Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences (original) (raw)
Abstract
It is thought that gene conversion (GCV) and somatic hypermutation (SHM) of immunoglobulin (Ig) genes occur in two steps: the generation of uracils in DNA by activation-induced cytidine deaminase, followed by their subsequent repair by various DNA repair pathways to generate sequence-diversified products. It is not known how either of the two steps is targeted specifically to Ig loci. Because of the tight link between transcription and SHM, we have investigated the role of endogenous Ig light chain (IgL) transcriptional control elements in GCV/SHM in the chicken B cell line DT40. Promoter substitution experiments led to identification of a strong RNA polymerase II promoter incapable of supporting efficient GCV/SHM. This surprising finding indicates that high levels of transcription are not sufficient for robust GCV/SHM in Ig loci. Deletion of the IgL enhancer in a context in which high-level transcription was not compromised showed that the enhancer is not necessary for GCV/SHM. Our results indicate that cis-acting elements are important for Ig gene diversification, and we propose that targeting specificity is achieved through the combined action of several Ig locus elements that include the promoter.
Antibody diversification in B cells is an important component of immune responses against foreign antigens. Ig gene diversity is first created by V(D)J recombination, which is responsible for assembling the primary functional repertoire, and this is followed by gene conversion (GCV) and somatic hypermutation (SHM), processes that further increase sequence diversity at the variable regions of Ig genes. GCV is achieved by copying patches of sequences from sequence donors into the variable regions, whereas SHM events are nontemplated and exist mainly as single point mutations. Both GCV and SHM are dependent on activation- induced cytidine deaminase (AID) (1–4), which is thought to initiate the reactions by deaminating deoxycytosine residues in Ig genes. The uracils thereby generated are then processed by uracil DNA glycosylase and mismatch repair proteins and channeled into homology-based repair for GCV or by error-prone repair for SHM (5–8).
GCV and SHM are thought to be restricted largely to Ig genes. The molecular basis of this restriction has been the subject of intensive investigation, with cis-acting DNA elements in Ig genes being the prime candidates for providing targeting specificity. Numerous studies indicate a tight link between AID-dependent diversification of Ig genes and transcription. The region that undergoes SHM is 1–2 kb downstream of Ig transcription start sites, which spans the variable region exon (9–11). Studies using knockout mice and transfected cell lines have shown that no SHM occurs in the absence of an active promoter (12, 13). Additional studies with transgenic mice demonstrated that promoter location dictates the region of SHM, as a newly inserted promoter creates a new window of mutation, and altered positioning of the promoter shifts the mutating region (10, 14). Nonetheless, the endogenous Ig promoters are thought not to be necessary because several non-Ig promoters in different model systems could support SHM when used to replace the Ig promoter (13, 15, 16). Similarly, studies of transgenes in mice and heterologous expression vectors in hypermutating B cell lines indicated that the Ig variable region exon itself is not required for SHM (17–19). Importantly, experiments performed in cell lines have indicated a positive correlation between transcription levels and SHM frequencies (12, 19). These results have led to the conclusion that rather than acting as targeting elements, Ig promoters simply mediate the transcription essential for SHM. However, very few experiments have been performed to examine the role of the promoter in the context of endogenous Ig genes (13, 15).
Another focus of interest has been Ig gene enhancers. Studies using mouse Ig light chain κ (Igκ) transgenes showed that deletion of the intronic or 3′ Igκ enhancer strongly reduced SHM, suggesting an important role of enhancers in SHM (16, 20, 21). However, when the mouse Igκ intronic enhancer, 3′κ enhancer, or Ig heavy chain (IgH) intronic enhancer were deleted in their endogenous context, SHM was not substantially affected (22–24). The discrepancy could reflect redundant functions provided by other cis-acting elements in the endogenous loci or limitations of transgenic studies related to low mutation rates and position effects coming from different integration sites. For its part, deletion of endogenous enhancers from the germline has the potential limitation that epigenetic alterations or selection might take place to compensate for the lack of the enhancer during earlier stages of B cell development. Recently, several reports have indicated positive effects of E2A proteins on AID-dependent diversification processes (25–27). As E2A binding sites are found in Ig enhancers, it is possible that the enhancers play an important role in Ig gene sequence diversification. In investigating such a role, it is important to distinguish a putative targeting activity of enhancers from their ability to facilitate transcription, because stronger transcription can itself contribute to higher levels of SHM.
To investigate the molecular mechanisms of targeting in Ig sequence diversification, we analyzed two regions in the Ig light chain (IgL) locus of DT40 cells, the promoter and the enhancer. DT40 is a chicken B cell line that undergoes continuous GCV and SHM in its IgL and IgH loci, and it performs homologous recombination efficiently, thereby enabling manipulation of DNA elements in their endogenous context (28–30). Deletion of the single known IgL enhancer element while maintaining high levels of transcription showed that the enhancer was not essential for GCV/SHM. Analysis of the promoter region revealed that an active promoter is crucial for GCV/SHM and that the endogenous promoter could be functionally replaced by a strong heterologous promoter. Surprisingly, we found that the human elongation factor 1-α (EF1-α) promoter supported GCV/SHM poorly, even though it drove transcription efficiently. This is the first time a strong non-Ig promoter has been shown to have a substantial defect in supporting Ig sequence diversification in an endogenous Ig locus, which argues that the function of the promoter in GCV/SHM must extend beyond simply driving transcription. Overall, our results indicate that cis-acting elements contribute to the targeting efficiency of Ig gene diversification and that high-level transcription in an Ig locus is not sufficient for efficient GCV/SHM.
