Predominant Mode of Human Immunodeficiency Virus Transfer between T Cells Is Mediated by Sustained Env-Dependent Neutralization-Resistant Virological Synapses (original) (raw)
Abstract
Cell-free human immunodeficiency virus type 1 (HIV-1) can initiate infections, but contact between infected and uninfected T cells can enhance viral spread through intercellular structures called virological synapses (VS). The relative contribution of VS to cell-free viral transfer has not been carefully measured. Using an ultrasensitive, fluorescent virus transfer assay, we estimate that when VS between HIV-expressing Jurkat T cells and primary CD4+ T cells are formed, cell-associated transfer of virus is 18,000-fold more efficient than uptake of cell-free virus. Furthermore, in contrast to cell-free virus uptake, the VS deposits virus rapidly into focal, trypsin-resistant compartments in target T cells. This massive virus internalization requires Env-CD4 receptor interactions but is resistant to inhibition by patient-derived neutralizing antisera that inhibit homologous cell-free virus. Deleting the Env cytoplasmic tail does not abrogate VS-mediated transfer, but it renders the VS sensitive to neutralizing antibodies, suggesting that the tail limits exposure of VS-neutralizing epitopes on the surface of infected cells. Dynamic live imaging of the VS reveals that HIV-expressing cells are polarized and make sustained, Env-dependent contacts with target cells through uropod-like structures. The polarized T-cell morphology, Env-CD4 coordinated adhesion, and viral transfer from HIV-infected to uninfected cells suggest that VS allows HIV-1 to evade antibody neutralization and to disseminate efficiently. Future studies will discern to what extent this massive viral transfer contributes to productive infection or viral dissemination through the migration of virus-carrying T cells.
Human immunodeficiency virus (HIV) produces billions of virus particles in infected patients each day (25, 48, 58, 62). Sensitive methods detect abundant cell-free virus in patient plasma at all disease stages (50), yet the efficiency with which virus can be cultured from cell-free fluids is low relative to that from cell-associated virus (11, 24). From viral preparations produced optimally in vitro, it is estimated that only 1 in 104 to approximately 107 virus particles is infectious (33, 36, 38). The production of defective particles may be attributable to numerous causes, many of which point to a paucity of functional Env on virus particles (9, 22, 23, 33, 39, 52, 67).
Levels of plasma virus can serve as a clinical indicator for disease progression (32, 43, 44) and viral transmission (60), but the extent to which cell-free plasma virus mediates viral dissemination still is unclear (21). The striking compartmentalization of HIV quasi-species in microdissected splenic white pulp suggests that the dissemination of HIV can be dominated by local, anatomically restricted viral transmission rather than by a systemic swarm of plasma virus (10).
The spread of HIV in cultured cells is known to be more rapid when cell-associated virus initiates an infection (14, 56), but the mechanisms for this enhancement are not well characterized. Pearce-Pratt et al. proposed that HIV transfer is activated by cell-to-cell contact (47) and found that viral budding was concentrated at subcellular locations where infected monocytes contacted epithelial cells (49). These early studies suggest that in HIV-infected cells, cellular adhesion and motility are enhanced.
To facilitate viral transfer, HIV-infected cells or cells that have endocytosed HIV can form intimate adhesive contacts, referred to as viral or infectious synapses (51). Direct transfer of HIV-1 between immune cells was first described for transfer between dendritic cells and T cells (7). In this type of infectious viral transfer, endocytic uptake of viral particles by dendritic cells occurs through CD4-independent mechanisms. The infectious virus is presented with high efficiency to target CD4+ T cells through a process sometimes referred to as trans infection.
Another mode of viral transfer involves transfer between infected T cells and uninfected epithelial cells. Infected T cells can adhere to the apical surface of intestinal epithelial cells and can induce the transcytosis of virus through the cell (4, 5). These studies demonstrate that infected cells can adhere to noninfected cells and induce them to endocytose virus in a cell-contact-dependent manner.
More recently, researchers revealed that stable adhesive contacts formed between T cells facilitate viral transfer from infected to uninfected cells (28, 29). In these virological synapses (VS), the viral Env protein and cellular coreceptors CD4 and CXCR4 were recruited toward the site of cell-to-cell contact. The relocalization of viral and cellular receptors required cytoskeletal rearrangements, suggesting the involvement of active cellular processes. Viral Gag antigen in the target T cells was detected following contact with infected donor cells. Others have reported that coreceptor-independent transfer of HIV is promoted by cell-to-cell contact (3) and that limiting cell contact in vitro by continuously agitating cells can reduce viral spread in culture (59). These studies all suggest that the process of cell-to-cell viral transfer is coordinated to maximize vectorial transfer of virus into uninfected cells.
In this study, we quantify and visualize highly efficient T-cell-to-T-cell viral transfer by directly tracking a highly fluorescent infectious clone of HIV-1. We find that the magnitude of VS-mediated viral antigen transfer is vastly greater than that mediated by high levels of cell-free virus. The viral transfer also is qualitatively distinct from that of cell-free infection, in that the viral particles are rapidly sequestered into a trypsin-resistant cellular compartment. The cell-cell adhesion and viral transfer is dependent upon the ectodomain of Env, is specific to CD4 T lymphocytes, and is resistant to neutralizing antisera that can inactivate cell-free virus. Using live-imaging methods, we find that productively infected cells are polarized and nonsessile and use Env to engage target cells with durable and high-multiplicity interactions. The high efficiency of VS-mediated transfer hijacks normal T-cell-adhesive and -interactive functions to disseminate large amounts of viral antigen from cell to cell.
MATERIALS AND METHODS
Viral constructs.
HIV Gag-iGFP is an NL4-3-based (1) HIV-1 molecular clone that carries green fluorescent protein (GFP) inserted internally into Gag between the MA and CA domains (27). A variant expressing the Env gene from molecular clone JRFL (45) was constructed carrying the NdeI to BamHI fragment in NL4-3 replaced with the corresponding sequence from JRFL. HIV Gag-iGFP ΔEnv carries a frameshift mutation in the Env gene generated by destroying a unique NdeI restriction site in the 5′ end. HIV Gag-iGFP ΔMA has a deletion of the globular head of MA that retains the N-terminal 28 amino acids of MA fused to the C-terminal 12 amino acids of MA (26). HIV Gag-iGFP ΔCT carries a premature termination codon that truncates the last 144 amino acids of the Env cytoplasmic tail (8).
Cells and tissue culture.
The CD4+ T-cell line Jurkat CE6.1 (ATCC) was maintained in RPMI 1640 with 100 U/ml penicillin, 100 g/ml streptomycin, and 10% fetal bovine serum (FBS). Cells were passaged regularly and were maintained at concentrations of less than 5 × 105/ml. HIV-1 proviral constructs were transduced into Jurkat cells using Amaxa nucleofection (Amaxa Biosystems). In brief, 3 μg of endotoxin-free HIV-1 proviral plasmids was nucleofected into 5 × 106 Jurkat cells by using Cell Line Nucleofector kit V, program S-18. Twenty-four hours after nucleofection, viable Jurkat cells were purified by centrifugation on a Ficoll-Hypaque density gradient. Forty-eight hours after nucleofection, cells were washed with complete buffer and enumerated for testing. Human peripheral blood mononuclear cells from healthy HIV/hepatitis B virus-seronegative donors were purified from leukocyte buffy coat by Ficoll gradient. CD4+ or D8+ T cells were enriched from peripheral blood mononuclear cells by negative selection using the CD4+ T-cell isolation kit II or by positive selection using the CD8 microbeads, according to the manufacturer's instructions (Miltenyi Biotec). The CD4+ or CD8+ T cells were uniformly above 95% purity. Unless indicated, primary cells were resuspended in RPMI medium and stimulated with phytohemagglutinin overnight.
Viral transfer assay.
CD4+ T cells were resuspended in RPMI 1640 and labeled with 1 μM CellTracker orange CMTMR fluorescent dye [(5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine); Molecular Probes] at 37°C for 15 to 45 min, followed by incubation with complete medium under growth conditions for 30 min. The nucleofected Jurkat cells (donor cells) or CMTMR-labeled CD4+ T cells (target cells) were extensively washed and resuspended in RPMI 1640 with 10% FBS to a final concentration of 1.25 × 106/ml. Virus transfer was carried out by mixing equal volumes (200 μl) of donor and target cells in a 5-ml round-bottom tube (BD Falcon), and the mixtures were incubated at 37°C for the indicated times. Virus transfer was terminated by washing the mixtures with 10 volumes of phosphate-buffered saline (PBS) and leaving them untreated or treating them with 0.05% trypsin-EDTA (Invitrogen) at 37°C for 5 to 10 min. Trypsin-treated cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 10 min before analysis by flow cytometry.
