Direct Involvement of Yeast Type I Myosins in Cdc42-Dependent Actin Polymerization (original) (raw)
Abstract
The generation of cortical actin filaments is necessary for processes such as cell motility and cell polarization. Several recent studies have demonstrated that Wiskott-Aldrich syndrome protein (WASP) family proteins and the actin-related protein (Arp) 2/3 complex are key factors in the nucleation of actin filaments in diverse eukaryotic organisms. To identify other factors involved in this process, we have isolated proteins that bind to Bee1p/Las17p, the yeast WASP-like protein, by affinity chromatography and mass spectroscopic analysis. The yeast type I myosins, Myo3p and Myo5p, have both been identified as Bee1p-interacting proteins. Like Bee1p, these myosins are essential for cortical actin assembly as assayed by in vitro reconstitution of actin nucleation sites in permeabilized yeast cells. Analysis using this assay further demonstrated that the motor activity of these myosins is required for the polymerization step, and that actin polymerization depends on phosphorylation of myosin motor domain by p21-activated kinases (PAKs), downstream effectors of the small guanosine triphosphatase, Cdc42p. The type I myosins also interact with the Arp2/3 complex through a sequence at the end of the tail domain homologous to the Arp2/3-activating region of WASP-like proteins. Combined deletions of the Arp2/3-interacting domains of Bee1p and the type I myosins abolish actin nucleation sites at the cortex, suggesting that these proteins function redundantly in the activation of the Arp2/3 complex.
Keywords: myosin, actin, Cdc42, actin-related protein (Arp) 2/3 complex, Wiskott-Aldrich syndrome protein (WASP)
Introduction
Type I myosins are highly conserved proteins that appear to play roles in numerous processes, such as endocytosis, membrane trafficking, contractility, and cell motility ( Mooseker and Cheney 1995). In some eukaryotic organisms, these proteins localize to the leading edge of motile cells and are speculated to power membrane protrusion ( Fukui et al. 1989; Wagner et al. 1992). The motor activity of type I myosins in amoebae depends on phosphorylation of the motor domain by p21-activated kinases (PAKs), downstream targets of Rho family small GTPases ( Maruta and Korn 1977; Brzeska et al. 1997, Brzeska et al. 1999). The same phosphorylation site, known as the TEDS-rule phosphorylation site, also exists in yeast type I myosins and type VI myosins of mammalian cells ( Wu et al. 1997; Buss et al. 1998), but the in vivo significance of this phosphorylation in Rho family GTPase-regulated cellular processes has not been demonstrated.
Budding yeast contains two type I myosins, Myo3p and Myo5p, which are functionally redundant in vivo ( Goodson et al. 1996). These myosins consist of an NH2-terminal motor domain with F-actin binding and ATPase activity and IQ motifs for binding to a light chain. The COOH-terminal tail of the type I myosins includes a putative lipid-binding domain, a second F-actin binding domain (TH2 domain), an Src homology domain 3 (SH3) domain, and an acidic tail motif (see Results). Therefore, the tail has the potential to interact with a variety of proteins and membrane lipids. Double knockout of MYO3 and MYO5 results in defects in polarized cell growth and endocytosis, but it is not known how the myosins participate in these actin-based processes. In vitro, these myosins are phosphorylated at the conserved site by Ste20p and Cla4p, two yeast PAKs ( Wu et al. 1997). Phosphorylation is required for function in vivo, as a nonphosphorylatable Myo3p mutant does not rescue the growth defect of the double myosin I–null ( Wu et al. 1997). Ste20p and Cla4p are effectors of the small GTPase Cdc42p (for review see Daniels and Bokoch 1999), which is required for polarized cell growth during bud formation ( Adams et al. 1990). Thus, yeast provides an excellent system for elucidating the in vivo function and regulation of type I myosins.
Cdc42 is required for cell polarity and actin polymerization in a number of systems. A requirement for Cdc42 activity in actin assembly has been shown in permeabilized yeast cells, and Acanthamoeba, Dictyostelium, Xenopus, and neutrophil extracts ( Li et al. 1995; Zigmond et al. 1997; Ma et al. 1998; Mullins and Pollard 1999). The mechanisms by which Cdc42 promotes actin assembly have been the subject of recent research. In both Dictyostelium and Xenopus egg extracts, the actin-related protein (Arp) 2/3 complex is required for Cdc42-induced actin assembly ( Ma et al. 1999; Mullins and Pollard 1999). This highly conserved seven-polypeptide complex is thought to catalyze the de novo nucleation of actin filaments, the rate-limiting step in actin polymerization ( Cooper et al. 1983; Tobacman and Korn 1983). It is also required for the actin polymerization-driven motility of pathogenic bacteria such as Listeria monocytogenes and Shigella flexneri ( Welch et al. 1997; Egile et al. 1999). The nucleation activity of purified Arp2/3 complex is low but can be greatly stimulated by the Listeria ActA protein ( Welch et al. 1998), and by members of the Wiskott-Aldrich syndrome protein (WASP) family, including WASP, neuronal (N)-WASP, WAVE/SCAR1, and Bee1p/Las17p ( Machesky et al. 1999; Rohatgi et al. 1999; Winter et al. 1999a; Yarar et al. 1999). This activation is dependent upon a COOH-terminal acidic domain conserved among all WASP family members ( Machesky and Insall 1998). Both WASP and N-WASP contain a Cdc42-binding motif ( Aspenstrom et al. 1996; Miki et al. 1996; Symons et al. 1996). It has been shown that the ability of N-WASP to activate the nucleation activity of the Arp2/3 complex is dependent upon binding to Cdc42p ( Rohatgi et al. 1999). It is believed that Cdc42 binding induces a conformational change in N-WASP, exposing its Arp2/3 activation domain. WAVE and Bee1p lack the Cdc42-interacting domain, and how they link signal transduction pathways to actin assembly is not yet known.
Our study started from an attempt to define functional components of cortical actin assembly sites using an in vitro reconstitution assay ( Lechler and Li 1997). In this assay, a Cdc42-dependent actin nucleation activity associated with cortical patches in permeabilized yeast cells was first eliminated by treatment with 2 M urea and reconstituted by incubation with a cytoplasmic extract. The extract was then washed away and actin polymerization at cortical patches was assayed after the addition of rhodamine-labeled actin monomers (G-actin). This approach led to the identification of Bee1p, a yeast member of the WASP family, as one of the proteins required for the first step of the reconstitution ( Lechler and Li 1997). However, the lack of a Cdc42-binding motif suggested that signaling to actin polymerization was occurring in a different manner in this system. Furthermore, deletion of the acidic Arp2/3-binding domain of Bee1p has little effect on actin assembly and cell growth, suggesting that Bee1p is unlikely to be the sole activator of the Arp2/3 complex ( Winter et al. 1999a). In this study, we first identified the yeast type I myosins as Bee1p-interacting proteins that are also required for cortical actin assembly in the permeabilized cells. Subsequent analysis provided evidence that these myosins have multiple roles in cortical actin assembly: they mediate Cdc42 regulation of actin polymerization through motor domain phosphorylation; and they function redundantly with Bee1p in the activation of the Arp2/3 complex.
Materials and Methods
Plasmid Construction
The Myo5ΔC-hemagglutinin (HA) construct was prepared by PCR of the promoter (−284) and the first 1169 codons of MYO5 using genomic DNA and the primers 5′-GCGCGCGCGGCCGCGGTGTGTTTTAACTCGTCGG-3′ and 5′-GCGCGCGGATCCTTGTACAGAATTCAGAACGGG-3′. The product was digested with NotI and BamHI and ligated into PRL129, an HA-tagging vector derived from PRS306. The Myo3ΔC-HA construct was generated by PCR using the primers 5′-GCGCGCGCGGCCGCCCTCGGGAAGAGCTTTACCG-3′ and 5′-GCGCGCGAATTCGTGGGGACGTTTAGCGACGG-3′. The product was digested with NotI and EcoRI and ligated into PRL129. The glutathione _S_-transferase (GST)-Myo3A–expressing plasmid was made by PCR of the acidic domain of MYO3 using the primers 5′-GCGCGCGAATTCGTGGGGACGTTTAGCGACGG-3′ and 5′-GCGCGCGAATTCACTCTGATTATCACTAGTGGG-3′, digesting the product with EcoRI, and ligating into the EcoRI site of pGEX4T-1. The GST-Myo5A construct was created by subcloning of the Myo5p acidic domain. Full-length MYO5 was subjected to PCR and ligated into NotI, BamHI sites of Bluescript. The EcoRI fragment containing the acidic domain was then cloned into pGEX4T-1. The rigor mutant of Myo3p was created by PCR-based mutagenesis of p1846 (a Myo3-HA–containing plasmid obtained from Charlie Boone, Queens University, Kingston, Ontario) using the primers 5′-GCTTCTGTCTTACCCGCTCTAGACTCACC-3′ and 5′-GGTGAGTCTAGAGCGGGTAAGACAGAAGC-3′.
