Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin (original) (raw)
Abstract
Aberrant expression of Jagged1 and Notch1 are associated with poor outcome in breast cancer. However, the reason that Jagged1 and/or Notch overexpression portends a poor prognosis is unknown. We identify Slug, a transcriptional repressor, as a novel Notch target and show that elevated levels of Slug correlate with increased expression of Jagged1 in various human cancers. Slug was essential for Notch-mediated repression of E-cadherin, which resulted in β-catenin activation and resistance to anoikis. Inhibition of ligand-induced Notch signaling in xenografted Slug-positive/E-cadherin–negative breast tumors promoted apoptosis and inhibited tumor growth and metastasis. This response was associated with down-regulated Slug expression, reexpression of E-cadherin, and suppression of active β-catenin. Our findings suggest that ligand-induced Notch activation, through the induction of Slug, promotes tumor growth and metastasis characterized by epithelial-to-mesenchymal transition and inhibition of anoikis.
Notch signaling is initiated when a Notch ligand interacts with a Notch transmembrane receptor expressed on an adjacent cell (1). This interaction triggers a series of proteolytic digestions that releases the Notch intracellular domain (NotchIC), allowing it to translocate into the nucleus. Within the nucleus, NotchIC binds to the transcriptional repressor CSL, resulting in derepression and coactivation of Notch downstream target genes and thereby regulating various cellular processes, including differentiation, proliferation, and apoptosis. Interestingly, in the development of cancer, Notch may act as either an oncogene or a tumor suppressor gene depending on the tumor type (2).
Mammary-specific overexpression of constitutively active Notch1IC, Notch3IC, or Notch4IC in mice leads to the formation of aggressive, metastatic breast tumors (3, 4). Recent studies have also highlighted a potential role for Notch signaling in human breast cancer development. Expression of all four Notch receptors has been reported in human breast tumors at varying frequencies (5). Poorly differentiated breast tumors are associated with elevated Notch1 protein levels and reduced patient survival (6). Interestingly, an association between increased mRNA expression of the Notch ligand Jagged1 and reduced survival in patients with breast cancer has recently been reported, with high-level coexpression of Jagged1 and Notch1 mRNA defining a subset of patients with very poor outcome (7). Notch has also been reported to be activated downstream of Ras and Wnt in the promotion of mammary tumors through the induction of Notch ligands and/or receptors (8, 9). Notch signaling may contribute to tumorigenesis by promoting mammary epithelial cell growth or inhibiting apoptosis (10, 11). However, much remains to be learned about the molecular mechanisms of Notch-mediated oncogenesis.
Numerous reports have indicated a role for epithelial-to-mesenchymal transition (EMT) in promoting the invasion and dissemination of malignant cells, particularly in breast cancer (12). Recent studies have suggested that Notch signaling induces a specialized type of EMT during normal heart development and that Notch up-regulates Snail in endothelial cells to promote mesenchymal transformation (13, 14). However, there is no direct or even correlative in vivo data that Notch regulates EMT in epithelial cancers.
In this paper, we identify Slug, a zinc-finger transcriptional repressor functionally linked to human breast cancer progression and metastasis (15), to be a direct downstream target gene of Notch that is up-regulated in Jagged1- and Notch1-positive human breast cancers. Jagged1-mediated activation of Notch in breast epithelial cells induces EMT through induction of Slug and subsequent repression of the cell–cell adhesion protein E-cadherin. Because Slug can be induced by factors other than Notch, we identify Notch downstream target genes of the HEY family as potential markers of primary human breast tumors that have activated the Jagged1–Notch–Slug signaling axis. In Slug-positive/E-cadherin–negative human breast cancer xenografts, inhibition of ligand-induced Notch signaling inhibits growth of the primary tumor and distant metastases, which correlates with reduced Slug expression and reexpression of E-cadherin. E-cadherin reexpression, either through Notch inhibition or enforced expression, is associated with relocalization of β-catenin from the nucleus to the plasma membrane and reversal of β-catenin activation in xenografted breast tumors. Our findings suggest a critical role for induction of EMT and inhibition of anoikis in promoting an aggressive phenotype in tumors exhibiting ligand-induced Notch signaling.
RESULTS
Notch activation inhibits E-cadherin expression in human breast epithelial cells through the induction of Slug
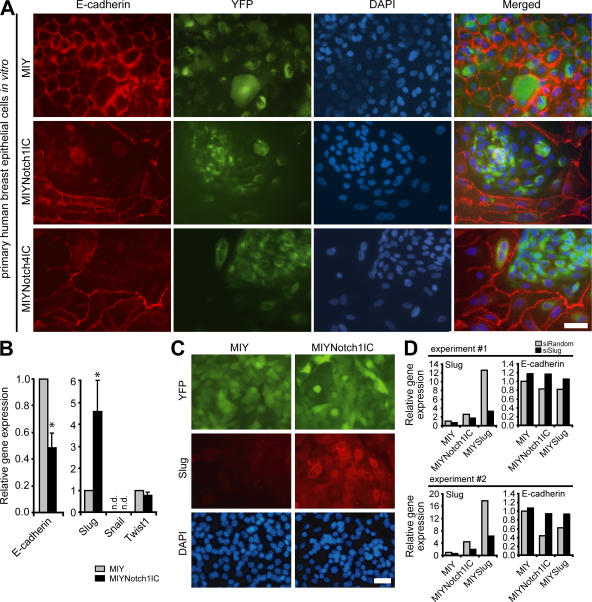
Down-regulation of E-cadherin is one of the best markers of EMT in human breast cancer (12). To determine whether Notch activation induces EMT in human breast epithelial cells as manifested by repression of E-cadherin, the E-cadherin–positive normal human breast epithelial cell line MCF-10A was transduced with a retroviral vector (MIY) linking yellow fluorescent protein (YFP) to activated Notch1 (Notch1IC) or activated Notch4 (Notch4IC). Hence, cells that express Notch1IC or Notch4IC also express YFP. Expression of either Notch1IC or Notch4IC caused this normal breast epithelial cell line to down-regulate E-cadherin, dissociate cell–cell contacts, and acquire a spindle-shaped morphology, consistent with mesenchymal transformation (Fig. S1, A–C, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1). A similar ability of activated Notch to down-regulate E-cadherin was demonstrated in primary human breast epithelial cells (Fig. 1 A).
Figure 1.
Notch activation inhibits E-cadherin expression in human breast epithelial cells through the induction of Slug. (A) Immunofluorescent staining for E-cadherin (red), YFP (green), and DAPI (blue) in primary human breast epithelial cells transduced with MIY, MIYNotch1IC, or MIYNotch4IC. Bar, 50 μm. (B) qPCR for expression of E-cadherin, Slug, Snail, and Twist1 in MCF-10A cells transduced with MIY or MIYNotch1IC. Data are expressed as the relative gene expression level, with MIY control as the comparator, and are from three independent experiments (mean + SEM). *, P ≤ 0.05. (C) Immunofluorescent staining for Slug (red), YFP (green), and DAPI (blue) in MCF-10A cells transduced with MIY or MIYNotch1IC. Bar, 50 μm. (D) qPCR for expression of Slug and E-cadherin in MCF-10A cell lines (MIY, MIYNotch1IC, or MIYSlug) transiently transfected with siRandom or siSlug. Data from two independent experiments are shown and are expressed as the relative gene expression level with MIY control as the comparator. n.d., not detectable.
To identify a potential mechanism of Notch-mediated E-cadherin silencing, expression of three known E-cadherin repressors that initiate EMT in breast cancer—Slug, Snail, and Twist1 (16)—was assessed in a normal breast epithelial cell line by quantitative RT-PCR (qPCR). In contrast to what has been reported in endothelial cells (13), Snail mRNA expression was not detected in either Notch1IC-expressing or control cells. Twist1 mRNA levels did not differ between control and Notch1IC-expressing cells (Fig. 1 B). Slug mRNA expression, however, was significantly increased in Notch1IC cells, which was associated with a decrease in E-cadherin mRNA expression (Fig. 1 B). Notch1IC-induced expression of Slug protein was confirmed by immunofluorescence microscopy (Fig. 1 C). Knockdown of Slug, achieved by delivering small interfering RNA (siRNA) targeting Slug, into cells transduced with either Notch1IC or Slug was sufficient to restore E-cadherin expression (Fig. 1 D).
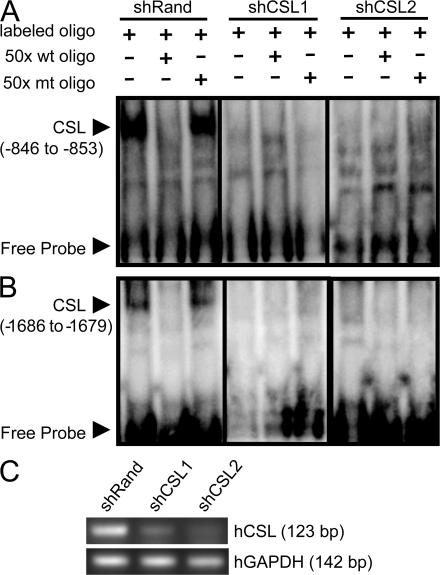
To determine whether Slug is a direct target of Notch/CSL, we examined the human Slug promoter and identified two potential CSL-binding consensus motifs (−846 to −853 and −1686 to −1679 relative to the transcriptional start site). EMSAs using double-stranded oligonucleotides spanning these sites showed a clear gel shift of MDA-MB-231 human breast carcinoma nuclear lysates (Fig. 2, A and B). The binding of each of the labeled oligonucleotides was competed away with unlabeled wild-type, but not mutated, double-stranded oligonucleotide (Fig. 2, A and B). In addition, lentiviral-delivered short hairpin RNAs (shRNAs) targeting two distinct sites of CSL (Fig. 2 C) also prevented the gel shift, indicating that these sites in the Slug promoter bind endogenous CSL and, thus, confirming that Slug is a direct target of Notch/CSL. These results, combined with our data demonstrating the ability of Slug alone (independent of Notch1IC) to induce a spindle-shaped morphology and down-regulate E-cadherin expression in normal breast epithelial cells (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1), identify Slug as a key player in the mechanism of Notch-induced EMT.