A control donor cell line, Jurkat GFP, was created by infecting Jurkat E6 with a murine stem cell virus enhanced GFP viral vector and was selected for with puromycin. When used as a control donor cell line, the cells were mock transduced with Amaxa nucleofection.
Inhibitors of viral transfer.
To test the ability of inhibitors to block VS-mediated viral transfer, donor and target cells were pretreated at 37°C for 30 to 60 min with inhibitors. CD4/gp120 blocking reagents were the following: soluble CD4 (Progenics Corporation); Leu 3a, an HIV-blocking anti-CD4 antibody (BD Biosciences); immunoglobulin G1b12 (IgG1b12), a gp120 CD4 binding site monoclonal antibody (from Dennis Burton and Paul Parren, through the AIDS Reference and Reagent Program [ARRP]); 2F5, a monoclonal antibody against gp41 (Hermann Katinger, through the ARRP); or AMD3100, a CXCR4 inhibitor (courtesy of Anormed, through the NIAID/ARRP). Actin inhibitors were the following: cytochalasin D (BD Biosciences) and latrunculin B (Axxora). HIV-1-positive patient sera and control patient sera (no. 2411, 1983, and 1984) were from Luba Vujcic through the ARRP. To test reagents for their ability to block cell-free infection, we preincubated virus with inhibitors for 60 min and then infected GHOST indicator cells (Vineet KewalRamani and Dan Littman, through the ARRP) with native HIV-1(NL4-3) (4 ng p24 per well) and measured infection at 24 h by flow cytometry.
Confocal and live imaging.
Live imaging was carried out in a sealed, gas-permeable microchamber (Ibidi Biosciences). Donor cells were mixed with CMTMR-labeled target cells at a ratio of 1:1.5 and were loaded onto a microchamber coated with 150 μg/ml fibronectin to provide the cells with a two-dimensional substrate for attachment and migration. The chamber was placed on a Zeiss Axiovert 200 microscope fitted with a laser-scanning confocal microscope 510 META detector. Differential interference contrast (DIC) imaging and confocal green (for GFP) and red (for CMTMR) fluorescence were acquired in a multitrack configuration to avoid cross-talk between fluorescence channels. Images were recorded at 10-s intervals continuously for 3 h. Confocal images and Quicktime movies were generated from laser-scanning confocal microscope file data using NIH ImageJ (http://rsb.info.nih.gov/ij/), Osirix (55), and Volocity (Improvision) software packages.
Conjugate analysis.
Image quantitation of the cell-to-cell conjugation was performed on a frame-by-frame basis. Composite three-color images were from manually inspected recording data for one cell at a time. For each donor cell (CMTMR-negative) interaction with a target cell, the initial and final frames of the interaction were recorded. From these data, a custom program was used to calculate a multiplicity of interaction for each cell in each frame. A spreadsheet with collective data enabled us to calculate the fraction of total cells that were engaged with 0, 1, 2, or 3 or more cells, the duration of interaction, and the frequency of new interaction. We directly compared data for HIV Gag-iGFP-expressing donor cells to those for internal control cells that were not GFP positive.
RESULTS
To understand the relative contribution of cell contact to viral dissemination, we set out to carefully measure how much virus is passed from T cell to T cell by comparing direct-contact routes to cell-free routes. Previous immunofluorescence microscopy studies have documented the relocalization of viral and cellular receptors to the site of infected-cell-to-uninfected-cell contact and revealed accumulated Gag in putative target T cells (28). To quantify viral uptake, we established an ultrasensitive flow cytometry assay that employed a novel infectious molecular clone of HIV, HIV Gag-iGFP (27). This infectious viral construct carries an internal insertion of GFP into the core structural protein, Gag, stoichiometrically loading each virus particle with an estimated 5,000 GFP molecules (6). This enables direct measurements of viral uptake with exceptional sensitivity.
Highly efficient transfer of fluorescent HIV-1 into target cells mediated by cell-to-cell contact.
To measure the efficiency of cell-to-cell viral transfer, we modified a previously described assay utilizing distinct donor and target CD4+ T cells (28). The donor cells were Jurkat CD4+ T cells expressing HIV Gag-iGFP, and the target cells were purified primary human CD4+ T cells labeled with the red fluorescent dye CMTMR. The red fluorescent dye was used to distinguish HIV-naïve target cells from the HIV Gag-iGFP-expressing donor cells. To synchronize our infections and to facilitate studies of viral mutants, we transduced Jurkat T cells with HIV Gag-iGFP by Amaxa nucleofection. Transducing the donor cells also ensured that the virus that we studied was produced in the donor cells and was not derived from carryover of high-dose, infectious inocula.
HIV-expressing donor cells were carefully washed to remove cell-free virus and were incubated with CMTMR-labeled target cells. Within 3 h, 22% of the CD4+ target T cells acquired high levels of GFP fluorescence (Fig. 1A). Analysis of the forward- and side-scatter plots of the red target cells that had acquired green fluorescence revealed that they were not simple aggregates with infected donor cells. When control GFP-expressing Jurkat donor cells were mixed with the same target cells, no transfer of GFP was observed (Fig. 1B). These control Jurkat cells express GFP fluorescence at an intensity comparable to that of the brighter HIV Gag-iGFP-expressing cells and were subjected to an Amaxa transduction protocol without proviral DNA.
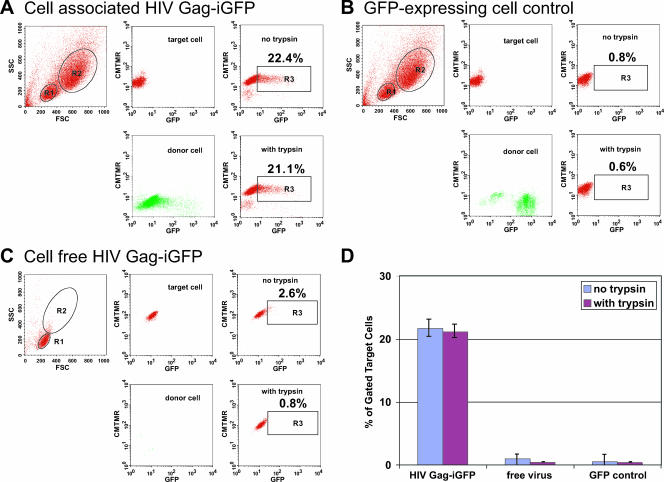
FIG. 1.
Massive VS-mediated transfer of HIV to target cells measured by flow-cytometry-based detection of HIV Gag-iGFP. (A) Target cells, which were CMTMR-labeled CD4+ primary T cells (R1 gate), were mixed with donor cells, which were HIV Gag-iGFP-expressing Jurkat cells (R2 gate), resulting in transfer of fluorescent virus into target cells (upper right). Middle panels display target and donor cell fluorescence prior to mixing. Trypsin-treated cells (lower right) did not decrease the signal. (B) Target cells mixed with control GFP-expressing cells did not show any viral transfer. (C) Target cells were mixed with 100 ng/ml of cell-free HIV Gag-iGFP fluorescent virus, resulting in weak fluorescence in a small percentage of cells. Numbers within the R3 gate indicate the percentage of target cells acquiring strong GFP fluorescence. (D) The average percentages of target cells acquiring GFP fluorescence were plotted from values obtained from at least three independent experiments with different donor cells, performed as described for panels A, B, and C. FSC, forward scatter; SSC, side scatter.
In contrast to the findings for GFP staining after exposure to infected cells, only faint GFP staining was observed when cells were exposed to cell-free viral stocks of HIV Gag-iGFP containing 100 ng/ml of p24 (Fig. 1C). Incubation of the target cells with supernatants collected over a 24-h period from HIV Gag-iGFP-nucleofected Jurkat cells (5 ng/ml p24) did not produce a detectable increase in cellular fluorescence (data not shown). These results strongly suggest that during coculture, the levels of cell-free viral transfer are very small relative to those of cell-to-cell transfer.