RESULTS
Generation of IgL promoter-substituted clones
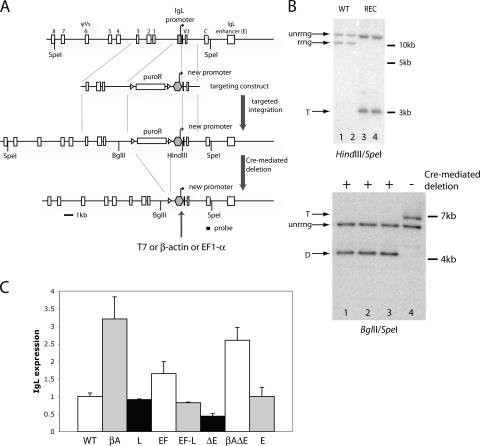
There is a well-established link between transcription and SHM in mammalian B cells, but this issue has not been investigated for GCV/SHM in chicken B cells. To address this, we replaced the endogenous IgL promoter in DT40 cells with three heterologous promoters: the bacteriophage T7 promoter, which should not be able to initiate transcription in eukaryotic cells, and two strong Pol II promoters, the chicken β-actin and the human EF1-α promoter. The IgL promoter region that was replaced extended from 206 bp upstream to 6 bp downstream of the transcription start site and contained the octamer binding motif and TATA-box essential for IgL promoter function (31, 32). The CL18 clone of DT40 cells was transfected with targeting constructs containing different promoters and seeded out at limiting dilution, and stable transfectants were identified after 6–8 d (see Materials and methods; Fig. 1 A) (30). Southern blots were performed to distinguish clones in which targeted integration of the constructs into the rearranged IgL allele had occurred (targeted integrants) from those in which random integration into other parts of the genome had taken place (random integrants). At least two independent targeted integrants were selected for subsequent analysis and treated with recombinant Cre protein to remove the puromycin selection cassette inserted upstream of the promoter as a result of targeted integration (Fig. 1 A). Cre-treated cells were single-cell seeded again, and clones that grew out were verified for successful Cre-mediated deletion by Southern blots (Fig. 1 B). Overall, the targeting efficiency of our promoter-swapping constructs was ∼45%, and the efficiency of Cre-mediated deletion was ∼90% (not depicted).
Figure 1.
Generation and analysis of promoter-substituted cells. (A) Schematic diagram of steps involved in replacing the endogenous IgL promoter with heterologous promoters. Relevant components of the IgL locus are indicated as rectangles, with the IgL promoter in gray, and the start site of transcription is indicated by the arrow. Only the eight most proximal ψV elements are shown. The puromycin resistance cassette (puroR; rectangle) is flanked by loxP sites (triangles) and the heterologous promoter (hexagon) is indicated. Relevant restriction enzyme sites and the constant region probe (solid bar) used in Southern and Northern blots are indicated. (B, top) A representative Southern blot for identifying targeted integrants (_Hind_III/_Spe_I digest) is shown; the higher molecular mass band in each lane derives from the unrearranged IgL allele (unrrng), whereas the lower band is derived from the rearranged allele (rrng; lanes 1 and 2) or the targeted rearranged allele (T; lanes 3 and 4). REC, two different targeted recombinants; WT, two randomly integrated clones that show wild-type band patterns. (B, bottom) A representative Southern blot for verification of successful Cre-mediated deletion (_Bgl_II/_Spe_I digest). Before Cre deletion, the rearranged IgL allele (T; lane 4) generates a larger fragment than the unrearranged allele (unrrng; lane 4); upon puro cassette removal, the band derived from the rearranged IgL allele becomes smaller in size than that of the unrearranged allele (D; lanes 1–3). (C) Quantitation of steady-state IgL transcript levels in various promoter-substituted or enhancer-deleted cell lines as assessed by Northern blots (a representative blot is shown in Fig. S1). Blots were hybridized with the IgL constant region probe and the GAPDH probe as an internal control and quantitated, and the IgL/GAPDH ratio was calculated. The ratio for wild-type CL18 cells was arbitrarily set to 1, and all other values are expressed relative to this. The height of each bar represents the average of two to four determinations with the standard deviation, as indicated by the error bars. WT, wild-type CL18 cells; βA, cells in which the IgL promoter was replaced with the β-actin promoter; L, cells in which the IgL promoter was replaced with itself; EF, cells in which the IgL promoter was replaced with the EF1-α promoter; EF-L, cells that have gone through two successive rounds of targeted replacement of the IgL promoter (the first to replace the IgL promoter with the EF1-α promoter and the second to revert the EF1-α promoter back to the IgL promoter); ΔE, cells in which the IgL enhancer was deleted; βAΔE, cells in which the IgL enhancer was deleted with the β-actin promoter driving IgL transcription; E, cells in which the IgL enhancer was replaced by itself.
IgL expression driven by the new promoters was measured by Northern blots. As expected, no transcription was detected from the T7 promoter (T7 cells), although removal of the normal V promoter appeared to activate cryptic promoters that generated low-level transcription initiating at the 3′ end of the V segment (not depicted). Conversely, both β-actin and EF1-α promoter-substituted cells (βA and EF cells, respectively) expressed the IgL locus at levels higher than the endogenous promoter (Fig. 1 C and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20061835/DC1).
The β-actin promoter supports GCV/SHM
Two established assays were used to measure levels of GCV/SHM. First, direct sequencing was performed on cloned PCR products of the 500-bp region downstream of the IgL transcription start site from genomic DNA of cells cultured for 28 d. Second, IgM reversion assays were performed, taking advantage of a frameshift mutation in the IgL variable region of CL18-derivative clones that can be corrected by certain GCV events to generate IgM+ cells (30). As only a few pseudogenes are able to correct the frameshift mutation and each culture begins as a single cell, the percentages of IgM+ cells in different clones of the same genotype exhibited considerable fluctuation. Consequently, multiple clones of a given genotype were analyzed, and the median of each group was used for comparison. In addition, successive rounds of subcloning were performed to ensure that GCV/SHM phenotypes were stable (cells that have undergone different numbers of subclonings will be referred to as belonging to different generations). Subcloning was performed when cells had been grown for 3 wk since they were last subcloned. Cells were maintained for a total of 4 wk for each round of subcloning, during which IgM reversion frequencies were assessed; at the end of this time, DNA was prepared for sequencing.