Figure 2.
Slug is a direct target of Notch signaling. EMSA of nuclear lysates from MDA-MB-231 human breast cancer cells transduced with nonspecific shRNA (shRand) or two different shRNAs targeting CSL (shCSL1 and shCSL2). CSL consensus binding sites in the human Slug promoter (A, −846 to −853 [TATGGGAA]; and B, −1686 to −1679 [TGTGGGAA]) relative to the transcriptional start site were used as the 32P-labeled probe, and either nonradioactive wild-type or mutated (mt) oligonucleotides were used as competitors in 50-fold excess. The CSL–DNA protein complex and the free DNA probe are identified by arrows. (C) shRNA-mediated knockdown of CSL in MDA-MB-231 cell lines was verified by RT-PCR analysis.
Jagged1/Notch1 correlates with Slug expression in human breast cancers
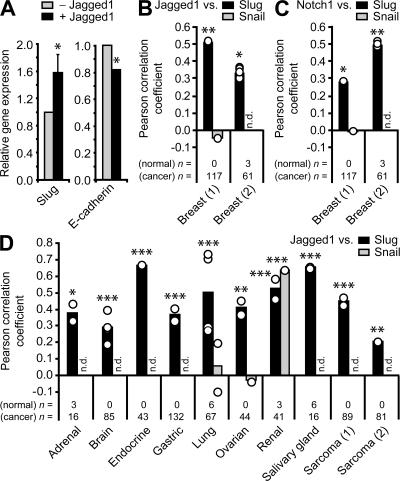
To determine whether Jagged1-triggered Notch activation could induce Slug expression, Jagged1-expressing mouse endothelial cells (from the SVEC 4-10 cell line) were co-cultured with normal parental MCF-10A cells, and qPCR using human-specific primers was used to measure Slug expression in the human breast epithelial cells. Jagged1-induced Notch activation, as demonstrated by induction of the target gene HEY1 (Fig. S2 E), resulted in increased levels of Slug mRNA levels with a concomitant decrease in E-cadherin mRNA expression in these normal breast epithelial cells (Fig. 3 A). These findings show that Jagged1 expression can activate Notch signaling in a juxtacrine manner to induce Slug expression and mesenchymal transformation of breast epithelial cells.
Figure 3.
Jagged1 and Slug expression are correlated in human breast epithelial cells and in primary human breast cancer. (A) qPCR for expression of Slug and E-cadherin in MCF-10A parental cells co-cultured with mouse endothelial cells transduced with MIY vector control (−Jagged1) or MIYJagged1 (+Jagged1). Human-specific primers were used to avoid amplification of mouse transcripts and limit analysis to MCF-10A cells. Data are expressed as the relative gene expression level, with the empty vector control co-culture (−Jagged1) as the comparator, and are from three independent experiments (mean + SEM). Slug: *, P ≤ 0.05; E-cadherin: *, P < 0.0001. (B–D) Expression correlations in primary human cancers. Pearson correlation coefficients were obtained from microarray datasets deposited in the Oncomine database. For each dataset, individual correlations between two genes of interest (as well as all possible correlations in cases where replicate probes were present in the microarray) are represented by open circles. Bars represent the mean Pearson correlation coefficient. The numbers of normal and cancer specimens included in the correlation analysis for each dataset are indicated. (B) Expression correlations between Jagged1 and Slug in primary human breast cancer. Jagged1 expression correlations with Snail are also shown. *, P < 0.01; **, P < 0.001. (C) Expression correlations between Notch1 and Slug in primary human breast cancer. Notch1 expression correlations with Snail are also shown. *, P < 0.01; **, P < 0.001. (D) Expression correlations between Jagged1 and Slug in various primary human cancers. Where available, expression correlations between Jagged1 and Snail are also shown. *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001. n.d., not determined.
Poor prognosis for breast cancer patients has been shown to correlate with elevated levels of expression of either Slug (16) or the Notch ligand Jagged1 (7). To examine whether increased Slug expression would be found in breast cancers showing increased Jagged1 or Notch1 expression, we accessed a compendium of 132 independent gene expression datasets representing >10,000 microarray experiments in the Oncomine database (17). Expression of both Jagged1 and Notch1 correlated positively with Slug in two independent breast cancer datasets (Fig. 3, B and C). Of significance, a positive correlation between the expression of Jagged1 and Slug, but not Snail, was observed in numerous cancers other than the breast, which was consistent with our finding that Jagged1 is capable of inducing Slug but not Snail (Fig. 3 D).
HEY genes are potential markers of human breast cancers that exhibit activation of the Jagged1–Notch–Slug signaling axis
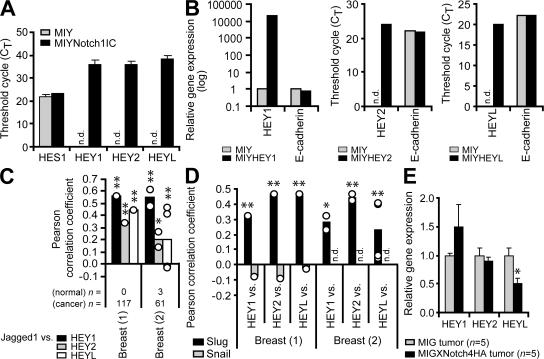
In response to Notch activation, transcriptional repressors of the HES and HEY families of basic helix-loop-helix proteins are induced (18). Cell type and context determine which members are induced (18). qPCR was used to analyze whether HES1 and HEY1/2/L were up-regulated in response to Notch activation in breast epithelial cells. Although HES1 levels did not differ between vector control and Notch1IC cells, all three HEY genes were absent in control cells and up-regulated in the presence of Notch1IC (Fig. 4 A).
Figure 4.
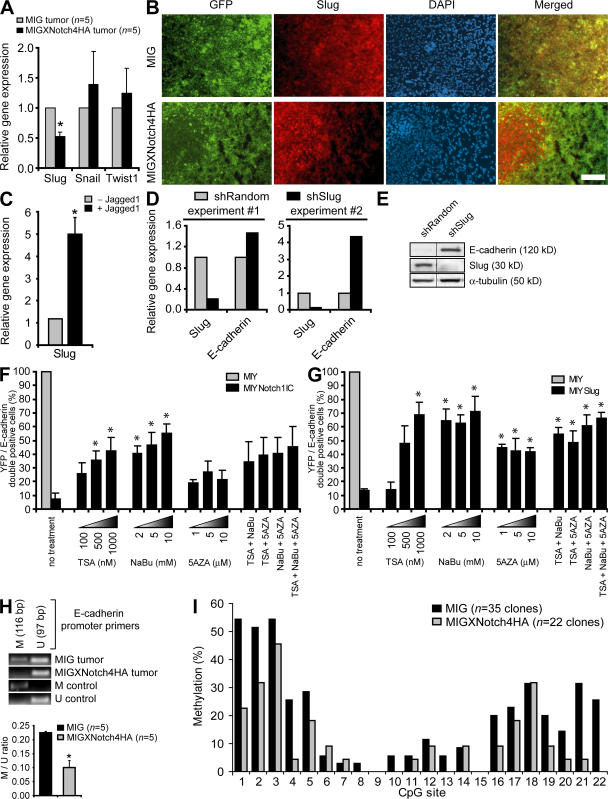
HEY genes are potential markers of human breast cancers that exhibit activation of the Jagged1–Notch–Slug signaling axis. (A) qPCR for Notch target genes (HES1, HEY1, HEY2, and HEYL) in MCF-10A MIY and MIYNotch1IC cell lines. Data shown are the mean threshold cycle number (CT) + SEM from three independent experiments. n.d., not detectable. (B) qPCR for gene expression in MCF-10A cells transduced with MIY, MIYHEY1, MIYHEY2, or MIYHEYL. Data show relative gene expression level or threshold cycle number (CT). n.d., not detectable. (C and D) Expression correlations in primary human breast cancers. Pearson correlation coefficients were obtained from microarray datasets deposited in the Oncomine database. For each dataset, correlations between two genes of interest (as well as all possible correlations in the event of replicate genes in the microarray) are represented by open circles. Bars represent the mean Pearson correlation coefficient. The number of normal and cancer specimens included in the correlation analysis for each dataset are indicated. n.d., not determined. (C) Expression correlations between Jagged1 and each of the HEY target genes in primary human breast cancer. *, P ≤ 0.05; **, P < 0.001. (D) Expression correlations between each of the HEY target genes and Slug in primary human breast cancer. Where available, expression correlations between each of the HEY target genes and Snail are also shown. *, P < 0.01; **, P < 0.001. (E) Semiquantitative RT-PCR for Notch target genes in MDA-MB-231 MIG and MIGXNotch4HA tumor xenografts. Data are expressed as the relative gene expression level with MIG control tumor as the comparator (mean + SEM). *, P ≤ 0.05. n.d.
