ZW10 Helps Recruit Dynactin and Dynein to the Kinetochore (original) (raw)
Abstract
Mutations in the Drosophila melanogaster zw10 gene, which encodes a conserved, essential kinetochore component, abolish the ability of dynein to localize to kinetochores. Several similarities between the behavior of ZW10 protein and dynein further support a role for ZW10 in the recruitment of dynein to the kinetochore: (a) in response to bipolar tension across the chromosomes, both proteins mostly leave the kinetochore at metaphase, when their association with the spindle becomes apparent; (b) ZW10 and dynein both bind to functional neocentromeres of structurally acentric minichromosomes; and (c) the localization of both ZW10 and dynein to the kinetochore is abolished in cells mutant for the gene rough deal. ZW10's role in the recruitment of dynein to the kinetochore is likely to be reasonably direct, because dynamitin, the p50 subunit of the dynactin complex, interacts with ZW10 in a yeast two-hybrid screen. Since in zw10 mutants no defects in chromosome behavior are observed before anaphase onset, our results suggest that dynein at the kinetochore is essential for neither microtubule capture nor congression to the metaphase plate. Instead, dynein's role at the kinetochore is more likely to be involved in the coordination of chromosome separation and/or poleward movement at anaphase onset.
Keywords: ZW10, dynein, dynamitin, rough deal, kinetochore
The kinetochores elaborated by the centromeres of eukaryotic chromosomes play three major roles during mitosis and meiosis (for review see Pluta et al., 1995). First, the kinetochores serve as the mechanical link allowing the chromosomes to attach to the dynamic plus ends of microtubules of the spindle apparatus. Second, the kinetochores contain microtubule motor activities that are probably responsible for poleward movements of the chromosomes during prometaphase (Rieder and Alexander, 1990), for at least some aspects of chromosomal movements accompanying congression to the metaphase plate (Rieder and Salmon, 1994), and for the poleward forces exerted on chromosomes during anaphase A (Nicklas, 1989). Finally, the kinetochores are intimately involved in the elaboration of a “wait anaphase” checkpoint control that ensures cells will not enter anaphase until all chromosomes are properly oriented at the metaphase plate (Li and Nicklas, 1995; Nicklas et al., 1995; Chen et al., 1996; Li and Benerza, 1996; Taylor and McKeon, 1997). These three kinetochore functions may not in fact be fundamentally distinct. For example, recent evidence suggests that the kinesin-related microtubule motor centromere-associated E protein (CENP-E) may act by tethering kinetochores to the plus ends of disassembling microtubules during chromosome congression (Yen et al., 1992; Lombillo et al., 1995; Duesbery et al., 1997; Wood et al., 1997; Yao et al., 1997).
Cytoplasmic dynein is one of three microtubule motor proteins currently known to localize to the kinetochore of mammalian chromosomes (Pfarr et al., 1990; Steuer et al., 1990; Wordeman et al., 1991); the two others are CENP-E (see above) and mitotic centromere-associated kinesin/ Xenopus kinesin-central motor 1 (MCAK/XKCM1), a member of the KIF2 subfamily of plus end–directed kinesins (Walczak et al., 1996; Wordeman and Mitchison, 1995). It has been extremely difficult to determine the importance of dynein's association with the kinetochore because dynein is required for many intracellular processes. For example, a complex of cytoplasmic dynein and the protein NuMA at the spindle poles has recently been demonstrated to be essential for proper assembly of the mitotic spindle (Merdes et al., 1996). Disruption of this activity would be particularly likely to mask possible effects of the perturbation of dynein at the kinetochore. Thus, microinjection of anti-dynein into cells induces spindle collapse (Vaisberg et al., 1993), whereas depletion of dynein from Xenopus or HeLa cell extracts disrupts aster formation or spindle pole assembly (Verde et al., 1991; Gaglio et al., 1996; Heald et al., 1996). Moreover, in Drosophila melanogaster, recent mutational analysis of dynein function has revealed defects in centrosome behavior and spindle morphogenesis during the nuclear divisions of the early syncytial embryo (Robinson, J.R., E.J. Wojcik, M. Sanders, M. McGrail, and T.S. Hays, manuscript in preparation). Some role for cytoplasmic dynein in mitotic chromosome movements has been inferred from studies of transfected tissue culture cells that overexpress dynamitin, the p50 component of the dynactin complex that may help target dynein to intracellular cargoes (Echeverri et al., 1996). In these cells with excess dynamitin, both dynein and dynactin are no longer associated with the kinetochores, and the chromosomes do not align properly at the metaphase plate (Echeverri et al., 1996). However, as these authors point out, the observed difficulties in chromosome behavior may be indirect effects of distortions of the spindle that also occur in these cells. Because of these complications, the significance of dynein's localization at the kinetochore remains highly controversial. Does this microtubule motor in fact play any role in attaching the chromosomes to spindle fibers, in moving the chromosomes along these microtubules, or in the wait anaphase checkpoint?
In this report, we establish a connection between dynein and ZW10, a kinetochore component conserved in most if not all multicellular eukaryotes (Starr et al., 1997). Null mutations in the Drosophila gene l(1)zw10 (hereafter abbreviated zw10) encoding the fly ZW10 protein disrupt chromosome segregation during mitosis and both meiotic divisions. Mitotic missegregation in zw10 mutants produces many aneuploid cells and consequent lethality to the organism (Smith et al., 1985; Williams et al., 1992). Although in zw10 mutants the chromosomes congress normally to the metaphase plate, defects are first detected during anaphase of the cell cycle where the separation and poleward movements of sister chromatids (during mitosis and meiosis II) or of homologous chromosomes (during meiosis I) occur asynchronously. As a result, some lagging chromatids or chromosomes remain behind in the vicinity of the former metaphase plate during anaphase. Related effects can be phenocopied in Caenorhabditis elegans embryos by injection of antisense RNA of the nematode ZW10 homologue into gonads (Starr et al., 1997).
ZW10 proteins in Drosophila and HeLa cells display a similar and intriguing cell cycle-dependent intracellular distribution. ZW10 protein first becomes localized to the kinetochore at prometaphase, but then appears to move onto the kinetochore microtubules of the spindle at metaphase, and then back to the kinetochore at anaphase (Williams et al., 1992; Williams and Goldberg, 1994; Williams et al., 1996; Starr et al., 1997). Interestingly, the pattern of ZW10 localization with respect to each chromosome's kinetochores is influenced by the presence or absence of tension across the centromere. During metaphase of the first meiotic division in Drosophila spermatocytes, ZW10 remains at the kinetochore of univalents that are attached only to a single spindle pole, but appears in the same cell to move from the kinetochores of bivalent chromosomes under bipolar tension onto the attached kinetochore microtubules (Williams et al., 1996). This observation suggests that ZW10 may act as part of, or immediately downstream of, the wait anaphase tension–sensing checkpoint. In further support of a possible relationship between ZW10 and the anaphase onset signaling mechanism, sister chromatids in zw10 mutants often separate precociously in the presence of microtubule-depolymerizing drugs, in contrast to their behavior in wild-type (Smith et al., 1985; Williams et al., 1992).
In this paper, we show that mutations in the Drosophila zw10 gene prevent the association of dynein heavy chain (Dhc)1 with the kinetochores of both meiotic and mitotic chromosomes. Interestingly, our studies also demonstrate that dynein's kinetochore localization is influenced by tension across the centromere. We further present evidence suggesting that the function of ZW10 in the targeting of dynein to the kinetochore is mediated by direct interactions of ZW10 with dynamitin, the p50 subunit of dynactin. Because zw10 mutations appear specifically to disrupt dynein at the kinetochore but not elsewhere in the cell, the phenotype caused by zw10 mutations provides information important to understanding dynein's role at the kinetochore.
Materials and Methods
Cytological Analysis of Meiosis and Mitosis in Drosophila
The Drosophila stocks used in these experiments have been previously described in Williams et al. (1996) and Murphy and Karpen (1995).
