifs (gray: bicoid motif, VVVBTAATCC) and miRNA motifs (black: miR-iab-4 motif, GTATACG), instance recovery increases until an optimal window size (500 and 400 nucleotides, respectively) and then decreases for larger movements, suggesting that tolerating motif movements increases overall discovery power. (Right panel) Performance across all TF motifs (black) and all miRNA motifs (gray) shows improved recovery until windows of 300–500 nucleotides (for 60%–80% of motifs) but reduced performance for larger window sizes. Performance for individual examples (left panel) shows a sharper peak than the overall performance across all motifs (right panel), as different window sizes are optimal for different motifs. (B) BLS measure leads to increased sensitivity. Number of motif instances recovered (_Y_-axis) at each confidence value (_X_-axis) for transcription factor (TF) motifs (left panel) and miRNA motifs (right panel). The BLS measure applied to the 12 fly genomes (blue) recovers more motif instances at each confidence, as compared to approaches requiring motif presence in all compared species (“full” conservation), applied to the five melanogaster species (red), the pairwise comparison of D. melanogaster and D. pseudoobscura (yellow), or the nine Sophophora species (green). (C) Additional species lead to increased specificity. Two measures of discovery power for the BLS measure applied to the five melanogaster group species (green), a pairwise comparison of D. melanogaster and D. pseudoobscura (gray), the nine Sophophora species (black), and all 12 Drosophila species (red). (Left panel) More TF and miRNA motifs reach 60% confidence for increasing number of genomes at larger evolutionary distances. (Right panel) Increasing numbers of genomes at larger evolutionary distances also lead to increased signal-to-noise ratio, measured as the conservation level of real motifs vs. control motifs at the most stringent BLS cutoff.
Overall, the single best window improved the recovery of 56% of TF motifs (20 nucleotides), and of 71% of miRNA motifs (50 nucleotides; both at 60% confidence). For 71% TF motifs, some window between 10 and 500 nucleotides improved sensitivity, and improvement was substantial for 11% (at 60% confidence; P ≤ 0.05 after Bonferroni correction to account for testing multiple windows). Similarly, 93% of miRNA motifs showed improved sensitivity, which was substantial for 13%. Improvements were observed over a wide range of confidence cutoffs, showing that tolerating motif movement is important at any desired confidence level for motif instance identification. These results confirm our intuition that indeed, many motif instances are offset considerably in the 12-species alignments, whether due to alignment artifacts or evolutionary plasticity of regulatory motifs.
BLS measure enables increased sensitivity
The confidence measure also enabled us to gauge the sensitivity of the BLS measure, measured as the number of instances recovered at a fixed specificity, compared to different methodological choices. In particular, we asked whether requiring perfect conservation across fewer species (the nine Sophophora subgroup species, the four melanogaster subgroup species, and D. pseudoobscura as the only informant) would lead to higher sensitivity/specificity levels, perhaps due to many lineage-specific motifs.
We found that the BLS measure across all 12 species recovered most instances for all TF and miRNA motifs, at all confidence levels (Fig. 3B). For TF motifs, our approach recovers more than 1.4-fold more instances than the second most sensitive of the other approaches at 60%, 1.5-fold more at 70%, and threefold more at 80% confidence (for miRNAs motifs, 1.8-fold more at 60%, twofold more at 70%, and 1.8-fold more at 80% confidence). When comparing the three other approaches for confidence thresholds <65%, we found that perfect conservation was indeed more sensitive across the four closely related species in the melanogaster subgroup and in D. pseudoobscura compared to perfect conservation across nine Sophophora species. However, very few motifs reached higher confidence levels, due to the high overall sequence similarity between these species, resulting in an apparent drop in motif recovery. We also found that the discovery power in D. pseudoobscura was comparable to the four melanogaster species, likely due to its position in the phylogenetic tree.
Lastly, the BLS and confidence measures allow us to gauge the effect of additional species. We found that evaluating motif conservation across all 12 species allowed more motifs to reach confidence levels of 60% than was possible with the other species combination and led to higher average signal-to-noise ratios than any other species combination for TFs and miRNAs (Fig. 3C).
These results show that the discovery power for target gene identification continues to increase even with more distantly related species. The usefulness of distant species only becomes effective by the use of the BLS measure, while inclusion of distantly related species resulted in lower performance when perfect conservation was required. Overall, the combination of additional species and a phylogenetic framework for evaluating motif conservation allowed high sensitivity and high specificity in motif-instance identification.
Conserved motif instances identify functional in vivo targets
We then compared our computationally determined conserved motif instances with experimentally determined in vivo targets of known regulators. To define in vivo targets, we used several large-scale experimental datasets: a set of high-confidence direct CrebA targets confirmed with a variety of reporter assays (Abrams and Andrew 2005), three genome-wide chromatin IP (ChIP) experiments for developmental TFs with known motifs (Snail, Mef-2, and Twist) (Sandmann et al. 2006, 2007; Zeitlinger et al. 2007), and a set of experimentally confirmed targets for different miRNAs (Stark et al. 2005; Sethupathy et al. 2006). We note that the experimentally validated miRNA sites were initially predicted based on conservation to D. pseudoobscura and thus are biased toward higher conservation (already showing BLS > 0.26). However, the CrebA and the three ChIP data sets were determined independently of any comparative information and thus provide an entirely independent evaluation of our methodology, allowing us to estimate both sensitivity and specificity of our predictions.