Because HEY proteins, similar to Slug, silence gene expression by binding to E-boxes in target gene promoters (18), we determined whether enforced expression of any one of the HEY proteins was sufficient to down-regulate E-cadherin. In contrast to Slug (Fig. S2, B-E), none of the three HEY proteins altered E-cadherin transcript levels in these cells (Fig. 4 B).
Although these findings suggest that repression of E-cadherin is mediated by Slug and not HES/HEY, HEY genes may represent surrogate markers of activation of the Jagged1–Notch–Slug signaling axis in human breast cancer. Examination of two independent breast cancer microarray datasets confirmed positive expression correlations between Jagged1 and HEY1, HEY2, and HEYL, respectively (Fig. 4 C). Moreover, the expression of HEY1, HEY2, and HEYL were all positively correlated with the expression of Slug but not Snail (Fig. 4 D). Thus, the HEY genes may be potential markers of human breast cancers that exhibit Notch activation and may, therefore, classify a subset of breast cancer patients that would benefit from therapeutics specifically designed to target the Jagged1–Notch–Slug pathway.
Inhibition of ligand-induced Notch activation blocks growth and metastasis of breast tumors in vivo
To determine whether blockade of Notch ligand–receptor interaction would reverse Slug-induced EMT and inhibit breast tumor invasion and metastasis, we used the Slug-positive/E-cadherin–negative MDA-MB-231 human breast carcinoma cell line. MDA-MB-231 cells possess a wild-type E-cadherin gene and, thus, exhibit reversible E-cadherin silencing (19), and they also express Notch4 (20). We confirmed Notch4 expression and also found that MDA-MB-231 cells expressed multiple other Notch receptors and ligands, thus providing these cells with the potential to activate Notch signaling through juxtacrine/autocrine ligand–receptor interactions (Fig. S3 A, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1).
To target the Notch pathway in vivo, we chose to use a soluble Notch receptor, which has been shown to block ligand-induced Notch signaling (21), rather than γ-secretase inhibitors that can directly increase the expression of E-cadherin by preventing its proteolysis (22). MDA-MB-231 cells were retrovirally transduced with the soluble ectodomain of human Notch4 (XNotch4) to block ligand-induced Notch activation. Secretion of the soluble protein was confirmed by immunoblotting the concentrated medium in which the cells were cultured (Fig. S3 B). The ability of XNotch4 to inhibit tumor growth in vivo was tested in a xenograft model by implanting XNotch4-secreting MDA-MB-231 cells subcutaneously into immunodeficient mice. Expression of XNotch4 protein in the xenografts was confirmed by immunohistochemistry (Fig. S3 C).
We assessed the mRNA levels of HEY genes in this model to determine whether any of the HEY genes were down-regulated and to confirm inhibition of the Notch pathway. Interestingly, of the three Notch target genes assessed, only HEYL exhibited a decrease in mRNA expression similar to Slug in XNotch4 tumors (Fig. 4 E and see Fig. 6 A). The reason for this is not clear, but given that some HEY genes are also downstream targets of other signaling pathways, such as TGF-β (23), it is possible that in our model HEYL is up-regulated solely by the Notch pathway, whereas other factors may also be up-regulating HEY1/2. Hence, inhibition of Notch would only block HEYL expression, but would not abrogate expression of HEY1/2.
Figure 6.
Restoration of E-cadherin expression by Notch inhibition is associated with Slug down-regulation and attenuation of E-cadherin promoter methylation. (A) qPCR for expression of Slug, Snail, and Twist1 in MDA-MB-231 MIG and MIGXNotch4HA tumors. Data are expressed as the relative gene expression level with MIG control tumors as the comparator (mean + SEM). *, P ≤ 0.01. (B) Immunofluorescent staining for Slug (red), GFP (green), and DAPI (blue) in MDA-MB-231 MIG and MIGXNotch4HA tumor xenografts. Yellow represents the overlap of GFP and Slug immunostaining. Bar, 100 μm. (C) qPCR for gene expression in MDA-MB-231 parental cells co-cultured with mouse endothelial cells transduced with MIY vector control (−Jagged1) or MIYJagged1 (+Jagged1). Human-specific primers were used to assay Slug specifically in MDA-MB-231 cells and avoid amplification of mouse transcripts. Data are expressed as the relative gene expression level, with the empty vector control co-culture (−Jagged1) as the comparator, and are from three independent experiments (mean + SEM). *, P ≤ 0.05. (D) qPCR for expression of Slug and E-cadherin in MDA-MB-231 cells transduced with shRandom or shSlug. Data from two independent experiments are shown and are expressed as the relative gene expression level with shRandom control as the comparator. (E) Immunoblot for expression of E-cadherin, Slug, and α-tubulin in MDA-MB-231 cells transduced with shRandom or shSlug. (F) Quantitation of the proportion of MCF-10A cells transduced with MIY or MIYNotch1IC that are positive for both YFP and E-cadherin. Cells were treated with TSA (100/500/1000 nM), NaBu (2/5/10 mM), or 5AZA (1/5/10 μM) alone or in combination, and E-cadherin expression was assessed 3 d after treatment by immunofluorescent microscopy. Data shown represent mean + SEM of at least three independent experiments. *, P ≤ 0.05. (G) Quantitation of the proportion of MCF-10A cells transduced with MIY or MIYSlug that are positive for both YFP and E-cadherin. Cells were treated with TSA (100/500/1000 nM), NaBu (2/5/10 mM), or 5AZA (1/5/10 μM) alone or in combination. Data shown represent mean + SEM of at least three independent experiments. *, P ≤ 0.05. (H) MSP to assess methylation status of the E-cadherin promoter in MDA-MB-231 tumors. Primers specific for methylated (M) or unmethylated (U) E-cadherin promoter were used. Amplified M and U products were quantitated by densitometry and expressed as the M/U ratio. Data shown represent mean + SEM. *, P < 0.001. (I) Genomic bisulfite sequencing to assess methylation status of the E-cadherin promoter in MDA-MB-231 tumors (MIG, n = 35 clones from five tumors; MIGXNotch4HA, n = 22 clones from three tumors). A total of 22 CpG sites within the E-cadherin proximal promoter (−104 to +118) were analyzed. Data shown represent the percentage of methylation observed at each CpG site.
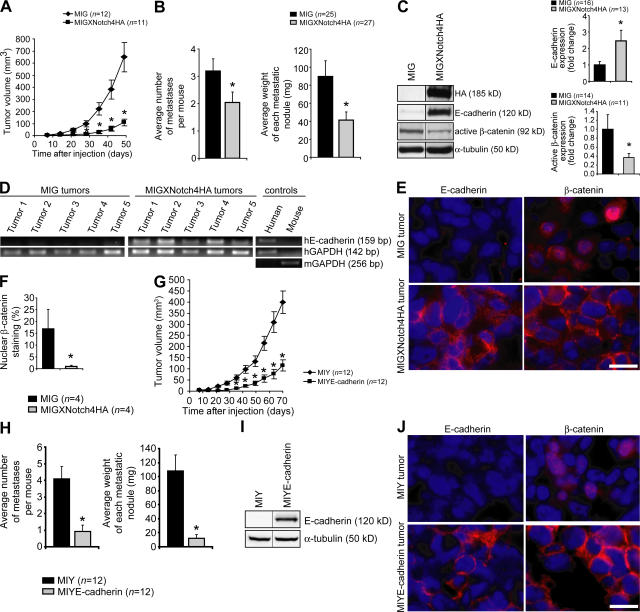
Tumor growth was significantly inhibited using XNotch4 to inhibit ligand-induced Notch signaling (Fig. 5 A). Inhibition of tumor growth was also achieved by enforced expression of soluble extracellular Notch1 or dominant-negative CSL, confirming that direct inhibition of Notch signaling within the tumor cells attenuates growth in MDA-MB-231 tumors (unpublished data). In addition to inhibiting the growth of the primary implant, attenuation of Notch signaling also reduced the number and size of metastases, as would be expected with inhibition of EMT (Fig. 5 B).
Figure 5.