The following techniques were used to localize various molecules within Drosophila spermatocytes. Larval, pupal, or adult testes were dissected in 0.7% NaCl and then placed in a small drop of PHEMT (60 mM Pipes, 25 mM HEPES, pH 7.0, 10 mM EGTA, 4 mM MgSO4, 0.5% Triton X-100) for 2 min. The testes were subsequently transferred to 4 μl of PHEMT + 3.7% formaldehyde on a coverslip and then immediately squashed on an inverted slide. The squashed testes were left on the slides for 10 min to allow fixation, after which the slide was immersed in liquid nitrogen and the coverslip was removed. The slide was then incubated in methanol for 20 min at −20°C, and the squash subsequently rehydrated in several changes of PBT (2.6 mM KCl, 1.5 mM KH2PO4, 137 mM NaCl, 8.1 mM Na2HPO4, 0.02% NaN3, 0.1% Triton-X100) at room temperature. To detect Dhc, either the anti-Dhc monoclonal antibody P1H4 (McGrail and Hays, 1997) at a 1:2,000 dilution in PBT or the rabbit anti-Dhc polyclonal antibody (Hays et al., 1994) at a dilution of 1:30 in PBT, was incubated with the squashed, fixed preparation overnight at 4°C. The samples were next washed in PBT 3 times for 5 min each at room temperature and then incubated overnight at 4°C with secondary antibody: either a 5-μg/ml dilution of TRITC-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc., Oak Grove, PA) if the primary reagent was the P1H4 monoclonal antibody, or with a 7.5-μg/ml dilution of TRITC-conjugated goat anti–rabbit IgG if the primary antibody was the polyclonal anti-Dhc antibody. In experiments where Dhc localization was examined in zw10 or rod mutants, wild-type control testes were placed side by side on the same slide as the mutant testes. For simultaneous localization of ZW10 and Dhc, affinity-purified rabbit anti-ZW10 polyclonal antibodies (Williams et al., 1992) at a dilution of 1:120 in PBT were mixed with the anti-Dhc P1H4 monoclonal antibody diluted as above. The secondary antibodies in this double-staining protocol were FITC-conjugated anti-rabbit IgG (The Jackson Laboratory, Bar Harbor, ME) or 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-_s_-indacene- 3- propionic acid (BODIPY-FL)– conjugated goat anti–rabbit antibody (both at 7.5 μg/ml; Molecular Probes Inc., Eugene, OR) to detect ZW10 antigen and the TRITC-conjugated anti-mouse IgG secondary antibody diluted as above to follow Dhc. After incubation with secondary antibody, all slides were washed in PBT for 2 h, stained with Hoechst 33258 (0.5 μg/ml in PBS; Sigma Chemical Co., St. Louis, MO) for 15 min, dried, and then mounted in glycerol + 2% _n_-propyl gallate to attenuate photobleaching.
To analyze mitotic figures in larval neuroblasts, larval brains were fixed, squashed, and then stained for immunofluorescence exactly according to Williams and Goldberg (1994), except that visualization of Dhc was performed as described above for testis preparations.
Images were collected using a charge-coupled device (CCD) camera (KAF1400 chip; 5 MgHz controller; Princeton Laboratories, Inc., Princeton, NJ) attached to a fluorescence microscope (model BX50; Olympus America, Lake Success, NY). Images were collected and processed with the Metamorph Imaging System (version 3.0; Universal Imaging Corporation, West Chester, PA). Alternatively, immunostained testes preparations were also observed using an ImagePointR CCD camera (Photometrics, Tucson, AZ) connected to a Zeiss Axioskop (Carl Zeiss, Inc., Oberkochen, Germany) using IPLab Spectrum software (Signal Analytics Co., Vienna, VA). All images were converted to Photoshop format (Adobe Systems Inc., Mountain View, CA). Final images were produced on a dye sublimation printer (Codonics NP1600; Cleveland, OH).
Two-hybrid Screen
A yeast two-hybrid interaction screen (Fields and Song, 1989) was preformed using the kit developed and provided to us by S. Elledge and colleagues (Baylor College of Medicine, Houston, TX), essentially following their published protocols (Bai and Elledge, 1996). The entire coding region of HZW10 was amplified by PCR using primers with 5′ NcoI and 3′ BamHI restriction site overhangs. The PCR product was then digested with NcoI and BamHI and cloned in frame and downstream of the galactose metabolism regulatory gene 4 (GAL4) DNA-binding domain (residues 1–147) in the pAS2 vector. This “bait” fusion construct (pAS2/ HZW10) was transformed into the host yeast strain Y190 (MAT a gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,-112 + URA3::GAL-lacZ, LYS2::GAL[UAS]-HIS3 cyhr). A human B cell cDNA library cloned downstream of the GAL4 transcription activation domain in the vector pACT1 (provided by S. Elledge) was transformed into Y190 + pAS2/ HZW10 as previously described (Bai and Elledge, 1996), and the transformed cells were plated onto synthetic minimal media (SD)-Trp, Leu, His + 25 mM 3-amino-1,2,4-triazole (3-AT; Sigma Chemical Co.). Transformation efficiency was determined by plating a small aliquot on SD-Trp, Leu plates. After 3–7 d, large Trp+ (presence of bait construct), Leu+ (presence of library prey construct), His+ (reporter turned on) colonies were streaked to fresh plates and colony filter lifts were made and tested for lacZ activity by a 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal) assay as previously described (Bai and Elledge, 1996).
Several criteria were used to screen against false positives. First, potential positives were streaked on SD-Leu, grown at 30°C for 2–3 d, and then streaked on SD-Leu + 2.5 mg/ml cycloheximide to select against pAS2/ HZW10. Once the bait plasmid was removed, a second X-Gal assay was preformed to identify false positives. Second, additional nonbait-specific false positives were identified by mating colonies with the potential positive pACT plasmids to the yeast strain Y187 (MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,-112 URA3::GAL-lacZ) with bait constructs encoding CDK2, SNF1, lamin, or p53 in pAS1. Diploids, selected for growth on SD-Trp, Leu, were assayed for X-Gal activity.
Plasmids containing prey constructs for pAS2/HZW10 bait-dependent positives were isolated from the host yeast as previously described (Bai and Elledge, 1996), and electroporated using an Escherichia coli pulser (Bio-Rad Laboratories, Hercules, CA) into E. coli XL1-blue (Stratagene, La Jolla, CA). Positives were sequenced using the pACT forward 5′ primer (Bai and Elledge, 1996) by the ddNTP chain termination method with the Sequenase kit (United States Biochemical, Cleveland, OH) and potential identity was determined by a BLAST search of GenBank (Altschul et al., 1990).
To exchange bait and prey, the NcoI/BamHI restriction fragment from the dynamitin/pACT plasmid was isolated and cloned into the bait vector pAS2, whereas the NcoI/BamHI PCR fragment of HZW10 was cloned into the prey vector pACTII. In addition, fragments of dynamitin were cloned by PCR with restriction site overhangs into pACTII, whereas fragments of HZW10 were cloned in a similar manner into pAS2 (see Fig. 6 for the exact size of each fragment). These constructs were tested in the two-hybrid system in the host strain Y190 by following the activity of lacZ in X-gal assays in the combinations described in the text and Fig. 6.
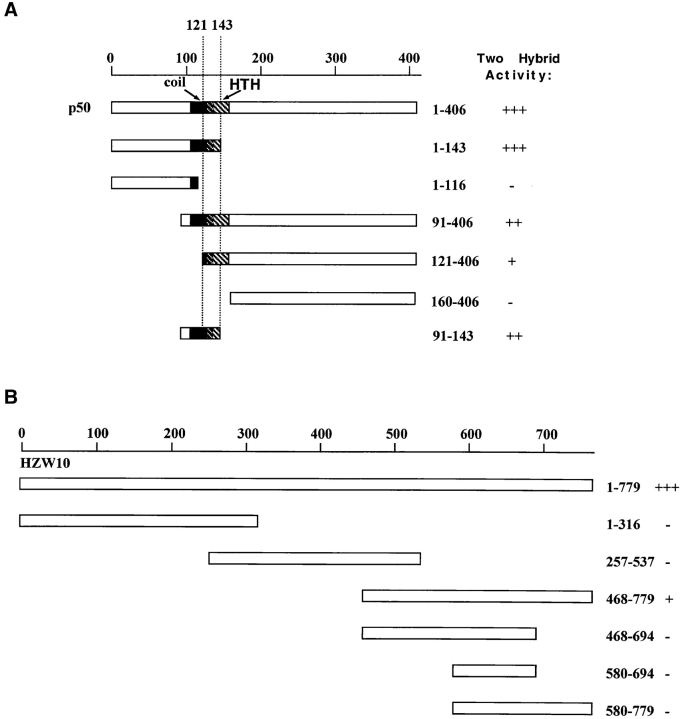
Figure 6.