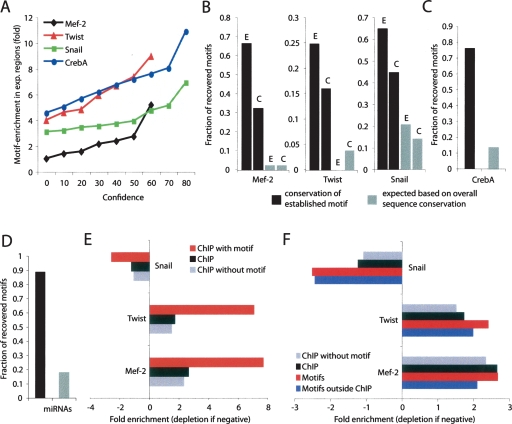
For each regulator, we compared motif instances at different confidence cutoffs with the experimentally derived in vivo targets. We found that motif instances at increasing confidence thresholds were strongly enriched for experimentally derived in vivo targets (Fig. 4A). In absence of any comparative information, Mef-2 motif instances in D. melanogaster showed no enrichment for experimentally derived targets, while conserved instances showed up to fivefold enrichment (at 60% confidence). Similarly, enrichment rose from threefold to sevenfold for Snail at increasing confidence levels, from fourfold to ninefold for Twist, and from 4.5-fold to 12-fold for CrebA (P = 4 × 10−11, 3 × 10−10, 2 × 10−6, and 1 × 10−7 at the highest confidence for the four factors). This illustrates the ability of evolutionary information to select for functional motif occurrences, experimentally shown to be bound and/or functional in vivo. In fact, the enrichment was most pronounced for CrebA (12-fold enrichment; P = 1.4 × 10−7), for which the targets had been shown to be direct transcriptional targets, while some of the ChIP-derived targets may reflect indirect binding or binding that is nonconsequential for transcription.
Figure 4.
Conserved motif instances identify functional in vivo targets. Functional in vivo targets were determined for Mef-2, Twist, and Snail using ChIP-chip (Sandmann et al. 2006, 2007; Zeitlinger et al. 2007), and direct transcriptional targets were determined for CrebA using various assays (Abrams and Andrew 2005). (A) Increasing confidence values show increased enrichment for in vivo sites. Fold enrichment in functional in vivo sites (_Y_-axis) for conserved motif instances at varying confidence values (_X_-axis). Hypergeometric _P_-values for max fold enrichments are 4 × 10−11 for Mef-2, 2 × 10−6 for Twist, 3 × 10−10 for Snail, and 1 × 10−7 for CrebA. Increasing confidence levels selected functional in vivo sites with increased enrichment for all four regulators, showing that high conservation selects for functional motif instances (X = 0% shows the enrichment in the absence of comparative information, i.e., without requiring conservation). Curves are truncated when motifs do not reach the respective confidence levels. (B_–_D) High-sensitivity recovery of in vivo targets for TF and miRNA regulators. Fraction of motifs in bound regions recovered at 60% confidence (black bars), compared to the fraction expected given the overall conservation of the respective regions, as assessed by control motifs using the same BLS cutoff (gray; suggesting preferential conservation of the corresponding TF motif instances). (B) Recovery of ChIP-bound motifs, across all ChIP-bound regions (lableled “C”), and only those instances overlapping known enhancers (labeled “E”). Recovery rates show high sensitivity for TF motif instances, especially when these overlap enhancer regions. (C) Recovery of experimentally validated direct CrebA targets shows even higher sensitivity, likely due to the multiple lines of experimental evidence establishing them as direct targets. (D) miRNA recovery at 80% confidence is very high. (E) Nonconserved ChIP sites show reduced functional enrichments. Enrichment in promoter regions of muscle genes for motif instances of activators Twist and Mef-2, and depletion for motif instances of repressor Snail are reduced for ChIP-bound regions for which motif instances are not conserved, suggesting they may contain a higher fraction of nonfunctional sites. The enrichment/depletion is even weaker for ChIP-bound regions without motif instances (all enrichments are significant with _P_-values between 1.1 × 10−4 and 5.1 × 10−13 except those for Snail). (F) Conservation-inferred targets and ChIP-inferred targets show comparable functional enrichments. Conservation-inferred motif targets at 60% confidence (red; all P < 10−4) show higher muscle-gene enrichment/depletion than ChIP-inferred targets (black). Even outside ChIP-bound regions, conserved motifs show comparable enrichment and depletion (blue; all P < 5 × 10−3).
We also found that even stringent confidence thresholds recovered a large fraction of motif instances in experimentally derived in vivo targets, illustrating the high sensitivity of our approach (Fig. 4B). When ChIP-bound motifs overlapped experimentally defined enhancer elements (Sandmann et al. 2006, 2007; Zeitlinger et al. 2007), 65% Mef-2, 65% Snail, and 25% Twist motif instances were recovered at our 60% confidence cutoff. The lower rate for Twist was possibly due to an overly specific Twist motif used (Markstein et al. 2004). Recovery was again highest for CrebA, for which 76% of motif instances were conserved, illustrating the high sensitivity of comparative genomics methods for validated direct targets (Fig. 4C).