Inhibition of ligand-induced Notch activation blocks breast tumor growth and metastasis, restores E-cadherin expression, and inactivates β-catenin in vivo. (A) Tumor growth curves for MDA-MB-231 cells transduced with MIG or MIGXNotch4HA grown as xenografts on the dorsa of immunodeficient mice. Data are presented as the mean ± SEM of the tumor volumes. *, P < 0.01. (B) Quantitation of metastases in MDA-MB-231 MIG and MIGXNotch4HA tumor-bearing mice. Data shown represent the mean number of metastases per mouse + SEM and the mean weight of each metastatic nodule + SEM. *, P ≤ 0.05. (C) Immunoblots for expression of XNotch4HA, E-cadherin, active β-catenin, and α-tubulin in MDA-MB-231 MIG and MIGXNotch4HA tumors. Protein expression was quantitated by densitometry and normalized to α-tubulin. Data shown represent mean + SEM. *, P ≤ 0.05. (D) RT-PCR for expression of human E-cadherin in MDA-MB-231 MIG and MIGXNotch4HA tumors. Human-specific primers were used to avoid amplification of mouse E-cadherin. (E) Immunofluorescent staining for E-cadherin (red), β-catenin (red), and DAPI (blue) in MDA-MB-231 MIG and MIGXNotch4HA tumors. Bar, 15 μm. (F) Quantitation of the proportion of cells exhibiting nuclear β-catenin staining in MDA-MB-231 MIG and MIGXNotch4HA tumors. Data shown represent mean + SEM. *, P ≤ 0.05. (G) Tumor growth curves for MDA-MB-231 cells transduced with MIY or MIYE-cadherin grown as xenografts on the dorsa of immunodeficient mice. Data are presented as the mean ± SEM of the tumor volumes. *, P < 0.001. (H) Quantitation of metastases in MDA-MB-231 MIY and MIYE-cadherin tumor-bearing mice. Data shown represent the mean number of metastases per mouse + SEM and the mean weight of each metastatic nodule + SEM. *, P < 0.001. (I) Immunoblots for expression of E-cadherin and α-tubulin in MDA-MB-231 MIY and MIYE-cadherin tumors. (J) Immunofluorescent staining for E-cadherin (red), β-catenin (red), and DAPI (blue) in MDA-MB-231 MIY and MIYE-cadherin tumors. Bar, 15 μm.
Because aberrant Notch signaling results in disrupted blood vessel development (24–26), we determined whether attenuated tumor growth could be explained by an antiangiogenic effect of XNotch4. We did not observe differences in vascular density in the implanted tumors, suggesting that angiogenesis inhibition is not the mechanism of tumor suppression in this model (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1). These findings show that inhibition of ligand-induced Notch signaling in breast tumor cells can inhibit tumor growth and metastasis.
Inhibition of ligand-induced Notch signaling restores E-cadherin expression and inactivates β-catenin in breast tumors in vivo
Having demonstrated an antitumor effect of XNotch4 on breast tumor growth in vivo, we sought to determine whether inhibition of Notch signaling would reinduce expression of E-cadherin in the tumor cells. Lysates from XNotch4 tumor xenografts exhibited E-cadherin protein expression in contrast to tumors lacking XNotch4, which remained E-cadherin negative (Fig. 5 C). Because the antibody used recognizes both human and mouse E-cadherin, RT-PCR was performed with human E-cadherin–specific primers to confirm E-cadherin reexpression in the XNotch4 tumor cells (Fig. 5 D). Functional reexpression of E-cadherin at the plasma membrane of the xenografted tumor cells in response to Notch inhibition was demonstrated by immunofluorescent microscopy (Fig. 5 E).
β-Catenin contributes to breast tumorigenesis by regulating the expression of genes involved in proliferation, invasion, and EMT (27). When β-catenin is bound to E-cadherin, signaling-competent nuclear β-catenin levels diminish, and cell proliferation and invasion are suppressed (28). To determine whether surface E-cadherin reexpression in XNotch4 tumors would affect β-catenin activity, we immunoblotted tumor cell lysates with an antibody specific for active β-catenin and found significantly reduced β-catenin activity in tumors where Notch activation was blocked (Fig. 5 C). Consistent with this finding, β-catenin relocated from the nucleus to the plasma membrane in tumors where Notch was inhibited (Fig. 5, E and F).
To verify that activated Notch is able to activate β-catenin function, we examined expression of the β-catenin target genes in Notch-activated MCF-10A cells by qPCR. Interestingly, Notch activation induced Axin2 and APCDD1 but not Lef1 (Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1). In parental MCF-10A cells, Wnt3a stimulation also induced Axin2, but not Lef1 (Fig. S5 B), suggesting that the active β-catenin levels correlate with transcriptional activation. However, conditioned medium from Notch-activated cells did not induce Axin2 (Fig. S5 C), suggesting that the observed β-catenin activation is independent of a secreted factor. Further, inhibition of Wnt activation by Dkk1 did not block activation of the β-catenin target Axin2 in Notch-activated cells, whereas Dkk1 did block Wnt3a-induced Axin2 (Fig. S5, C and D), thereby confirming Wnt-independent activation of β-catenin by Notch.
To determine whether enforced expression of E-cadherin was sufficient to reproduce the phenotype induced by blockade of Notch signaling, we transduced MDA-MB-231 cells with E-cadherin complementary DNA (cDNA) and implanted vector- or E-cadherin–expressing cells onto the backs of immunodeficient mice. Enforced expression of E-cadherin (independent of Notch inhibition) was sufficient to inhibit tumor growth and metastasis with concomitant relocalization of β-catenin to the plasma membrane (Fig. 5, G–J). These data suggest that inhibition of ligand-induced Notch activation is tumor suppressive, in part because of reinduction of surface E-cadherin expression. This in turn retains β-catenin at the plasma membrane, thereby attenuating β-catenin nuclear activity and inhibiting tumor growth and metastasis.
Restoration of E-cadherin expression by Notch inhibition is caused by Slug down-regulation and attenuation of E-cadherin promoter methylation
To determine whether inhibition of ligand-induced Notch signaling restored E-cadherin expression secondary to repression of Slug, expression of the genes encoding Slug, Snail, and Twist1 was assessed by qPCR in the breast tumor xenografts. Although transcript levels of Snail and Twist1 did not differ between control and XNotch4 tumors, Slug expression was significantly reduced when Notch signaling was inhibited (Fig. 6 A). XNotch4 also blocked expression of Slug protein, because Slug was only present in areas of the tumor where XNotch4 was absent, as determined by immunofluorescent staining of breast tumor xenografts (Fig. 6 B).
To directly demonstrate that Jagged1 could induce Slug in breast tumor cells, MDA-MB-231 parental cells were co-cultured with mouse stromal cells transduced with either Jagged1 or empty vector, and human Slug transcript levels were quantitated by RT-PCR using human-specific Slug primers. In response to Jagged1-induced Notch signaling, Slug transcripts in MDA-MB-231 cells were increased approximately fivefold (Fig. 6 C). To prove that Slug was responsible for E-cadherin repression in these cells, Slug was targeted using lentiviral-delivered shRNA in breast tumor cells. Knockdown of Slug was confirmed by qPCR and immunoblotting, and was found to be sufficient to restore expression of E-cadherin (Fig. 6, D and E).
Because Slug recruits histone deacetylase (HDAC) complexes to mediate transcriptional silencing (29), we determined whether HDAC activity was required for the ability of Notch-induced Slug to down-regulate E-cadherin expression. Treatment of NotchIC- or Slug-expressing breast epithelial cells with the HDAC inhibitors trichostatin A (TSA) and sodium butyrate (NaBu), either alone or together, was sufficient to reverse the down-regulation of E-cadherin expression by Notch1IC or Slug (Fig. 6, F and G). A current molecular model for transcriptional gene silencing suggests that histone deacetylation is a primary event involved in the initiation of chromatin compaction, and that DNA methylation functions subsequent to histone deacetylation to establish a permanent state of gene inactivation (30). NotchIC- or Slug-expressing cells were thus treated with the DNA methyltransferase inhibitor 5-azacytidine (5AZA), which also elicited reinduction of E-cadherin expression. However, 5AZA and TSA/NaBu together did not produce an additive effect on E-cadherin reexpression, suggesting that DNA methylation may occur secondary to histone deacetylation, as previously suggested (30).
The E-cadherin promoter contains numerous cytosine-phosphate-guanine (CpG) sites, which result in E-cadherin silencing when methylated on the corresponding cytosine residue (31). Given that DNA methylation has been reported to be dominant over histone deacetylation in mediating gene silencing (32), as well as our data suggesting that methylation may occur as a later step in silencing the E-cadherin promoter, we sought to determine whether reduced Slug expression and reexpression of E-cadherin in our breast tumor xenografts correlated with reduced DNA methylation at the E-cadherin promoter. Using two independent methods, methylation-specific PCR (MSP) and genomic bisulfite sequencing, E-cadherin promoter methylation in XNotch4 tumors was found to be reduced compared with control tumors (Fig. 6, H and I). Because XNotch4 tumors did not display generalized hypomethylation of the genome (Fig. S6, A and B, available at http://www.jem.org/cgi/content/full/jem.20071082/DC1), Notch inhibition likely results in demethylation at specific promoters regulated by Notch/Slug. These results suggest that inhibition of Notch signaling reinduces E-cadherin by repressing Slug and reversing E-cadherin promoter methylation.
The Notch–Slug signaling axis inhibits anoikis of human breast cells
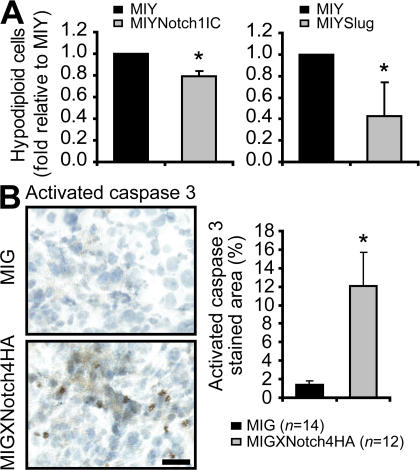
During cancer progression and dissemination, malignant cancer cells enter the circulation or lymphatic system and then must survive in the vasculature until they successfully extravasate into a tissue site. Cell death associated with abolished matrix-initiated integrin signaling has been termed anoikis. For a tumor cell to metastasize to a distant site, it needs to overcome anoikis. Activated Notch inhibits apoptosis in response to various triggers in different cell types, but it has also been reported to induce apoptosis in other situations (11, 33, 34). We thus asked whether breast epithelial cells expressing Notch1IC or Slug would exhibit a survival advantage compared with control cells when maintained under conditions that prevented adhesion. For this assay, cells were placed in suspension cultures, rather than soft agar (11), to mimic transit through the bloodstream. Both NotchIC- and Slug-expressing breast epithelial cells exhibited protection against anoikis compared with control cells (Fig. 7 A). Conversely, breast tumor xenografts in which Notch signaling was inhibited showed reduced Slug expression (Fig. 6, A and B), with a concomitant increase in cell death as measured by activated Caspase 3 levels (Fig. 7 B).