Determination of the interaction domains of dynamitin and HZW10. (A) Various NH2- and COOH-terminal deletions of dynamitin (the p50 subunit of dynactin) were constructed and put into the prey vector and tested for their ability to interact with HZW10 bait in the two-hybrid system. Two-hybrid activity was measured by β-galactosidase activity. +++ was arbitrarily set as the strength of the interaction (intensity of blue) between the entire dynamitin protein and HZW10. The coil-coil (coil) and helix-turn-helix (HTH) domains of dynamitin as identified by Echeverri et al. (1996) are marked. (B) Various parts of the HZW10 protein were cloned into the prey vector and tested in the two- hybrid system with dynamitin as bait. Interaction of the entire HZW10 protein with dynamitin is set as +++.
Cytological Analysis of Human Tissue Culture Cells
HeLa cells (gift of E. Keller, Cornell University, Ithaca, NY) were grown in DME supplemented with 10% fetal bovine serum, 1,000 units/ml penicillin G sodium, and 1 mg/ml streptomycin sulfate at 37°C in 5% CO2 (all tissue culture media were from Life Technologies, Grand Island, NY). Metaphase-arrested chromosome spreads were made and fixed as previously described (Starr et al., 1997). Unsynchronized HeLa cells were grown on coverslips, further attached by centrifugation at 500 g for 1 min, and then preextracted in 0.5% Triton X-100 as described by Echeverri et al. (1996). Cells were then fixed in 4% paraformaldehyde in PBS for 10 min. Primary antibodies (affinity-purified anti-HZW10 at a dilution of 1:100) (Starr et al., 1997) and anti-dynamitin monoclonal Ab 50-1 at a 1:500 dilution (Echeverri et al., 1996), were added in PBS to fixed cells for 1 h. After washing for 15 min in PBS, the secondary antibodies (FITC-conjugated goat anti–rabbit IgG and TRITC-conjugated goat anti–mouse IgG [both from Jackson ImmunoResearch Laboratories, Inc.]), were added at a dilution of 1:100 in PBS for 1 h and then the slides were subsequently washed for 30 min in PBS. DNA was stained with 0.05 μg/ml Hoechst 33258 (Sigma Chemical Co.) for 5 min. Coverslips were mounted in 2% _N_-propyl gallate, 80% glycerol, and examined on a Zeiss Axioskop attached to a MC100 camera (Carl Zeiss, Inc., Oberkochen, Germany). Negatives were digitized using a SprintScan 35 slide scanner (Polaroid, Cambridge, MA). Anti-HZW10 signals were pseudocolored green, whereas anti-dynamitin signals were pseudocolored red using Adobe Photoshop (Adobe Systems, Inc.).
Results
Dynein Localizes to Kinetochores during Meiosis in Drosophila Males
Despite ample evidence for the association of dynein with the kinetochores of mammalian mitotic chromosomes (Pfarr et al., 1990; Steuer et al., 1990; Wordeman et al., 1991), previous attempts to visualize dynein at the kinetochores of mitotic chromosomes in Drosophila have been unsuccessful (Hays et al., 1994). We reasoned that this failure could be due to the use of dynein at other intracellular locations such as the spindle microtubules or spindle poles, which might have obscured the observation of dynein at the kinetochores in Drosophila mitotic cells. Because Drosophila primary spermatocytes are much larger than all nonsyncytial mitotic cells, with spindles over four times the size of those of other cell types, we thought it possible that the association of dynein with the kinetochores in spermatocytes might be apparent if other dynein signals were dispersed in the large volume of these cells. Meiosis in Drosophila males is easily observed cytologically, and the behaviors of chromosomes, kinetochores, and microtubules throughout both meiotic divisions have been extensively characterized (Cenci et al., 1994; Williams et al., 1996). We thus used monoclonal and polyclonal antibodies against the Dhc to examine the localization of cytoplasmic dynein through the male meiotic cell divisions by immunofluorescence. To further lower backgrounds due to cytoplasmic Dhc, we preextracted the testes with the nonionic detergent Triton X-100 before fixation, as was done by the investigators who described the association of Dhc with mammalian kinetochores (e.g., Pfarr et al., 1990; Steuer et al., 1990; Echeverri et al., 1996).
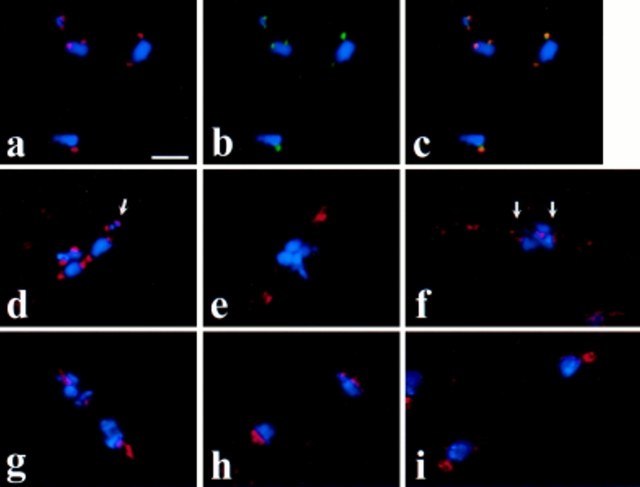
The results of this analysis, which were consistent for two different anti-Drosophila Dhc preparations (refer to Materials and Methods), are shown in Fig. 1. Dhc did not localize to discrete intracellular structures in mature primary spermatocytes before prophase I (stages M1a–M1b according to the stage designations of Cenci et al. [1994]; data not shown). However, as the bivalents condense during prometaphase I (stage M2), bright Dhc staining appeared at two separate sites on each bivalent, at the positions of the kinetochores (Fig. 1, a and d). At this stage, each kinetochore is shared by the two sister centromeres in each dyad comprising the bivalent (Goldstein, 1981; Church and Lin, 1982). The Dhc staining often assumed a hemispherical character (Fig. 1 d, arrow), reflecting the shape of the kinetochore visible in electron micrographs (Lin and Church, 1982). Dhc exactly colocalizes with the kinetochore component ZW10 during prometaphase I (Fig. 1, a–c). Further evidence that Dhc indeed associates with the kinetochores during prometaphase is presented below, where we show that Dhc association with chromosomes is correlated with the ability of DNA sequences in those chromosomes to assemble functional kinetochores, and that Dhc is bound to the kinetochore in Drosophila mitotic cells arrested in prometaphase.
Figure 1.
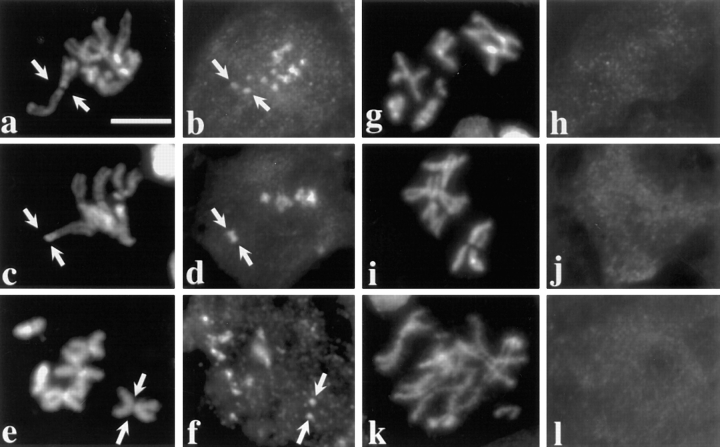
Dhc localization in Drosophila spermatocytes. Blue, DNA; red, Dhc; green, ZW10 protein. (a–c) A prometaphase I figure illustrating the presence of Dhc at the kinetochores of the bivalents. In addition, the colocalization of Dhc and ZW10 is indicated by the yellow signals in c, representing overlap of the sites of Dhc (seen alone in a) and ZW10 staining (seen alone in b). (d) Another prometaphase I figure with Dhc at the kinetochores of the chromosomes in the bivalents. Arrow, a hemispherical zone of Dhc staining on one of the kinetochores of the small fourth chromosome bivalent. (e and f) Metaphase I figures with decreased Dhc staining at the kinetochores (arrows) and with Dhc staining near the poles (e) or along kinetochore microtubules (f). (g and h) Anaphase I figures with Dhc concentrated near the poles. (i) Telophase I figure with Dhc excluded from the reforming nuclei but present at the centrosomes associated with the daughter nuclei. Bar, 10 μm.