Recovery was much lower when all ChIP-bound regions were considered, regardless of enhancer information, suggesting that some of the ChIP-derived targets may be due to noise and that conservation is able to pinpoint functional enhancers within ChIP-bound regions. Lastly, we recovered 90% of miRNA motif instances in experimentally confirmed targets at 80% confidence (Stark et al. 2005; Sethupathy et al. 2006) (Fig. 4D), showing that despite the added branch length (from BLS > 0.26 for D. pseudoobscura to BLS > 0.60 at 80% confidence across the 12 genomes on average), our methods maintain high sensitivity.
In contrast to evaluating conservation by the BLS methodology, requiring perfect conservation across all 12 Drosophila species or across the nine Sophophora species recovered significantly fewer experimentally validated motif instances for TF and miRNA motifs (see above and Supplementary Fig. S2).
Nonconserved binding events show decreased functional enrichment
Although the overlap between conservation derived motif instances and in vivo binding was highly significant and we recovered a substantial fraction of instances in ChIP-bound enhancers, CrebA targets, and miRNA targets, we noted that numerous motif instances in ChIP-bound regions were not conserved above 60% confidence, especially for regions that had not previously been shown to be enhancers (Fig. 4B). Nonconserved sites might be functional but missed due to unusually large motif movements or sequencing and alignment errors. Alternatively, they may play roles with only lineage-specific selection (and thus not meeting our 60% confidence threshold) or represent largely nonconsequential binding, without a specific biological role subject to evolutionary selection. To distinguish the two possibilities, we studied the enrichment of conserved and nonconserved motif instances of the mesodermal factors Mef-2, Twist, and Snail in muscle genes.
We found that ChIP-bound motif instances that were evolutionarily conserved showed enrichment or depletion in promoters of muscle genes for all three factors: The transcriptional activators Mef-2 and Twist showed eightfold and sevenfold enrichment, respectively, and Snail, a mesodermal repressor, showed threefold depletion in muscle genes. In contrast, ChIP-bound motif instances that were not conserved showed only one- to twofold enrichment or depletion for all three factors (Fig. 4E). This suggests that potential lineage-specific roles corresponding to nonconserved ChIP-bound sites may lie outside the regulators’ conserved functions in core development processes (e.g., mesoderm/muscle development). Alternatively, these sites may be of decreased biological significance, perhaps representing nonconsequential binding sites with no role in gene-expression regulation, which are known to be recovered in ChIP experiments (Boyer et al. 2005; Lee et al. 2006). In either case, our results show that nonconserved sites are not simply due to low sensitivity of comparative methods but are functionally distinct from conserved sites.
ChIP-derived and conservation-derived targets show comparable functional significance
Interestingly, evolutionary conservation identified many high-confidence motif instances outside ChIP-bound regions. These may be functional sites reflecting higher coverage for conservation-derived targets or spurious sites reflecting noise in the methodology. To distinguish the two possibilities, we used the correlation of these additional motif instances with muscle genes, providing an independent assessment of the overall quality of our predictions.
We found that conservation-derived targets outside ChIP regions were enriched in the same categories in which the factors are known to act. In fact, even outside ChIP regions, conserved sites showed comparable or higher enrichment or depletion in muscle genes than those identified by the ChIP methodology (Fig. 4F), suggesting they may be of similar overall quality. For Twist, enrichment was 1.3-fold higher; for Snail, depletion was 2.5-fold higher; and for Mef-2, enrichment was slightly lower (0.9-fold). Overall, when assessing ChIP- and conservation-derived targets independently (i.e., considering all ChIP targets and all conservation-derived targets), our approach showed a consistently higher enrichment or depletion in muscle genes than ChIP-chip (1.4-fold for Twist, twofold for Snail, and 1.01-fold for Mef-2; Fig. 4F).
Our results suggest that the additional sites outside ChIP-bound regions are likely functional and reflect the higher coverage of conservation-derived targets as compared to experimentally derived targets. Indeed, while ChIP-derived targets are constrained by the developmental stages or cell types surveyed, comparative approaches capture all conserved gene targets regardless of their spatial or temporal constraints. Moreover, comparative approaches are not constrained by the abundance of TFs at bound sites, but only by the strength of evolutionary selection; they can thus identify important sites even when these are bound more rarely (or in few cell types). Lastly, comparative genomics enables us to capture additional functional targets that may be missed due to experimental limitations of ChIP technology, for which reported false-negative rates are up to 30% (Boyer et al. 2005; Lee et al. 2006).
Regulatory network of D. melanogaster at 60% confidence
We conclude that comparative genomics provides a powerful methodology for identifying functional targets showing high sensitivity and high specificity. For factors with experimentally determined in vivo binding sites, we showed that evolutionary conservation provides comparable discover power as ChIP and importantly reveals additional functional sites that potentially function at stages or tissues not surveyed. More generally, even when ChIP studies are not available, comparative genomics can provide a first overview of the regulatory connections across a complete genome.