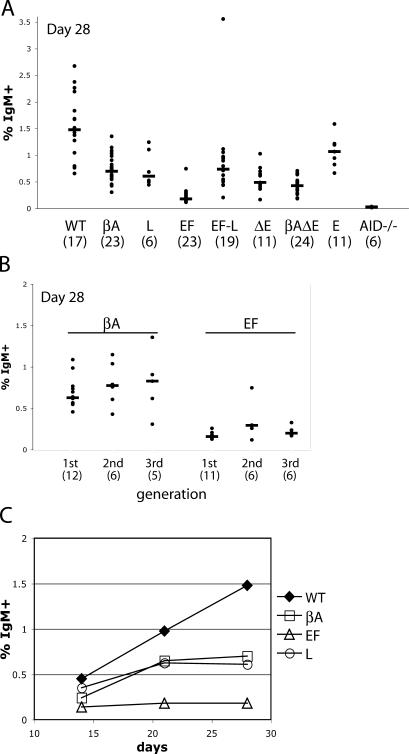
After 4 wk of culture, a significant percentage of βA cells had become surface IgM+ (sIgM+) compared with AID−/− controls (combined dataset from three generations of βA cells; P ≤ 0.0001; Fig. 2 A) (2). When the IgM reversion results were separated based on the number of subclonings the cells had gone through, no difference was found between different generations, indicating that the GCV/SHM phenotype was stable over time (Fig. 2 B). Moreover, weekly examination of the cells showed that the percentages of IgM+ cells increased over time (Fig. 2 C shows median values; data for all clones are shown in Fig. S2 B, available at http://www.jem.org/cgi/content/full/jem.20061835/DC1), confirming the ability of the β-actin promoter to support GCV in the IgL locus. Although we noted a slowing of accumulation of sIgM+ cells in the βA cell cultures between days 21 and 28 as compared with between days 14 and 21, the data from successive subclonings of these cells indicate no loss of GCV/SHM activity with the age of the cells (Fig. 2 B).
Figure 2.
IgM reversion assay of various promoter-substituted and enhancer-deleted cell lines. (A) The percentages of surface IgM+ cells at day 28 for the cell types indicated along the x axis; cells of all generations are included. Each dot represents a subclone of the indicated cell type, and the black bar marks the median of each group. The number in parentheses below the name of each cell type is the total number of subclones analyzed for that cell type. (B) The percentages of surface IgM+ cells in βA and EF cells at day 28, separated according to the different generations of subclones analyzed. The black bar marks the median of each group. (C) Time course analysis of IgM+ cell accumulation. Each data point represents the median of all clones of the given genotype at the indicated time point. Cell lines are abbreviated as in Fig. 1.
Sequence analysis corroborated the findings from the IgM reversion assays. At day 28, we found 43 total events (long-track GCV, templated mutations, and nontemplated events combined) out of 274 sequences analyzed for the βA cells (combined dataset over three generations; Table I). In comparison, T7 cells generated no diversification events in the 138 sequences obtained (βA vs. T7 cells; P ≤ 0.0001). These results indicate that GCV/SHM in DT40 cells relies on a functional promoter and that the endogenous promoter is not essential.
Table I.
Sequence diversification in the IgL locus
| Cell typea | Totaleventsb | Totalsequences | Frequency(×10−4 events/bp)c | Event typesd | ||
|---|---|---|---|---|---|---|
| Long-trackGCV | Templatedmutations | Nontemplatedeventse | ||||
| WT | 86 | 276 | 6.2 | 31 | 32 | 14 (23) |
| T7 | 0 | 138 | 0 | 0 | 0 | 0 (0) |
| βA | 43 | 274 | 3.1 | 18 | 13 | 12 (12) |
| EF | 25 | 396 | 1.3 | 10 | 6 | 6 (8) |
| EF-L | 15 | 114 | 2.6 | 5 | 8 | 2 (2) |
| ΔE | 11 | 174 | 1.3 | 5 | 3 | 3 (3) |
| βAΔE | 33 | 208 | 3.2 | 11 | 13 | 6 (8) |
Manipulation of the IgL locus around the promoter interferes with optimal GCV/SHM
Although the βA cells were able to perform GCV/SHM, we noticed that they did so only half as efficiently as the wild-type CL18 cells (WT cells; Fig. 2, A–C; and Table I). This was not an artifact resulting from transfection, subcloning, selection, or phenotypic fluctuation of the cells, as random-integrant clones (unaltered in the IgL locus) derived from transfections of different targeting constructs performed GCV/SHM in the endogenous IgL locus at levels comparable to those of untransfected CL18 cells (not depicted). Sequencing and IgM reversion results from random integrants and untransfected CL18 cells have thus been pooled together as the wild-type dataset.
We therefore considered the possibility that the promoter-exchange scheme altered the IgL locus in a manner that impaired GCV/SHM. To address this, a targeting vector similar to that used to generate βA and EF cells was created to replace the IgL promoter with an exact copy of itself. We expected that cells arising from targeted integration of this construct (L cells, derived from one parental clone) would be identical to WT cells, except that they would have undergone the same recombination events in the IgL locus as other promoter-substituted cells and retained a loxP site upstream of the promoter. Northern blots demonstrated that IgL expression was not affected by this synonymous substitution (compare WT with L; Fig. 1 C).
Interestingly, L cells exhibited a twofold reduction in IgM reversion relative to WT cells (P = 0.0016; Fig. 2 A and Fig. S2 C), indicating that the promoter replacement scheme reduced the efficiency of GCV/SHM. Furthermore, comparison of βA and L cells (Fig. 2 A) indicates that the β-actin promoter supports the same frequency of GCV/SHM as the normal IgL promoter, and that the reduction in GCV/SHM seen in βA cells relative to WT cells was most likely caused by locus manipulations (see Discussion).