At metaphase I (stage M3), after the bivalents have congressed to the metaphase plate, the intensity of Dhc staining in the kinetochore regions was significantly decreased (Fig. 1, e and f). In many metaphase I figures, Dhc signals could be also seen on the spindle, between the kinetochores and the poles, presumably on the kinetochore microtubules, as well as near the poles themselves (Fig. 1, e and f). It is possible that these apparent movements from the kinetochore at prometaphase to the spindle at metaphase may reflect stretching of the kinetochore's fibrous corona along kinetochore microtubules as previously observed by Rieder (1982); alternatively, the Dhc on the spindle could be recruited independently from a different intracellular pool. During anaphase I (stages M4a–c), Dhc was mostly in the vicinity of the poles, though it is difficult to distinguish what fraction of this staining represents localization on the microtubules near the poles as opposed to residual signal on the kinetochores of each dyad (Fig. 1, g and h). By telophase I (stages M4c–M5), Dhc was excluded from the reforming nuclei, but remained associated with polar regions juxtaposed to these nuclei (Fig. 1 i).
The pattern of Dhc localization during the second meiotic division was very similar to that observed during the first division. Dhc occupied the sister kinetochores of each prometaphase II chromosome (data not shown). During metaphase II, some kinetochore staining was visible, concomitant with increased staining along the spindle. At anaphase II, staining in the vicinity of the poles, possibly including some residual kinetochore signals, was visible.
Localization of Dynein to the Kinetochore Correlates with Centromere Activity but Not with Specific Centromeric Sequences
In an effort to determine which sequences at the centromere are required for the localization of Dhc to the kinetochore, we asked whether Dhc would associate with Drosophila minichromosomes containing relatively short, defined DNA sequences. G. Karpen and colleagues (Molecular Biology and Virology Laboratory, The Salk Institute, La Jolla, CA) have described the minichromosome Dp(1;f)1187 (Dp1187) and its derivatives Dp8-23 and γ238, all of which are deleted for most of the X chromosome, but nonetheless retain a functional centromere within 1 Mb of X chromosome centric heterochromatin. These minichromosomes also contain 290 kb of noncentromeric sequences from the tip of the X, including subtelomeric heterochromatin and euchromatin (collectively referred to as subtelomeric DNA). As shown in Fig. 2, Dhc is targeted to these minichromosomes during prometaphase I in primary spermatocytes.
Figure 2.
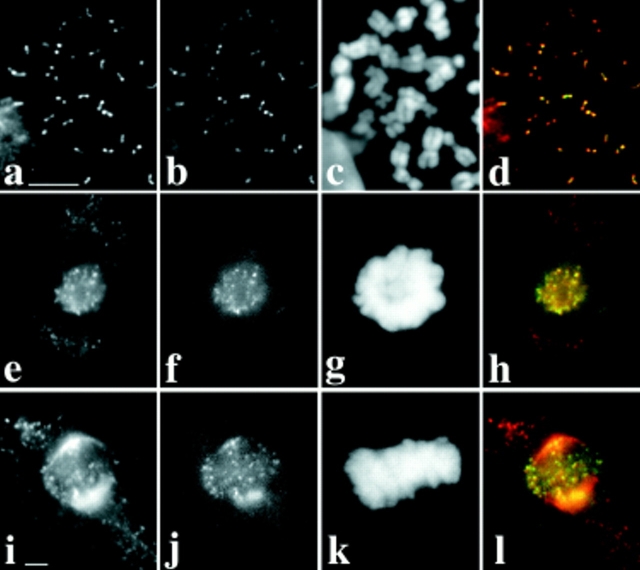
Dhc binds to acentric minichromosomes. Two views of the same field are presented to show the relationship between the distribution of DNA (a, c, e, g, and i) and Dhc (b, d, f, h, and j) in primary spermatocytes containing minichromosomes or their deleted derivatives. In all panels, arrows point to the position of the minichromosome(s). (a and b) Dp1187. Other full-length derivatives of Dp1187 (Dp8-23 and γ238) also bind Dhc (data not shown). (c and d) 31E. This deleted minichromosome contains all centromeric heterochromatin, but none of the subtelomeric DNA in Dp1187. (e and f) J21A. This deleted minichromosome contains all the subtelomeric DNA, but only approximately half the centromeric heterochromatin of Dp1187. As the stocks containing the various minichromosomes have not been continuously selected for maintenance of minichromosome number, and because transmission of smaller minichromosomes such as J21A through the male and female is imperfect, some spermatocytes in some individuals contain more than one copy of the minichromosome. (g and h) 19C. This deleted minichromosome is similar to J21A, but has lost ∼150 kb more centromeric heterochromatin. (i and j) 26C. This deleted minichromosome is only 285 kb in length and contains only the subtelomeric DNA, and thus none of the centromeric heterochromatin, of Dp1187. Details on the physical maps of all these minichromosomes can be found in Murphy and Karpen (1995) and in Williams et al. (1998). Bar, 10 μm.
After irradiation of the γ238 minichromosome, the Karpen laboratory subsequently recovered deleted minichromosomes, some less than 300 kb in length (Murphy and Karpen, 1995). We found that Dhc could associate with all of the deleted minichromosomes tested (Fig. 2). Two of these, 31E and 26C, do not contain any sequences in common. In addition, the structurally acentric minichromosome 26C completely lacks detectable centromeric DNA. Nonetheless, the levels of Dhc staining on minichromosome were comparable to those seen at the kinetochores of full-length endogenous chromosomes in the same cell (Fig. 2). All of these deleted minichromosomes appear to have centromeres that function in the male germline to organize kinetochores: as they are efficiently transmitted between generations through the male germline, they migrate toward the spindle poles during anaphase and they bind the kinetochore protein ZW10 (Williams et al., 1998). We have previously argued that the transmission of structurally acentric minichromosome deletions results from the acquisition of centromere function by the normally noncentromeric DNA from the tip of the X chromosome that is retained in these deleted minichromosomes. Several other examples of such neocentromere activity have been reported in the literature (e.g., Cancilla et al., 1998), and may involve the generation of a self-propagating centromeric chromatin structure. Regardless of the underlying mechanism, the association of Dhc with the acentric minichromosomes has at least two implications. First, Dhc cannot simply be considered a component of centromeric heterochromatin. Second, the targeting of Dhc to the kinetochores does not depend upon specific DNA sequences, but rather reflects the ability of a chromosome to organize a functional kinetochore regardless of sequence.
Dhc Localization Responds to Bipolar Orientation of Bivalents
For bivalents to achieve a stable bipolar orientation during the first meiotic metaphase, tension must be exerted across the bivalent from opposite poles of the spindle. In at least some if not all cell types, all chromosomes must be subjected to this tension before the cell progresses into anaphase (Nicklas et al., 1995). Because the Dhc signal at the kinetochores appeared to decrease in metaphase spermatocytes (refer to Fig. 1, e and f), we entertained the possibility that the association of Dhc with the kinetochores might be lessened by the presence of spindle tension. If this were the case, it might then be imagined that Dhc acts either to help measure bipolar tension, or as part of the system that transduces the measurement of tension to the eventual disjunction of homologous chromosomes at onset of anaphase I. To investigate these hypotheses, we analyzed the distribution of Dhc in primary spermatocytes containing monooriented chromosomes (univalents). In these studies, we used two compound chromosomes, the attached X-Y (X̂Y) and the compound 4th (C[4]RM), as univalents. Such compound chromosomes, in which homologues are attached to a single centromere, behave as univalents because they do not possess a pairing partner (Yamamoto, 1979; Church and Lin, 1982). As univalents can attach to only a single pole during prometaphase I and metaphase I, they are not subject to normal forces of bipolar tension (Ault and Lin, 1984; Ault and Nicklas, 1989). As a result, they cannot attain a stable metaphase orientation, and oscillate along the spindle from one pole to the other, eventually becoming randomly incorporated into daughter nuclei (Church and Lin, 1982).
Fig. 3 displays the staining of the kinetochores of these univalent chromosomes with anti-Dhc relative to the kinetochores of the second and third chromosomes in the same spermatocytes, which pair normally as bivalents. During prometaphase I, levels of kinetochore staining on univalents and bivalents were essentially identical (138 univalents scored in 13 testes; Fig. 3, a and b). However, clear differences were observed at metaphase I (Fig. 3, c–f). Although staining of the bivalent kinetochores was quite weak, the kinetochores of the univalents (when clearly separated from the bivalents) in the same cells showed intense anti-Dhc signals (44 univalents scored in 5 testes). By comparing prometaphase and metaphase figures in the same preparation, it appears that this difference is due to the loss of signal from bivalent kinetochores between prometaphase and metaphase, whereas the levels of Dhc association with univalent kinetochores seem little changed between these points of the cell cycle (Fig. 3). In summary, these observations indicate that although bipolar orientation and/or forces are not needed for the initial localization of Dhc to the kinetochore, they are necessary for the redistribution of Dhc at metaphase. We presume that this redistribution involves in part the movement of Dhc from the kinetochores to the kinetochores microtubules of chromosomes subjected to bipolar tension.