We used our comparative approach to present an initial regulatory network of D. melanogaster at 60% confidence for both pre- and post-transcriptional regulators (Fig. 5). Overall, 49 of 57 miRNA motifs (86%) and 67 of 83 TF motifs (81%) had instances with confidence values of 60% or higher and were considered (Supplemental Tables S1, S2). The remaining motifs may have too few physiologically relevant and conserved target sites to discern them reliably from background, or they may not accurately reflect the factors’ binding properties, potentially being overly specific or degenerate.
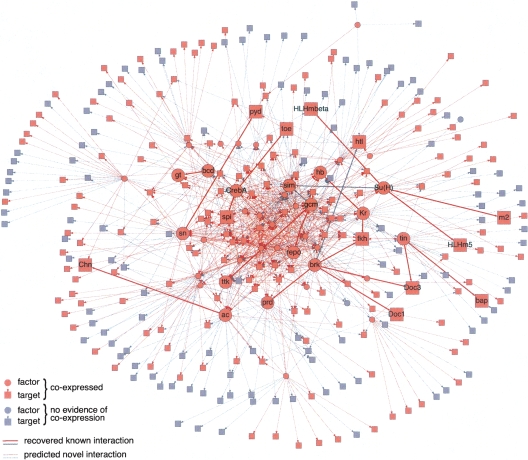
Figure 5.
An initial regulatory network in Drosophila. Regulatory network with 46,525 connections between 83 TF and 57 miRNA motifs (circles) and their target genes (squares) at 60% confidence. If the regulator and its target are co-expressed in at least one tissue according to ImaGO (Tomancak et al. 2002), the corresponding edges (lines) and nodes (circles or squares) are colored red; otherwise they are gray. The high fraction of red edges (46%, P = 2 × 10−3) highlights the quality of the network. Nodes with gene names and connected by bold edges indicate examples of regulatory connections with evidence in the literature (see Supplemental Table S4).
We find a total of 46,525 regulatory connections for TF motifs and 3662 for miRNA motifs, targeting 8287 genes and 2003 genes, respectively. The distribution of targets is highly asymmetric: While we find on average 123 targets per TF motif and 41 targets per miRNA motif, some TF motifs have up to 4129 targets (homeobox factors), and some miRNA motifs more than 150 targets (miR-4, miR-92, and miR-1). We note, that some motifs (e.g., the homeobox TF motif or the K-box miRNA motif) correspond to multiple TFs or miRNAs, and thus the numbers likely represent combined targets for all individual factors. The distribution of target sites per gene (indegree) is also highly imbalanced: While a typical gene is regulated by six different TF motifs and two different miRNA motifs on average, some genes are targeted by up to 33 different TF and up to 14 different miRNA motifs. Genes with high indegree were enriched in morphogenesis, organogenesis, neurogenesis, and a variety of tissues, while genes with small indegree were enriched in ubiquitously expressed or maternal genes with functions in DNA, RNA, or protein metabolism for both TF and miRNA motifs (Supplemental Table S3). Many genes with high indegree were TFs (P < 10−9 for TF and miRNA motifs), and transcriptional regulators were indeed more densely targeted than other genes, by both TF (10.1 vs. 5.5, P < 10−20) and miRNA motifs (2.3 vs. 1.8, P < 5 × 10−5). The similarity between the TF and miRNA motif network was further illustrated by mutual enrichment: Genes with high TF indegree are enriched in genes with high miRNA indegree (P = 8 × 10−5), as are genes with low indegree for both types of regulators (P = 2 × 10−7).
This initial network contained many connections with independent support in the literature (Fig. 5; Supplemental Table S4). For example, we identified the direct regulation of achaete by Hairy (Van Doren et al. 1994), several direct targets of Suppressor of Hairless Su(H) in the Enhancer of split E(spl) complex (Bailey and Posakony 1995), and direct regulation of the gap gene giant by Bicoid (Kraut and Levine 1991). In addition, the network proposed many novel connections supported by experimental evidence, including direct regulation of bagpipe by Tinman, which both cooperate in mesoderm induction and heart specification (Yin and Frasch 1998). More generally, when tissue-specific expression data was available, we found that on average 46% of all targets were co-expressed with their factor in at least one tissue (Fig. 5), which is significantly higher than expected by chance (P = 2 × 10−3).
Discussion
We showed that comparative analysis of many related genomes allows us to identify functional motif instances with very high confidence. Overall, 86% miRNA motifs and 81% TF motifs had instances with confidence values of ≥60. The remaining factors may have too few physiologically relevant and conserved target sites to discern them reliably from background, or may contain inaccuracies in their binding site motifs might be artificially specific or degenerate.