Strain Construction
The myo5ΔA Δmyo3 Δmyo5 strain was constructed by transforming a _myo5ΔA_-carrying plasmid (a COOH-terminal 50 amino acid [aa] deletion) into the double mutant background. The bee1ΔA strain was created by deletion of the COOH-terminal 33 codons of the BEE1 gene by homologous recombination. Deletion was confirmed by PCR and Western blot analysis. The bee1ΔA myo5ΔA Δmyo3 strain was made by crossing the bee1ΔA strain with the myo5ΔA Δmyo3 Δmyo5 strain and subsequently selecting haploids with the desired genotype. All yeast strains used in this study are listed in Table .
Table 1.
Yeast Strains
| Name | Genotype | Source |
|---|---|---|
| RLY1 | MATa his3-Δ200 leu2-3 lys2-801 ura3-52 | Drubin lab |
| RLY171 | MATa leu2-3 ura3-52 cdc42-1 | Drubin lab |
| RLY446 | MATa ade2 his3-Δ200 leu2-3 trp1-1 ura3-52 bar1 Δmyo3::HIS3 Δmyo5::TRP1 pMYO3-HA (p1846) | This work |
| RLY454 | MATa his3-Δ200 leu2-3 lys2-801 ura3-52 trp1-1 Δvrp1::TRP1 pMYO3-HA (p1846) | This work |
| RLY650 | MATa his3-Δ_200 leu2-3 lys2-801 ura3-52 Δ_bee1::LEU2 pBEE1-PrA (pTL84) | Winter et al., 1999 |
| RLY802 | MATa ade2 can1 his3-Δ200 leu2-3 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 pmyo3-S357A (pVL97) | This work |
| RLY803 | MATa ade2 can1 his3-Δ200 leu2-3 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 pMYO3-S357D (pVL98) | This work |
| RLY804 | MATa ade2 can1 his3-Δ200 leu2-3 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 pMyo5_Δ_C-HA (pTL88) | This work |
| RLY805 | MATa ade2 can1 his3-Δ200 leu2-3 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 pMyo3ΔC-HA (pTL91) | This work |
| RLY817 | MATa his3-Δ200 3 lys2-801 trp1-1 ura3-52 Δmyo3::HIS3 | This work |
| RLY819 | MATa his3-Δ200 leu2-3 lys2-801 trp1-1 ura3-52 Δmyo5::TRP1 | This work |
| RLY822 | MATa his3-Δ200 lys2-801 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 | This work |
| RLY865 | MATa his3-Δ200 leu2-3 lys2-801 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 pmyo3-G132R-HA (pTL103) | This work |
| RLY871 | MATa his3-Δ200 leu2-3 lys2-801 trp1-1 ura3-52 Δmyo3::HIS3 Δmyo5::TRP1 URA3::myo5ΔC bee1ΔC::LEU2 | This work |
| RLY889 | MATa leu2-3 ura3-52 cdc42-1 pmyo3-S357A (pVL97) | This work |
| RLY890 | MATa leu2-3 ura3-52 cdc42-1 pMYO3-S357D (pVL98) | This work |
Isolation of Bee1p-Interacting Proteins
Extracts were prepared from RLY650 cells by passing cells suspended in UB (50 mM Hepes, pH 7.5, 100 mM KCl, 3 mM MgCl2, 1 mM EGTA, 1 mM DTT, supplemented with protease inhibitors as described [ Li et al. 1995]) through a French pressure cell at 2,300 psi. A high-speed supernatant was obtained by centrifugation at 250,000 g for 1 h. Ammonium sulfate was added to the resulting extract to bring it to 30% saturation (the protein A–tagged Bee1 is efficiently precipitated at this concentration). Precipitated protein was collected by centrifugation, resuspended in UB, and clarified. IgG–Sepharose beads (Amersham Pharmacia Biotech) were added to this solution and subsequently washed with UB plus 0.5% Tween 20, and then UB plus 1 M KCl. Protein was then eluted with 0.5 M acetic acid, pH 3.6. Bee1p-associated proteins were separated by one-dimensional SDS-PAGE and identified by high mass accuracy matrix-assisted laser desorption/ionization (MALDI) peptide mapping as described ( Shevchenko et al. 1996a). In brief, protein bands visualized by staining with Coomassie were excised from the gel. Proteins were reduced by dithiothreitol, alkylated with iodoacetamide, and in-gel digested with trypsin (unmodified, sequencing grade; Boehringer Mannheim) at 37°C overnight ( Shevchenko et al. 1996b). 0.5-μl aliquots of the supernatant were withdrawn from the digests and analyzed on a REFLEX MALDI TOF mass spectrometer (Bruker Daltonics). Mass spectra were internally calibrated using known m/z of ions originating from the matrix and autolysis products of trypsin. Database searches were performed using PeptideSearch 3.0 software developed in European Molecular Biology Laboratory. No limitations on protein molecular weights and species of origin were imposed. 18 (38) peptides were matched to the sequence of Myo3p (Myo5p) with the mass accuracy better than 75 ppm. These peptides covered 17 and 39% of the sequence of Myo3p and Myo5p, respectively.
Protein Purification and Interaction Assays
GST-Myo3A contained the COOH-terminal 38 aa of Myo3p, and GST-Myo5A and GST-Bee1A contained the COOH-terminal 54 aa of each protein. Extracts were prepared from bacteria that were induced to express these proteins. Fusion proteins were purified on glutathione-agarose, washed extensively with PBS plus 0.5% Tween 20, eluted with reduced glutathione, and then equilibrated into UB. Arp2/3 complex was purified as described ( Winter et al. 1999a). Purified Arp2/3 complex at 50 nM was incubated with fusion protein coupled beads, washed with UB plus 0.4% Tween 20, and then eluted into SDS-PAGE sample buffer. Immunoprecipitations were performed from 20 mg/ml, 250,000 g yeast extracts prepared by the liquid nitrogen grinding technique. In brief, anti-HA (monoclonal 12CA5 ascites; BABCO) or anti-myc ( Evan et al. 1985) coupled protein A beads were incubated with extract at 4°C for 1 h. After washing extensively with UB plus 0.4% Tween 20, proteins were eluted into SDS-PAGE sample buffer. For immunodepletions, concentrated extracts were treated as for immunoprecipitations, but the supernatant was removed for analysis.
Actin Assembly Assays
Small-budded cells and rhodamine-labeled actin were prepared as described ( Li et al. 1995). The actin assembly and reconstitution assays in permeabilized cells was performed as described ( Li et al. 1995; Lechler and Li 1997). In brief, frozen small budded cells were permeabilized by incubation in 0.5 mg/ml saponin for 30 min, treated with 2 M urea for 25 min to inactivate actin assembly activity, and then incubated with concentrated extracts for 25 min. Extract was washed away and rhodamine-labeled actin was added to the permeabilized cells in polymerization buffer (UB plus 1 mM ATP). After an 8-min incubation, cells were fixed with formaldehyde and observed by fluorescence microscopy. All incubations were at 27°C. For _cdc42-1_–containing strains, the cells were grown at 34°C for 2 h before freezing. For pharmacologic experiments, 2,3-butanedione 2-monoxime (BDM) (gift of T.J. Mitchison, Harvard Medical School, Boston, MA) was used at 20 mM, and adenosine 5′-[beta, gamma-imido]triphosphate (AMPPNP) (Sigma Chemical Co.) at 1 mM. Inhibitors were added either with concentrated extracts during the reconstitution, or with labeled actin during the polymerization. For the dephosphorylation experiments, concentrated extracts were treated with 10 U alkaline phosphatase (Boehringer Mannheim) for 20 min at 37°C before incubation with cells.
