Mdv1p Is a Wd Repeat Protein That Interacts with the Dynamin-Related Gtpase, Dnm1p, to Trigger Mitochondrial Division (original) (raw)
Abstract
Mitochondrial fission is mediated by the dynamin-related GTPase, Dnm1p, which assembles on the mitochondrial outer membrane into punctate structures associated with sites of membrane constriction and fission. We have identified additional nuclear genes required for mitochondrial fission, termed MDV (for mitochondrial division). MDV1 encodes a predicted soluble protein, containing a coiled-coil motif and seven COOH-terminal WD repeats. Genetic and two-hybrid analyses indicate that Mdv1p interacts with Dnm1p to mediate mitochondrial fission. In addition, Mdv1p colocalizes with Dnm1p in fission-mediating punctate structures on the mitochondrial outer membrane. Whereas localization of Mdv1p to these structures requires Dnm1p, localization of Mdv1p to mitochondrial membranes does not. This indicates that Mdv1p possesses a Dnm1p-independent mitochondrial targeting signal. Dnm1p-independent targeting of Mdv1p to mitochondria requires MDV2. Our data indicate that MDV2 also functions separately to regulate the assembly of Dnm1p into punctate structures. In contrast, Mdv1p is not required for the assembly of Dnm1p, but Dnm1p-containing punctate structures lacking Mdv1p are not able to complete division. Our studies suggest that mitochondrial fission is a multi-step process in which Mdv2p regulates the assembly of Dnm1p into punctate structures and together with Mdv1p functions later during fission to facilitate Dnm1p-dependent mitochondrial membrane constriction and/or division.
Keywords: fission, membrane remodeling, morphology, outer membrane, organelle
Introduction
The steady state copy number and shape of mitochondria varies dramatically in different cell types, ranging from multiple spherical organelles to low copy number branched structures (Johnson et al. 1980). Thus, not surprisingly, the regulation of mitochondrial copy number, morphology, and distribution is important for cellular differentiation and function. For example, mitochondrial fusion is required in flies for sperm development and fertility and in yeast for mitochondrial DNA (mtDNA) recombination and inheritance (Hales and Fuller 1997; Hermann et al. 1998; Rapaport et al. 1998), and in worms mitochondrial fission is required for embryonic development (Labrousse et al. 1999).
In Saccharomyces cerevisiae, mitochondria form a continuous reticulum evenly distributed at the cell cortex (Hoffman and Avers 1973; Stevens and White 1979; Nunnari et al. 1997). The establishment and maintenance of this reticulum requires a balanced frequency of mitochondrial fusion and fission events (Nunnari et al. 1997). The molecular components and mechanisms involved in mitochondrial fusion and fission are just beginning to be described. The coordinate fusion of the mitochondrial outer and inner membranes is mediated by the evolutionarily conserved Fzo family of mitochondrial outer membrane GTPases (Hales and Fuller 1997; Hermann et al. 1998; Rapaport et al. 1998). In Drosophila melanogaster, mutations in Fzo block a developmentally regulated mitochondrial fusion event during spermatogenesis (Hales and Fuller 1997). In yeast, the conditional fzo1-1 mutation causes mitochondrial reticuli to fragment in cells, a phenotype consistent with a block in mitochondrial fusion and ongoing mitochondrial fission, and mitochondrial DNA loss (Hermann et al. 1998). In addition, mitochondrial fusion is blocked during mating in fzo1-1 cells (Hermann et al. 1998).
Mitochondrial fission in yeast is mediated by Dnm1p, which is localized on the mitochondrial outer membrane in punctate structures associated with sites of mitochondrial constriction and fission (Otsuga et al. 1998; Bleazard et al. 1999; Sesaki and Jensen 1999). Dnm1p is a GTPase structurally related to dynamin, a protein required during endocytosis for the formation and scission of clathrin-coated vesicles from the plasma membrane (Sever et al. 2000). Dnm1p homologues in higher eucaryotes also have been shown to control mitochondrial fission, indicating that the mechanism of this process is evolutionarily conserved (Smirnova et al. 1998; Labrousse et al. 1999). Deletion of DNM1 causes mitochondria to form net-like structures of interconnected mitochondrial tubules in cells, but has no effect on mtDNA inheritance (Bleazard et al. 1999; Sesaki and Jensen 1999). These net-like mitochondrial structures arise in Δ_dnm1_ cells because the tips of mitochondrial tubules fuse with tubule sides, and new tubule ends cannot be generated by mitochondrial division. In addition, deletion of DNM1 blocks mitochondrial fragmentation in fzo1 cells, consistent with the respective antagonistic roles of these genes in fission and fusion (Bleazard et al. 1999; Sesaki and Jensen 1999).