We found that the availability of many genomes allowed for very high signal-to-noise levels for many motifs at the most stringent settings. However, more importantly, we showed that the BLS measure allowed us to use the increased number of species to strongly increase sensitivity at any given specificity compared to requiring perfect motif conservation in arbitrary subsets of species. While requiring perfect conservation across many genomes is of limited use, the increased power enables approaches that account for artifacts in sequencing, assembly and alignment, and tolerate diverged, missing, or moved motif instances. Our BLS measure is more generally applicable to PWMs (Stormo 2000), to more complex models of regulatory motifs that account for dependencies between individual motif positions (Yada et al. 1998; Naughton et al. 2006), and to more advanced rules for miRNA-target recognition that for example score the contribution of the 3′pairing energy (Stark et al. 2003; Brennecke et al. 2005).
We found that comparative genomics and ChIP-chip showed similar power for functional target identification. The two approaches are complementary, each with unique advantages: Conservation helps pinpoint evolutionarily selected functional targets across all conditions, while ChIP-chip reveals stage- and tissue-specific binding in vivo, as well as species-specific sites which may play important evolutionary roles in the emergence of new functions. As motifs of additional regulators are derived by experimental (e.g., by SELEX, in Tuerk and Gold [1990] or protein-binding microarrays [Mukherjee et al. 2004]) or computational approaches (e.g., by motif-overrepresentation [Tompa et al. 2005] or genome-wide motif-instance conservation [Kellis et al. 2003; Xie et al. 2005]), and tissue-specific binding becomes available for dozens of factors (e.g., through the ENCODE and modENCODE projects), comparative studies can help establish and refine their genome-wide targets. Indeed, we found that motif instances identified by both approaches had the highest functional enrichments, suggesting that combined approaches may prove useful in the future. Although the regulatory network we present likely lacks many true regulatory relationships that could not be reliably recovered, our comparison with ChIP-chip data and other validated targets showed that the network is of high overall quality. We anticipate that the network and the predicted regulatory connections prove to be a useful resource for the fly community working on the biology of TFs or miRNAs and their target genes and their roles in development. The methodology to assess motif conservation across many genomes and predict functional motif instances with high sensitivity is more generally applicable for the study of any genome.
Methods
Regulatory motifs
We obtained TF motifs from Transfac (Matys et al. 2003), Jaspar (Sandelin et al. 2004), FlyReg (Bergman et al. 2005), and the literature. To remove redundancy for global statements about motif targets, we clustered TF motifs using centroid-linkage hierarchical clustering with a Pearson correlation coefficient cutoff of 0.8 (calculated on the columns of the equivalent PWM) at the best alignment offset (Pietrokovski 1996; Schones et al. 2005; Xie et al. 2005; Gupta et al. 2007). To avoid the creation of artificial motifs by averaging, we chose the original motif from each cluster that is closest to the cluster average as the cluster representative. We defined miRNA motifs as the nonredundant set of 7mers reverse complementary to miRNA 5′ end positions 2–8 (seeds after Lewis et al. 2003) for all Drosophila miRNAs in Rfam release 9.2 (Griffiths-Jones et al. 2006). We represent all motifs as consensus sequences over an alphabet of 15 characters (IUPAC code, http://www.chem.qmul.ac.uk/iupac/) consisting of the four nucleotides A,C,G,T, the six twofold degenerate characters S = (CG), W = (AT), Y = (CT), R = (AG), M = (AC), and K = (GT), the four threefold degenerate characters H = (ACT), B = (GCT), V = (G,A,C), and D = (G,A,T), and the fourfold degenerate character N = (ACGT). A motif instance (or motif occurrence) is a sequence that matches the motif at each position, i.e., containing one of the allowed characters at that position.
We translate consensus sequences to PWMs given the definition of the degenerate characters. We translate PWMs to consensus sequences by choosing the character with the highest sum of the PWM column entries corresponding to that character minus a correction for character degeneracy (1/2 for ACGT, 2/3 for SYRMK, 5/6 for HBVD, and 1 for N).
Genome alignments and annotation
For all analyses, we used whole genome MULTIZ alignments of 12 Drosophila genomes (Stark et al. 2007), available from UCSC (Kent et al. 2002). We used the D. melanogaster genome-annotations from FlyBase (Release 4.3), and excluded simple repeats, repeat masked regions obtained from UCSC, and noncoding exons according to FlyBase 4.3.
Motif matching and BLS measure
We searched all motif instances in the D. melanogaster genome and evaluated their conservation in the 12 species using the whole-genome alignments. For each motif instance in D. melanogaster, we recorded all instances in the other genomes that were aligned, allowing for motif movements (see below). We prevented double counting of motif instances by assigning each instance in an informant species to the closest instance in D. melanogaster. We evaluated the conservation of all motif instances by summing the branch-lengths of the subtree of the species with conserved motif instances (BLS). This procedure implicitly assumes that all instances are potentially ancestral, such that an instance conserved in a remote informant species would score more highly than instances in closely related informants. One disadvantage of this approach is therefore that chance occurrences or gains in distant species may contribute false positives. The phylogenetic tree branch lengths were obtained from a whole-genome alignment of all 12 species (Dewey et al. 2006; Stark et al. 2007).
_P_-values
All _P_-values are calculated based on the hypergeometric distribution, and correction for multiple-testing was done with the Bonferroni correction.