To examine whether the virus acquired by cells through cell-to-cell or cell-free exposure were bound to the cell surface, we digested the cells with trypsin to strip away cell surface viral proteins after the 3-h exposure to cell-free virus or HIV-expressing cells. The VS-mediated transfer was resistant to trypsin treatment (Fig. 1A), whereas the small amount of fluorescence acquired by cell-free virus was largely trypsin sensitive (Fig. 1C). To ensure that the trypsin treatment was effective, we conducted immunostaining of target cells treated with trypsin and found a total loss of cell surface CD4 (data not shown). We conclude that VS-mediated viral transfer results in rapid sequestration of viral particles into a trypsin-resistant compartment. Both the magnitude of fluorescence and the resistance to trypsin distinguish the VS-mediated transfer from cell-free viral transfer (Fig. 1D).
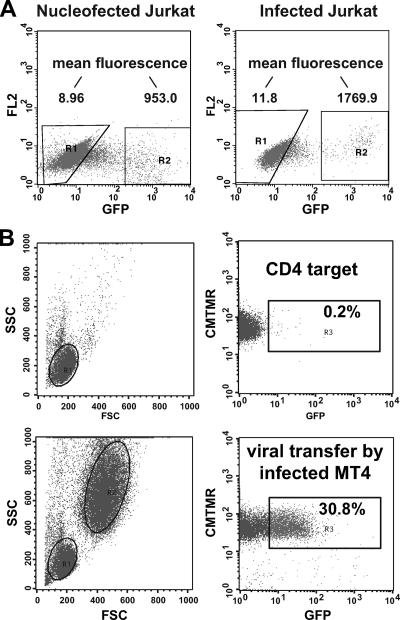
Because Amaxa nucleofection could lead to extraordinarily high levels of viral gene expression, we compared the fluorescence of Amaxa-nucleofected Jurkat cells to that of the Jurkat cells subjected to spinoculation by infectious HIV Gag-iGFP. The levels of HIV Gag-iGFP fluorescence in the two conditions were not dramatically different (Fig. 2A), yet the Amaxa nucleofection allowed us to avoid using large input of cell-free virus to initiate the infections. We have found that MT4 cells are highly permissive for replication of HIV Gag-iGFP (27). With infected MT4 cells as donor cells, they were able to engage in efficient VS-mediated viral transfer when coincubated briefly with primary CD4+ T cells (Fig. 2B).
FIG. 2.
HIV Gag-iGFP-infected cells can engage in VS-mediated transfer. (A) Levels of HIV Gag-iGFP expressed by Amaxa-nucleofected Jurkat cells are comparable to those of HIV Gag-iGFP-infected Jurkat cells at 48 h after nucleofection/infection. Infection of Jurkat cells employed a modified spinoculation method (46) to achieve maximal efficiency. (B) HIV Gag-iGFP-infected MT4 cells can function as donor cells in VS-mediated viral transfer into primary CD4+ T cells. Target cells alone (top panels) or target cells plus infected MT4 cells (bottom panels) are shown. FSC, forward scatter; SSC, side scatter; FL2, fluorescence channel 2.
To quantify the overall magnitude of viral transfer into the population of recipient T cells, we calculated the relative fluorescence transfer (RFT) index as the product of the average increase in fluorescence intensity of the target cells and the percentage of GFP+ cells (Table 1). The relative fluorescence intensity of the GFP-positive fraction was normalized to the background fluorescence of the GFP-negative population. VS-mediated viral transfer resulted in a large fraction (20%) of target cells that acquired a 10-fold-greater average fluorescence than nonexposed cells (Table 1). Exposure of target cells to high concentrations of cell-free virus (100 ng/ml of p24 antigen) resulted in a very low percentage of cells (1.0%) with a weak increase in fluorescence over background. Decreasing the input of cell-free virus resulted in small decreases in the RFT index (Table 1). The RFT index from cell-to-cell conditions was 178, which was 93-fold higher than that obtained with 100 ng/ml of cell-free virus (RFT of 1.91). The large quantities of cell-free virus tested were well beyond the levels of p24 obtained from infected Jurkat cells in vitro, but they were required to observe small, dose-dependent shifts in fluorescence. Jurkat cell viral supernatants harvested more than 24 h posttransfection, which contained 5 ng of p24 antigen, did not result in a detectable shift in fluorescence when exposed to target cells (data not shown).
TABLE 1.
Quantitative comparison of cell-to-cell viral transfer to cell-free viral transfer
| Transfer type | Amt examined (ng) | Avg mean fluorescence of population: | %GFP+ | RFT indexa | |
|---|---|---|---|---|---|
| GFP+ MFd | GFP− MFd | ||||
| Cell to cell | 59.42 | 6.17 | 20.67 | 178.39 | |
| Cell free | 100 | 15.77 | 5.49 | 1.02 | 1.91 |
| 33 | 10.38 | 5.40 | 0.68 | 0.63 | |
| 11b | 16.81 | 5.18 | 0.26 | 0.58 | |
| 0c | 25.57 | 8.90 | 0.13 | 0.24 |
Under VS-forming conditions, we found that the amount of viral p24 released by the donor cells during the 3-h coculture period was 0.5 ng, a value 200-fold lower than the concentration of cell-free virus tested. Assuming that dilution of these lower levels of cell-free virus is likely to result, at best, in a linear decrease in cell-free uptake, we can estimate the difference between the efficiency of cell-free transfer and that of cell-to-cell transfer to be 200-fold greater than the calculated difference in the RFT index, or 18,600-fold (93-fold × 200-fold). We conclude that the efficiency of viral transfer from a cell-cell mode is at least 92- to 18,600-fold more efficient than that of a cell-free mode of uptake.
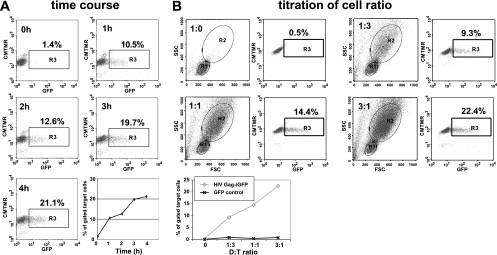
We next examined the time course of VS-mediated viral transfer. The rate of transfer was rapid and efficient, with the onset of transfer occurring after 1 h and increasing steadily over a 4-h time course. When mixed at a fixed ratio of donor Jurkat cells to acceptor CD4+ T cells of 1:1, a 4-h incubation resulted in more than 20% of the target cells exhibiting strong green fluorescence (Fig. 3A). In addition, we altered the ratio of infected donor cells to target cells to test its effect on the efficiency of viral transfer. At donor-to-target ratios of 1:3, 1:1, and 3:1, the efficiency of transfer increased as the donor-to-target ratio was increased (Fig. 3B). We thus observed a rapid, cell-ratio-dependent increase in virus-associated fluorescence in target cells.
FIG. 3.
VS-mediated viral transfer is rapid and dependent on donor target ratio. (A) A 1:1 ratio of HIV Gag-iGFP-expressing donor cells and CD4+ target cells was mixed and incubated for 0, 1, 2, 3, or 4 h. The percentage of GFP+ cells is plotted over time (lower right). (B) Cells were mixed at donor/target ratios of 0:1, 1:3, 1:1, or 3:1 and were assayed after 3 h of transfer. The percentage of GFP+ cells following exposure to HIV Gag-iGFP-expressing cells is compared to that after exposure to control cells expressing GFP alone (bottom panel).
Dependence of VS-mediated HIV-1 transfer upon Env and CD4 interactions.