The EF1-α promoter is defective in supporting GCV/SHM
To our surprise, the EF1-α promoter yielded results quite different from those of the β-actin promoter. EF cells generated very few sIgM+ cells over the course of 28 d (three- to fourfold less than L cells; Fig. 2 A). This was a stable phenotype, observed in three successive generations of EF cells (Fig. 2 B). When the medians of the percentages of IgM+ cells of clones at days 14, 21, and 28 were plotted, minimal increases were observed with increased culture times (Fig. 2 C and Fig. S2 D). Sequencing results pooled from EF cell clones from different generations confirmed that their GCV/SHM activity was significantly reduced compared with WT and βA cells (P ≤ 0.0001 and P = 0.024, respectively; Table I). Though the defect in GCV/SHM of EF cells as assayed by sequencing analysis is less pronounced than by IgM reversion assay, the latter is more quantitative, as it monitors hundreds of thousands of cells, whereas the sequencing assay sampled only a couple of hundred alleles for each cell type.
As the EF1-α promoter was able to drive higher levels of IgL transcription than the endogenous IgL promoter (Fig. 1 C), we were intrigued by its inability to support efficient GCV/SHM. To ascertain whether EF clones might have some unrelated defect that could explain the IgL GCV/SHM results, we first sequenced the IgH locus in the EF cells. Comparable levels of IgH GCV/SHM were found in EF and WT cells (Table II); therefore, the GCV/SHM defect was specific to the IgL locus. We next tested whether the IgL locus itself had been inactivated for GCV/SHM by reinserting the endogenous IgL promoter into the rearranged IgL locus of EF cells (with attendant deletion of the EF1-α promoter) to generate EF-L cells. As expected, levels of IgL expression in EF-L cells were similar to those seen in WT and L cells and lower than in EF cells (Fig. 1 C). Analysis of IgM reversion in EF-L cells showed levels of IgM reversion fourfold higher than those in EF cells (P = 0.0006; Fig. 2 A and Table I); sequence analysis also confirmed the GCV/SHM frequencies in EF-L cells to be higher than those in EF cells (Table I). Our ability to rescue the GCV/SHM defect in the IgL locus of EF cells by reinsertion of the endogenous promoter strongly suggests that the EF cell defect was specific to the EF1-α promoter and was not caused by other factors or alterations. In addition, EF-L cells were only able to perform GCV/SHM at about half the efficiency of wild-type cells (Fig. 2 A and Table I), consistent with observations in L cells and demonstrating again the effect of the targeting scheme on GCV/SHM.
Table II.
Sequence diversification in the IgH locus
| Cell typea | Diversifiedsequencesb | Totalsequences | % diversifiedsequences |
|---|---|---|---|
| WT | 14 | 65 | 22 |
| EF | 24 | 114 | 21 |
| AID−/− | 0 | 38 | 0 |
Deletion of the IgL enhancer
The only known transcriptional enhancer in the chicken IgL locus was identified in a transgenic mouse study as a 1.7-kb fragment 2 kb downstream of the constant region (33). Transient transfection assays were subsequently used to localize the enhancer to a ∼500-bp region (32). To test the involvement of the enhancer in IgL GCV/SHM, this region was deleted in CL18 and βA cells (see Materials and methods; Fig. 3 A). Deletion of the IgL enhancer in the βA cells was of particular interest, because we anticipated that transcription driven by the β-actin promoter would be minimally dependent on the enhancer, thereby allowing us to identify potential roles of the enhancer in GCV/SHM beyond facilitation of transcription. Stable transfectants of the enhancer deletion construct into WT and βA cells were screened by Southern blots to identify targeted integrants (Fig. 3 B), and for each at least two independent clones were selected for further characterization. As a control, a cell line in which the enhancer was retargeted into the rearranged IgL allele of CL18 cells was also generated (E cells); the only genotypic difference between E cells and CL18 cells should be an 80-bp fragment including the loxP site inserted immediately upstream of the enhancer. Targeted integration efficiency of the enhancer deletion construct was ∼24% (not depicted). Clones were subsequently treated with Cre protein and seeded out as described in Generation of IgL promoter…, and deletion of the puromycin selection cassette was verified by Southern blotting (Fig. 3 B).
Figure 3.
Analyses of enhancer-deleted cells. (A) Stepwise schematic diagram of the generation of IgL enhancer-deleted cells. Components of the IgL locus are as detailed in Fig. 1. Relevant restriction enzyme sites and the constant region probe (solid bar) used in Southern and Northern blots are indicated, as are the puromycin resistance cassette (puroR; rectangle) and its flanking loxP sites (triangles). (B) Representative Southern blots for identifying targeted integrants of the enhancer deletion construct (_Bcl_I/_Hind_III digest, top) and verifying successful Cre-mediated deletion (_Bcl_I digest, bottom). WT, random integrants that show wild-type band patterns; REC, targeted recombinant clones. In both blots, the higher molecular mass bands in each sample come from the unrearranged IgL allele (unrrng). The lower bands are derived from the rearranged allele (rrng; lanes 3 and 4, top), the targeted allele (T; lanes 1 and 2, top; lane 1, bottom), or the Cre-deleted allele (D; lanes 2–4, bottom). (C) Time course analysis of IgM+ cell accumulation in different cell types. The median of each cell type, including all generations of subclones, is plotted at various time points.