Allowing for motif movements
When assessing motif conservation, we allowed motif instances in the informant species to be offset relative to the alignment position of the D. melanogaster instances within a given window (counted as distance in either direction in characters excluding gaps). We did not use a prior for a cutoff on maximal tolerable motif movement, as we are not aware of a systematic experimental study that assessed typical movements of functionally equivalent motifs in related species nor systematically assessed of the maximum movement tolerable while maintaining function. We consequently used the window that maximized signal over noise.
While it is clear that increasing tolerated windows may capture additional equivalent instances across genomes, thereby increasing sensitivity, they also increase the number of spurious motif instances that are recovered by chance. We account for the increased background conservation by the use of control motifs (see above), and determine the optimal allowable motif movement window (the one that recovered most motif instances) out of 32 windows between 0 and 500 nucleotides (0, 5, 10, 20, 30, . . . , 90, 100, 120, 140, . . . , 480, 500). For Figure 4B and for analyzing the correlation of optimal window size with different motif properties, we assessed 119 windows between 0 and 10,000 nucleotides (0, 10, 20, . . . , 190, 200, 300, . . . , 9900, 10,000). Similarly, we allow for strand reversals of TF motif instances in informant species, when they help instance recovery in the respective windows. The significance of sensitivity improvement for individual windows and for allowing windows in general was assessed by hypergeometric _P_-values compared to motif instances identified with a window of 0 nucleotides, i.e., perfect alignment of instances.
Estimation of confidence levels of motif instances
For each motif and type of genomic region (promoter, 5′ UTR, 3′ UTR, intron, etc.), we created 100 shuffled control motifs and selected those that had a similar number of matches to the region in the D. melanogaster genome (±20%). By requiring the control motifs to have occurrence rates similar to real motifs in the respective genomic regions in D. melanogaster (i.e., without conservation), we corrected for biases in di- or trinucleotide frequencies (see discussion in Lewis et al. 2003). To remove possible redundancy, we clustered the control motifs (cutoff 0.8) and selected only one representative per cluster, limiting to 10 motifs total that were least similar to known motifs. For each real motif and its controls, we computed the conservation rate (the number of conserved instances at a given BLS cutoff divided by the total number of instances in the D. melanogaster genome) in each region and at each BLS cutoff. We determined the confidence at each BLS as the fraction of conserved motif instances above background conservation, where the latter was estimated using the conservation ratio of the control motifs. This provided a BLS-to-confidence mapping for each motif and region. The variation between the control motifs lead to an average standard-error of 5% for TF motifs, and 4% for miRNA motifs at 60% confidence, indicating an accurate assessment of background conservation.
Comparison with experimental data sets
We obtained all experimentally validated miRNA target gene pairs from TarBase (Sethupathy et al. 2006) and our previous study (Stark et al. 2005). We obtained ChIP-chip regions and the subset that overlapped known enhancers from (Sandmann et al. 2006, 2007; Zeitlinger et al. 2007) and CrebA target genes from Abrams and Andrew (2005). We calculated the enrichment of sites at different confidence cutoffs between 3′UTRs of validated miRNA/target pairs and all 3′UTRs, and between ChIP regions within 2 kb upstream regions and the union of all 2 kb upstream regions. As CrebA targets were originally defined through mostly 5′ UTR instances (Abrams and Andrew 2005) and Mef-2 showed considerable overlap with 5′ UTR regions, we included the 5′ UTR and restricted the upstream region to 500 bp instead. We assessed the recovery of motif instances as the fraction of motif instances in the functional regions (with the same restrictions) that reached the indicated confidence. To assess the fraction of these that are expected by putatively increased overall conservation in these regions, we assess the recovery of control motifs at the same BLS (not confidence, as the control motifs, by definition, would not reach high confidence levels).
Evaluation of experimental and motif instances by correlation with muscle genes
We used correlation with expression patterns to independently evaluate ChIP-regions and predicted motif instances. Muscle genes were 616 genes annotated as “muscle system (13-16)” by the manually curated BDGP in situ database (ImaGO) (Tomancak et al. 2002). To obtain a unique assignment of regions to genes, we restricted our analysis to the 5′ UTR and 500 bases upstream of each gene. We calculated functional enrichments as the fraction of nucleotides covered by motif instances (at 60% confidence) or ChIP regions in muscle genes divided by the corresponding number in all genes present in ImaGO. Hypergeometric _P_-values were computed for motif instances using control motifs at the same BLS and window and for ChIP regions using the fraction of muscle genes matched versus the fraction of all genes matched (note that individual nucleotides are correlated, such that nucleotide _P_-values would overestimate the significance).
Assessing the indegree distribution
We assessed the nonrandomness of the indegree distribution against a control Erdos–Renyi random network (Bollobás 2001) with the same number of edges. To construct this network, we added edges by selecting a source and target node with probability 1/m and 1/n, where m and n were the number of source and target nodes in the true network, respectively. We assessed the difference of indegree distributions between the true and control network with a Wilcoxon rank-sum test. We also assessed the difference in indegree distribution between all transcription factors (as defined by Adryan and Teichmann 2006) and all other genes also with a Wilcoxon rank-sum test.