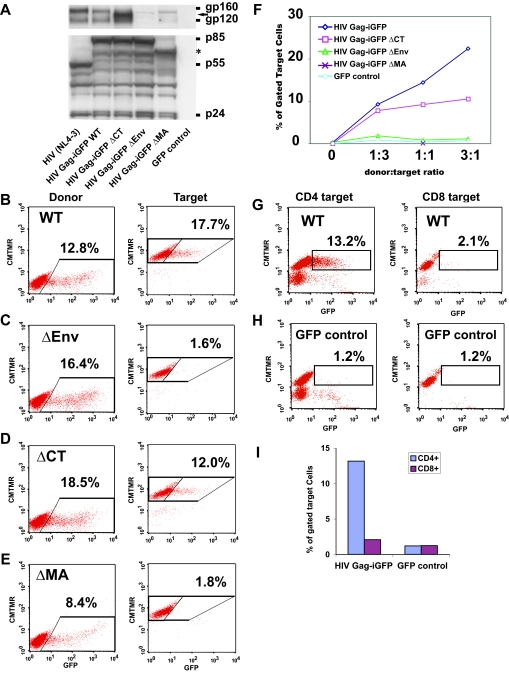
Previous studies have illustrated that the viral Env protein is relocalized to the site of cell-cell contact during VS formation (28). We therefore examined mutant HIV clones to determine the dependence of viral transfer upon Env. An Env-deficient HIV-1 clone, HIV Gag-iGFP ΔEnv, was nucleofected to generate donor cells that abundantly express all HIV-1 genes except for Env (Fig. 4A). Jurkat cells were nucleofected with the Env-deleted construct with an efficiency comparable to that of native HIV Gag-iGFP (Fig. 4B and C). However, when HIV Gag-iGFP ΔEnv-expressing donor cells were mixed with target cells, only a background level of HIV Gag-iGFP transfer to target cells was observed (Fig. 4C). The massive transfer of virus to target cells therefore is completely dependent on expression of Env in the donor cell.
FIG. 4.
Efficient VS-mediated viral transfer is dependent upon Env and Gag. (A) Western blot of cell lysates of viral mutants nucleofected into Jurkat T cells and probed with anti-Env antibodies (top) or anti-HIV antisera (bottom). (B) HIV Gag-iGFP fluorescence in the donor cells (left) and in the target cells (right) after 3 h of coincubation. (C) Viral transfer assay on the Env-deleted clone ΔEnv. (D) Viral transfer assay on the cytoplasmic-tail-deleted clone ΔCT. (E) Viral transfer assay on the globular head of the MA deletion construct ΔMA. (F) Titration of the donor/target ratios of the various mutant clones. (G) CD8+ T cells do not engage in viral transfer with HIV Gag-iGFP-expressing donor cells. (H) Incubation of CD4+ or CD8+ cells with control GFP-expressing cells. (I) Comparison of CD4 to CD8 cells as target cells, graphing the percentage of GFP+ cells from experiments depicted in G and H. WT, wild type.
Prior studies suggest that interactions between Env and Gag enhance the cell-to-cell spread of HIV-1 (13). We therefore tested viral clones carrying deletion mutations in either the cytoplasmic tail of Env (2, 41) or the globular head of the MA domain of Gag (54), both of which attenuate viral replication but are dispensable for viral production. These viral mutants, called HIV Gag-iGFP ΔCT and HIV Gag-iGFP ΔMA, were efficiently nucleofected into donor cells (Fig. 4A, D, and E). The levels of supernatant p24 released from cells nucleofected with the HIV Gag-iGFP ΔCT construct were close to wild-type levels (about 2 ng/ml), while HIV Gag-iGFP ΔMA produced roughly threefold less virus than wild-type HIV Gag-iGFP (data not shown). Surprisingly, deletion of the cytoplasmic domain of Env resulted in only a small reduction in viral transfer (Fig. 4D and F), while the MA domain mutation abolished viral transfer completely (Fig. 4E and F). The ΔMA mutation illustrates that the viral transfer is not driven simply by an Env-mediated adhesion or fusion event but that it may require proper targeting of Gag to the synapse. VS-mediated transfer therefore is likely to involve coordinated interactions between Gag and Env, yet the presence of the full cytoplasmic tail is not essential for the process.
It has been shown that human intestinal epithelial cells can transcytose HIV-1 in a CD4-independent manner following contact with HIV-infected T cells (4, 37). We therefore tested whether VS-mediated viral transfer occurred specifically only with CD4+ lymphocytes. Purified primary CD8+ T cells were CMTMR labeled and tested for their ability to capture HIV Gag-iGFP when cocultured with HIV Gag-iGFP-expressing donor cells. CD8+ T cells did not engage in viral transfer with the donor cells (Fig. 4G). The level of transfer of HIV Gag-iGFP into the CD8 cells was slightly higher than that of the background observed for GFP-expressing cell controls (Fig. 4H and I). These data showed that VS-mediated transfer was specific to CD4+ T cells.
To further examine the role of CD4 in VS-mediated viral transfer, we incubated the donor cells with the HIV-blocking, anti-CD4 antibody Leu 3a. The pretreatment of target cells with Leu 3a at 0.25 μg/ml blocked more than 80% of the VS-mediated transfer (Table 2). Similarly, treatment of the cells with monoclonal antibody against the CD4 binding site on gp120, IgG1b12, inhibited 50% of VS-mediated transfer (Table 2). Incubation with soluble CD4 blocked more than 60% of the viral transfer (Table 2). Thus, three inhibitors of the interaction between Env and CD4 all had inhibitory effects on VS-mediated viral transfer. We conclude that VS-mediated transfer requires the engagement of target cell CD4 by Env on the surface of HIV-expressing donor cells.
TABLE 2.
Inhibition of VS-mediated transfer
| Target and inhibitor | Concnb | % VS-mediated viral transfer fora: | Cell-free infectionc | |
|---|---|---|---|---|
| HIV Gag-iGFP | HIV Gag- iGFP ΔCT | |||
| Control | 100 | 100 | 100 | |
| CD4 inhibitors | ||||
| sCD4 | 50 μg/ml | 37 | 46 | 6 ± 0.5 |
| 50 μg/ml | 23 | 11 | ||
| 100 μg/ml | 18 | 9 | ||
| Leu 3a | 0.25 μg/ml | 15 | 10 | |
| 0.5 μg/ml | 13 | 8 | 31 ± 2.5 | |
| IgG1b12 | 10 μg/ml | 47 | 38 | |
| 20 μg/ml | ND | 23 | 19 ± 1.2 | |
| gp41 inhibitors | ||||
| T20 | 100 μg/ml | 108 | 104 | 11 ± 1.8 |
| 2F5 | 100 μg/ml | 104 | 109 | |
| 20 μg/ml | 22 ± 2.2 | |||
| CXCR4 antagonist | ||||
| AMD3100 | 10 μg/ml | 79 | 90 | 14 ± 2.6 |
| Actin inhibitors | ||||
| Cytochalasin D | 1.5 μM | 53 | 47 | 86 ± 10.2 |
| 1.5 μM | 39 | 86 | 91 ± 16.4 | |
| Latrunculin B | 1.0 μM | 13 | 27 | 82 ± 8.4 |
HIV-1 transfer using an R5-tropic Env does not require CCR5 on target cells.
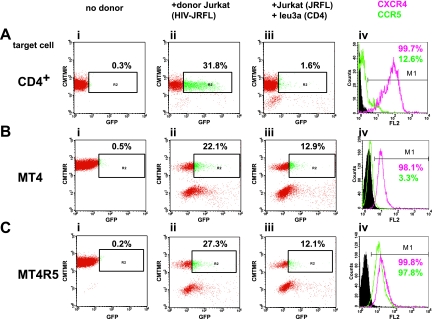
To examine the efficiency of R5-tropic Env in mediating cell-to-cell transfer, we nucleofected Jurkat cells with an HIV Gag-iGFP clone carrying the Env from the R5 molecular clone, JRFL. When the HIV Gag-iGFP(JRFL)-expressing Jurkat cells were mixed with primary CD4+ T cells, a large fraction (31.8%) became highly GFP positive (Fig. 5A, panel ii). This fraction of cells was higher than the fraction of CD4+ T cells that expressed high levels of CCR5 (12.6%) (Fig. 5A, panel iv). The transfer also could be blocked efficiently with the anti-CD4 antibody Leu 3a. This suggests that CCR5 expression is not required for VS-mediated transfer of an R5-tropic virus into primary CD4+ T cells. Viral transfer mediated by the R5 Env also was observed in CCR5-negative MT4 cells (Fig. 5B) and was not greatly enhanced by the stable expression of CCR5 in these cells (Fig. 5C). While R5-tropic Env was very efficient at directing cell-to-cell transfer of virus, the presence of the CCR5 coreceptor was not required for VS-mediated viral transfer.
FIG. 5.