Fluorescent Microscopy
Rhodamine-phalloidin (Molecular Probes) staining was performed as described ( Pringle et al. 1989). Rhodamine-labeled actin ( Kellogg et al. 1988) and phalloidin were imaged with a Zeiss Axiophot microscope with a HB100 W/Z high pressure mercury lamp and a Zeiss 100× Plan Neofluar oil immersion objective. Image acquisition was carried out using Northern Exposure (Phase 3 Imaging Systems).
Results
Bee1p Interacts with Type I Myosins
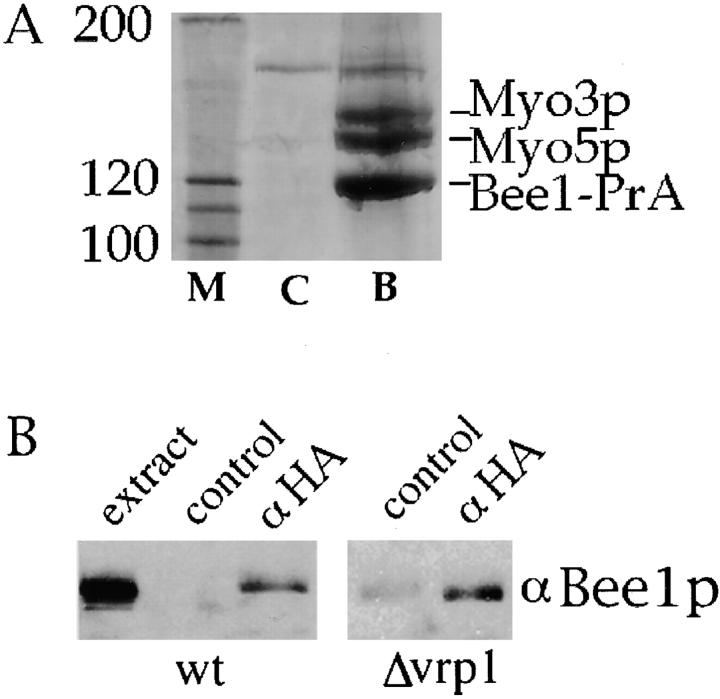
We demonstrated previously that Bee1p is required for actin assembly activity in permeabilized yeast cells. In part, Bee1p may function through activation of the Arp2/3 complex's nucleation activity. However, Bee1p contains several putative protein–protein interaction motifs, and appears to function with other proteins in the reconstitution of actin assembly ( Lechler and Li 1997). Therefore, an affinity chromatography strategy was used to identify Bee1p-interacting proteins. Bee1p was tagged at its COOH terminus with four repeats of the IgG binding domain of protein A (Bee1-PrA). Under the endogenous BEE1 promoter, this construct rescued the growth defect of the Δbee1 mutation (data not shown). Extracts were prepared from this strain, precipitated with ammonium sulfate to enrich for Bee1p, and then subjected to affinity chromatography on IgG–Sepharose beads. The ammonium sulfate treatment concentrates Bee1p, and possibly its binding partners as well, from the extract by ∼28-fold. Bound proteins were eluted with acid, separated by SDS-PAGE, and individual protein bands were identified by mass spectroscopy. Two proteins that bound specifically to Bee1-PrA and were present in approximately stoichiometric amounts were identified as Myo3p and Myo5p, the two type I myosins of budding yeast ( Fig. 1 A). Characterization of other proteins will be published elsewhere. Reciprocal immunoprecipitation was carried out to verify the Bee1–myosin interaction in a strain that expresses HA-tagged Myo3p at an endogenous level. Although Bee1p was not detected in a control immunoprecipitate, the anti-HA immunoprecipitate specifically and reproducibly brought down Bee1p ( Fig. 1 B). However, only a small fraction (<1%) of the total Bee1p was immunoprecipitated under these conditions, suggesting that the interaction is of low affinity. Concentration of the proteins through ammonium sulfate precipitation, as in Fig. 1 A, increases the level of interaction.
Figure 1.
Type I myosins associate with Bee1p. (A) High speed extracts were prepared from a wild-type strain and a strain carrying Bee1–protein A under the control of the BEE1 promoter. Ammonium sulfate was added to 30% saturation, and precipitated proteins were collected, resuspended, and bound to IgG–Sepharose. After extensive washing, proteins were eluted with 0.5 M acetic acid, pH 3.6. Shown is part of a Coomassie blue–stained gel that contains the indicated proteins: M, protein molecular weight markers; C, control (untagged wild-type extract); B, Bee1–protein A extract. The identity of proteins determined by mass spectroscopy analysis is indicated. (B) Extracts were prepared from strains carrying Myo3-HA under the control of the MYO3 promoter in either wild-type or Δvrp1 backgrounds. Immunoprecipitation was performed with anti-HA or anti-myc (control immunoprecipitate) coupled protein A beads. After washing and elution, proteins were subjected to SDS-PAGE, transferred to nitrocellulose, and blotted with the anti-Bee1p antibody ( Winter et al. 1999a). The extract lane is loaded with 5% of the input extract.
The yeast protein Vrp1p has been shown previously to interact with both Bee1p and Myo5p ( Anderson et al. 1998; Naqvi et al. 1998). To determine whether the Bee1p–type I myosin interaction was mediated by Vrp1p, we performed immunoprecipitations of Myo3-HA from Δvrp1 cells. Consistently, we find that Bee1p is still present in the anti-HA immunoprecipitates, albeit at lower levels than in wild-type cells, suggesting that at least some fraction of the Bee1p interacts with type I myosins in a Vrp1p-independent manner ( Fig. 1 B). These results, combined with the fact that both Bee1p and type I myosins localize to cortical patches ( Goodson et al. 1996; Li 1997) suggest that these proteins interact in vivo.
Type I Myosins Are Required for the Reconstitution of Actin Assembly Sites
Type I myosins have traditionally been thought of as molecular motors that transport cargo along actin filaments ( Mooseker and Cheney 1995). Therefore, their interaction with a protein involved in regulating actin dynamics was somewhat surprising. To determine whether the type I myosins have a direct role in the production of actin filaments, extracts were prepared from single or double myosin knockout yeast strains and tested for their ability to reconstitute actin assembly in urea-inactivated permeabilized cells. Typical positive and negative results of this assay are shown in Fig. 2 A. Extracts prepared from Δmyo3 cells or Δmyo5 cells were able to restore actin assembly activity to the urea-inactivated permeabilized yeast cells to the same extent as wild-type extracts ( Fig. 2 B). However, extracts from Δmyo3 Δmyo5 double mutant showed no more activity than addition of buffer alone. Therefore, the type I myosins are required for the actin assembly activity and appear to be functionally redundant in this assay as they are in vivo.
Figure 2.