Figure 7.
The Notch–Slug signaling axis inhibits anoikis of human breast cells. (A) Anoikis assay to assess cell death in MCF-10A cell lines (MIY, MIYNotch1IC, and MIYSlug). The fraction of hypodiploid cells was determined by flow cytometry, and the data representing the mean + SEM of three independent experiments are expressed as the fold change in hypodiploid cells relative to MIY control. MIYNotch1IC: *, P < 0.01; MIYSlug: *, P ≤ 0.05. (B) Immunohistochemical staining for activated caspase 3 in implanted MDA-MB-231 MIG and MIGXNotch4HA tumors. Data representing the mean + SEM are shown as the proportion of the activated caspase 3–stained area compared with the total tumor area. *, P = 0.012. Bar, 25 μm.
DISCUSSION
Although mouse models have indicated that mammary-specific overexpression of a truncated constitutively active Notch can result in breast tumors, human data suggest that both Notch ligands and receptors are up-regulated in a proportion of breast cancers and that this expression is correlated with poor outcome. The implication is that juxtacrine or autocrine Notch activation, rather than activating Notch mutations, may be responsible for an aggressive tumor phenotype. This is in marked contrast to T cell acute lymphoblastic leukemia, where more than half of the cases have activating mutations of Notch1 (35). In this paper, we have shown that Jagged1 activation of Notch up-regulates the transcriptional repressor Slug to promote carcinogenesis by two cellular mechanisms: (a) by facilitating cancer cell metastasis through initiation of EMT, and (b) by enhancing cell survival in the absence of cell matrix adhesion. Importantly, we have demonstrated that, in human breast cancers, expression of Jagged1 and Notch1 correlates positively with Slug expression.
In breast cancer patients, increased expression of either Jagged1 or Notch1 is predictive of poor overall survival (7). If both Jagged1 and Notch1 are increased, there is a further substantial reduction in overall survival (7). Our data provide an explanation for the reported dose-dependent relationship of Jagged1 expression and negative outcome in breast cancer (7), suggesting that juxtacrine or autocrine Jagged1–Notch interactions induce Slug, which initiates EMT and inhibits anoikis, thereby promoting tumor invasion and metastasis. Interestingly, Jagged1 expression has also been associated with prostate cancer metastasis and recurrence (36), and our demonstration of positive correlations between Jagged1 and Slug expression in a wide variety of tumors raises the possibility that our findings may be generalized to other tumors in which Notch is activated by ligand.
Expression of the Notch target gene HEYL has previously been reported to be absent in the normal breast epithelium but present in the tumor cell compartment of invasive breast cancers (37). These findings fit with our results, which show that HEYL mRNA is undetectable in control breast epithelial cells but is up-regulated in cells exhibiting activation of the Notch–Slug signaling axis. Importantly, our analysis of primary human breast cancers has identified HEY genes, in particular HEYL, as potential markers of human breast cancers that exhibit activation of the Jagged1–Notch–Slug signaling axis.
The Snail gene has previously been reported to be a direct target gene of Notch in endothelial cells (13). However, our data suggest that this is not the case in epithelial cells, because we did not observe a positive correlation between activated Notch signaling and Snail induction. Rather, we observed a positive correlation between Notch activation and Slug expression both in human breast epithelial cells in vitro and in primary human breast cancers. Hence, our findings indicate that Slug, but not Snail, is a downstream target gene of Notch in epithelial and other tumors. Interestingly, increased levels of Slug, but not Snail, have been associated with tumors from patients with metastatic disease or disease recurrence (16).
Although it has been reported that Slug has antiapoptotic activity in some cell types, the current study is the first demonstration that Slug can inhibit anoikis. To initiate EMT, repression of E-cadherin permits loss of cell–cell cohesion, thereby disrupting apical-basal polarity and promoting cell migration. However, given that the loss of E-cadherin renders cells susceptible to anoikis, Slug must independently promote cell survival in this context to allow local tumor invasion and distant metastases. Thus, our findings that Notch blockade inhibits growth of the primary tumor by triggering apoptosis and reduces distant metastases is concordant with this model of Slug-dependent EMT and cell survival.
Both Slug and Notch have been shown to inhibit p53 function, in part through the inhibition of Puma expression, which may be one mechanism of the antianoikis effect (11, 38). Notch has also been shown to activate the phosphatidylinositol 3′-kinase–Akt antiapoptotic pathway, but whether there is cross talk with Slug remains to be seen (39, 40). The ability of Notch to suppress c-Jun N-terminal kinase activation and activate phosphatidylinositol 3′-kinase–Akt may explain the inhibition of p53 by Notch (11, 33, 39). Both Notch and Slug have also been shown to induce Bcl-2 (33, 41); thus, there are multiple potential anoikis–apoptosis pathways that Notch may regulate through Slug.
It is also possible that Slug-mediated repression of E-cadherin, which we have shown results in activation of β-catenin, may be responsible for an antianoikis effect. Indeed, even modest overexpression of active β-catenin has been shown to prevent anoikis (42). Of interest, β-catenin has been reported to induce Slug promoter activity (43). Hence, Notch induction of Slug is potentially enhanced by a feed-forward loop. Initially, Notch/CSL would directly induce Slug. Subsequent Slug-mediated E-cadherin repression, via interaction with E2-boxes in the E-cadherin promoter (12), would then release β-catenin from the plasma membrane, which would accumulate in the nucleus and further activate the Slug promoter. However, the human Slug promoter also contains several cis elements predicted to bind the HEY/HES family of Notch-induced transcriptional repressors (unpublished data). Thus, these factors may act to attenuate the positive feedback loop described. Of note, our data showed that enforced expression of E-cadherin mimicked the effects of blocking Notch signaling in vivo, suggesting that a major function of Notch-induced Slug in this model may be to repress E-cadherin.
Recent evidence has demonstrated that breast cancers may be initiated in and propagated by a minority cell population that have been designated tumor stem cells (44). Notch signaling appears to play an important role in normal mammary stem cell self-renewal (44). Thus, the Notch pathway may be selectively activated in more primitive cancer stem cells. A mouse transgenic model of activated Notch4-driven breast cancer indicates that these tumors are highly metastatic (3). Interestingly, Slug has been shown to have antiapoptotic activity in hematopoietic progenitor cells (38); thus, the Notch–Slug axis may play a role in maintaining the tumor stem cell, as well as in permitting these cells to survive the metastatic process.
Tumor cell metastasis is the primary cause of mortality in a vast majority of cancer patients. As such, there is a compelling need to elucidate the mechanisms of tumor cell metastasis and to develop rationally designed therapeutics to specifically target the metastatic process. Our findings highlight the potential use of inhibitors of ligand-induced Notch signaling as viable and effective agents to block EMT and tumor metastasis in the treatment of human cancers that exhibit activation of the Jagged1–Notch–Slug signaling axis. By specifically targeting ligand-induced activation, there is the potential of reducing the more widespread side effects potentially evoked by less-specific therapeutics, such as the γ-secretase inhibitors.
MATERIALS AND METHODS
Cell lines.
The human breast epithelial cell line MCF-10A was cultured in a 1:1 mixture of DMEM/F12 (Sigma-Aldrich) supplemented with 5% horse serum (Sigma-Aldrich), 2 mM glutamine (Sigma-Aldrich), 20 ng/ml of epidermal growth factor (Sigma-Aldrich), 100 ng/ml cholera toxin (Cedarlane), 10 μg/ml insulin (Sigma-Aldrich), 500 ng/ml hydrocortisone (Sigma-Aldrich), and 100 U/ml each of penicillin and streptomycin (Invitrogen). The human breast carcinoma cell lines MDA-MB-231 and T47D and the mouse endothelial cell line SVEC4-10 were cultured in DMEM supplemented with 10% heat-inactivated calf serum (HyClone), 2 mM glutamine, and 100 U/ml each of penicillin and streptomycin. All cells were maintained at 37°C in an atmosphere of 5% CO2.
Isolation of primary human breast epithelial cells.
Normal human breast tissue was obtained in accordance with guidelines approved by the University of British Columbia from anonymized discarded material from normal premenopausal women undergoing reduction mammoplasty surgeries. A crude epithelial cell–enriched cell suspension was obtained enzymatically and cryopreserved. As required, cells were thawed, and single-cell suspensions were prepared as previously described (45). Cells were co-cultured with 1.2 × 106 X-irradiated NIH3T3 mouse fibroblasts for 1 d in Epicult-B medium (StemCell Technologies Inc.) supplemented with 5% FCS. Cells were harvested, and epithelial cell adhesion molecule (EpCAM)–positive breast epithelial cells were magnetically separated using the human EpCAM selection cocktail EasySep (StemCell Technologies Inc.). These selected cells were cultured in Epicult-B medium at a density of 2 × 105 cells per dish in 35-mm dishes precoated with Vitrogen (Cohesion Technologies). To precoat the dishes, 2–3 ml of Vitrogen (67 μg/ml in PBS) was used for 1 h at 37°C and washed with PBS before use.