Viral transfer mediated through R5-tropic Env from molecular clone JRFL occurs efficiently in CCR5+ or CCR5− CD4+ T cells. (A) VS-mediated transfer from HIV Gag-iGFP(JRFL)-expressing Jurkat cells into primary CD4+ T cells shows efficient transfer into target cells. A parallel analysis of coreceptor expression in the target cells shows a small fraction of CCR5+ T cells. (B) VS-mediated transfer into MT4 does not require CCR5. (C) VS-mediated transfer into CCR5-expressing MT4R5 cells is similar to that for CCR5− MT4. Panel i, target cells alone; panel ii, target cells plus HIV Gag-iGFP(JRFL)-expressing Jurkat donor cells; panel iii, donor and target cells inhibited with 0.5 μg/ml of the anti-CD4 blocking antibody Leu 3a; panel iv, chemokine receptor staining of the target cells, illustrating the percentage of cells expressing CXCR4 (red) or CCR5 (green). FL2, fluorescence channel 2.
HIV-1 transfer to CD4+ T cells requires cytoskeletal rearrangements but is not blocked by inhibitors of viral membrane fusion.
HIV-1 VS formation is thought to involve active cytoskeletal rearrangements that stabilize adhesive interactions and promote the concentration of viral and cellular receptors at the VS (28). We therefore examined the extent to which viral transfer was sensitive to the actin inhibitor cytochalasin D. Cytochalasin D treatment of the donor cells resulted in a 50 to 60% decrease in viral transfer, suggesting that active cytoskeletal rearrangements are necessary for the process (Table 2). Another inhibitor of actin polymerization, latrunculin B, reduced VS-mediated viral transfer by 87% (Table 2). These results confirm that cytoskeletal rearrangements are required for VS-mediated transfer. Interestingly, the same cytoskeletal inhibitors had little effect on cell-free infection (Table 2), showing that the requirements for cell-to-cell transfer are distinct from those of cell-free infection.
To examine the dependence of viral transfer upon viral membrane fusion, we tested the ability of a peptide inhibitor of Env-mediated membrane fusion, T20, to block VS-mediated viral transfer. T20 had no measurable effect on VS-mediated transfer, suggesting that the uptake of virus into target cells does not require triggering of viral membrane fusion (Table 2). Similarly, treatment of cells with the broadly neutralizing antibody 2F5 did not block VS-mediated transfer (Table 2). Control experiments using these inhibitors at the same or lower concentrations showed that they were effective at blocking cell-free infection (Table 2). Since T20 and 2F5 both target fusion intermediates in gp41, we conclude that the highly efficient VS-mediated transfer is not dependent on activation of viral membrane fusion, or that it may occur in a manner that blocks the accessibility of these inhibitors. Because these inhibitors typically are effective at blocking the formation of syncytia, the results also suggest that syncytia do not explain the massive transfer of HIV-1 into target cells. In a previous study, treatment of cells with the CXCR4 antagonist AMD3100 did not block conjugate formation, but it did inhibit CD4 polarization during VS formation (28). We found that high concentrations of AMD3100 had little effect on viral transfer (Table 2). Based on our tests with AMD3100, engagement of CXCR4 by gp120 is not required for highly efficient VS-mediated viral transfer.
Resistance of VS-mediated viral transfer to patient-derived neutralizing antisera.
Viral transmission in the dendritic cell to T-cell-infectious synapses has been shown to mediate efficient infection of T cells by a mechanism that is resistant to neutralizing antibodies (18). We therefore examined whether the VS-mediated viral transfer in our system is sensitive to antibody-mediated neutralization. We tested two well-studied neutralizing antisera against a control nonneutralizing serum (61) and found that viral transfer was not affected at a 1:50 dilution, a concentration that effectively blocked infection by homologous HIV-1(NL4-3) under cell-free conditions (Table 3). Interestingly, the neutralizing serum did partially block the viral mutant HIV Gag-iGFP ΔCT, reducing the percentage of viral transfer by 40% relative to the level of transfer of the nonimmune control serum (Table 3). In recent studies, it has been found that deletions or mutations in the cytoplasmic tail of Env can expose neutralizing epitopes in its ectodomain that are otherwise masked (31, 64). These results suggest that the patient sera can recognize Env with a truncated cytoplasmic tail on the surface of cells and can block its ability to mediate viral transfer, but they do not recognize native HIV Env on the surface of cells. The cytoplasmic tail therefore may play a role in limiting the exposure of neutralizing epitopes on the cell surface.
TABLE 3.
Resistance of VS-mediated viral transfer to patient neutralizing antibodies
| Serum and concnb | % VS-mediated viral transfer fora: | % Cell-free infectionc | |
|---|---|---|---|
| HIV Gag-iGFP | HIV Gag-iGFP ΔCT | ||
| None | 100 | 100 | 100 |
| Negative serum | |||
| 1:50 | 100 | 100 | 105 |
| 1:50 | 100 | 100 | 124 |
| 1:50 | 123 | 109 | |
| Avgd | 108 ± 13 | 103 ± 5 | 115 |
| Neutralizing serum 1 | |||
| 1:50 | 89 | 56 | 22 |
| 1:50 | 98 | 57 | 16 |
| 1:50 | 146 | 64 | |
| Avgd | 111 ± 31 | 59 ± 4 | 19 |
| Neutralizing serum 2 | |||
| 1:50 | 100 | 59 | 10 |
| 1:50 | 113 | 61 | 1 |
| 1:50 | 144 | 61 | |
| Avgd | 119 ± 23 | 60 ± 1 | 6 |
Dynamic VS imaging reveals enhanced T-cell adhesion, polarity, and chemokinesis.
The rapid and high level of HIV Gag-iGFP fluorescence transferred into target cells measured by flow cytometry suggested that we should be able to visualize the virus as it is being transferred into target cells and to capture dynamic aspects of the living VS. Red-fluorescent-dye-labeled target cells were mixed with HIV Gag-iGFP-expressing donor cells and were imaged with two-color, time-lapse, confocal microscopy. To perform long-term imaging of live samples, we use a sealed, gas-permeable microchamber (Ibidi Biosciences) maintained at 37°C. A fibronectin-coated microchamber provided the T cells with a two-dimensional substrate for attachment and migration. Simultaneous DIC and confocal green and red fluorescence images were acquired. Confocal images were acquired every 10 s for 3 h.
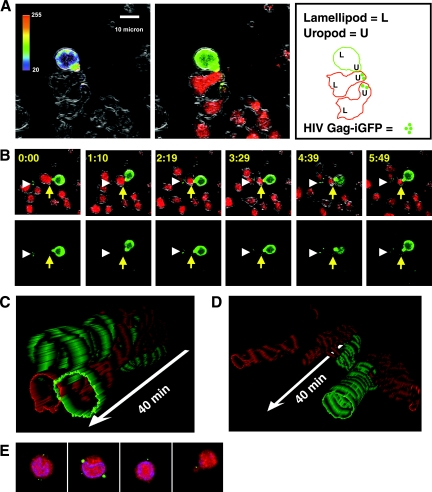
A large majority of infected donor cells was stably associated with one or more target cells (Fig. 6; also see movies S1 and S2 in the supplemental material). T cells engaged in synapses assumed a polarized morphology and interacted with target cells through uropod-like structures (Fig. 6A). A polarized morphology is characteristic of motile T cells (53) and is distinct from the morphology of T cells engaged in immunological synapses (15). In the donor cells, higher concentrations of Gag-iGFP were observed at the site of cell-cell contact (Fig. 6A, left). When threshold settings were set to measure low-intensity green signals in target cells, we readily observed accumulations of green fluorescence on the target cells engaged in synapses (Fig. 6B, bottom panels). We also observed spots of green fluorescence on target cells that were not engaged with donor cells, which likely represented cells that already had participated in a VS. These spots also localized asymmetrically in the cell in a location that appeared to be the cell uropod (Fig. 6A and B).
FIG. 6.