Northern blotting demonstrated that deletion of the enhancer in the context of the wild-type promoter (ΔE cells) reduced transcription twofold compared with WT cells, whereas deletion in the context of the β-actin promoter (βAΔE cells) reduced transcription by only 15% compared with βA cells, with transcription levels remaining 2.6-fold higher than in WT cells (Fig. 1 C). These results indicate that the enhancer only contributes moderately to transcription in the DT40 IgL locus. In the E cells, IgL expression was unaltered compared with WT cells (Fig. 1 C), showing that synonymous replacement of the enhancer had no measurable effect on IgL transcription.
The IgL enhancer is dispensable for GCV/SHM
We analyzed the ΔE, βAΔE, and E cells for IgL GCV/SHM and included the same subcloning procedure as described in The β-actin promoter… to test the stability of GCV/SHM phenotypes. Sequencing results from two generations of ΔE cells indicated that GCV/SHM levels were reduced by almost fivefold compared with WT cells (Table I). IgM reversion assays showed reductions of about four- and twofold relative to WT and E cells, respectively (two generations of cells analyzed; Fig. 2 A, Fig. 3 C, and Fig. S2 E). Both assays indicated that some GCV/SHM activity remained in the absence of the enhancer. In addition, much of the decrease in GCV/SHM observed in ΔE cells compared with E cells was likely caused by the twofold decrease in IgL transcription caused by deletion of the enhancer. In the βAΔE cells, IgL transcription was substantially stronger than in WT or L cells (Fig. 1 C); thus, effects on GCV/SHM are unlikely to be results of reduced transcription. Analysis of three generations of βAΔE cells demonstrated that they were able to produce some sIgM+ cells, confirming the finding from ΔE cells that the enhancer is not essential for GCV/SHM. Nevertheless, the median was reduced by 30% compared with βA cells (P ≤ 0.0001; Fig. 2 A, Fig. 3 C, and Fig. S2 F). Interestingly, by IgM reversion assay, E cells also showed a ∼30% reduction in levels of GCV/SHM compared with WT cells (Fig. 2 A), which was similar in magnitude to the reduction observed in βAΔE cells compared with βA cells. This suggests that the IgL locus manipulations that accompanied enhancer deletion cause a measurable reduction in GCV/SHM and that the drop in GCV/SHM activity seen in βAΔE cells compared with βA cells was the result of the manipulations and not the absence of the enhancer sequences. This was supported by sequencing analyses of two generations of βAΔE cells, as the GCV/SHM levels were indistinguishable from that observed in βA cells (Table I). Collectively, our results indicate that the IgL enhancer is not required for GCV/SHM of the locus and that when transcription is driven at high levels, the contribution of the enhancer to GCV/SHM is quite modest.
DISCUSSION
In this study, the roles of two cis-acting elements in GCV/SHM of the chicken DT40 IgL locus were investigated: the promoter and the enhancer. Replacement of the endogenous IgL promoter with the inactive bacteriophage T7 promoter led to complete abrogation of GCV/SHM, indicating an essential role for an active promoter in chicken Ig gene diversification. Two other non-Ig promoters were also tested for their ability to substitute functionally for the IgL promoter. Unexpectedly, the EF1-α promoter supported GCV/SHM poorly despite driving IgL expression at levels ∼1.5-fold higher than that obtained with the endogenous IgL promoter. Given the proposal that a positive correlation exists between transcription levels and Ig gene diversification frequencies (12, 19), the GCV/SHM defect in EF cells could be as much as sixfold compared with L and EF-L cells. To our knowledge, this is the first time a strong promoter has been shown to have a significant defect in supporting SHM or GCV of an endogenous Ig gene. These results indicate, in contrast to currently prevailing models, that there are roles of the promoter in GCV/SHM beyond driving high levels of transcription.
Although the β-actin promoter was able to substitute for the endogenous IgL promoter to support GCV/SHM, it did so at levels similar to those supported by the endogenous IgL promoter (L and EF-L cells), despite driving more than threefold higher steady-state IgL transcript levels than the endogenous promoter. That a higher GCV/SHM activity was not observed in the βA cells could reflect saturating levels of transcription driven by the endogenous IgL promoter for GCV/SHM. Alternatively, the β-actin promoter, like the EF1-α promoter, could be suboptimal for recruitment of AID or other aspects of GCV/SHM, which in turn would suggest that the endogenous IgL promoter has features that render it particularly efficient in mediating GCV/SHM. A third possibility is that the positive correlation between transcription and diversification levels inferred from studies of transgenes and artificial SHM substrates does not apply to the endogenous Ig loci. Our experiments only assessed steady-state mRNA levels, but because the transcribed region is virtually identical in all of the cell lines examined in this paper, the results are likely to reflect relative transcription rates as well.
Most previous analyses of the role of promoters in SHM have been performed using transgenes in mice and cell lines, and the ability of non-Ig promoters to support SHM has led to the proposal that endogenous Ig promoters are entirely replaceable (12, 16, 18, 19, 34, 35). These results should be interpreted cautiously, because transgenes typically mutate at substantially lower frequencies than endogenous Ig genes and are subject to integration site effects. In the two previous studies in which an endogenous Ig promoter was manipulated (13, 15), mutation frequencies were reduced but so was transcription. In our study, targeted manipulation of the endogenous IgL promoter yielded loci with high-level transcription, which revealed previously unappreciated roles of the promoter region in the targeting of GCV/SHM.
Cell line–based studies, as well as bacterial and in vitro assays, have led to the model that transcription is important for generating single-stranded DNA suitable for AID deamination (12, 36, 37), implying that stronger promoter activity translates into more AID substrates being available, thereby leading to higher levels of mutation. There is also evidence to indicate that AID interacts directly with RNA polymerases, supporting the idea that AID could traverse the transcribed region with the transcription elongation complexes (14, 38, 39). Though our results do not contradict the role of transcription in such models, they demonstrate that an active promoter is important for GCV/SHM for reasons beyond driving high levels of transcription. One possibility is that the EF1-α promoter fails to recruit certain transcription factors to its transcription initiation or elongation complexes that are important for AID recruitment or for locus interactions involved in GCV/SHM.