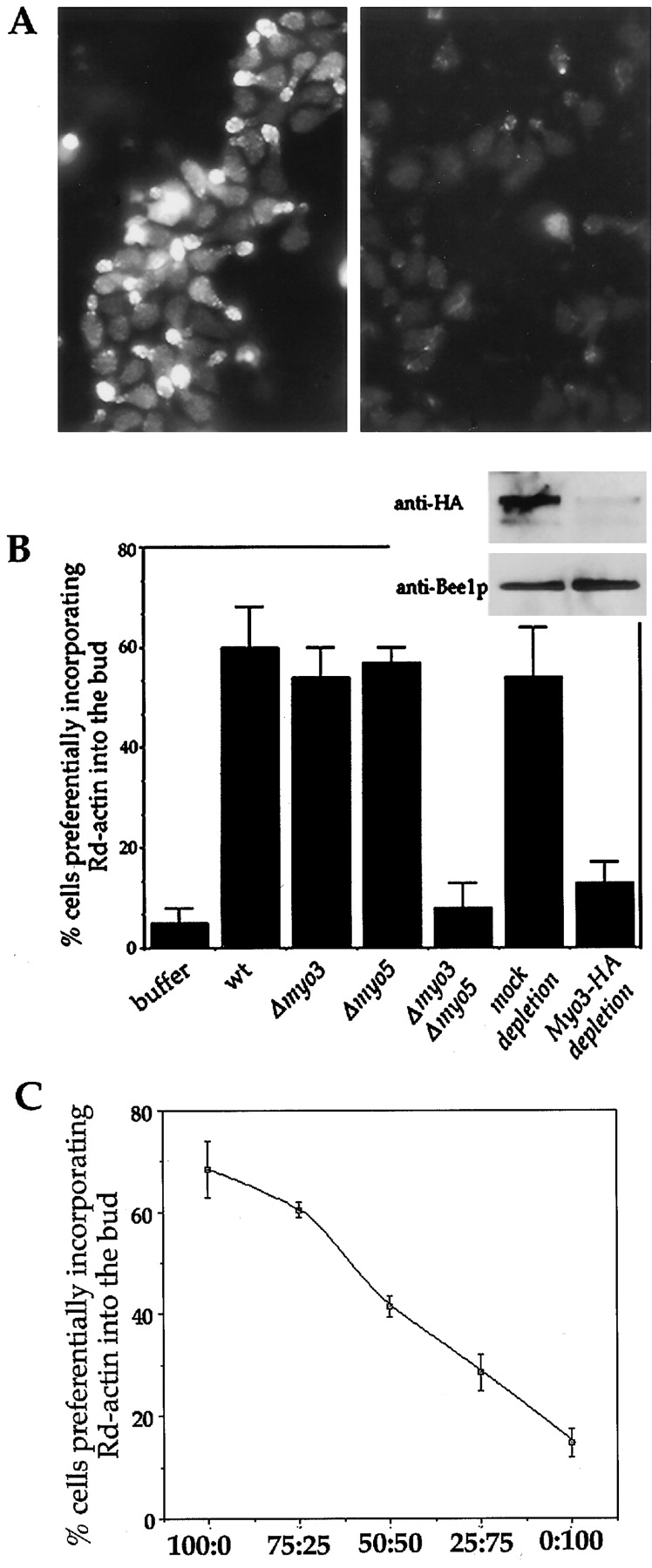
Type I myosins are necessary for cortical actin polymerization. (A) Examples of positive (left panel, wild-type extract), and negative (right panel, myo3G132R mutant extract) results for the reconstitution of actin assembly sites in permeabilized yeast cells. Assay conditions are described in Materials and Methods. (B) Concentrated extracts were prepared from various strains as indicated below the histograms and assayed for their ability to restore actin assembly activity to urea-treated permeabilized yeast cells. Quantitation of cells that assembled rhodamine actin into the bud was as described ( Lechler and Li 1997). For immunodepletion, anti-myc (mock depletion) or anti-HA coupled protein A beads were added to concentrated extracts prepared from a strain that expresses Myo3-HA in the Δmyo3 Δmyo5 background. After incubation, beads were pelleted and supernatants were removed for analysis. Gel inset above corresponding lanes of the histogram, shows depletion of Myo3-HA, but not Bee1p by the anti-HA beads. (C) Extracts from wild-type and Δmyo3 Δmyo5 cells were mixed at the indicated ratios and then assayed for their ability to restore actin assembly in urea-treated permeabilized cells. The complementation efficiency was quantified and plotted against the extract ratios. Each data point is an average of duplicate reactions.
To demonstrate that the type I myosins are required for actin polymerization in a dose-dependent manner, wild-type and Δmyo3 Δmyo5 extracts were mixed at varying ratios to vary the concentration of the type I myosin at the same time as maintaining the concentration of other active components ( Fig. 2 C). We find that as type I myosin concentration decreases, the ability to assemble actin also decreases. This closely resembles the results seen by dilution of wild-type extract with buffer alone ( Lechler and Li 1997), suggesting that the type I myosins are limiting factors in the wild-type extract.
To ensure that the lack of activity in the Δmyo3 Δmyo5 extract was not due to unhealthiness of the double mutant cells, we tested whether biochemical depletion of the type I myosins also led to a loss of ability to reconstitute actin assembly. To do so, a strain was generated that contained the HA-tagged Myo3p in the Δmyo3 Δmyo5 background. Therefore, Myo3-HA was the only source of type I myosins in this strain. The Myo3-HA rescued the growth defect of the double deletion strain (data not shown). Extracts were prepared from this strain and then mixed with protein A–Sepharose beads conjugated to either anti-HA or anti-myc (control) antibodies. Immunodepletion of the Myo3-HA is demonstrated in the immunoblot as shown in Fig. 2 B. Bee1p as well as the actin nucleator Arp2/3 complex (data not shown) were not significantly depleted from extracts by this treatment. The Myo3-depleted extract lacked the ability to restore actin assembly in urea-inactivated cells, whereas mock depletion had little effect ( Fig. 2 B). These results support a direct involvement of the type I myosins in cortical actin assembly.
Motor Activity of Type I Myosins Is Necessary for Cortical Actin Polymerization
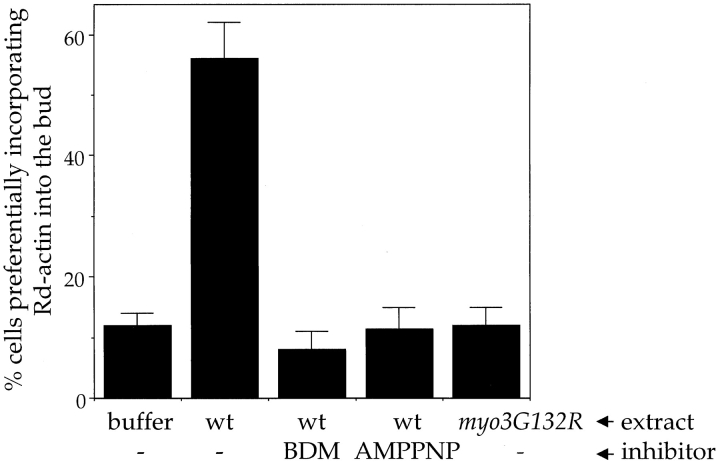
Type I myosins consist of an NH2-terminal motor domain harboring the ATP and actin binding sites, followed by a tail domain that is thought to interact with cargos and other cytoskeletal proteins. We first took a pharmacological approach to determine whether the motor activity of myosins is required for actin polymerization. BDM is a low affinity inhibitor of myosins ( Backx et al. 1994; Cramer and Mitchison 1995). To separate the effect of the drug on the reconstitution of actin assembly sites from that on actin polymerization, BDM was either added during the incubation with the extract but washed out before the polymerization step, or was only present during the polymerization step. Addition of 20 mM BDM to extracts during reconstitution had no effect on actin assembly (data not shown), but when BDM was added with G-actin at the time of polymerization, actin assembly into the permeabilized cells was inhibited ( Fig. 3). Similarly, addition of AMPPNP to extracts during the reconstitution had no effect on actin assembly activity (data not shown); however, when added with G-actin at the time of polymerization, it inhibited actin polymerization ( Fig. 3). Because neither the same concentration of BDM nor AMPPNP blocked spontaneous assembly of pyrene-labeled actin (data not shown), these results suggest that myosin motor domain activity is not required during the reconstitution (e.g., to localize the proteins), but is required for actin polymerization at the cell cortex.
Figure 3.
Myosin motor activity is required for actin assembly. Reconstitution was performed with buffer or extracts as listed in the first line under the histogram. Polymerization was performed in the presence of 1 mM ATP (−), 1 mM ATP plus 20 mM BDM, or with 1 mM AMPPNP (with no ATP), as indicated below the histogram. Reconstitution procedure and quantitation of the results were performed as described ( Lechler and Li 1997).
These pharmacological experiments support a role for myosins and ATP hydrolysis in actin assembly. However, care must be taken in interpreting these results, as the specificities of these inhibitors are broad. To further test the requirement of myosin motor activity in actin polymerization, Glycine132, within the P-loop required for nucleotide binding of Myo3p, was mutated to an arginine residue. Similar mutations in this loop of other myosins lead to an inability to bind nucleotide ( Bejsovec and Anderson 1990). Since nucleotide-free myosin binds tightly to actin filaments, these mutations are known as rigor mutations. The myo3G132R mutation results in a complete loss of function: it did not rescue the growth defect of the Δmyo3 Δmyo5 strain (data not shown). Extracts prepared from a strain carrying the rigor Myo3p as its sole source of type I myosins (i.e., in the Δmyo3 Δmyo5 background) were unable to restore actin assembly activity to the permeabilized cells ( Fig. 3). This result further suggests that type I myosins require a functional motor domain in order to promote actin polymerization.