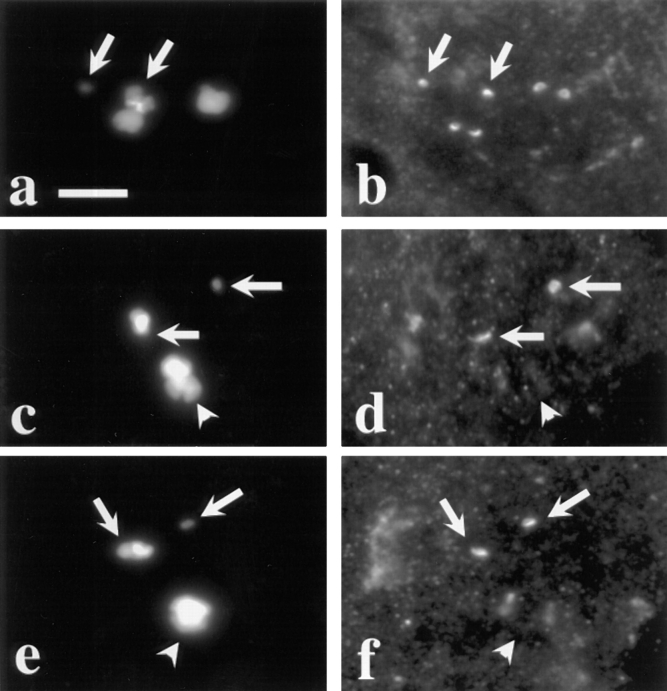
Figure 3.
Dhc localization is tension dependent. Two views of the same field are presented to show the relationship between the distribution of DNA (a, c, and e) and Dhc (b, d, and f) in primary spermatocytes containing two univalents: X^Y and C(4)RM (refer to text). Arrows, positions of these univalents; the univalent with less DNA is C(4)RM, the univalent with more DNA in the same panel is X^Y. The remaining DNA staining is due to the large autosomal bivalents that are present in the same spermatocytes. (a and b) Prometaphase I. All kinetochores, including those on the bivalents and those on the univalents, stain with equal intensity. Note that there is only a single spot of Dhc localization associated with the univalents but two spots associated with each bivalent, consistent with the number of kinetochores they contain. (c–f) Metaphase I. Univalents have a much brighter intensity of Dhc staining. Arrowheads, bivalent chromosomes situated on the metaphase plate. Bar, 10 μm.
Dhc Does Not Associate with Kinetochores in zw10 Mutants
Our observations on the intracellular distribution of Dhc through meiotic cell cycles were strongly reminiscent of the behavior of the kinetochore component ZW10, which we had previously characterized in Drosophila spermatocytes (Williams et al., 1996). ZW10 protein also associates with kinetochores starting in prometaphase, and appears to move to the kinetochore microtubules during metaphase in a tension-sensitive manner. Indeed, in both meiotic and mitotic cells stained simultaneously for ZW10 and Dhc, ZW10 signals strongly colocalized with Dhc signals at the kinetochore during prometaphase (see below). The similarities in the intracellular positions of these two proteins, along with evidence presented below pointing to a direct interaction between ZW10 and a component of the dynein-associated dynactin complex in human cells, together impelled us to determine if ZW10 protein was required for Dhc association with Drosophila kinetochores. To test this possibility, we examined Dhc protein localization in zw10 mutant spermatocytes. We used two null mutations, zw10S1 and zw10S2M, both of which abolish production of ZW10 protein (Williams et al., 1992). These mutants disrupt chromosome segregation during anaphase but do not obviously alter chromosome condensation or congression to the metaphase plate (Williams and Goldberg, 1994; Williams et al., 1996).
The results of these experiments, shown in Fig. 4, clearly demonstrate that the ZW10 protein is needed for the association of Dhc with kinetochores. In all zw10 mutant prometaphase I figures examined (n = 104 from 15 testes), the Dhc localization at the kinetochores seen in wild-type (as in Fig. 1, a and d) was eliminated (Fig. 4, a–d). Additionally, no Dhc staining was visualized on kinetochores or kinetochore microtubules at metaphase I and metaphase II (Fig. 4, e–h). The elimination of Dhc staining in zw10 mutants appears to be specific to the kinetochore, since Dhc still inhabited the polar areas during metaphase (Fig. 4, e–h) and anaphase (data not shown). Thus, the absence of the ZW10 protein affected Dhc localization to kinetochores but not to the spindle poles during metaphase and anaphase.
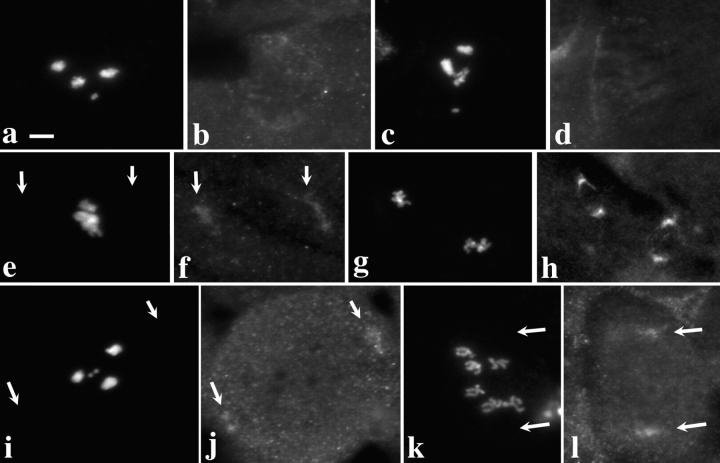
Figure 4.
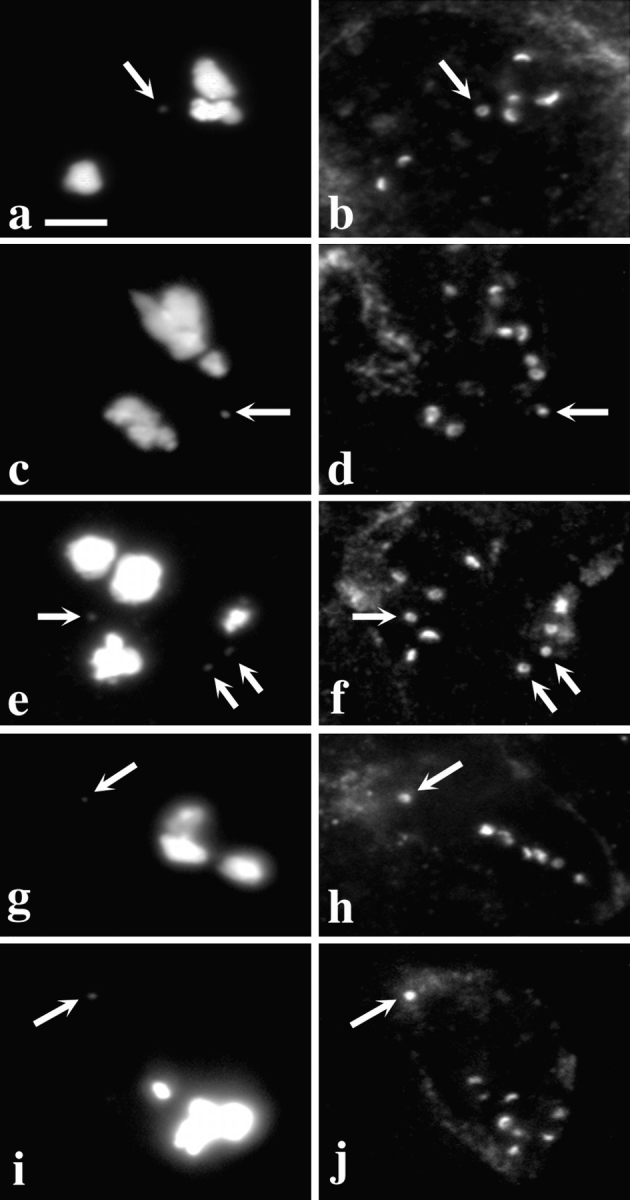
Dhc fails to localize to kinetochores in zw10 or rod mutant spermatocytes. Two views of the same field are presented to show the relationship between the distribution of DNA (a, c, e, g, i, and k) and Dhc (b, d, f, h, j, and l) in zw10S1/Y mutant testes (a–h) or in rod X−5 mutant testes (i–l). (a–d; i and j) Prometaphase I; (e and f) Metaphase I; (k and l) Anaphase I; (g and h) Metaphase II. Arrows, residual Dhc staining near the poles. Kinetochore localization of Dhc is absent at all stages. Bar, 10 μm.