Like dynamin-mediated endocytosis, mitochondrial fission is likely a multi-step process regulated by the Dnm1p GTPase cycle, which in turn is both influenced by and dependent on interactions with a variety of binding partners (Schmid et al. 1998; Sever et al. 2000). However, to date, no additional mitochondrial fission components in any organism have been described. Here, we report the identification and characterization of two novel nuclear genes required for mitochondrial fission, termed MDV1 and MDV2, for mitochondrial division. Our studies indicate that Mdv2p regulates the assembly of Dnm1p into prefission structures, and Mdv1p functions later in the fission reaction, possibly with Mdv2p, to stimulate Dnm1p-dependent mitochondrial membrane constriction and/or division.
Materials and Methods
Media and Yeast Genetic Techniques
Yeast strains used in this study are listed in Table . Standard genetic techniques and yeast media, including YPD (2% glucose), YPG (3% glycerol), SD, SRaf (2% raffinose), and SGal (2% galactose), were prepared as described (Guthrie and Fink 1991). Yeast transformations were performed as described (Gietz and Schiestl 1994).
Table 1.
Yeast Strains
| Strain | Genotype | Source |
|---|---|---|
| JSY1826 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Mata | Hermann et al. 1998 |
| JSY2977 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, fzo1-1, Mata | Hermann et al. 1998 |
| JSY2788 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_fzo1::HIS3_, pRS314-fzo1-1, Mata | Hermann et al. 1998 |
| JSY2793 | JSY2788, except _Mat_α | Hermann et al. 1998 |
| JSY1371 | leu2_Δ_1, his3_Δ_200, ura3-52, Δ_dnm1::HIS3_, _Mat_α | Otsuga et al. 1998 |
| ADM378 | leu2_Δ_1, his3_Δ_200, ura3-52, fzo1-1, Δ_dnm1::HIS3_, Mata | Bleazard et al. 1999 |
| JNY546 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_fzo1::HIS3,_ pRS314-fzo1-1, mdv1-1, Mata | This study |
| JNY547 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, fzo1-1, mdv1-1, Mata | This study |
| JNY548 | JNY547, except pVT100GFP, Mata | This study |
| JNY549 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_fzo1::HIS3,_ pRS314-fzo1-1, mdv2-1, _Mat_α | This study |
| JNY550 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, fzo1-1, mdv2-1, _Mat_α | This study |
| JNY551 | JNY550, except pVT100GFP, _Mat_α | This study |
| JNY552 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_mdv1::his5_+, Mata | This study |
| JNY553 | JNY552, except pVT100GFP, Mata | This study |
| JNY554 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, fzo1-1 Δ_mdv1::his5_+, Mata | This study |
| JNY555 | JNY 554, except pVT100GFP, Mata | This study |
| JNY556 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, TRP1GAL1GFP-MDV1, Mata | This study |
| JNY558 | JNY556_,_ except pRS315-_DNM1-_DsRED | This study |
| JNY559 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, fzo1-1, _Mat_α | This study |
| JNY560 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_mdv1::his5_+, pRS315-DNM1_-GFP, Mata_ | This study |
| JNY561 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, mdv2-1, pRS315-_DNM1_-GFP, Mata | This study |
| JNY562 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, Δ_mdv1::his5_+, mdv2-1, pRS315-DNM1_-GFP, Mata_ | This study |
| JNY563 | PJ69-4A, except pGAD-DNM1 and pGBDU-C1 | This study |
| JNY564 | PJ69-4A, except pGAD-C1 and pGBDU-MDV1 | This study |
| JNY565 | PJ69-4A, except pGAD-DNM1 and pGBDU-MDV1 | This study |
| JNY566 | JSY1826, except pRS315-_DNM1_-GFP, Mata | This study |
| JNY567 | leu2_Δ_1, his3_Δ_200, trp1_Δ_6, ura3-52, mdv2-1, Mata | This study |
| JNY568 | JNY567, except pVT100GFP, Mata | This study |
| JNY569 | leu2_Δ_1, his3_Δ_200, ura3-52, Δ_dnm1::HIS3, mdv2-1_, Mata | This study |
| JNY570 | JNY556, except Δ_dnm1::HIS3_ | This study |
| JNY571 | JNY556, except mdv2-1 | This study |
| JNY572 | JNY556, except Δ_dnm1::HIS3, mdv2-1_ | This study |
| JNY573 | JSY1826, except mdv1-1 | This study |
| JNY574 | JNY573, except pVT100GFP | This study |
| JNY575 | JSY1371, except pVT100GFP | This study |
Isolation of mdv Mutants
Extragenic mutations that suppress the glycerol growth defects of fzo1-1 cells were selected by plating 106 cells of JSY2788 and JSY2793 haploid cells on solid YPG media at the nonpermissive temperature of 37°C. Recessive nuclear fzo1-1 extragenic suppressor mutations were identified by crossing cells from colonies that formed under these conditions to naive fzo1-1 cells. Recessive extragenic fzo1-1 suppressor mutations were characterized by complementation analysis, which revealed three groups: dnm1, mdv1, and mdv2. Mutations allelic to DNM1 were identified by complementation analysis after crossing to fzo1-1 Δ_dnm1_ cells (ADM378). Sporulation of these diploids and tetrad analysis were used to determine whether these mutations were linked to the DNM1 locus.