Type I Myosins Mediate Cdc42p Regulation of Cortical Actin Assembly
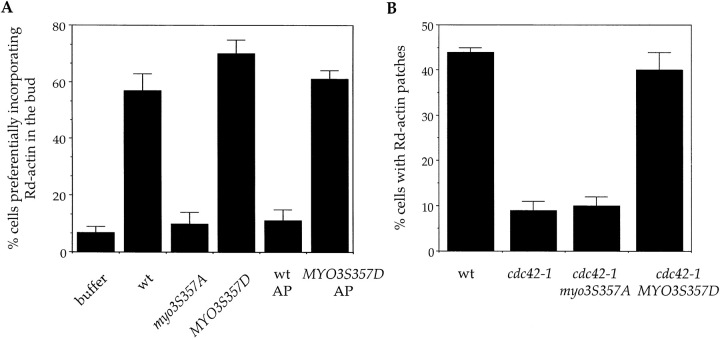
The distribution of actin patches in vivo as well as actin polymerization in the permeabilized cells are dependent on the small GTPase Cdc42 ( Adams et al. 1990; Li et al. 1995), but the mechanism by which Cdc42 regulates actin assembly in yeast was not understood. Because the motor activity of type I myosins, which appears to be required for cortical actin polymerization, is stimulated through phosphorylation by PAKs ( Maruta and Korn 1977; Wu et al. 1996; Brzeska et al. 1997, Brzeska et al. 1999), it is possible that the type I myosins mediate Cdc42 regulation of actin assembly through motor domain phosphorylation. Mutation of the conserved serine residue (ser357) to alanine in Myo3 leads to a complete loss of function, whereas mutation to aspartate has no phenotype ( Wu et al. 1997). To test the effects of these mutations on actin assembly, extracts were prepared from strains carrying the myo3S357A and MYO3S357D mutants as their sole source of type I myosins. The myo3S357A mutant extract was unable to restore actin assembly in urea-treated cells, whereas the MYO3S357D mutant extract reproducibly showed slightly higher levels of activity than wild-type extracts ( Fig. 4 A).
Figure 4.
Phosphorylation of type I myosins is an important step in the stimulation of actin polymerization by Cdc42p. (A) Reconstitution of actin assembly sites was carried out with concentrated extracts prepared from strains carrying the myo3S357A and MYO3S357D mutants as their only source of type I myosins. Reconstitution procedure and quantitation of the results were performed as described ( Lechler and Li 1997). AP: alkaline phosphatase used for treating wild-type or MYO3S357D mutant extracts (see text). (B) A mutant Myo3p mimicking constitutive phosphorylation rescues the actin assembly defect of a temperature-sensitive CDC42 allele. Wild-type, cdc42-1, and cdc42-1 cells expressing MYO3S357D or myo3S357A under the endogenous MYO3 promoter were grown at room temperature, shifted to 34°C for 2 h, permeabilized, and subjected to rhodamine actin assembly assays as described ( Li et al. 1995).
To further establish a requirement of myosin I phosphorylation in cortical actin assembly, we reasoned that if this were true then the extract used for reconstitution of actin assembly should be sensitive to phosphatase treatment. In fact, incubation of extracts with alkaline phosphatase caused almost a complete loss of actin assembly activity ( Fig. 4 A). Treatment of the MYO3S357D extract with phosphatase, on the other hand, had very little effect on the activity of the extract. These data confirm the requirement for myosin I phosphorylation in actin assembly, and suggest that it is the only phosphorylation required for the reconstitution of actin assembly sites.
We showed previously that the actin assembly activity in permeabilized yeast cells was abolished by the cdc42-1 mutation ( Li et al. 1995). If phosphorylation of the type I myosins is a key downstream event in the activation of actin assembly, then the MYO3S357D mutation may rescue the actin assembly defect of cdc42-1 cells. To test this possibility, the MYO3S357D and myo3S357A alleles were transformed into cdc42-1 cells, and actin polymerization was assayed in the permeabilized cells from the resulting strains. As shown in Fig. 4 B, permeabilized cdc42-1 cells expressing MYO3S357D regained actin polymerization activity, whereas those expressing the myo3S357A allele were as defective as the untransformed cdc42-1 cells. This result supports the hypothesis that type I myosins are a key target of active Cdc42p in promoting actin polymerization.
Involvement of Type I Myosins in the Activation of the Arp2/3 Complex
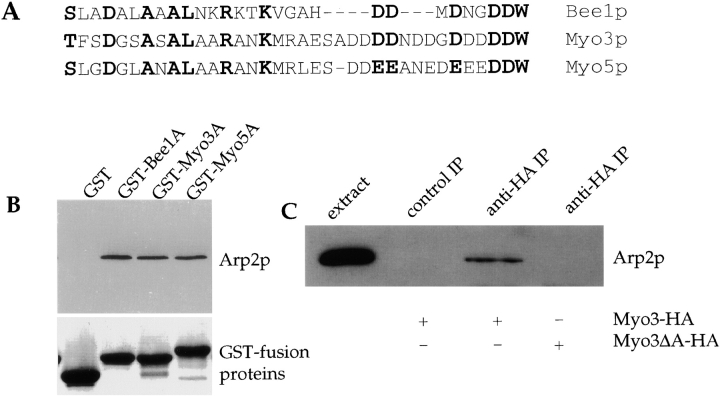
A unique feature of the yeast type I myosins that is not shared by known type I myosins of other organisms is the presence of an acidic region at the COOH-terminus. This domain shows significant similarity to the acidic COOH-terminal tails of Bee1p and other members of WASP family proteins. Over ∼35 aa, the Myo3p and Myo5p tails are 45% conserved with the Bee1p tail motif ( Fig. 5 A). This motif mediates the interaction of WASP family proteins with the Arp2/3 complex and is essential for activation of the Arp2/3 complex's nucleation activity in vitro ( Machesky and Insall 1998; Winter et al. 1999a). To determine if the myosins could interact with the Arp2/3 complex through the acidic motif, beads coupled to GST fusions of Myo3p or Myo5p COOH-terminal acidic fragments (GST-Myo3A and GST-Myo5A) were incubated with purified Arp2/3 complex, washed, and then examined for the presence of Arp2p. Arp2p was found associated with these fusion proteins to the same extent as with the Bee1p acidic domain (GST-Bee1A) ( Fig. 5 B). To determine whether the interaction between the type I myosins and Arp2/3 complex occurs in vivo, we immunoprecipitated HA-tagged Myo3p expressed at an endogenous level from cell extracts, and blotted with an antibody against Arp2p. Arp2p was detected in Myo3-HA immunoprecipitates but not in control immunoprecipitates ( Fig. 5 C). As we have found previously with Bee1p, this interaction is of low affinity, and <1% of total Arp2p is immunoprecipitated with the type I myosins (data not shown). The COOH-terminal domain of Myo3p was required for this interaction, as a deletion mutant lacking the COOH-terminal 34 aa no longer immunoprecipitated Arp2p ( Fig. 5 C). This result, together with the observation that Myo3p, Myo5p, Arp2p, and Arp3p all localize to cortical actin patches, suggest that the type I myosins and the Arp2/3 complex interact in vivo ( Goodson et al. 1996; Moreau et al. 1996; Winter et al. 1997).
Figure 5.
Type I myosins interact with the Arp2/3 complex. (A) Alignment of the COOH-terminal acidic domains of Myo3p, Myo5p, and Bee1p. Residues conserved in all three are in bold. (B) GST fusions to the COOH-terminal 38 aa of Myo3p and the COOH-terminal 54 aa of Myo5p and Bee1p were purified from bacteria. Fusion proteins bound to glutathione-agarose beads were incubated with 50 nM Arp2/3 complex purified as described ( Winter et al. 1999b). After washing, bound proteins were eluted into SDS-PAGE sample buffer. Upper panel, anti-Arp2p immunoblot; lower panel, Coomassie-stained gel showing that equal amounts of GST fusion proteins were added. (C) Extracts at 15 mg/ml were prepared from strains carrying Myo3-HA or Myo3ΔA-HA (Myo3p lacking the COOH-terminal acidic domain) in the Δmyo3 Δmyo5 background. Anti-myc (control immunoprecipitate) and anti-HA immunoprecipitations were performed, and bound proteins were analyzed by immunoblotting with anti-Arp2p antibodies. Extracts used are listed below the gel. The extract lane shows the presence of Arp2p in preimmunoprecipitate extracts.