Importantly, it is not simply the presence of ZW10 in spermatocytes that is required for Dhc association with kinetochores; the intracellular position of ZW10 in these cells is also critical. We have previously established that mutations in the gene rough deal (rod; Karess and Glover, 1989) prevent the localization of ZW10 to either the kinetochores or kinetochore microtubules at any meiotic stage (Williams and Goldberg, 1994; Williams et al., 1996), but these mutations do not affect the intracellular levels of ZW10 protein. When we examined the testes of rod mutants, it was clear that Dhc did not associate with the kinetochores during meiosis, leaving only residual spindle staining of microtubules at the poles (Fig. 4, i–l). It thus appears that the targeting of Dhc to kinetochores demands that ZW10 protein must also localize to the kinetochores.
To see if ZW10 was also required for Dhc localization at the kinetochores of mitotic chromosomes, we examined the localization of Dhc in Drosophila brain neuroblast cells. Dhc localization in these mitotic cells appeared to closely reflected its behavior in meiotic cells, including its localization to kinetochores and the spindle (data not shown), but a high cytoplasmic background hampered our observations. To improve the cytology, we therefore examined Dhc in wild-type larval brain neuroblasts arrested in a prometaphase-like state with the microtubule poison colchicine, and swollen with hypotonic solution (Gatti and Goldberg, 1991). In these cells, Dhc was clearly localized at the sister kinetochores of the duplicated chromosomes (Fig. 5, a–f). Note that because these preparations were not preextracted with Triton X-100 before fixation, the kinetochore localization of Dhc cannot be only a detergent-induced artifact. When zw10 mutant brains were stained under exactly the same conditions, however, Dhc protein was absent from the kinetochore, and was only uniformly dispersed throughout the cell (Fig. 5, g–j). Similar results were observed in the neuroblasts of animals carrying mutations in rod (Fig. 5, k and l). Thus, ZW10 (as well as the product of the rod gene) is required to recruit Dhc to the kinetochore in both meiosis and mitosis.
Figure 5.
Dhc fails to localize to the kinetochore in zw10 or rod mutant larval brain neuroblasts. Two views of the same field are presented to show the relationship between the distribution of DNA (a, c, e, g, i, and k) and Dhc (b, d, f, h, j, and l) in neuroblasts from brain preparations treated with colchicine and swelled in hypotonic solution as described in Materials and Methods. (a–f) Wild-type (Oregon-R). (g–j) zw10S1/Y. (k and l) rod X −5. Arrows, clear Dhc staining at the kinetochore regions in wild-type neuroblasts. Dhc in zw10 and rod mutant brains is absent at the kinetochore. Bar, 10 μm.
Human ZW10 and Dynamitin Interact in the Two-hybrid System
To find potential molecular interactors of ZW10, we performed a two-hybrid screen (Fields and Song, 1989) using the GAL4 system (Bai and Elledge, 1996). As Drosophila ZW10 protein in a bait construct activates transcription of the reporter genes by itself, we attempted these experiments using human ZW10 protein (HZW10; Starr et al., 1997). The HZW10 open reading frame was cloned downstream and in frame to sequences encoding the DNA binding domain of GAL4 to make the bait construct (pAS2/HZW10), which was transformed into the host yeast strain (refer to Materials and Methods). Synthesis of the GAL4/HZW10 fusion protein in the transformed yeast cells was confirmed by Western blot analysis (data not shown). The GAL4/HZW10 bait was not able to turn on the reporter genes (HIS3 or lacZ), allowing its use to screen a human B cell cDNA library fused to the GAL4 transcription activation domain. Positives were defined as being able to turn on both reporter genes only in the presence of the HZW10 bait, but not with other baits (CDK2, SNF1, lamin, or p53). DNA sequences were determined from the 21 positives obtained; two independent positives proved to be dynamitin, the p50 subunit of the dynactin complex, a known component of the kinetochore (Table I; Echeverri et al., 1996). Other results from this two-hybrid screen will be presented elsewhere (Starr, D.A., and M.L. Goldberg, manuscript in preparation). To verify the interaction between HZW10 and dynamitin, the constructs were then interchanged. The entire coding region of dynamitin was cloned into the bait vector whereas HZW10 was cloned into the prey vector; this combination also activated the lacZ reporter gene (Table I).
Table I.
Two-hybrid β-galactosidase Assay Results
| Bait construct (in pAS2) | Prey construct (in pACT) | β-galactosidase activity |
|---|---|---|
| HZW10 | p50 (isolate 1) | +++ |
| HZW10 | p50 (isolate 2) | +++ |
| p50 (isolate 1) | HZW10 | ++ |
| CDK2 | p50 (isolate 1) | — |
| SNF1 | p50 (isolate 1) | — |
| lamin | p50 (isolate 1) | — |
| p53 | p50 (isolate 1) | — |
| CDK2 | HZW10 | — |
| SNF1 | HZW10 | — |
| lamin | HZW10 | — |
| p53 | HZW10 | — |
| HZW10 | — | — |
| p50 (isolate 1) | — | — |
| — | p50 (isolate 1) | — |
| — | p50 (isolate 2) | — |
| — | HZW10 | — |
| HZW10 | HZW10 | — |
We also used the two-hybrid system to map the regions in both HZW10 and dynamitin that were responsible for this interaction. Within dynamitin, the region participating in the interaction is quite small. The minimal binding domain allowing a weak interaction can be roughly defined as extending from amino acids 121–143, but for optimal interaction, additional amino acids toward the NH2 terminus of dynamitin are also required, especially amino acids 105– 120 (Fig. 6). This region of dynamitin includes what has previously been considered on the basis of structural considerations to be a predicted coil-coil domain (amino acids 105–135; Echeverri et al., 1996). Within HZW10, the region involved in the two-hybrid interaction appeared to be restricted to the COOH-terminal 300 amino acids of the protein, the part of ZW10 proteins that is best conserved during evolution (Starr et al., 1997). However, expression of the β-galactosidase reporter was considerably lower using a construct containing only this region relative to the entire HZW10 protein, and attempts to map the binding region more precisely within the COOH-terminal part of the protein were unsuccessful (Fig. 6). It is therefore possible that HZW10's ability to recognize dynamitin in the two-hybrid system involves sequences dispersed throughout the COOH-terminal 300 amino acids, as well as one or more domains closer to the NH2-terminus for optimal interaction. Another possibility, that the association between HZW10 and dynamitin requires multimerization of HZW10 to form a binding surface for dynamitin seems unlikely, since at least in two-hybrid analysis, HZW10 appears not to interact with itself (Table I).
Dhc and Dynamitin Colocalize with ZW10 at the Kinetochore
As argued more fully in the Discussion below, the results reported in this paper support a model in which ZW10 targets the dynactin complex to the kinetochore through a direct interaction with dynamitin, and that the dynactin complex in turn recruits dynein to the kinetochore. If this hypothesis is valid, one would expect that all three proteins would be found in close association at the kinetochore. We have already shown in Fig. 1, a–c that in Drosophila, Dhc and ZW10 colocalize at the kinetochores during prometaphase of meiosis I; this is also true during mitosis and meiosis II in the fly (data not shown). A similar finding was also obtained for human cells: Fig. 7, a–d depicts the results of immunofluorescence experiments demonstrating that dynamitin and HZW10 also completely colocalize to the kinetochores in HeLa cell chromosome spreads from cells arrested by nocodazole treatment. Echeverri et al. (1996) have previously shown that several components of dynactin and dynein complexes colocalize at the kinetochore of similarly treated HeLa cells. It should be noted that substantial evidence shows that the location of these three molecules is in fact at the kinetochore (or its fibrous corona) rather than in centromeric heterochromatin (Wordemann et al., 1991; Starr et al., 1997).
Figure 7.
Colocalization of dynamitin and HZW10. (a–d) A chromosome spread from a HeLa cell arrested in mitosis by nocodazole. (e–h) A HeLa cell in prometaphase. (i–l) A HeLa cell in metaphase. The panels in the first column of each row show staining with anti-dynamitin (a, e, and i); in the second column, staining with anti HZW10 (b, f, and j); and in the third column staining with Hoechst 33258 to visualize DNA (c, g, and k). In the column at the right (d, h, and l), dynamitin staining is shown in red, and HZW10 staining in green; the overlap between these signals is yellow. Bars, 5 μm.