Live imaging of VS formation reveals polarized HIV-infected and uninfected cells engaging in long-lived contacts. (A) An HIV Gag-iGFP-expressing donor cell bound to a target cell through uropod-like structures. (Left) At the VS, increased concentrations of Gag-iGFP in the donor cell at the site of cell-cell contact were observed, with GFP fluorescence in a rainbow intensity scale overlaid on a DIC edge image to show the cell boundaries. (Middle) Three-color image with Gag-iGFP (green), a CMTMR-labeled target cell (red), and DIC edges (gray). (Right) Outlines of a donor and target cell and one unconjugated target cell that has accumulations of HIV Gag-iGFP in its uropod. (B) An image series following the same VS that focuses on one pair of cells for 6 min. The time stamp is shown in upper left corner. Three colors are merged so that the cell edges defined by the DIC channel are represented in grayscale, while the red and green show target and donor cells, respectively. A yellow arrow indicates a target cell that is stably bound to a green HIV Gag-iGFP-expressing donor cell. Near the yellow arrow in the green channel (lower images), small accumulations of green signal on the target cell are indicative of viral transfer. The DIC channel in this series highlights the polarity of the cells by showing extended lamellipodia in both the donor and target cells (see movie S1 in the supplemental material). (C) Kymograph representations of donor and target cell interactions. A three-dimensional reconstruction illustrating the movements of an HIV Gag-iGFP-infected T cell (green) and stable conjugates with two uninfected target cells (red) over time is shown. An HIV-Gag-GFP-expressing cell is represented as a green outline, and the CD4 target cell is represented as a red outline, with successive x-y sections stacked along a temporal axis (white arrow) encompassing 40 min of imaging. The donor and target cells remain attached throughout the entire image sequence. (D) Kymograph illustrating transient adhesive interactions between an uninfected Jurkat cell (green outline) and two uninfected target cells (red). Two red target cells transiently interact with the uninfected green donor cell during the time frame. (E) Three-dimensional reconstruction of confocal images of flow-sorted double-positive target cells. Strong dots of green fluorescence resemble the dots seen in live images.
Neighboring Jurkat cells not expressing HIV-1 were observed to interact with target cells; however, these interactions usually were relatively short lived. To illustrate the interactions of representative donor cells over the duration of the movie, we converted the temporal sequence of images into a three-dimensional kymograph. In the kymograph, a green donor cell outline (or control cell) is plotted in an _x_-y plane with the outlines of all target cells with which it interacts during a 40-min imaging period. Time is plotted on a third z plane. HIV-1-infected cells typically stayed engaged with target cells for most of the imaging period (Fig. 6C), whereas uninfected control cells engaged in multiple short-term interactions with target T cells during the same time frame (Fig. 6D).
To determine whether the small green spots observed microscopically in the target cells were representative of the same cells that had acquired massive viral fluorescence as measured by flow cytometry, we acquired images of the GFP-positive, CMTMR-positive target cells after they were flow sorted. Confocal images revealed the presence of similar intense green spots localizing to the perimeter of the cells in dots (Fig. 6E; also see movie S3 in the supplemental material). Most cells appeared to contain more than one spot. Importantly, in flow-sorted target cells, we did not find multicell aggregates or syncytia. By comparing the images of the fixed, flow-sorted target cells to the live images of cells undergoing VS, we confirmed that the target cells that had acquired bright dots of green fluorescence resembled those measured by flow cytometry.
Measurement of Env-dependent adhesive interactions between infected and uninfected T cells.
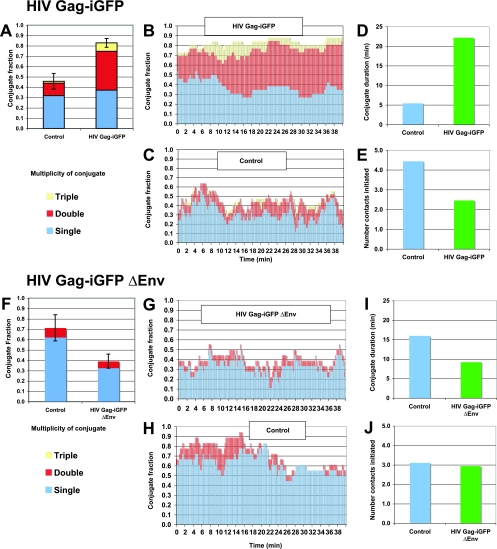
To quantify the number of cell-to-cell conjugates formed by HIV-infected versus noninfected donor cells, we analyzed the live images on a frame-by-frame basis. In every frame, each infected cell was classified by the number of target cells that it had engaged. An equal number of uninfected, non-GFP-expressing Jurkat cells were analyzed as controls. The frequency of HIV-expressing cells that had formed stable cell conjugates was roughly twofold higher than that of nonexpressing cells (Fig. 7A). Infected cells therefore were twice as likely to be engaged with one or more target cells. In addition, a large fraction of HIV-expressing donor cells interacted with two or three cells at a time (Fig. 7B). In contrast, control cells that were not expressing HIV were much less likely to interact with more than one target cell (Fig. 7C). The average duration of interaction with the target cells also was fourfold greater for the infected cells (Fig. 7D), although the number of cell contacts initiated was lower than that of uninfected controls (Fig. 7E). Infected cells engaged fewer target cells than uninfected cells over a period of time, but the interactions with HIV-expressing cells were much more durable.
FIG. 7.
Durable VS adhesion is driven by Env. (A to E) HIV Gag-iGFP-expressing donor cells were mixed with purified target CD4+ T cells and imaged for 40 min. (A) The conjugate fraction of control or HIV Gag-iGFP-expressing cells engaged with one, two, three, or more target cells was averaged over the entire imaging series. The interactions of 25 HIV-expressing and 25 control cells were analyzed for 200 frames at 12-s intervals. (B) The conjugate fraction of HIV Gag-iGFP-expressing cells represented over time. (C) The conjugate fraction of non-HIV-expressing control cells. (D) The average duration of each cell-to-cell contact between HIV-expressing and nonexpressing control cells. (E) Average number of new contacts initiated between HIV-expressing and control, nonexpressing cells. (F to J) HIV Gag-iGFP ΔEnv-expressing donor cells were mixed with purified CD4+ primary target T cells and were imaged for 40 min. (F) Time-averaged conjugate fractions of HIV Gag-iGFP ΔEnv-expressing cells compared to those of noninfected cells. (G) The conjugate fraction of HIV Gag-iGFP ΔEnv-expressing cells represented over time. (H) The conjugate fraction of non-HIV-expressing control cells. (I) Average duration of contacts of control and HIV Gag-iGFP ΔEnv-expressing cells. (J) Average number of new contacts initiated by HIV Gag-iGFP ΔEnv-expressing cells compared to that of control cells. Live-imaging results are indicative of experiments with two or more independent donors.
We conducted the same experiment with an Env-deficient viral clone, HIV Gag-iGFP ΔEnv, to determine the role of Env in mediating these adhesive contacts. The HIV Gag-iGFP ΔEnv-expressing cells bound to target cells with a lower level of efficiency than uninfected controls (Fig. 7F). This suggests that the expression of all HIV-1 genes in the absence of Env does not enhance cell-to-cell adhesion (Fig. 7G and H). The duration of the typical interaction of the HIV ΔEnv-expressing cells was less than that of neighboring control cells (Fig. 7I), and the average number of interactions per donor cell was not affected by expression of HIV ΔEnv (Fig. 7J). These results demonstrate that cell surface Env functions as an adhesion molecule to upregulate the duration of contact between infected and uninfected cells.
DISCUSSION
We have used a novel infectious HIV-1 clone that is stoichiometrically labeled with GFP to quantify the efficiency of and to visualize the dynamics of T-cell-to-T-cell viral transfer. The results reveal a remarkable capacity of primary human CD4+ T cells to internalize large quantities of viral particles through Env-directed contact with HIV-expressing T cells. It is critical to understand how VS-mediated transfer differs from cell-free viral adsorption and internalization. We illustrate that both the quantity and quality of this transfer of virus to CD4+ T cells is distinct from those of cell-free viral infection. With direct measurements of viral fluorescence in a coculture system, we estimate that cell-to-cell contact is 18,000-fold more efficient in transferring viral particles into target cells during a relatively brief coculture period. We find that the transfer process produces target cells carrying large amounts of Gag in an internal, trypsin-resistant compartment. Cell-to-cell transfer therefore generates a reservoir of virus that is held stably within T cells, protected from serum-mediated inactivation or mechanical clearance. The great disparity in the efficiency of virus internalization suggests that the VS is the dominant mechanism for viral spread.