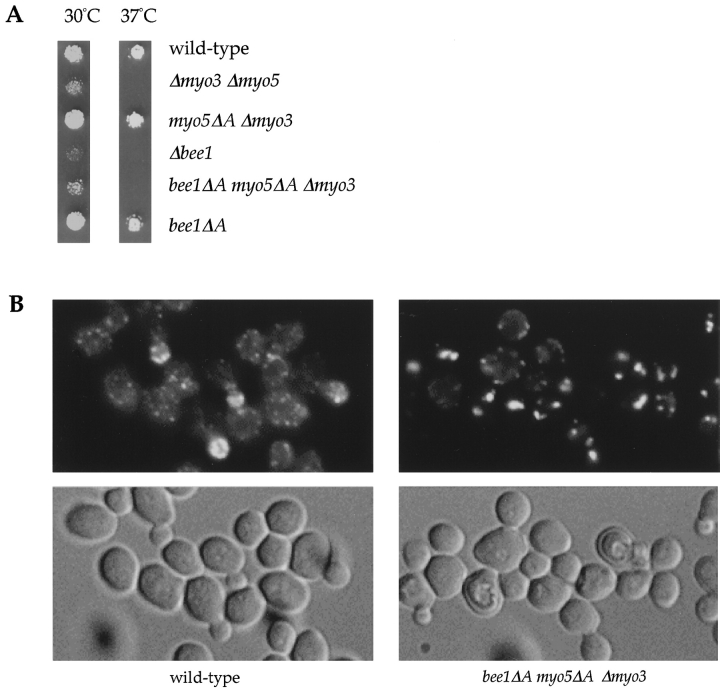
We showed previously that deletion of the Arp2/3-interacting domain of Bee1p does not cause severe defects in cortical actin organization, as opposed to disruption of the entire BEE1 gene or subunits of the Arp2/3 complex ( Winter et al. 1999a). To determine whether this lack of phenotype was due to redundant functions of the homologous domains in the type I myosins, strains were created that contained deletions of these domains in combination. The growth defect of a Δmyo3 Δmyo5 double mutant is fully rescued by an allele of Myo5p in which the COOH-terminal Arp2/3-interacting domain (34 aa) was deleted (myo5ΔA) ( Fig. 6 A). However, although neither the bee1ΔA mutation alone nor the myo5ΔA truncation in the Δmyo3 Δmyo5 double mutant background showed any severe phenotypes, a combination of the two in the Δmyo3 Δmyo5 double mutant background had a clear synthetic effect: the mutant cells grew almost as poorly as _BEE1_-null cells at 30°C and did not grow at 37°C. The double mutant cells also show severe defects in actin organization and accumulate abnormal F-actin aggregates, characteristic of null alleles of BEE1 and genes encoding subunits of the Arp2/3 complex ( Fig. 6 B) ( Li 1997; Winter et al. 1999b). These data suggest that the homologous COOH-terminal domains of Bee1p and type I myosins have a redundant but important role in regulation of the actin in vivo.
Figure 6.
The type I myosin and Bee1p acidic tails have redundant functions. (A) Equal concentration of cells from strains as indicated were spotted onto yeast extract, peptone, dextrose (YPD) plates, grown for 36 h, and then photographed. myo5ΔA Δmyo3: a strain expressing myo5ΔA in the Δmyo3 Δmyo5 double mutant background; bee1ΔA myo5ΔA Δmyo3: a strain expressing bee1ΔA and myo5ΔA in the Δmyo3 Δmyo5 double mutant background. (B) Rhodamine-phalloidin staining (upper panels) and differential interference contrast (DIC) images (lower panels) of wild-type and bee1ΔA myo5ΔA Δmyo3 cells grown at room temperature.
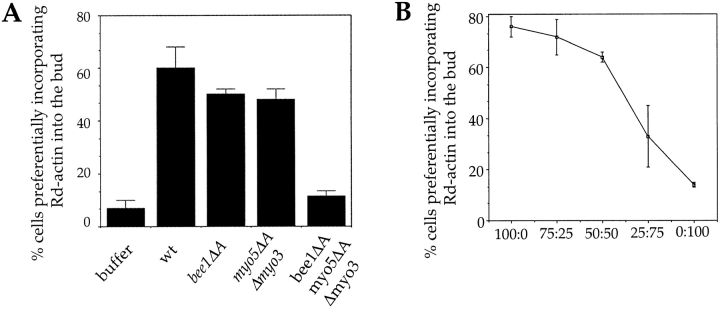
This synthetic effect is also evident in vitro. Extracts were prepared from the bee1ΔA, the myo5ΔA Δmyo3, and the bee1ΔA myo5ΔA Δmyo3 strains and tested for their ability to reconstitute actin polymerization in the urea-inactivated permeabilized cells. Whereas the actin assembly activity was intact in the former two extracts, an extract from the strain lacking both Bee1p and Myo5p COOH-terminal acidic domains (in the Δmyo3 Δmyo5 double mutant background) had essentially no activity ( Fig. 7 A). These results suggest that the COOH-terminal domains of Bee1p and Myo5p share a redundant function both in vivo and in vitro. Finally, to address how the concentration of these activation domains affects actin assembly activity, we titrated wild-type extract with the bee1ΔA myo5ΔA Δmyo3 extract ( Fig. 7 B). Actin assembly decreases at low levels of the activation domain; however, actin assembly is less sensitive to loss of activation domains than to loss of type I myosins (compare Fig. 2 C and Fig. 7 B).
Figure 7.
Bee1p and type I myosin acidic tails function redundantly in actin assembly. (A) Concentrated extracts were prepared from the strains as indicated and tested for their ability to restore the actin assembly activity of urea-treated permeabilized yeast cells. Reconstitution procedure and quantitation of the results were performed as described ( Lechler and Li 1997). (B) Extracts from wild-type and bee1ΔA myo5ΔA Δmyo3 cells were mixed at the indicated ratios on the horizontal axis and then assayed for their ability to restore actin assembly in urea-treated permeabilized cells. The complementation efficiency was quantified and plotted against the extract ratios. Each data point is an average of duplicate reactions.
Discussion
Type I myosins are a highly conserved family of molecular motors, first identified almost 30 years ago (for review see Mooseker and Cheney 1995). In spite of this, precise in vivo function of these myosins have not been defined. Genetic and biochemical evidence in Acanthamoeba, Dictyostelium, and yeast have implicated these proteins in actin-based processes such as cell motility, phagocytosis, endocytosis, and contractile activity ( Mooseker and Cheney 1995). Type I myosins have been hypothesized to transport membranous vesicles or to provide contractile force on actin filaments in order to function in these processes. Here we have presented evidence that type I myosins are directly involved in the promotion of actin assembly at the cell cortex.
Myo3p and Myo5p physically interact with Bee1p, the yeast WASP homologue. This interaction appears to be of low affinity, as only a small fraction coimmunoprecipitates. However, we consistently find that increasing extract concentration increases the amount of bound proteins. In the case where extracts were first precipitated with 30% ammonium sulfate, we achieved a 28-fold concentration over the starting extract, leading to an almost stoichiometric interaction between the proteins. This suggests that with the high in vivo concentrations of these proteins in cortical patches, the majority of these proteins may be in complex. The interaction between Bee1p and the myosins is likely to occur through interaction of the SH3 domain of the myosins and the proline-rich repeats of Bee1p (M. Evangelista and C. Boone, personal communication). The only other identified ligand for the SH3 domain of type I myosins is Acan125, a protein purified from Acanthamoeba which shows no homology to Bee1p ( Xu et al. 1997). Interestingly, the Bee1p binding protein, Vrp1p, also interacts with the SH3 domain of type I myosins ( Anderson et al. 1998; Naqvi et al. 1998). Since the SH3 domain of the yeast type I myosins was shown to be important for their localization to actin patches ( Anderson et al. 1998), it is possible that Bee1p and Vrp1p function redundantly in targeting the myosins to regions of actin assembly. Another possibility is that Bee1p and Vrp1p are cargo for the myosins and are transported to the barbed ends of filaments by these molecular motors (see below). Regardless, since all these proteins have mammalian counterparts (Vrp1p is a homologue of the mammalian WASP-interacting protein [WIP] protein [ Ramesh et al. 1997]), it would be interesting to test if their complex formation is also conserved.