Plasmid constructs and gene transfer.
Retroviral vectors (MIY and MIG) containing an internal ribosomal entry site and either YFP (MIY) or GFP (MIG) were used to facilitate the sorting of transduced cells. cDNA constructs encoding the human Notch1IC, C-terminal hemagglutinin (HA)-tagged human Notch4IC, full-length C-terminal Flag-tagged human Slug (a gift of E. Fearon, University of Michigan, Ann Arbor, MI), full-length human Jagged1, full-length human E-cadherin (a gift of B.M. Gumbiner, University of Virginia, Charlottesville, VA), and N-terminal myc-tagged full-length human HEY1/2/L (a gift of D. Srivastava, University of California, San Francisco, San Francisco, CA) were subcloned into MIY. The cDNA construct encoding the entire extracellular domain of human Notch4 (XNotch4; amino acids 1–1,443) tagged with a C-terminal HA epitope was subcloned into MIG. The pLentilox3.7-shRandom and pLentilox3.7-shSlug constructs were generated by inserting shRNAs targeting the sequences 5′-GTTGCTTGCCACGTCCTAGAT-3′ (Random) and 5′-GCATTTGCAGACAGGTCAAAT-3′ (Slug) into pLentilox3.7 (a gift of L. Van Parijs, Massachusetts Institute of Technology, Cambridge, MA). Cells were transduced with empty vector control or vector containing cDNA inserts, and transduced cells were sorted based on YFP or GFP expression using a cell sorter (FACS 440; Becton Dickinson).
Immunoblotting.
Cultured cells or tumor tissue were lysed and analyzed by SDS-PAGE and immunoblotting with rabbit polyclonal antibody to Slug (Santa Cruz Biotechnology, Inc.), or mouse monoclonal antibodies to HA (Sigma-Aldrich), E-cadherin (BD Biosciences), β-catenin (BD Biosciences), active β-catenin (Millipore), and α-tubulin (Sigma-Aldrich). Protein expression was quantitated by densitometry.
RNA isolation, RT-PCR, and qPCR.
Total RNA isolation was performed using TRIzol reagent (Invitrogen) or an RNeasy kit (QIAGEN), according to the manufacturers' recommendations. First-strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen). After ribonuclease H treatment (Invitrogen), PCR was performed. Control reactions omitting reverse transcriptase were performed in each experiment. Primer sequences and annealing temperatures are described in Tables S1 and S2 (available at http://www.jem.org/cgi/content/full/jem.20071082/DC1). For RT-PCR, entire PCR samples were assessed in 1.5% TAE-agarose gels containing ethidium bromide. Control human cDNA was generated from pooled total RNA isolated from the following human cells: human mammary epithelial cells; vascular smooth muscle cells; cervical cancer cells, SiHa; colon cancer cells, WiDr; and kidney epithelial cells, 293T. Control mouse cDNA was generated from pooled total RNA isolated from the following mouse cells: Lewis lung carcinoma cells; endothelial cells, SVEC4-10; and fibroblasts, NIH3T3. For qPCR, reactions were run on a real-time PCR system (ABI Prism 7900; Applied Biosystems). Gene expression was detected with SYBR green (Applied Biosystems), and relative gene expression was determined by normalizing to GAPDH using the ΔCT method.
EMSA.
Nuclear lysates were collected from shRandom, shCSL1, and shCSL2 overexpressing MDA-MB-231 cells for the CSL EMSA assays. The binding reaction (10 mM TrisHCl, 50 mM NaCl, 1 mM EDTA [pH 8], 4% glycerol, 2 μg PolydI-dC binding buffer, and 10 μg of nuclear protein) was performed by preincubating with either 50-fold excess wild-type (Slug 850, [forward] GGGCCCTTTTTCCCATAAAAAAAAAG and [reverse] GGGAAAAAGGGTATTTTTTTTTCGGG; Slug 1680, [forward] TGTGTGTTTTGTGGGAAATGGAG and [reverse] CTCCATTTCCCACAAAA) or mutant (Slug 800, [forward] GGGCCCTTTGCAGCATAAAAAAAAAAG and [reverse] GGGAAACGTCGTATTTTTTTTTTCGGG; Slug 1600, [forward] TGTGTGTTTTGTGCTGCATGGAG and [reverse] CTCCATGCAGCACAAAA) nonradioactive duplex oligos for 15 min on ice, and then adding a 150,000-cpm 32P-labeled double-stranded probe and incubating for 30 min at room temperature. Binding reactions were run on 5% Tris-Borate EDTA gels and exposed to a phosphorimager plate for 12–16 h.
Inhibition of histone acetylation and DNA methylation.
MCF-10A cell lines were flow sorted, and YFP-positive cells were plated into fourwell chamber slides at 7 × 105 cells per well and allowed to adhere and grow for 48 h. Cells were then treated with TSA (100/500/1000 nM), NaBu (2/5/10 mM), and/or 5AZA (1/5/10 μM) for 72 h. After immunofluorescent staining for E-cadherin, at least six random fields at 200× magnification were analyzed per well using Northern Eclipse software (Empix Imaging). Data are presented as the percentage of YFP/E-cadherin double-positive cells + SEM.
Methylation assays.
Genomic DNA was isolated from cultured cells or tumor tissue using a DNeasy tissue kit (QIAGEN) according to the manufacturer's recommendations. 1 μg of genomic DNA was bisulfite modified using a CpGenome DNA modification kit (Chemicon) and eluted in 25 μl Tris-EDTA buffer, according to the manufacturer's recommendations. MSP was performed using ∼120 ng of bisulfite-modified DNA, 400 nM of 5′ and 3′ primers, 0.2 mM of 2′-deoxynucleoside 5′-triphosphates (Invitrogen), 1× PCR buffer, and 0.625 U of HotStarTaq DNA polymerase (QIAGEN), according to the manufacturer's recommendations. Primer sequences and annealing temperatures are described in Tables S1 and S2. Entire MSP reactions were assessed in 2% TAE-agarose gels containing ethidium bromide. Bands corresponding to methylated and unmethylated PCR products were quantitated by densitometry. Data are expressed as a ratio of methylated over unmethylated PCR products (M/U ratio) and represent the mean ratio + SE from five control tumors and five XNotch4 tumors. For genomic bisulfite sequencing, bisulfite-modified genomic DNA isolated from tumor tissue was amplified by PCR using primers spanning the E-cadherin proximal promoter. Primer sequences and annealing temperatures are described in Tables S1 and S2. PCR products were purified and cloned into the pDrive cloning vector (QIAGEN), and individual clones were sequenced. Five control tumors (a total of 35 MIG clones) and three XNotch4 tumors (a total of 22 XNotch4 clones) were analyzed, and data were expressed as the percentage of methylation per CpG site (total number of methylated clones/total number of clones).
Immunostaining.
Breast epithelial cells were stained with mouse monoclonal antibody to extracellular E-cadherin (Chemicon) or rabbit polyclonal antibody to Slug (Santa Cruz Biotechnology, Inc.). 7-μm-thick tumor cryosections were stained with rabbit polyclonal antibody to Slug (Santa Cruz Biotechnology, Inc.), mouse monoclonal antibodies to E-cadherin (BD Biosciences) or β-catenin (BD Biosciences), and rabbit monoclonal antibody to activated caspase-3 (BD Biosciences). For immunohistochemistry, a biotinylated goat anti–rabbit antibody (Vector Laboratories) followed by horseradish peroxidase–conjugated streptavidin (Vector Laboratories) were used, and nuclei were counterstained with hematoxylin. For immunofluorescence, the fluorochrome-conjugated secondary antibodies goat anti–rat Alexa Fluor 594 (Invitrogen) and goat anti–mouse Alexa Fluor 594 (Invitrogen) were used, and nuclei were counterstained with DAPI (Sigma-Aldrich). Immunofluorescence was detected with an imaging microscope (Axioplan II; Carl Zeiss, Inc.), and images were captured with a digital camera (1350EX; QImaging). To quantify the proportion of cells exhibiting nuclear β-catenin staining, at least six random fields at 200× magnification were analyzed per tumor using Northern Eclipse software. Data are presented as the mean percentage of nuclear β-catenin staining (total number of nuclear β-catenin–positive cells/total number of cells) + SEM from four control tumors and four XNotch4 tumors. To quantify the proportional area of tumors showing activated caspase 3, entire tumor sections were analyzed using Northern Eclipse software. Data are presented as the mean percentage of the activated caspase 3–stained area (total area positive for activated caspase 3/total area of tumor section) + SEM from 14 control tumors and 12 XNotch4 tumors.
siRNA transfection.
MCF-10A MIY and MIYNotch1IC cells were flow sorted, and YFP-positive cells were transiently transfected with 100 nM siRNA targeting the sequences 5′-GTTGCTTGCCACGTCCTAGAT-3′ (siRandom) or 5′-GCATTTGCAGACAGGTCAAAT-3′ (siSlug) using DharmaFECT 1 transfection reagent (Dharmacon Inc.). Cells were harvested 4 d after transfection.
Co-culture.