Our experiments also revealed that the targeted integration scheme used interfered with GCV/SHM. After targeted integration of the designated DNA elements and Cre-mediated deletion of the selection cassette, 59- and 80-bp regions including the loxP site are left in the IgL locus immediately upstream of the promoter and in place of the enhancer with the respective targeting constructs (Fig. 1 A and Fig. 3 A). These insertions might interrupt DNA elements needed for optimal GCV/SHM. Alternatively, sequences within the insertions or the targeted integration process itself could have interfered with GCV/SHM.
A recent study reported that deletion of the enhancer in the DT40 IgL locus leads to a 1,000-fold reduction in IgL transcription (25). However, additional Southern blot analyses of the single enhancer-deleted clone generated by Conlon and Meyer indicated that Cre-mediated removal of the selection cassette integrated at the site of the deleted enhancer was unsuccessful (not depicted; Meyer, K.B., personal communication), and thus those cells and that study are not informative as to the phenotype caused by enhancer deletion. The ability of our ΔE and βAΔE cells to perform some levels of GCV/SHM indicates that the enhancer is not necessary for IgL GCV/SHM. Nonetheless, GCV/SHM efficiencies are moderately compromised in both ΔE and βAΔE cells. Although the reduction in GCV/SHM of ΔE cells could be caused by reduced transcription compared with WT cells, the small decrease in GCV/SHM seen in βAΔE cells relative to βA cells is likely caused by the targeted integration scheme. Overexpression of the E2A-encoded protein E47 stimulates GCV/SHM in DT40 cells (25), whereas deletion of the E2A gene substantially reduces GCV/SHM in DT40 (27), leading to the proposal that E2A proteins facilitate GCV/SHM through direct binding to the IgL enhancer (25, 27). Our data, however, suggest that the IgL enhancer, which contains six E2A binding motifs, is not the E2A-responsive element involved in the potentiation of IgL GCV/SHM.
Based on our findings, we favor the model that Ig loci are organized into higher order structures through interactions between multiple cis-acting elements, including the promoter, thereby creating an interaction surface that is important for targeting of GCV/SHM. As none of the Ig elements manipulated in this study led to significant changes in the GCV to SHM ratio or altered SHM patterns (χ2 analyses; Table I and not depicted), the effects we observed are most likely mediated at the level of recruitment of AID. Although deletion or alteration of one of the interacting elements would perturb targeting efficiency, it does not necessarily lead to the collapse of the entire structure. Further examination of the IgL locus will be important for identifying other DNA elements that contribute to GCV/SHM. It will be interesting to determine if any single IgL element is necessary for GCV/SHM, or if the targeting specificity of GCV/SHM is established through many interactions that in combination impose a high degree of specificity on Ig gene diversification processes.
MATERIALS AND METHODS
Constructs.
Homology arms used in the promoter-exchange targeting construct were PCR-amplified from DT40 genomic DNA. A 3.5-kb left homology arm (5LEFT1-5′-gatctatagcagcggccgcagaccctcaaacatcccttca-3′, 3LEFT-5′-gccaggccttaattaagggacacgtg-3′) and a 1-kb right homology arm (CVLF1-5′-cagtaagcttaccatggcctgggctcctctcctcctg-3′, CLA2X-5′-gactctcgaggacagcacttacctggacagctg-3′) flanked the promoters of interest and a puromycin selection cassette. The bacteriophage T7 promoter was cloned by annealing two complementary oligos (T7BHT-5′-gatcttaattaaggatcctaatacgactcactataggga-3′ and T7BHBN-5′-agcttccctatagtgagtcgtattaggatccttaattaa-3′), and the chicken β-actin promoter was derived as an _Nhe_I_-Hinf_I fragment from the pLoxPuro plasmid (40). Two constructs carrying the human EF1-α promoter were generated through PCR amplification of the pEBB plasmid (41): one included the region up to the end of the small first exon of the human EF1-α gene (5BAMEF-5′-cttgaaaggaggatccattggctccg-3′, EFHIN3-5′-aaccacacacgaagcttacctgtg-3′), whereas the other ended immediately downstream of the transcription start site of the EF1-α promoter (5BAMEF, EFHINR2-5′-ccatggtaagctttgggatgttctggcggcaaacccgttg-3′). The first construct led to bypass of the leader exon during mRNA splicing, and the target-integrated clones derived with this construct were assayed for GCV/SHM by sequencing and not IgM reversion; the second construct preserved the original leader to V region splice pattern (not depicted). To generate the construct for reinserting the IgL promoter into its endogenous location, the right homology arm was rederived using a different primer set (VLP5BF-5′-gctgccacgggatcccgctgca-3′, CLA2X). The puromycin selection cassettes were flanked by loxP sites to allow removal by Cre recombinase with the resulting single loxP site containing mutations that render it inactive (40).
The enhancer deletion construct contained homology arms 2.2 kb upstream and downstream of the enhancer flanking a Cre-removable puromycin selection cassette. The arms were PCR-amplified from DT40 genomic DNA (upstream arm: CLEUF2-5′-ggcgcatacacacatattgg-3′, CLEUPR-5′-gcctgctaattaattgccatgtc-3′; downstream arm: CLEDHF-5′-ccacgcataagcttggccccac-3′, CLEDXR-5′-gaatatctcgagaggagac-3′). The targeting construct for reinsertion of the IgL enhancer contained a different downstream homology arm (CLE-HF-5′-ctggcgacaaagcttcagcagagc-3′, CLEDXR).
Cell culture, transfection, and Cre-mediated deletion.