Functional/ImaGO enrichment of high and low indegree genes
We considered all genes with a GO (Ashburner et al. 2000) and ImaGO (Tomancak et al. 2002) functional annotation (n = 7495 and 5996, respectively) and computed the indegree (number of incoming edges) for each gene in the transcription factor (TF) and miRNA networks. For both networks we defined high-indegree nodes as the 1% with the highest indegree (≥20 for the TF network and ≥4 for the miRNA network) and low-indegree nodes as miRNA antitargets (indegree = 0) and the same fraction of nodes with lowest indegree in the TF network (80%; ≤7 edges). For each GO/ImaGO category, we assessed over-representation and depletion with a hypergeometric _P_-value.
Mutual enrichment between high indegree transcriptional and miRNA targets
We considered all genes that were either a target or a regulator in the TF and microRNA networks resulting in a total of 8760 nodes and defined high- and low-indegree sets as above. We then evaluated if nodes in the miRNA network with high indegree were enriched high-indegree nodes of the transcriptional network (or vice versa) using a hypergeometric _P_-value.
Tissue co-expression
For each TF with available expression information (n = 42; ImaGO; see Tomancak et al. 2002), we counted the number of targets that were co-expressed with the TF in any of the annotated tissues and the number of targets that were not annotated to be co-expressed. The statistical significance of co-expression of a TF with its target was estimated using the hypergeometric distribution given the number of co-expressed targets, and the total number of targets of the TF with known tissue expression, and the corresponding counts for all genes.
Network figure
The network figure was drawn in Cytoscape (Shannon et al. 2003) to display genes (nodes) and regulatory connections (edges) of the 60% confidence network. We colored edges and nodes if genes were expressed in the same tissue according to ImaGO (Tomancak et al. 2002). For clarity, we only show 20 randomly picked targets per transcription factor, i.e., without influencing the fraction of colored edges.
Acknowledgments
We thank Matt Rasmussen, Mike Lin (CSAIL, Broad), and other members of the Kellis laboratory for helpful discussions and for sharing unpublished data. A.S. thanks the Human Frontier Science Program Organization (HFSPO) for a postdoctoral fellowship (LT00495/2006-L). P.K. was supported in part by a National Science Foundation Graduate Research Fellowship. S.R. thanks Terran Lane and Maggie Werner-Washburne (University of New Mexico) for their support.
Footnotes
References
- Abrams E.W., Andrew D.J., Andrew D.J. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. doi: 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- Adryan B., Teichmann S.A., Teichmann S.A. FlyTF: A systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.M., Posakony J.W., Posakony J.W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bergman C.M., Carlson J.W., Celniker S.E., Carlson J.W., Celniker S.E., Celniker S.E. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Levine M., Rubin G.M., Eisen M.B., Rubin G.M., Eisen M.B., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko I., Pachter L., Rubin E.M., Pachter L., Rubin E.M., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Bollobás B. Random graphs. Cambridge University Press; Cambridge, UK: 2001. [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Elemento O., Tavazoie S., Elemento O., Tavazoie S., Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Green E.D., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H., Erwin D.H., Erwin D.H. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Dewey C.N., Huggins P.M., Woods K., Sturmfels B., Pachter L., Huggins P.M., Woods K., Sturmfels B., Pachter L., Woods K., Sturmfels B., Pachter L., Sturmfels B., Pachter L., Pachter L. Parametric alignment of Drosophila genomes. PLoS Comput. Biol. 2006;2:e73. doi: 10.1371/journal.pcbi.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007 doi: 10.1038/nature06341. (in press) [DOI] [PubMed] [Google Scholar]
- Eddy S.R. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Schwartz S., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwiller L., Paten B., Souren M., Loosli F., Wittbrodt J., Birney E., Paten B., Souren M., Loosli F., Wittbrodt J., Birney E., Souren M., Loosli F., Wittbrodt J., Birney E., Loosli F., Wittbrodt J., Birney E., Wittbrodt J., Birney E., Birney E. The discovery, positioning and verification of a set of transcription-associated motifs in vertebrates. Genome Biol. 2005;6:R104. doi: 10.1186/gb-2005-6-12-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J., Grocock R.J., van Dongen S., Bateman A., Enright A.J., van Dongen S., Bateman A., Enright A.J., Bateman A., Enright A.J., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Langenberger D., Gunsalus K.C., Rajewsky N., Gunsalus K.C., Rajewsky N., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S., Bailey T.L., Noble W.S., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R., Levine M., Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991;111:601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- Lai E.C. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C., Burks C., Posakony J.W., Burks C., Posakony J.W., Posakony J.W. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Johnstone S.E., Cole M.F., Isono K., Cole M.F., Isono K., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M., Bergman C., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Bergman C.M., Nathan J., Kreitman M., Nathan J., Kreitman M., Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., Haussler D., Green E.D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M., et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Yee K.P., Erives A., Stathopoulos A., Levine M., Erives A., Stathopoulos A., Levine M., Stathopoulos A., Levine M., Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Karas D., Kel A.E., Kel-Margoulis O.V., Kel A.E., Kel-Margoulis O.V., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A.P., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Zanet J., Srinivasan D.G., Payre F., Stern D.L., Srinivasan D.G., Payre F., Stern D.L., Payre F., Stern D.L., Stern D.L. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Miller W., Makova K.D., Nekrutenko A., Hardison R.C., Makova K.D., Nekrutenko A., Hardison R.C., Nekrutenko A., Hardison R.C., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- Moses A.M., Chiang D.Y., Pollard D.A., Iyer V.N., Eisen M.B., Chiang D.Y., Pollard D.A., Iyer V.N., Eisen M.B., Pollard D.A., Iyer V.N., Eisen M.B., Iyer V.N., Eisen M.B., Eisen M.B. MONKEY: Identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Snyder M., Young R.A., Bulyk M.L., Young R.A., Bulyk M.L., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton B.T., Fratkin E., Batzoglou S., Brutlag D.L., Fratkin E., Batzoglou S., Brutlag D.L., Batzoglou S., Brutlag D.L., Brutlag D.L. A graph-based motif detection algorithm models complex nucleotide dependencies in transcription factor binding sites. Nucleic Acids Res. 2006;34:5730–5739. doi: 10.1093/nar/gkl585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis A.A., Busser B.W., Gisselbrecht S.S., He F.S., Estrada B., Michelson A.M., Bulyk M.L., Busser B.W., Gisselbrecht S.S., He F.S., Estrada B., Michelson A.M., Bulyk M.L., Gisselbrecht S.S., He F.S., Estrada B., Michelson A.M., Bulyk M.L., He F.S., Estrada B., Michelson A.M., Bulyk M.L., Estrada B., Michelson A.M., Bulyk M.L., Michelson A.M., Bulyk M.L., Bulyk M.L. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Comput. Biol. 2006;2:e53. doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3846. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Yeh S.D., True J.R., Carroll S.B., True J.R., Carroll S.B., Carroll S.B. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. MicroRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Engstrom P., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E.E., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E.E., Brehme M., Tongprasit W., Stolc V., Furlong E.E., Tongprasit W., Stolc V., Furlong E.E., Stolc V., Furlong E.E., Furlong E.E. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes & Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Eichenlaub M.P., Bork P., Furlong E.E., Bork P., Furlong E.E., Furlong E.E. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Schones D.E., Sumazin P., Zhang M.Q., Sumazin P., Zhang M.Q., Zhang M.Q. Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics. 2005;21:307–313. doi: 10.1093/bioinformatics/bth480. [DOI] [PubMed] [Google Scholar]
- Schroeder M.D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Rajewsky N., Siggia E.D., Gaul U., Siggia E.D., Gaul U., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Corda B., Hatzigeorgiou A.G., Corda B., Hatzigeorgiou A.G., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Ramage D., Amin N., Schwikowski B., Ideker T., Amin N., Schwikowski B., Ideker T., Schwikowski B., Ideker T., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M., Brennecke J., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., Rasmussen M.D., Roy S., Deoras A.N., Roy S., Deoras A.N., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007 doi: 10.1038/nature06340. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G.D. DNA binding sites: Representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., King D.C., Hardison R.C., Miller W., Chiaromonte F., Hardison R.C., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Siepel A.C., Thomas P.J., McDowell J.C., Thomas P.J., McDowell J.C., McDowell J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Weiszmann R., Kwan E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Kwan E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., Ashburner M., Hartenstein V., Celniker S.E., Hartenstein V., Celniker S.E., Celniker S.E., et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Frith M.C., Fu Y., Kent W.J., Fu Y., Kent W.J., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A., Ettwiller L., Birney E., Ettwiller L., Birney E., Birney E. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Van Doren M., Bailey A.M., Esnayra J., Ede K., Posakony J.W., Bailey A.M., Esnayra J., Ede K., Posakony J.W., Esnayra J., Ede K., Posakony J.W., Ede K., Posakony J.W., Posakony J.W. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes & Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Wasserman W.W., Sandelin A., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Thompson W., Fickett J.W., Lawrence C.E., Fickett J.W., Lawrence C.E., Lawrence C.E. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A., Pizer M., Rockman M.V., Romano L.A., Rockman M.V., Romano L.A., Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada T., Totoki Y., Ishikawa M., Asai K., Nakai K., Totoki Y., Ishikawa M., Asai K., Nakai K., Ishikawa M., Asai K., Nakai K., Asai K., Nakai K., Nakai K. Automatic extraction of motifs represented in the hidden Markov model from a number of DNA sequences. Bioinformatics. 1998;14:317–325. doi: 10.1093/bioinformatics/14.4.317. [DOI] [PubMed] [Google Scholar]
- Yin Z., Frasch M., Frasch M. Regulation and function of tinman during dorsal mesoderm induction and heart specification in Drosophila. Dev. Genet. 1998;22:187–200. doi: 10.1002/(SICI)1520-6408(1998)22:3<187::AID-DVG2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R.P., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Zinzen R.P., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Kellis M., Zhang H., Young R.A., Levine M., Zhang H., Young R.A., Levine M., Young R.A., Levine M., Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes & Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wong W.H., Wong W.H. CisModule: De novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]