106 parental human cells (MCF-10A or MDA-MB-231) were co-cultured with 106 mouse SVEC4-10 endothelial cells transduced with MIY control vector or MIYJagged1 and plated in 100-mm tissue culture dishes. Cells were harvested after 3 d of co-culture. qPCR was performed with human-specific primers to avoid amplification of mouse transcripts.
Anoikis assay.
MCF-10A cell lines were flow sorted, and YFP-positive cells were plated into 60-mm tissue culture plates (8 × 105 cells per plate) coated with 1% agarose. After 24 h of incubation at 37°C, cells were fixed/permeabilized in 70% ethanol and stained with propidium iodide (Sigma-Aldrich), and the proportion of cells with hypodiploid DNA content was determined by flow cytometry. Data are expressed as the proportion of hypodiploid cells relative to MIY control.
Tumorigenicity assays.
Female nonobese diabetic/severe combined immunodeficient mice were obtained from the Animal Resource Centre of the British Columbia Cancer Research Centre. For MDA-MB-231 tumor implantation, 5 × 106 cells were injected subcutaneously into the dorsa of mice. Once tumors were palpable, tumor volume (0.523 × length × width × height) was measured weekly using calipers. For each time point, MDA-MB-231 tumor data are presented as the mean tumor volume ± SEM from (a) 12 MIG tumors and 11 MIGXNotch4HA tumors, and (b) 12 MIY tumors and 12 MIYE-cadherin tumors. Tumor-growth curves for one experiment are presented and are representative of three independent experiments, with 8–16 mice per tumor group for each experiment. Mice bearing MDA-MB-231 tumors (MIG and MIGXNotch4HA, or MIY and MIYE-cadherin) were killed at the same time, and the total number and total weight (in milligrams) of metastases were determined. Data are presented as the mean number of metastases per mouse + SEM and the mean weight of each metastatic nodule (in milligrams) + SEM. Metastasis data were determined by analyzing (a) 25 MIG tumors and 27 MIGXNotch4HA tumors, and (b) 12 MIY tumors and 12 MIYE-cadherin tumors. Animal experiments were approved by the University of British Columbia Institutional Animal Care and Ethics Committee, and all animals were handled according to institutional animal care procedures.
Microarray data analysis.
Pearson correlation coefficients were obtained from publicly available microarray datasets deposited in the Oncomine database (available at www.oncomine.org): breast 1 (46), breast 2 (47), adrenal (48), brain (49), endocrine (50), gastric (51), lung (52), ovarian (53), renal (54), salivary gland (55), sarcoma 1 (56), and sarcoma 2 (57). For each dataset, individual correlations between two genes of interest (as well as all possible correlations in the event of replicate genes in the microarray) were determined, as well as the mean Pearson correlation coefficient. Microarray data accession numbers are shown in Table S3 (available at http://www.jem.org/cgi/content/full/jem.20071082/DC1).
Statistical analysis.
To determine statistical significance, a one-way analysis of variance with a Tukey test for multiple comparisons was performed using the statistics program Statistical Package for Social Scientists (version 11.0; SPSS Inc.). Statistical significance was taken at P ≤ 0.05.
Online supplemental material.
Tables S1 and S2 provide all primer sequences used in this study. Table S3 provides microarray accession number information. Fig. S1 shows Notch-induced E-cadherin repression in MCF-10A cells. Fig. S2 shows Notch-induced Slug expression in MCF-10A cells. Fig. S3 demonstrates the expression of Notch ligands and receptors in MDA-MB-231 cells, and secreted XNotch4HA protein. Fig. S4 shows a lack of a vascular effect by XNotch4HA. Fig. S5 demonstrates Notch-induced β-catenin activation independent of Wnt activation. Fig. S6 reveals that inhibition of Notch does not cause a generalized hypomethylation of the genome. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071082/DC1.
Supplemental Material
[Supplemental Material Index]
[Related biobytes podcast]
Acknowledgments
We thank F. Wong and D. McDougal for assistance with flow cytometry and cell sorting. We also thank P.L. Olive for critical review of the manuscript.
This research was supported by grants to A. Karsan from the National Cancer Institute of Canada with funds from the Canadian Cancer Society and the Cancer Research Society, and to C. Eaves from Genome Canada. K.G. Leong was supported by a Doctoral Research Award from the Canadian Institutes of Health Research and a Predoctoral Fellowship Award from the Department of the Army (DAMD17-01-1-0164). The U.S. Army Medical Research Acquisition Activity was the awarding and administering acquisition office. K. Niessen is the recipient of a Senior Graduate Studentship from the Michael Smith Foundation for Health Research, and I. Kulic is supported by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and a Junior Graduate Studentship from the Michael Smith Foundation for Health Research. A. Raouf is supported by a Post-doctoral Research Award from the Canadian Institutes of Health Research. A. Karsan is a Senior Scholar of the Michael Smith Foundation for Health Research.
The authors declare that they have no competing financial interests.
Abbreviations used: 5AZA, 5-azacytidine; cDNA, complementary DNA; CpG, cytosine-phosphate-guanine; EMT, epithelial-to-mesenchymal transition; HA, hemagglutinin; HDAC, histone deacetylase; MSP, methylation-specific PCR; NaBu, sodium butyrate; NotchIC, Notch intracellular domain; qPCR, quantitative RT-PCR; shRNA, short hairpin RNA; siRNA, small interfering RNA; TSA, trichostatin A; YFP, yellow fluorescent protein.
K.G. Leong, K. Niessen, and I. Kulic contributed equally to this work.
References
- 1.Mumm, J.S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151–165. [DOI] [PubMed] [Google Scholar]
- 2.Leong, K.G., and A. Karsan. 2006. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 107:2223–2233. [DOI] [PubMed] [Google Scholar]
- 3.Gallahan, D., C. Jhappan, G. Robinson, L. Hennighausen, R. Sharp, E. Kordon, R. Callahan, G. Merlino, and G.H. Smith. 1996. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56:1775–1785. [PubMed] [Google Scholar]
- 4.Hu, C., A. Dievart, M. Lupien, E. Calvo, G. Tremblay, and P. Jolicoeur. 2006. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am. J. Pathol. 168:973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan, R., and S.E. Egan. 2004. Notch signaling in mammary development and oncogenesis. J. Mammary Gland Biol. Neoplasia. 9:145–163. [DOI] [PubMed] [Google Scholar]
- 6.Parr, C., G. Watkins, and W.G. Jiang. 2004. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int. J. Mol. Med. 14:779–786. [DOI] [PubMed] [Google Scholar]
- 7.Reedijk, M., S. Odorcic, L. Chang, H. Zhang, N. Miller, D.R. McCready, G. Lockwood, and S.E. Egan. 2005. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 65:8530–8537. [DOI] [PubMed] [Google Scholar]
- 8.Ayyanan, A., G. Civenni, L. Ciarloni, C. Morel, N. Mueller, K. Lefort, A. Mandinova, W. Raffoul, M. Fiche, G.P. Dotto, and C. Brisken. 2006. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc. Natl. Acad. Sci. USA. 103:3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S.M. Jonkheer, A. Zlobin, B.A. Osborne, S. Gottipati, J.C. Aster, W.C. Hahn, et al. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8:979–986. [DOI] [PubMed] [Google Scholar]
- 10.Pece, S., M. Serresi, E. Santolini, M. Capra, E. Hulleman, V. Galimberti, S. Zurrida, P. Maisonneuve, G. Viale, and P.P. Di Fiore. 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stylianou, S., R.B. Clarke, and K. Brennan. 2006. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66:1517–1525. [DOI] [PubMed] [Google Scholar]
- 12.Vincent-Salomon, A., and J.P. Thiery. 2003. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 5:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmerman, L.A., J. Grego-Bessa, A. Raya, E. Bertran, J.M. Perez-Pomares, J. Diez, S. Aranda, S. Palomo, F. McCormick, J.C. Izpisua-Belmonte, and J.L. de la Pompa. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noseda, M., G. McLean, K. Niessen, L. Chang, I. Pollet, R. Montpetit, R. Shahidi, K. Dorovini-Zis, L. Li, B. Beckstead, et al. 2004. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ. Res. 94:910–917. [DOI] [PubMed] [Google Scholar]
- 15.Come, C., V. Arnoux, F. Bibeau, and P. Savagner. 2004. Roles of the transcription factors snail and slug during mammary morphogenesis and breast carcinoma progression. J. Mammary Gland Biol. Neoplasia. 9:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, T.A., A. Goyal, G. Watkins, and W.G. Jiang. 2005. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann. Surg. Oncol. 12:488–496. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes, D.R., J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, and A.M. Chinnaiyan. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. [DOI] [PubMed] [Google Scholar]
- 19.van de Wetering, M., N. Barker, I.C. Harkes, M. van der Heyden, N.J. Dijk, A. Hollestelle, J.G. Klijn, H. Clevers, and M. Schutte. 2001. Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res. 61:278–284. [PubMed] [Google Scholar]