All DT40 clones were grown in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Benchmark), 1% heat-inactivated chicken serum (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), 100 U/L penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) at 41°C. The CL18 clone of DT40 cells and the AID−/− DT40 cells were generously provided by J.-M. Buerstedde (GSF-National Research Center for Environment and Health, Neuherberg, Germany) (2, 30). Transfections were performed by electroporating 107 cells with 25 μg linearized targeting vectors at 580 V and 25 μF in PBS. Transfected cells were seeded out at limited dilution in 0.5 μg/ml puromycin 10–14 h after electroporation, and stable transfectants were isolated 6–8 d later. Cre-mediated deletions were performed 14 d after transfection. 3 × 105 cells were treated with 6 μM his-Tat-NLS-Cre (HTNC; provided by F. Edenhofer, University of Bonn, Bonn, Germany) recombinant protein (42) for 1–6 h in 300 μl serum-free media. Cells were single-cell seeded after the incubation, and clones were identified 6–8 d later.
Southern and Northern blotting.
To screen clones for targeted integration and successful Cre deletion, genomic DNA was purified, digested with restriction enzymes, separated on 0.8% agarose gels, transferred onto membranes (GeneScreen Plus; PerkinElmer), and blotted with random hexamer-labeled DNA probes (Roche) against the IgL constant region (CCLF1-5′-cccaccgtcaaaggaggagctg-3′ and CCLR2-5′-gacagtgacaggtagctgctggccatatac-3′) and puromycin region (_Hind_III-_Bam_HI or _Hind_III-_Acc_I fragments of pLoxPuro) (40). To analyze expression, RNA was extracted from various clones using RNA-Bee (Tel-Test, Inc.), separated on formaldehyde agarose gels, and transferred onto GeneScreen Plus membranes. Blots were hybridized with random hexamer-labeled probes against the IgL constant region and GAPDH (CHGAPDHF-5′-accagggctgccgtcctctc-3′ and CHGAPDHR-5′-ttctccatggtggtgaagac-3′).
IgM reversion assay.
Clones were stained weekly with a PE–α-chicken IgM antibody (clone M-1; Southern Biotechnology Associates, Inc.) and analyzed by a flow cytometer (FACScan; BD Biosciences).
Sequencing analysis.
IgL and IgH variable regions were PCR amplified using high-fidelity Pfu (Stratagene) or Phusion (New England Biolabs, Inc.) polymerases (IgL: CVLF1-5′-ccatggcctgggctcctctcctcctg-3′, CLA2-5′-gacagcacttacctggacagctg-3′; heavy chain: CVH1F1-5′-cgggagctccgtcagcgctctctgtcc-3′, CJH1R1-5′-ggggtacccggaggagacgatgacttcgg-3′) (43), TA-cloned (Invitrogen), and sequenced with the forward PCR primer or universal primers at the W.M. Keck Facility at the Yale University School of Medicine.
Sequences were aligned using CLUSTALW (http://align.genome.jp), and sequence variations in the IgL gene were categorized as long-track GCV, templated mutations, and nontemplated events. Long-track GCV events were those consisting of at least two nucleotide changes that were perfect matches to the same pseudoV (ψV) gene. Single nucleotide changes were scored as templated mutations when there was perfect identity between a ψV gene and the nucleotide change plus at least five nucleotides upstream and downstream of it. All other changes were classified as nontemplated events. Sequence diversification in IgH was not subcategorized, because the IgH ψV gene database is incomplete. Instead, the percentages of sequences that contained at least one nucleotide change relative to the germline sequence were calculated and used for comparisons.
Statistics.
Two-sample t tests were used to determine the statistical significance of differences in IgM reversion levels and mutation frequencies by sequencing using Data Desk software (version 6.2; Data Description, Inc.). χ2 analyses were performed to determine if the distribution of GCV/SHM events in different cell types were statistically significant.
Online supplemental material.
Fig. S1 is a Northern blot analysis of IgL expression in promoter-substituted cells. Fig. S2 is a time course analysis of IgM+ reversion in individual subclones of various promoter-substituted cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061835/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We wish to thank the entire Schatz Laboratory, M.J. Shlomchik, E. Ullu, and A.J. Koleske for critical discussions. We also wish to thank F. Edenhofer for his generous gift of the HTNC protein and K. Meyer for generously sharing reagents and information. All DNA sequencing was carried out at the W.M. Keck Facility at the Yale University School of Medicine.
This work was supported by grant AI066130 from the National Institutes of Health to D.G. Schatz. S.D. Fugmann was supported by the Irvington Institute for Immunological Research. D.G. Schatz is an investigator of the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; EF1-α, elongation factor 1-α; GCV, gene conversion; IgH, Ig heavy chain; Igκ, Ig light chain κ; IgL, Ig light chain; SHM, somatic hypermutation.
S.D. Fugmann's present address is National Institute on Aging, National Institutes of Health, Baltimore, MD 21224.