The type I myosins also interact with the Arp2/3 complex. The COOH-terminal tails of Myo3p and Myo5p show significant homology to the COOH-terminal tails of Bee1p and other WASP family proteins. These acidic tails are actually more similar to WASP and N-WASP than they are to Bee1p, as they have a large net negative charge, as opposed to Bee1p which has a more neutral charge. These motifs mediate a direct physical interaction with the Arp2/3 complex. This interaction is likely to occur in vivo, as Arp2/3 complex can be immunoprecipitated with the type I myosins, an association that is dependent upon the COOH-terminal tail of the myosins. There are several possible functions for this interaction. First, it could be involved in localization of either the myosins or the Arp2/3 complex. This is unlikely, as Arp2/3 complex localization is not affected by deletion of the myosin acidic tail motif (data not shown). Also, deletion of the myosin acidic tails causes no obvious defects in the presence of wild-type Bee1p, suggesting that myosin localization is unlikely to be abolished. Another possible function for the myosin–Arp2/3 interaction could be to activate the nucleation activity of the Arp2/3 complex. This possibility is supported by three findings. First, homologous motifs in Bee1p and WASP proteins have been shown to be required for the activation of the Arp2/3 complex ( Machesky et al. 1999; Rohatgi et al. 1999; Winter et al. 1999a; Yarar et al. 1999). Second, we found a specific synthetic interaction due to loss of the Bee1p and myosin I acidic motifs: loss of the Arp2/3-activating acidic domain of either Bee1p or the type I myosins does not cause a dramatic phenotype; however, in combination, these deletions lead to drastic growth and actin organization defects. Third, by using an in vitro reconstitution assay, we showed that loss of the acidic motifs of Bee1p and type I myosins causes a defect in actin assembly activity. These data support redundant functions of the myosin and Bee1p acidic domains in activation of the Arp2/3 complex.
It was surprising to us that an involvement in actin polymerization is not limited to the tail domain of type I myosin, but that motor activity is also required. The requirement for myosin motor activity in actin polymerization may be explained by one of the following models. First, the type I myosins may function by transporting nucleation or elongation machinery to the barbed ends of existing filaments. If the cargo is a WASP family member or the Arp2/3 complex, this would ensure that the nucleation machinery remains near the barbed end of filaments, promoting the generation of dendritic actin filaments at the membrane cortex. Alternatively, if the cargo is an elongation machinery ( Zigmond et al. 1998), it could serve to maintain this machinery at the growing barbed end of actin filaments. Type I myosins may also function in cortical filament assembly as a molecular ratchet. In this model, type I myosins associate with the membrane through the lipid-binding domain within their tails ( Adams and Pollard 1989); movement of the myosin head along actin towards the barbed end could then generate a membrane protrusion, allowing addition of G-actin onto the barbed end. This would explain the requirement for myosin motor activity during polymerization. In filopodia, it has been proposed that such a function may be important for coupling polymerization and leading edge protrusion ( Sheetz et al. 1992). One caveat is that all of these models imply processivity of the myosin motor. In vitro experiments suggested that individual myosin I molecules are unlikely to remain bound to F-actin long enough to translocate through any significant distance ( Ostap and Pollard 1996). However, other experiments have shown that myosin I can transport vesicles or F-actin filaments ( Adams and Pollard 1986; Zot et al. 1992), possibly due to a second F-actin binding site in the myosin I tail that can cross-link actin filaments ( Fujisaki et al. 1985). An alternative possibility is that the type I myosins do not function like conventional motors in promoting actin assembly. For example, members of the Kin I kinesin family of microtubule motors have been shown to use their microtubule-dependent ATPase activity to catalyze the depolymerization of microtubules ( Desai et al. 1999).
The mammalian N-WASP protein has been shown to mediate Cdc42-dependent actin polymerization by interacting directly with Cdc42 ( Miki et al. 1998a; Rohatgi et al. 1999). However, WASP-family proteins such as Bee1p and WAVE, do not contain the Cdc42-binding motif ( Li 1997; Miki et al. 1998b). In yeast, PAKs are known Cdc42 effectors. The involvement of yeast PAKs in actin polymerization was first suggested by the result that overexpression of Ste20p, a yeast PAK, suppresses both the polarization and actin assembly defects of cdc42-1 mutant ( Eby et al. 1998). We have shown here that a mutation in Myo3p mimicking constitutive phosphorylation of the site known to be acted upon by PAKs rescues the actin assembly defects in cdc42-1 mutant cells. This finding suggests that Myo3p and probably also Myo5p are major targets of Cdc42p/PAK in the regulation of actin assembly at the cell cortex. A function in mediating polarized actin assembly may explain the phenotype of myosin I mutant Dictyostelium cells that exhibit defects in maintaining direction of movement in chemotactic streaming assays ( Jung et al. 1996; Titus et al. 1993). Since the PAK phosphorylation site is conserved in amoeba type I myosins, but not in those of higher cells, it is possible that this pathway of actin regulation by small GTPases only operates in protozoan organisms. However, the mammalian type VI myosins, which were recently shown to move toward the pointed ends of actin filaments ( Wells et al. 1999), have the same PAK phosphorylation sites ( Buss et al. 1998). It will be interesting to define the role of these myosins in actin polymerization and actin-based motility.
Acknowledgments
We thank B. Winsor, E. Leberer, C. Boone, L. Pon, and P. Silver for providing antibodies, strains, and plasmids; D. Winter for purified Arp2/3 complex; T. Mitchison, J. Yarrow, D. Winter, and S. Schuyler for comments on the manuscript; and members of the Li lab for support and encouragement.
This work was supported by grant GM57063 from the National Institutes of Health to R. Li.
Footnotes
Abbreviations used in this paper: aa, amino acid(s); AMPPNP, adenosine 5′-[beta, gamma-imido]triphosphate; Arp, actin-related protein; BDM, 2,3-butanedione 2-monoxime; GST, glutathione _S_-transferase; HA, hemagglutinin; N-WASP, neuronal WASP; PAK, p21-activated kinase; SH3, Src homology domain 3; WASP, Wiskott-Aldrich syndrome protein.