- 20.Imatani, A., and R. Callahan. 2000. Identification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell lines. Oncogene. 19:223–231. [DOI] [PubMed] [Google Scholar]
- 21.Small, D., D. Kovalenko, D. Kacer, L. Liaw, M. Landriscina, C. Di Serio, I. Prudovsky, and T. Maciag. 2001. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J. Biol. Chem. 276:32022–32030. [DOI] [PubMed] [Google Scholar]
- 22.Marambaud, P., J. Shioi, G. Serban, A. Georgakopoulos, S. Sarner, V. Nagy, L. Baki, P. Wen, S. Efthimiopoulos, Z. Shao, et al. 2002. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21:1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavadil, J., L. Cermak, N. Soto-Nieves, and E.P. Bottinger. 2004. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 23:1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krebs, L.T., Y. Xue, C.R. Norton, J.R. Shutter, M. Maguire, J.P. Sundberg, D. Gallahan, V. Closson, J. Kitajewski, R. Callahan, et al. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 25.Leong, K.G., X. Hu, L. Li, M. Noseda, B. Larrivee, C. Hull, L. Hood, F. Wong, and A. Karsan. 2002. Activated Notch4 inhibits angiogenesis: role of beta 1-integrin activation. Mol. Cell. Biol. 22:2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng, Q., S. Li, D.B. Chepeha, T.J. Giordano, J. Li, H. Zhang, P.J. Polverini, J. Nor, J. Kitajewski, and C.Y. Wang. 2005. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 8:13–23. [DOI] [PubMed] [Google Scholar]
- 27.Hatsell, S., T. Rowlands, M. Hiremath, and P. Cowin. 2003. Beta-catenin and Tcfs in mammary development and cancer. J. Mammary Gland Biol. Neoplasia. 8:145–158. [DOI] [PubMed] [Google Scholar]
- 28.Wong, A.S., and B.M. Gumbiner. 2003. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J. Cell Biol. 161:1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi, M.K., S. Misra, S.V. Khedkar, N. Hamilton, C. Irvin-Wilson, C. Sharan, L. Sealy, and G. Chaudhuri. 2005. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J. Biol. Chem. 280:17163–17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachman, K.E., B.H. Park, I. Rhee, H. Rajagopalan, J.G. Herman, S.B. Baylin, K.W. Kinzler, and B. Vogelstein. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 3:89–95. [DOI] [PubMed] [Google Scholar]
- 31.Berx, G., K. Staes, J. van Hengel, F. Molemans, M.J. Bussemakers, A. van Bokhoven, and F. van Roy. 1995. Cloning and characterization of the human invasion suppressor gene E-cadherin (CDH1). Genomics. 26:281–289. [DOI] [PubMed] [Google Scholar]
- 32.Cameron, E.E., K.E. Bachman, S. Myohanen, J.G. Herman, and S.B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103–107. [DOI] [PubMed] [Google Scholar]
- 33.MacKenzie, F., P. Duriez, F. Wong, M. Noseda, and A. Karsan. 2004. Notch4 inhibits endothelial apoptosis via RBP-Jkappa-dependent and -independent pathways. J. Biol. Chem. 279:11657–11663. [DOI] [PubMed] [Google Scholar]
- 34.Zweidler-McKay, P.A., Y. He, L. Xu, C.G. Rodriguez, F.G. Karnell, A.C. Carpenter, J.C. Aster, D. Allman, and W.S. Pear. 2005. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B cell malignancies. Blood. 106:3898–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng, A.P., A.A. Ferrando, W. Lee, J.P. Morris IV, L.B. Silverman, C. Sanchez-Irizarry, S.C. Blacklow, A.T. Look, and J.C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271. [DOI] [PubMed] [Google Scholar]
- 36.Santagata, S., F. Demichelis, A. Riva, S. Varambally, M.D. Hofer, J.L. Kutok, R. Kim, J. Tang, J.E. Montie, A.M. Chinnaiyan, et al. 2004. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 64:6854–6857. [DOI] [PubMed] [Google Scholar]
- 37.Parker, B.S., P. Argani, B.P. Cook, H. Liangfeng, S.D. Chartrand, M. Zhang, S. Saha, A. Bardelli, Y. Jiang, T.B. St Martin, et al. 2004. Alterations in vascular gene expression in invasive breast carcinoma. Cancer Res. 64:7857–7866. [DOI] [PubMed] [Google Scholar]
- 38.Wu, W.S., S. Heinrichs, D. Xu, S.P. Garrison, G.P. Zambetti, J.M. Adams, and A.T. Look. 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 123:641–653. [DOI] [PubMed] [Google Scholar]
- 39.Mungamuri, S.K., X. Yang, A.D. Thor, and K. Somasundaram. 2006. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 66:4715–4724. [DOI] [PubMed] [Google Scholar]
- 40.Androutsellis-Theotokis, A., R.R. Leker, F. Soldner, D.J. Hoeppner, R. Ravin, S.W. Poser, M.A. Rueger, S.K. Bae, R. Kittappa, and R.D. McKay. 2006. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 442:823–826. [DOI] [PubMed] [Google Scholar]
- 41.Bermejo-Rodriguez, C., M. Perez-Caro, P.A. Perez-Mancera, M. Sanchez-Beato, M.A. Piris, and I. Sanchez-Garcia. 2006. Mouse cDNA microarray analysis uncovers Slug targets in mouse embryonic fibroblasts. Genomics. 87:113–118. [DOI] [PubMed] [Google Scholar]
- 42.Weng, Z., M. Xin, L. Pablo, D. Grueneberg, M. Hagel, G. Bain, T. Muller, and J. Papkoff. 2002. Protection against anoikis and down-regulation of cadherin expression by a regulatable beta-catenin protein. J. Biol. Chem. 277:18677–18686. [DOI] [PubMed] [Google Scholar]
- 43.Conacci-Sorrell, M., I. Simcha, T. Ben-Yedidia, J. Blechman, P. Savagner, and A. Ben-Ze'ev. 2003. Autoregulation of E-cadherin expression by cadherin–cadherin interactions: the roles of β-catenin signaling, Slug, and MAPK. J. Cell Biol. 163:847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, S., G. Dontu, and M.S. Wicha. 2005. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 7:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stingl, J., C.J. Eaves, I. Zandieh, and J.T. Emerman. 2001. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res. Treat. 67:93–109. [DOI] [PubMed] [Google Scholar]
- 46.van ‘t Veer, L.J., H. Dai, M.J. van de Vijver, Y.D. He, A.A. Hart, M. Mao, H.L. Peterse, K. van der Kooy, M.J. Marton, A.T. Witteveen, et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415:530–536. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, H., A. Langerod, Y. Ji, K.W. Nowels, J.M. Nesland, R. Tibshirani, I.K. Bukholm, R. Karesen, D. Botstein, A.L. Borresen-Dale, and S.S. Jeffrey. 2004. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell. 15:2523–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giordano, T.J., D.G. Thomas, R. Kuick, M. Lizyness, D.E. Misek, A.L. Smith, D. Sanders, R.T. Aljundi, P.G. Gauger, N.W. Thompson, et al. 2003. Distinct transcriptional profiles of adrenocortical tumors uncovered by DNA microarray analysis. Am. J. Pathol. 162:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freije, W.A., F.E. Castro-Vargas, Z. Fang, S. Horvath, T. Cloughesy, L.M. Liau, P.S. Mischel, and S.F. Nelson. 2004. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64:6503–6510. [DOI] [PubMed] [Google Scholar]
- 50.Jain, S., M.A. Watson, M.K. DeBenedetti, Y. Hiraki, J.F. Moley, and J. Milbrandt. 2004. Expression profiles provide insights into early malignant potential and skeletal abnormalities in multiple endocrine neoplasia type 2B syndrome tumors. Cancer Res. 64:3907–3913. [DOI] [PubMed] [Google Scholar]
- 51.Chen, X., S.Y. Leung, S.T. Yuen, K.M. Chu, J. Ji, R. Li, A.S. Chan, S. Law, O.G. Troyanskaya, J. Wong, et al. 2003. Variation in gene expression patterns in human gastric cancers. Mol. Biol. Cell. 14:3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garber, M.E., O.G. Troyanskaya, K. Schluens, S. Petersen, Z. Thaesler, M. Pacyna-Gengelbach, M. van de Rijn, G.D. Rosen, C.M. Perou, R.I. Whyte, et al. 2001. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. USA. 98:13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaner, M.E., D.T. Ross, G. Ciaravino, T. Sorlie, O. Troyanskaya, M. Diehn, Y.C. Wang, G.E. Duran, T.L. Sikic, S. Caldeira, et al. 2003. Gene expression patterns in ovarian carcinomas. Mol. Biol. Cell. 14:4376–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higgins, J.P., R. Shinghal, H. Gill, J.H. Reese, M. Terris, R.J. Cohen, M. Fero, J.R. Pollack, M. van de Rijn, and J.D. Brooks. 2003. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am. J. Pathol. 162:925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frierson, H.F., Jr., A.K. El-Naggar, J.B. Welsh, L.M. Sapinoso, A.I. Su, J. Cheng, T. Saku, C.A. Moskaluk, and G.M. Hampton. 2002. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am. J. Pathol. 161:1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linn, S.C., R.B. West, J.R. Pollack, S. Zhu, T. Hernandez-Boussard, T.O. Nielsen, B.P. Rubin, R. Patel, J.R. Goldblum, D. Siegmund, et al. 2003. Gene expression patterns and gene copy number changes in dermatofibrosarcoma protuberans. Am. J. Pathol. 163:2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Segal, N.H., P. Pavlidis, W.S. Noble, C.R. Antonescu, A. Viale, U.V. Wesley, K. Busam, H. Gallardo, D. DeSantis, M.F. Brennan, et al. 2003. Classification of clear-cell sarcoma as a subtype of melanoma by genomic profiling. J. Clin. Oncol. 21:1775–1781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]
[Related biobytes podcast]