We also show partial colocalization of HZW10 and dynamitin in cycling HeLa cells. At prometaphase both antibodies colocalize to punctate dots in the chromatin (Fig. 7, e–h) presumed to be kinetochores based on previously published results (Echeverri et al., 1996; Starr et al., 1997). At metaphase, dynamitin is primarily on the spindle, where a large amount of HZW10 also localizes (Fig. 7, i–l). Although HZW10 and dynamitin do not completely colocalize on the spindle, there is considerable overlap.
Discussion
ZW10 Is Required for the Association of Dynein with the Kinetochore
Several previous publications have documented the presence of dynein at the kinetochores of chromosomes in mammalian tissue culture cells (Pfarr et al., 1990; Steuer et al., 1990; Wordeman et al., 1991; Echeverri et al., 1996; Dujardin et al., 1998). In this paper, we have found that Dhc also associates with the kinetochores of mitotic and meiotic chromosomes in Drosophila. All previous work has used detergent preextraction to remove background staining due to high concentrations of dynein in the cytoplasm in order to visualize dynein at the kinetochore. Although we have used a similar procedure to see Dhc at the kinetochore in Drosophila primary spermatocytes (Figs. 1–4), we have also been able to detect Dhc at the kinetochore of mitotic chromosomes that have not been subjected to detergent preextraction, demonstrating that the association of dynein with the kinetochore is not artefactually induced by this method of preparation.
We have presented a number of lines of evidence supporting the hypothesis that one role played by the wild-type zw10 gene product is to recruit dynein to the kinetochore. ZW10 and Dhc colocalize to the kinetochores of fly and human chromosomes (refer to Fig. 1 and Fig. 7) in a fashion that is identically influenced by tension across the chromosomes (refer to Fig. 3). Both proteins are found at the kinetochores elaborated by minichromosome derivatives that lack centromeric sequences but that display neocentric activity (refer to Fig. 2). Both proteins fail to associate with the kinetochore in rod mutant spermatocytes (refer to Fig. 4, i–l) and neuroblasts (refer to Fig. 5, k and l). Yeast two-hybrid experiments show that human ZW10 protein and dynamitin, the p50 subunit of dynactin complex, can interact with each other directly (refer to Table I and Fig. 6).
The most compelling evidence pointing to a role for the ZW10 protein in dynein localization to the kinetochore is documented in Fig. 4, a–d and Fig. 5, g–j, which show that in zw10 mutant spermatocytes and neuroblasts, Dhc fails to localize to the kinetochore at levels detectable by immunofluorescence. Of course, because of significant backgrounds of anti-Dhc staining (presumably reflecting the use of Dhc in many intracellular contexts), we cannot exclude the possibility that an undetectable small fraction of Dhc is retained at the prometaphase kinetochore in zw10 mutants. The same caveat also applies in our studies demonstrating that Dhc is not kinetochore associated in rod mutant testes and brains (refer to Fig. 4, i–l and Fig. 5, k and l). These latter results indicate that the large majority of the binding of Dhc to the kinetochore is dependent upon the arrival of ZW10 protein at the same location. This is because the ZW10 protein is present in rod mutant cells in normal amounts but is not present at the kinetochore (Williams and Goldberg, 1994). The requirement for both ZW10 and ROD proteins would be most easily explained if these two polypeptides were complexed with each other. Indeed, in collaboration with F. Scaerou and R. Karess (CNRS Centre de Génétique Moléculaire, Gif-sur-Yvette, France), we have recently obtained evidence for the existence of such a complex, which requires the participation of both proteins for its localization to the kinetochore (our manuscript in preparation).
It could be argued that the depletion of Dhc from the kinetochore in zw10 or rod mutant spermatocytes is only an indirect effect of major disruptions in kinetochore structure caused by these mutations. Three observations indicate that this is unlikely to be true. First, many aspects of kinetochore function are relatively untouched by these mutations. In mutant zw10 and rod meiotic and mitotic cells, the chromosomes appear to condense appropriately and congress to the metaphase plate. At anaphase, most sister chromatids (mitosis and meiosis II) or homologous chromosomes (meiosis I) separate from each other and migrate toward the poles during anaphase. Second, other kinetochore proteins, like the those recognized by the 3F3/2 antibody (Bousbaa et al., 1997; Gorbsky and Ricketts, 1993) and Drosophila Bub1, are properly localized in zw10 and rod mutants (Basu, J., B.C. Williams, and M.L. Goldberg, manuscript in preparation). Third, we have two- hybrid evidence that the interaction between ZW10 and dynein is reasonably direct, being mediated by contacts between ZW10 and dynamitin, the p50 subunit of the dynactin complex (refer to Table I). This scenario is in accord with previous work suggesting that the dynactin complex helps target cytoplasmic dynein to appropriate intracellular sites (for review see Vallee and Sheetz, 1996).
In the yeast two-hybrid system, human ZW10 and dynamitin associate with each other to activate the transcription of two different reporter genes. We have mapped the interaction domain of dynamitin to a 22-amino acid region including part of a conserved coil-coil domain and the region immediately downstream (refer to Fig. 6). This same region has been shown to be important for dynamitin function (Echeverri, C., and R. Vallee, personal communication). Echeverri et al. (1996) demonstrated that overexpression of wild-type dynamitin disrupts the dynactin complex, leading to a number of phenotypes. However, when expressed at even low levels, dynamitin, with a small deletion including the human ZW10-interacting domain, causes the same phenotypes, including a lack of dynein at the kinetochore (Echeverri, C., and R. Vallee, personal communication). The idea that ZW10 and dynamitin interact directly with each other in the cell must nonetheless be approached with some caution. We have thus far been unable to show a direct interaction between ZW10 and dynamitin by methods other than the yeast two-hybrid system, such as coimmunoprecipitation or binding assays using in vitro–translated proteins. We believe this is because the two proteins are normally able to interact only in the context of the kinetochore, an insoluble structure. In addition, though we know that the region of dynamitin that interacts with ZW10 in the two-hybrid system is necessary for dynamitin function, it is not yet clear whether this region alone can target dynactin to the kinetochore.
Our results taken together strongly imply a model in which a complex including ZW10 and ROD arrives at the kinetochore early in prometaphase, that this complex then attracts the dynactin complex to the kinetochore by virtue of direct contacts between ZW10 and dynamitin, and finally, that dynactin in turn targets dynein to the kinetochore (Fig. 8).
Figure 8.
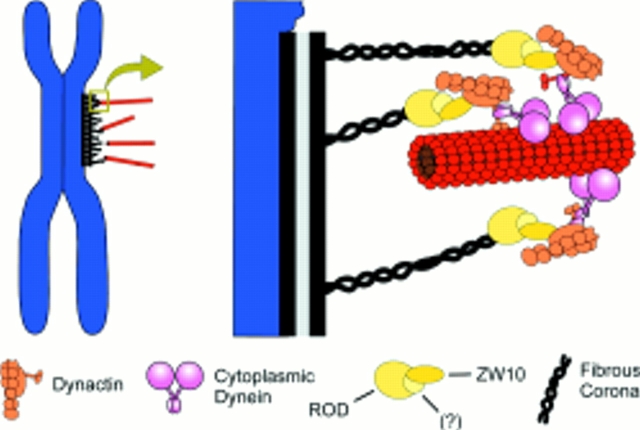
Model for the ZW10/ROD-dependent targeting of dynein to the kinetochore. In this model, a complex containing ZW10 and ROD proteins, as well as potential unknown additional components (?), is associated with the fibrous corona of the prometaphase kinetochore. Direct interactions between ZW10 and the p50 subunit of the dynactin complex then bring dynactin to the kinetochore. Dynactin in turn recruits cytoplasmic dynein to the kinetochore, providing one possible contact between the kinetochore and microtubules. Our results suggest that this contact is sensitive to bipolar tension exerted across the chromosome. As described in the Introduction, additional kinetochore/microtubule interactions are undoubtedly mediated by other microtubule motors such as CENP-E (data not shown). This figure is modified from Vallee et al. (1995).
What Can the zw10 and rod Mutant Phenotypes Tell Us about the Function of Dynein at the Kinetochore?
As stated in the Introduction, it has been difficult to assess the exact role of dynein at the kinetochore because of the complications presented by dynein function at other locations, particularly in the organization of the spindle (Heald et al., 1996). It is tempting, however, to ascribe the chromosomal missegregation phenotypes seen in Drosophila zw10 or rod mutants (Karess and Glover, 1989; Williams et al., 1992, 1996), or in C. elegans zw10 antisense RNA-treated embryos (Starr et al., 1997) to the failure of dynein to localize to the kinetochore. There are some dangers with this assumption. ZW10 and ROD proteins may play additional roles at the kinetochore beyond the targeting of dynein, and it is possible that a small amount of dynein remains at the kinetochore in zw10 or rod mutant spermatocytes. We nonetheless believe that it is a useful exercise to assume that the zw10 and rod phenotypes reflect the loss of dynein from the kinetochore.