References
- 1.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, H., J. Hauschild, and J.M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 3.Harris, R.S., J.E. Sale, S.K. Petersen-Mahrt, and M.S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12:435–438. [DOI] [PubMed] [Google Scholar]
- 4.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 5.Saribasak, H., N.N. Saribasak, F.M. Ipek, J.W. Ellwart, H. Arakawa, and J.M. Buerstedde. 2006. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 176:365–371. [DOI] [PubMed] [Google Scholar]
- 6.Di Noia, J.M., and M.S. Neuberger. 2004. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34:504–508. [DOI] [PubMed] [Google Scholar]
- 7.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa, H., H. Saribasak, and J.M. Buerstedde. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rada, C., A. Gonzalez-Fernandez, J.M. Jarvis, and C. Milstein. 1994. The 5′ boundary of somatic hypermutation in a V kappa gene is in the leader intron. Eur. J. Immunol. 24:1453–1457. [DOI] [PubMed] [Google Scholar]
- 10.Winter, D.B., N. Sattar, J.J. Mai, and P.J. Gearhart. 1997. Insertion of 2 kb of bacteriophage DNA between an immunoglobulin promoter and leader exon stops somatic hypermutation in a kappa transgene. Mol. Immunol. 34:359–366. [DOI] [PubMed] [Google Scholar]
- 11.Lebecque, S.G., and P.J. Gearhart. 1990. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is ∼1 kb from V(D)J gene. J. Exp. Med. 172:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa, K., I.M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036. [DOI] [PubMed] [Google Scholar]
- 13.Fukita, Y., H. Jacobs, and K. Rajewsky. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 9:105–114. [DOI] [PubMed] [Google Scholar]
- 14.Peters, A., and U. Storb. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 4:57–65. [DOI] [PubMed] [Google Scholar]
- 15.Tumas-Brundage, K., and T. Manser. 1997. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J. Exp. Med. 185:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betz, A.G., C. Milstein, A. Gonzalez-Fernandez, R. Pannell, T. Larson, and M.S. Neuberger. 1994. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 77:239–248. [DOI] [PubMed] [Google Scholar]
- 17.Yelamos, J., N. Klix, B. Goyenechea, F. Lozano, Y.L. Chui, A. Gonzalez Fernandez, R. Pannell, M.S. Neuberger, and C. Milstein. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 376:225–229. [DOI] [PubMed] [Google Scholar]
- 18.Martin, A., and M.D. Scharff. 2002. Somatic hypermutation of the AID transgene in B and non-B cells. Proc. Natl. Acad. Sci. USA. 99:12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachl, J., C. Carlson, V. Gray-Schopfer, M. Dessing, and C. Olsson. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051–5057. [DOI] [PubMed] [Google Scholar]
- 20.Goyenechea, B., N. Klix, J. Yelamos, G.T. Williams, A. Riddell, M.S. Neuberger, and C. Milstein. 1997. Cells strongly expressing Ig(kappa) transgenes show clonal recruitment of hypermutation: a role for both MAR and the enhancers. EMBO J. 16:3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klix, N., C.J. Jolly, S.L. Davies, M. Bruggemann, G.T. Williams, and M.S. Neuberger. 1998. Multiple sequences from downstream of the J kappa cluster can combine to recruit somatic hypermutation to a heterologous, upstream mutation domain. Eur. J. Immunol. 28:317–326. [DOI] [PubMed] [Google Scholar]
- 22.Perlot, T., F.W. Alt, C.H. Bassing, H. Suh, and E. Pinaud. 2005. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 102:14362–14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Stoep, N., J.R. Gorman, and F.W. Alt. 1998. Reevaluation of 3′Ekappa function in stage- and lineage-specific rearrangement and somatic hypermutation. Immunity. 8:743–750. [DOI] [PubMed] [Google Scholar]
- 24.Inlay, M.A., H.H. Gao, V.H. Odegard, T. Lin, D.G. Schatz, and Y. Xu. 2006. Roles of the Ig kappa light chain intronic and 3′ enhancers in Igk somatic hypermutation. J. Immunol. 177:1146–1151. [DOI] [PubMed] [Google Scholar]
- 25.Conlon, T.M., and K.B. Meyer. 2006. The chicken Ig light chain 3′-enhancer is essential for gene expression and regulates gene conversion via the transcription factor E2A. Eur. J. Immunol. 36:139–148. [DOI] [PubMed] [Google Scholar]
- 26.Michael, N., H.M. Shen, S. Longerich, N. Kim, A. Longacre, and U. Storb. 2003. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 19:235–242. [DOI] [PubMed] [Google Scholar]
- 27.Schoetz, U., M. Cervelli, Y.D. Wang, P. Fiedler, and J.M. Buerstedde. 2006. E2A expression stimulates Ig hypermutation. J. Immunol. 177:395–400. [DOI] [PubMed] [Google Scholar]
- 28.Buerstedde, J.M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 67:179–188. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S., E.H. Humphries, L. Tjoelker, L. Carlson, and C.B. Thompson. 1990. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol. 10:3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buerstedde, J.M., C.A. Reynaud, E.H. Humphries, W. Olson, D.L. Ewert, and J.C. Weill. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heltemes, L.M., C.K. Tuggle, and S.J. Lamont. 1997. Octamer function in the chicken lambda immunoglobulin light chain promoter. Immunogenetics. 47:73–76. [DOI] [PubMed] [Google Scholar]
- 32.Bulfone-Paus, S., L. Reiners-Schramm, and R. Lauster. 1995. The chicken immunoglobulin lambda light chain gene is transcriptionally controlled by a modularly organized enhancer and an octamer-dependent silencer. Nucleic Acids Res. 23:1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauster, R., C.A. Reynaud, I.L. Martensson, A. Peter, D. Bucchini, J. Jami, and J.C. Weill. 1993. Promoter, enhancer and silencer elements regulate rearrangement of an immunoglobulin transgene. EMBO J. 12:4615–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachl, J., C. Olsson, N. Chitkara, and M. Wabl. 1998. The Ig mutator is dependent on the presence, position, and orientation of the large intron enhancer. Proc. Natl. Acad. Sci. USA. 95:2396–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachl, J., and M. Wabl. 1996. Enhancers of hypermutation. Immunogenetics. 45:59–64. [DOI] [PubMed] [Google Scholar]
- 36.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 38.Besmer, E., E. Market, and F.N. Papavasiliou. 2006. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol. Cell. Biol. 26:4378–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nambu, Y., M. Sugai, H. Gonda, C.G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 302:2137–2140. [DOI] [PubMed] [Google Scholar]
- 40.Arakawa, H., D. Lodygin, and J.M. Buerstedde. 2001. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peitz, M., K. Pfannkuche, K. Rajewsky, and F. Edenhofer. 2002. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA. 99:4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sale, J.E., D.M. Calandrini, M. Takata, S. Takeda, and M.S. Neuberger. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 412:921–926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]