References
- Adams A.E., Johnson D.I., Longnecker R.M., Sloat B.F., Pringle J.R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae . J. Cell Biol. 1990;111:131–142 . doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.J., Pollard T.D. Propulsion of organelles isolated from Acanthamoeba along actin filaments by myosin-I. Nature. 1986;322:754–756 . doi: 10.1038/322754a0. [DOI] [PubMed] [Google Scholar]
- Adams R.J., Pollard T.D. Binding of myosin I to membrane lipids. Nature. 1989;340:565–568 . doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- Anderson B.L., Boldogh I., Evangelista M., Boone C., Greene L.A., Pon L.A. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polymerization. J. Cell Biol. 1998;141:1357–1370 . doi: 10.1083/jcb.141.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenstrom P., Lindberg U., Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr. Biol. 1996;6:70–75 . doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Backx P.H., Gao W.D., Azan-Backx M.C., Marban E. Mechanism of force inhibition by 2,3-butanedione monoxime in rat cardiac muscleroles of [Ca2+] and cross-bridge kinetics. J. Physiol. 1994;476:487–500 . doi: 10.1113/jphysiol.1994.sp020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A., Anderson P. Functions of the myosin ATP and actin binding sites are required for C. elegans thick filament assembly. Cell. 1990;60:133–140 . doi: 10.1016/0092-8674(90)90723-r. [DOI] [PubMed] [Google Scholar]
- Brzeska H., Knaus U.G., Wang Z.Y., Bokoch G.M., Korn E.D. p21-activated kinase has substrate specificity similar to Acanthamoeba myosin I heavy chain kinase and activates Acanthamoeba myosin I. Proc. Natl. Acad. Sci. USA. 1997;18:1092–1095 . doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H., Young R., Knaus U., Korn E.D. Myosin I heavy chain kinasecloning of the full-length gene and acidic lipid-dependent activation by Rac and Cdc42. Proc. Natl. Acad. Sci. USA. 1999;96:394–399 . doi: 10.1073/pnas.96.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F., Kendrick-Jones J., Lionne C., Knight A.E., Cote G.P., Paul Luzio J. The localization of myosin VI at the Golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J. Cell Biol. 1998;143:1535–1545 . doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A., Buhle E.L.J., Walker S.B., Tsong T.Y., Pollard T.D. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22:2193–2202 . doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- Cramer L.P., Mitchison T.J. Myosin is involved in postmitotic cell spreading. J. Cell Biol. 1995;131:179–189 . doi: 10.1083/jcb.131.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R.H., Bokoch G.M. p21-activated protein kinasea crucial component of morphological signaling? Trends Biochem. Sci. 1999;24:350–355 . doi: 10.1016/s0968-0004(99)01442-5. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78 . doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Eby J.J., Holly S.P., van Drogen F., Grishin A.V., Peter M., Drubin D.G., Blumer K.J. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr. Biol. 1998;8:967–970 . doi: 10.1016/s0960-9822(98)00398-4. [DOI] [PubMed] [Google Scholar]
- Egile C., Loisel T.P., Laurent V., Li R., Pantaloni D., Sansonetti P.J., Carlier M.F. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999;146:1319–1332 . doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G.I., Lewis G.K., Ramsay G., Bishop J.M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616 . doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki H., Albanesi J.P., Korn E.D. Experimental evidence for the contractile activities of Acanthamoeba myosins IA and IB. J. Biol. Chem. 1985;260:11183–11189 . [PubMed] [Google Scholar]
- Fukui Y., Lynch T.J., Brzeska H., Korn E.D. Myosin I is located at the leading edge of locomoting Dictyostelium amoeba. Nature. 1989;341:328–331 . doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- Goodson H.V., Anderson B.L., Warrick H.M., Pon L.A., Spudich J.A. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5)myosin proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 1996;133:1277–1291 . doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Wu X., Hammer J.A., III. Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol. 1996;133:305–323 . doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Mitchison T.J., Alberts B.M. Behavior of microtubules and actin filaments in living Drosophila embryos. Development. 1988;103:675–686 . doi: 10.1242/dev.103.4.675. [DOI] [PubMed] [Google Scholar]
- Lechler T., Li R. In vitro reconstitution of cortical actin assembly sites in budding yeast. J. Cell Biol. 1997;138:95–103 . doi: 10.1083/jcb.138.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiskott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 1997;136:649–658 . doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zheng Y., Drubin D. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J. Cell Biol. 1995;128:599–615 . doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Cantley L.C., Janmey P.A., Kirschner M.W. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J. Cell Biol. 1998;140:1125–1136 . doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Rohatgi R., Kirschner M.W. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc. Natl. Acad. Sci. USA. 1999;95:15362–15367 . doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Insall R.H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 1998;8:1347–1356 . doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky L.M., Mullins R.D., Higgs H.N., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., Pollard T.D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 1999;96:3739–3744 . doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H., Korn E.D. Acanthamoeba cofactor protein is a heavy chain kinase required for actin activation of the Mg2+-ATPase activity of Acanthamoeba myosin I. J. Biol. Chem. 1977;252:8329–8332 . [PubMed] [Google Scholar]
- Miki H., Miura K., Takenawa T. N-WASP, a novel actin depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5326–5335 . [PMC free article] [PubMed] [Google Scholar]
- Miki H., Sasaki T., Takai Y., Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP Nature. 391 1998. 93 96a [DOI] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac EMBO (Eur. Mol. Biol. Organ.) J. 17 1998. 6932 6941b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker M.S., Cheney R.E. Unconventional myosins. Annu. Rev. Cell Dev. Biol. 1995;11:633–675 . doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Moreau V., Madania A., Martin R.P., Winsor B. The Saccharomyces cerevisiae actin-related protein, Arp2, is involved in the actin cytoskeleton. J. Cell Biol. 1996;134:117–132 . doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D., Pollard T.D. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr. Biol. 1999;9:405–415 . doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- Naqvi S.N., Zahn R., Mitchell D.A., Stevenson B.J., Munn A.L. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 1998;8:959–962 . doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- Ostap E.M., Pollard T.D. Biochemical kinetic characterization of the Acanthamoeba myosin-I ATPase. J. Cell Biol. 1996;132:1053–1060 . doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R., Preston R.A., Adams A.E., Stearns T., Drubin D.G., Haarer B.K., Jones E.W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435 . doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Anton I.M., Hartwig J.H., Geha R.S. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. USA. 1997;94:14671–14676 . doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M.W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231 . doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Sheetz M.P., Wayne D.B., Pearlman A.L. Extension of filopodia by motor-dependent actin assembly. Cell Motil. Cytoskelet. 1992;22:160–169 . doi: 10.1002/cm.970220303. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Jensen O.N., Podtelejnikov A.V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. Linking genome and proteome by mass spectrometrylarge-scale identification of yeast proteins from two dimensional gels Proc. Natl. Acad. Sci. USA. 93 1996. 14440 14445a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels Anal. Chem 68 1996. 850 858b [DOI] [PubMed] [Google Scholar]
- Symons M., Derry J.M.J., Karlak B., Jiang S., Lemahieu V., McCormick F., Francke U., Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734 . doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Titus M.A., Wessels D., Spudich J.A., Soll D. The unconventional myosin encoded by the myoA gene plays a role in Dictyostelium motility. Mol. Biol. Cell. 1993;4:233–246 . doi: 10.1091/mbc.4.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacman L.S., Korn E.D. The kinetics of actin nucleation and polymerization. J. Biol. Chem. 1983;258:3207–3214 . [PubMed] [Google Scholar]
- Wagner M.C., Barylko B., Albanesi J.P. Tissue distribution and subcellular localization of mammalian myosin I. J. Cell Biol. 1992;119:163–170 . doi: 10.1083/jcb.119.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., Iwamatsu A., Mitchison T.J. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes . Nature. 1997;385:265–268 . doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch M.D., Rosenblatt J., Skoble J., Portnoy D.A., Mitchison T.J. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108 . doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Wells A.L., Lin A.W., Chen L.Q., Safer D., Cain S.M., Hasson T., Carragher B.O., Milligan R.A., Sweeney H.L. Myosin VI is an actin-based motor that moves backward. Nature. 1999;401:505–508 . doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Winter D., Podtelejnikov A.V., Mann M., Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 1997;7:519–529 . doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Winter D., Lechler T., Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP family protein Curr. Biol. 9 1999. 501 504a [DOI] [PubMed] [Google Scholar]
- Winter D.C., Chau E.C., Li R. Genetic dissection of the yeast Arp2/3 complexa comparison of the in vivo and structural roles of individual subunits Proc. Natl. Acad. Sci. USA. 96 1999. 7288 7293b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Lee S.F., Furmaniak-Kazmierczak E., Cote G.P., Thomas D.Y., Leberer E. Activation of myosin-I by members of the Ste20p protein kinase family. J. Biol. Chem. 1996;271:31787–31790 . doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- Wu C., Lytvyn V., Thomas D.Y., Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 1997;272:30623–30626 . doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- Xu P., Mitchelhill K.I., Kobe B., Kemp B.E., Zot H.G. The myosin-I binding protein Acan125 binds the SH3 domain and belongs to the superfamily of leucine-rich repeat proteins. Proc. Natl. Acad. Sci. USA. 1997;94:3685–3690 . doi: 10.1073/pnas.94.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D., To W., Abo A., Welch M.D. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr. Biol. 1999;9:555–558 . doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zigmond S.H., Joyce M., Borleis J., Bokoch G.M., Devreotes P.N. Regulation of actin polymerization in cell-free systems by GTPγS and Cdc42. J. Cell Biol. 1997;138:363–374 . doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H., Joyce M., Yang C., Brown K., Huang M., Pring M. Mechanism of Cdc42-induced actin polymerization in neutrophil extracts. J. Cell Biol. 1998;142:1001–1012 . doi: 10.1083/jcb.142.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zot H.G., Doberstein S.K., Pollard T.D. Myosin-I moves actin filaments on a phospholipid substrateimplications for membrane targeting. J. Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]