Although we have not fully demonstrated that viral transfer is an endocytic process, our current imaging studies (Fig. 6) (W. Hübner and B. K. Chen, unpublished data) suggest that the virus is translocated into an internal compartment in the target cells. Because the endocytic pathways often lead to lysosomal degradation of the vesicular contents, it is not clear to what extent this pathway contributes to productive infection in T cells. Recent studies on HIV have found that the endocytic route does lead to productive infection at efficiencies much greater than previously recognized (12, 16, 17, 35, 57). Because studies of endocytic pathways of viral internalization largely have been performed with cell-free virus, we now must consider that transfer of virus into cells by VS may drive HIV into endocytic routes at levels that are orders of magnitude higher than previously appreciated.
The highly efficient transfer of virus between T cells is driven by engagement of target cell CD4 with donor cell Env. Antibodies or soluble CD4 that disrupts this interaction effectively blocks cell-to-cell transfer of virus. In contrast, neutralizing antibodies or peptides that block viral membrane fusion by binding to gp41 prehairpin structures were unable to block cell-to-cell transmission. Therefore, cell-to-cell transfer is a process in which the virus internalization is triggered by CD4 engagement, but it is unlikely to involve the activation of viral membrane fusion. Our model therefore resembles the coreceptor-independent transfer of HIV-1 from T cell to T cell described by Blanco et al. (3). We find that R5-tropic Env can mediate highly efficient viral transfer, which does not require the presence of CCR5 on the cells. The Blanco group demonstrated that following cell-to-cell contact, internalized virus could serve as a source of infectious virus when exposed to fresh cells. Other recent studies suggest that upon exposure to cell-free virus, HIV-1 internalized through endocytic routes can contribute from 40 to 80% of the productive infection (12). Because our measurements indicate that cell-cell contact enhances viral uptake by up to four orders of magnitude, it is possible that VS-enhanced endocytic routes contribute to a major fraction of productive infections. To definitively determine the efficiency of infection following VS-mediated transfer, experiments using high-purity sorting of cells immediately after VS-mediated transfer need to be conducted.
Studies of dendritic-cell-to-T-cell viral transfer have demonstrated that synapse-mediated transfer is resistant to serum neutralization (18). Importantly, we found that patient sera were unable to block the VS-mediated transfer from a wild-type virus but could partly block transfer mediated by a virus with a truncated cytoplasmic tail of Env. These data suggest that immunogenic epitopes on the surface of cells are hidden but can be exposed in a manner that allows neutralizing antisera to interfere with cell-to-cell transfer. Recent studies have found that the interactions between Env and unprocessed Gag that occur during assembly and in immature virus particles may hold Env in a prefusogenic conformation that is distinct from fusogenic conformations found on mature virus particles (40, 63). If humoral responses are largely directed against epitopes present on mature cell-free virus, they may be less effective at binding to cell surface epitopes capable of blocking cell-to-cell transmission of HIV-1. Our current results suggest that vaccines expressing Gag and Env at the surfaces of cells may be better at eliciting antibodies that inhibit cell-to-cell transmission.
Recent immunological studies have revealed dynamic live images of T cells migrating rapidly within lymph nodes (19). As part of their normal biological functions, T cells are highly migratory and constantly engage in adhesive interactions with other immune cells. The migratory and adhesive behavior of HIV-infected T cells has not been examined under conditions in which we can monitor both infected donor cells and uninfected target cells and measure the transfer of virus between these cells. Under conditions in which donor and target T cells are able to move on a two-dimensional fibronectin substrate, we have found that infected cells engage in long-lived, Env-dependent adhesion with target CD4 cells. We formally demonstrate that HIV Env functions as a cell surface adhesion molecule in addition to its role in viral adsorption and membrane fusion. Furthermore, infected cells frequently were found to interact with target cells through the uropod, a structure indicative of a motile cell. Importantly, we find that HIV-1 infection does not diminish the intrinsic ability of the infected T cell to move. A key to understanding the pathogenesis of HIV-1 infection may therefore be to understand how viral dissemination is influenced by infected T-cell migration and adhesion.
At a subcellular level, we observed that high focal concentrations of virus are asymmetrically deposited on target cells in the vicinity of the uropod, a cellular structure that is appropriately enriched in cell adhesion molecules and cytoskeletal elements that are known to be concentrated in HIV-1 particles. ICAM-1, ICAM-3, CD44, CD43, ezrin, moesin, and radixin all are packaged into virus particles, suggesting that the uropod is a major site for coordinated assembly and transmission between T cells (42).
Our studies show that primary unstimulated or stimulated CD4 cells can take up virus in a highly efficient manner after they contact infected cells. Indeed, recent studies looking at simian immunodeficiency virus in vivo have found that nonactivated T cells frequently are found to be positive for viral RNA and capsid antigen (34, 65, 66). Although the authors suggested that the nonactivated cells produce HIV-1 at low levels, it also is plausible that these cells have undergone VS-mediated uptake and are not yet synthesizing new virus. Other studies of HIV-1 sequences in the spleen have found that infected cells are found in clusters with unique viral sequences, suggesting a fine spatial segregation of viral quasi-species within lymphoid tissue (10, 20, 30). If dissemination of HIV-1 were to occur mostly through cell-to-cell contact, one might expect to observe such a localized trail of virus-positive CD4+ cells.
Our studies suggest that a plausible mechanism for HIV spread primarily involves VS-mediated transfer, whereby physical contact between motile infected cells is the key parameter. Physical contact between T cells may promote local spread of the virus within microcompartments in the body. Distal spread within the body also may occur when productively infected or virus-laden target cells migrate from one organ to the next. Further study of T-cell-to-T-cell transmission will be essential to understanding AIDS pathogenesis and may provide new avenues to more efficiently block viral transmission and dissemination.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Jay Unkeless and David Baltimore for helpful comments on the manuscript. We thank James E. Robinson for MT4-R5 cells.
This work was supported in part by National Institutes of Health, Institute of Allergy and Infectious Diseases, grant AI-055321 to B.K.C. and a grant from the American Foundation for AIDS Research to B.K.C. Confocal laser-scanning microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported with funding from an NIH-NCI shared resources grant (CA095823), an NSF Major Research Instrumentation grant (DBI-9724504), and an NIH shared instrumentation grant (RR0 9145).