Since chromosomes in a zw10 or rod mutant cell congress to the metaphase plate (Karess et al., 1989; Williams et al., 1992), wild-type levels of dynein at the kinetochore can not be uniquely required for chromosome microtubule attachments or movements before anaphase onset. This conclusion is somewhat surprising, since it has been previously proposed that dynein at the kinetochore is involved in the initial capture of a microtubule and the rapid poleward movement observed by Rieder and Alexander (1990) (Pfarr et al., 1990; Steuer et al., 1990). The kinetics of this rapid minus end–directed movement along the side of a single microtubule match those of dynein (Rieder and Alexander, 1990). This initial poleward movement is thought to eventually facilitate the ability of kinetochores near the poles to capture the plus ends of additional microtubules. Our results do not disprove such a role for dynein in chromosome congression, but they do suggest that other microtubule motors are able to supplant dynein function in this regard.
Phenotypic analysis suggests that zw10 and rod mutations mostly interfere with the fidelity and coordination of events at anaphase onset. Sister chromatids (mitosis and meiosis II) or homologous chromosomes (meiosis I) for the most part separate at anaphase onset, and migrate towards the spindle pole in anaphase. However, some chromatids or chromosomes appear to separate from each other later than normal, and often remain in the vicinity of the metaphase plate even late in anaphase. To the extent that these phenotypes reflect the role of dynein at the kinetochore, they suggest two possibilities for dynein's function at this location. Dynein might participate in the checkpoint mechanisms that sense bipolar tension across the centromere, delaying anaphase onset until all the chromosomes are properly aligned on the metaphase plate. In this light, it is of interest that in zw10 and rod mutant, but not wild-type neuroblasts, sister chromatids separate precociously when the cells are treated with the microtubule poison colchicine. One interpretation consistent with our observations is that the lack of dynein at the kinetochore allows cells to bypass the wait anaphase checkpoint. Alternatively, dynein might be required at the kinetochore to supplement and/or coordinate other microtubule motors in moving chromosomes to the poles during anaphase. The resolution to this question may lie in a more detailed analysis of the zw10 and rod mutant phenotypes.
Abbreviations used in this paper
CCD
charge-coupled device
Dhc
dynein heavy chain
GAL4
galactose metabolism regulatory gene 4
rod
rough deal gene
SD
synthetic minimal media
X-Gal
5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside
Footnotes
We would like to dedicate this paper to the memory of B. Keller (Cornell University, Ithaca, NY). We thank Z. Li and E. Williams (both from Cornell University) for technical help, M. Serr and S. O'Rourke (both from University of Minnesota, St. Paul, MN) for the Dhc antibodies, T. Murphy and G. Karpen (both from The Salk Institute, La Jolla, CA) for the minichromosome stocks, J. Lis and C. Bayles (both from Cornell University) for assistance with CCD microscopy, and F. Scaerou and R. Karess (both from CNRS Centre de Génétique Moléculaire, Gif-sur-Yvette, France) for helpful discussion. We are indebted to C. Echeverri and R. Vallee (both from University of Massachusetts Medical Center, Boston, MA) for insightful discussions and artistic help in preparing Fig. 8.
This research was supported by a grant from the National Institutes of Health (NIH) to M.L. Goldberg (GM 48430) and by an NIH training grant (GM 07617) to the Field of Genetics and Development at Cornell University (Ithaca, NY).
D.A. Starr and B.C. Williams contributed equally to this work.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ault JG, Lin HP. Bivalent behavior in Drosophila melanogastermales containing the In(1)sc4Lsc8RX chromosome. Chromosoma. 1984;90:222–228. doi: 10.1007/BF00292400. [DOI] [PubMed] [Google Scholar]
- Ault JG, Nicklas RB. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma. 1989;98:33–39. doi: 10.1007/BF00293332. [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ. Gene identification using the two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Bousbaa H, Correia L, Gorbsky GJ, Sunkel CE. Mitotic phosphoepitopes are expressed in Kc cells, neuroblasts and isolated chromosomes of Drosophila melanogaster. . J Cell Sci. 1997;110:1979–1988. doi: 10.1242/jcs.110.17.1979. [DOI] [PubMed] [Google Scholar]
- Cancilla MR, Tainton KM, Barry AE, Larionov V, Kouprina N, Resnick MA, Sart DD, Choo KHA. Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast. Genomics. 1998;47:399–404. doi: 10.1006/geno.1997.5129. [DOI] [PubMed] [Google Scholar]
- Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogasterspermatogenesis. J Cell Sci. 1994;107:3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- Chen R, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Church K, Lin HP. Meiosis in Drosophila melanogaster. II. The prometaphase-I kinetochore microtubule bundle and kinetochore orientation in males. J Cell Biol. 1982;93:365–373. doi: 10.1083/jcb.93.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin D, Wacker UI, Moreau A, Schroer TA, Rickard JE, De Mey JR. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J Cell Biol. 1998;141:849–862. doi: 10.1083/jcb.141.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery NS, Choi T, Brown KD, Wood KW, Resau J, Fukasawa K, Cleveland DW, Van de Woude GF. CENP-E is an essential kinetochore motor in maturing oocytes and is masked during mos-dependent, cell cycle arrest at metaphase II. Proc Natl Acad Sci USA. 1997;94:9165–9170. doi: 10.1073/pnas.94.17.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Goldberg ML. Mutations affecting cell division in Drosophila. . Methods Cell Biol. 1991;35:543–586. doi: 10.1016/s0091-679x(08)60587-7. [DOI] [PubMed] [Google Scholar]
- Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. . Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- Gorbsky G, Ricketts WA. Differential expression of phosphoepitope at the kinetochore of moving chromosomes. J Cell Biol. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays TS, Porter ME, McGrail M, Grissom P, Gosch P, Fuller MT, McIntosh JR. A cytoplasmic dynein motor in Drosophila: identification and localization during embryogenesis. J Cell Sci. 1994;107:1557–1569. doi: 10.1242/jcs.107.6.1557. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopusegg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Karess RE, Glover DM. rough deal, a gene required for proper mitotic segregation in Drosophila. . J Cell Biol. 1989;109:2951–2961. doi: 10.1083/jcb.109.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Benerza R. Identification of a human mitotic checkpoint gene: hsMAD2. . Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Lin HP, Church K. Meiosis in Drosophila melanogaster, III. The effect of orientation disruptor (ord)on gonial mitotic and the meiotic divisions in males. Genetics. 1982;102:751–770. doi: 10.1093/genetics/102.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail M, Hays TS. The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila. . Development (Camb) 1997;124:2409–2419. doi: 10.1242/dev.124.12.2409. [DOI] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Location of centromere function in a Drosophilaminichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J Cell Biol. 1989;109:2245–2255. doi: 10.1083/jcb.109.5.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Baker BS, Gatti M. Mutations in genes controlling essential mitotic functions in Drosophila melanogaster. . Genetics. 1985;110:647–670. doi: 10.1093/genetics/110.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Li Z, Etemad-Moghadam B, Dawe RK, Goldberg ML. Conservation of the centromere/kinetochore protein ZW10. J Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Echeverri CJ, Vaughn KT. Targeting of cytoplasmic dynein to membranous organelles and kinetochores via dynactin. Cold Spr Harb Symp Quant Biol. 1995;60:803–811. doi: 10.1101/sqb.1995.060.01.086. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Sheetz MP. Targeting of motor proteins. Science. 1996;271:1539–1544. doi: 10.1126/science.271.5255.1539. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopuseggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopuskinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Williams BC, Gatti M, Goldberg ML. Bipolar spindle attachments affect redistributions of ZW10, a Drosophilacentromere/kinetochore component required for accurate chromosome segregation. J Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Goldberg ML. Determinants of Drosophilazw10 protein localization and function. J Cell Sci. 1994;107:785–798. doi: 10.1242/jcs.107.4.785. [DOI] [PubMed] [Google Scholar]
- Williams BC, Karr TL, Montgomery JM, Goldberg ML. The Drosophila l(1)zw10gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. . Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–105. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Steuer ER, Sheetz MP, Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M. Cytological studies of heterochromatin function in the Drosophila melanogastermale: autosomal meiotic paring. Chromosoma. 1979;72:293–328. doi: 10.1007/BF00331091. [DOI] [PubMed] [Google Scholar]
- Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochores to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]