Footnotes
▿
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59**:**284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., T. Fukumori, and A. Adachi. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74**:**4891-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, J., B. Bosch, M. T. Fernandez-Figueras, J. Barretina, B. Clotet, and J. A. Este. 2004. High level of coreceptor-independent HIV transfer induced by contacts between primary CD4 T cells. J. Biol. Chem. 279**:**51305-51314. [DOI] [PubMed] [Google Scholar]
- 4.Bomsel, M. 1997. Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat. Med. 3**:**42-47. [DOI] [PubMed] [Google Scholar]
- 5.Bomsel, M., M. Heyman, H. Hocini, S. Lagaye, L. Belec, C. Dupont, and C. Desgranges. 1998. Intracellular neutralization of HIV transcytosis across tight epithelial barriers by anti-HIV envelope protein dIgA or IgM. Immunity 9**:**277-287. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11**:**672-675. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, P. U., P. S. Freudenthal, J. M. Barker, S. Gezelter, K. Inaba, and R. M. Steinman. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257**:**383-387. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., W. Hubner, K. Riviere, Y. X. Liu, and B. K. Chen. 2006. Chimeric HIV-1 containing SIV matrix exhibit enhanced assembly in murine cells and replicate in a cell-type-dependent manner in human T cells. Virology 349**:**1-12. [DOI] [PubMed] [Google Scholar]
- 9.Chertova, E., J. W. Bess, Jr., B. J. Crise, I. R. Sowder, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76**:**5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheynier, R., S. Henrichwark, F. Hadida, E. Pelletier, E. Oksenhendler, B. Autran, and S. Wain-Hobson. 1994. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphocytes. Cell 78**:**373-387. [DOI] [PubMed] [Google Scholar]
- 11.Coombs, R. W., A. C. Collier, J. P. Allain, B. Nikora, M. Leuther, G. F. Gjerset, and L. Corey. 1989. Plasma viremia in human immunodeficiency virus infection. N. Engl. J. Med. 321**:**1626-1631. [DOI] [PubMed] [Google Scholar]
- 12.Daecke, J., O. T. Fackler, M. T. Dittmar, and H. G. Krausslich. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79**:**1581-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deschambeault, J., J. P. Lalonde, G. Cervantes-Acosta, R. Lodge, E. A. Cohen, and G. Lemay. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J. Virol. 73**:**5010-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov, D. S., R. L. Willey, H. Sato, L. J. Chang, R. Blumenthal, and M. A. Martin. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67**:**2182-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dustin, M. L. 2004. Stop and go traffic to tune T cell responses. Immunity 21**:**305-314. [DOI] [PubMed] [Google Scholar]
- 16.Fackler, O. T., and B. M. Peterlin. 2000. Endocytic entry of HIV-1. Curr. Biol. 10**:**1005-1008. [DOI] [PubMed] [Google Scholar]
- 17.Fredericksen, B. L., B. L. Wei, J. Yao, T. Luo, and D. Van Ryk. 2002. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 76**:**11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh, L., K. Leung, K. Lore, R. Levin, A. Panet, O. Schwartz, R. A. Koup, and G. J. Nabel. 2004. Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J. Virol. 78**:**11980-11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Germain, R. N., M. J. Miller, M. L. Dustin, and M. C. Nussenzweig. 2006. Dynamic imaging of the immune system: progress, pitfalls and promise. Nat. Rev. Immunol. 6**:**497-507. [DOI] [PubMed] [Google Scholar]
- 20.Gratton, S., R. Cheynier, M. J. Dumaurier, E. Oksenhendler, and S. Wain-Hobson. 2000. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. USA 97**:**14566-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossman, Z., M. B. Feinberg, and W. E. Paul. 1998. Multiple modes of cellular activation and virus transmission in HIV infection: a role for chronically and latently infected cells in sustaining viral replication. Proc. Natl. Acad. Sci. USA 95**:**6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart, T. K., A. M. Klinkner, J. Ventre, and P. J. Bugelski. 1993. Morphometric analysis of envelope glycoprotein gp120 distribution on HIV-1 virions. J. Histochem. Cytochem. 41**:**265-271. [DOI] [PubMed] [Google Scholar]
- 23.Herrera, C., C. Spenlehauer, M. S. Fung, D. R. Burton, S. Beddows, and J. P. Moore. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77**:**1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, D. D., T. Moudgil, and M. Alam. 1989. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N. Engl. J. Med. 321**:**1621-1625. [DOI] [PubMed] [Google Scholar]
- 25.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373**:**123-126. [DOI] [PubMed] [Google Scholar]
- 26.Hübner, W., and B. K. Chen. 2006. Inhibition of viral assembly in murine cells by HIV-1 matrix. Virology 352**:**27-38. [DOI] [PubMed] [Google Scholar]
- 27.Hübner, W., P. Chen, A. Del Portillo, Y. Liu, R. Gordon, and B. K. Chen. 2007. Sequence of human immunodeficiency virus (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81**:**12596-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolly, C., K. Kashefi, M. Hollinshead, and Q. J. Sattentau. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199**:**283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolly, C., and Q. J. Sattentau. 2005. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. J. Virol. 79**:**12088-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418**:**144. [DOI] [PubMed] [Google Scholar]
- 31.Kalia, V., S. Sarkar, P. Gupta, and R. C. Montelaro. 2005. Antibody neutralization escape mediated by point mutations in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41. J. Virol. 79**:**2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katzenstein, D. A., S. M. Hammer, M. D. Hughes, H. Gundacker, J. B. Jackson, S. Fiscus, S. Rasheed, T. Elbeik, R. Reichman, A. Japour, T. C. Merigan, M. S. Hirsch, et al. 1996. The relation of virologic and immunologic markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. N. Engl. J. Med. 335**:**1091-1098. [DOI] [PubMed] [Google Scholar]
- 33.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189**:**695-714. [DOI] [PubMed] [Google Scholar]
- 34.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434**:**1148-1152. [DOI] [PubMed] [Google Scholar]
- 35.Marchant, D., S. J. Neil, K. Aubin, C. Schmitz, and A. McKnight. 2005. An envelope-determined, pH-independent endocytic route of viral entry determines the susceptibility of human immunodeficiency virus type 1 (HIV-1) and HIV-2 to Lv2 restriction. J. Virol. 79**:**9410-9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marozsan, A. J., E. Fraundorf, A. Abraha, H. Baird, D. Moore, R. Troyer, I. Nankja, and E. J. Arts. 2004. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J. Virol. 78**:**11130-11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng, G., X. Wei, X. Wu, M. T. Sellers, J. M. Decker, Z. Moldoveanu, J. M. Orenstein, M. F. Graham, J. C. Kappes, J. Mestecky, G. M. Shaw, and P. D. Smith. 2002. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat. Med. 8**:**150-156. [DOI] [PubMed] [Google Scholar]
- 38.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37**:**2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250**:**1139-1142. [DOI] [PubMed] [Google Scholar]
- 40.Murakami, T., S. Ablan, E. O. Freed, and Y. Tanaka. 2004. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J. Virol. 78**:**1026-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97**:**343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74**:**3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien, W. A., P. M. Hartigan, E. S. Daar, M. S. Simberkoff, J. D. Hamilton, et al. 1997. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann. Intern. Med. 126**:**939-945. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien, W. A., P. M. Hartigan, D. Martin, J. Esinhart, A. Hill, S. Benoit, M. Rubin, M. S. Simberkoff, J. D. Hamilton, et al. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N. Engl. J. Med. 334**:**426-431. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien, W. A., Y. Koyanagi, A. Namazie, J. Q. Zhao, A. Diagne, K. Idler, J. A. Zack, and I. S. Chen. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348**:**69-73. [DOI] [PubMed] [Google Scholar]
- 46.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74**:**10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce-Pratt, R., D. Malamud, and D. M. Phillips. 1994. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J. Virol. 68**:**2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271**:**1582-1586. [DOI] [PubMed] [Google Scholar]
- 49.Perotti, M. E., X. Tan, and D. M. Phillips. 1996. Directional budding of human immunodeficiency virus from monocytes. J. Virol. 70**:**5916-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piatak, M., Jr., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K. C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259**:**1749-1754. [DOI] [PubMed] [Google Scholar]
- 51.Piguet, V., and Q. Sattentau. 2004. Dangerous liaisons at the virological synapse. J. Clin. Investig. 114**:**605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77**:**353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratner, S., M. P. Piechocki, and A. Galy. 2003. Role of Rho-family GTPase Cdc42 in polarized expression of lymphocyte appendages. J. Leukoc. Biol. 73**:**830-840. [DOI] [PubMed] [Google Scholar]
- 54.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17**:**2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosset, A., L. Spadola, and O. Ratib. 2004. OsiriX: an open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 17**:**205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, H., J. Orenstein, D. Dimitrov, and M. Martin. 1992. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology 186**:**712-724. [DOI] [PubMed] [Google Scholar]
- 57.Schaeffer, E., V. B. Soros, and W. C. Greene. 2004. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 78**:**1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, V., and D. D. Ho. 2003. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 1**:**181-190. [DOI] [PubMed] [Google Scholar]
- 59.Sourisseau, M., N. Sol-Foulon, F. Porrot, F. Blanchet, and O. Schwartz. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81**:**1000-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sperling, R. S., D. E. Shapiro, R. W. Coombs, J. A. Todd, S. A. Herman, G. D. McSherry, M. J. O'Sullivan, R. B. Van Dyke, E. Jimenez, C. Rouzioux, P. M. Flynn, J. L. Sullivan, et al. 1996. Maternal viral load, zidovudine treatment, and the risk of transmission of human immunodeficiency virus type 1 from mother to infant. N. Engl. J. Med. 335**:**1621-1629. [DOI] [PubMed] [Google Scholar]
- 61.Vujcic, L. K., and G. V. Quinnan, Jr. 1995. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res. Hum. Retrovir. 11**:**783-787. [DOI] [PubMed] [Google Scholar]
- 62.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373**:**117-122. [DOI] [PubMed] [Google Scholar]
- 63.Wyma, D. J., J. Jiang, J. Shi, J. Zhou, J. E. Lineberger, M. D. Miller, and C. Aiken. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78**:**3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79**:**12231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286**:**1353-1357. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, Z. Q., S. W. Wietgrefe, Q. Li, M. D. Shore, L. Duan, C. Reilly, J. D. Lifson, and A. T. Haase. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 101**:**5640-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, P., E. Chertova, J. Bess, Jr., J. D. Lifson, L. O. Arthur, J. Liu, K. A. Taylor, and K. H. Roux. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 100**:**15812-15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]