MDV1 Cloning
MDV1 was cloned by screening for yeast genomic library inserts that would restore temperature-sensitive growth on glycerol to JNY547 (mdv1-1 fzo1-1) cells (i.e., restore the fzo1-1 phenotype) (Rose et al. 1987; Rose and Broach 1991). Strains that were inviable at 37°C on YPG were identified by replica plating. One complimenting plasmid, pECJN231, was identified and recovered by transformation into Escherichia coli, as described (Strathern and Higgins 1991). When retransformed into naive JNY547 cells, pECJN231 conferred temperature-sensitive growth on YPG. Yeast open reading frames (ORFs) on pECJN231 were identified by DNA sequence analysis (Davis Sequencing Facility, Davis, CA) and subcloning revealed that the complimenting ORF was YJL112W. Sequence analysis of YJL112W indicates that the nucleotide at position 48 is an A and not a C, as reported (Cziepluch et al. 1996).
Strain and Plasmid Construction
A ΔYJL112W::his5 + mutation was generated as described (Longtine et al. 1998) by transforming JSY1826 and JSY2977 with a PCR product containing the Schizosaccharomyces pombe his5 + gene sandwiched between 40 bp of YJL112W 5′ and 3′ flanking sequences, and selecting for his5 + transformants (JNY552 and JNY554, respectively). Replacement of YJL112W with his5 + was confirmed by PCR analysis of the YJL112W locus. Sporulation of diploids and tetrad analysis from a cross between JNY547 and JNY554 cells confirmed that the his5 + was linked to mdv1-1 (n = 24, 4:0).
A yeast strain harboring a _GAL1_-regulated NH2-terminal GFP-tagged version of Mdv1p was created by homologous recombination between the MDV1 locus in JSY1826 cells and a PCR product generated using the plasmid pFA6a-TRP1-pGAL1-GFP and the primers: 5′-CGGCGTAAACAAGAGAAGAAATTAACTTTCTACAGAAAGTACGAATTCGAGC-TCGTTTAAAC-3′ and 5′-CGTGGTGGACAATGTTTTTCCTATATGAGTTATTTGGTCGTTCACTTTGTATAGTTATCCATGC-3′ (Longtine et al. 1998). This strain, JNY556, was confirmed by PCR of the MDV1 locus and Western blotting using anti-Mdv1p (see below) and anti-GFP antibodies (Covance Research).
The pRS315-_DNM1_-GFP plasmid was kindly provided by H. Sesaki and R. Jensen (Johns Hopkins University, Baltimore, MD). DNM1-DsRED was created by replacing GFP in the pRS315-_DNM1_-GFP plasmid with DsRED (Clontech) at the NotI and SacII sites to generate the plasmid pECJN233. 5′ NotI and 3′ SacII restriction sites were generated in the DsRED gene by PCR using Vent Polymerase (New England Biolabs, Inc.).
Generation of Affinity-purified Anti-Mdv1p Antibodies
For the production of anti-Mdv1p antibodies, a truncated form of MDV1, from the 5′ ATG initiator codon to +214 nucleotide, was cloned in frame with maltose-binding protein (MBP) into pMAL-C2 (New England Biolabs, Inc.) using a 5′ EcoRI site and a 3′ SalI site introduced into MDV1 by PCR. Mdv1–MBP fusion protein was expressed in E. coli (DH5α) at 37°C and purified by amylose affinity chromatography (New England Biolabs, Inc.). Anti-Mdv1p polyclonal antibodies were produced in rabbits by injection of the Mdv1–MBP fusion protein by Covance Research, Inc. An Mdv1–MBP fusion protein affinity column was created by coupling purified Mdv1–MBP fusion protein to CNBr-activated Sepharose (Amersham Pharmacia Biotech) and was used to purify anti-Mdv1p antibodies as described (Harlow and Lane 1998)
Biochemical Analyses
Cell extracts were prepared and fractionated by differential centrifugation from JSY1826, JSY1371, JNY567, and JNY569 cells and protease protection experiments were conducted on enriched mitochondrial fractions from JSY1826 as described (Nunnari et al. 1993; Meeusen et al. 1999). Samples were analyzed using 12.5% SDS-PAGE, prepared as described (Anderson et al. 1973). For Western blotting, affinity-purified anti-Mdv1p (see above), antiporin (Molecular Probes), anti-KDH (C. Koehler, UCLA, Los Angeles, CA), and anti-3PGK (Molecular Probes) were used at dilutions of 1:500, 1:2,000, 1:1,000, 1:1,000, and 1:125, respectively. Western blots were developed using ECL reagents (Amersham Pharmacia Biotech).
Yeast Two Hybrid Analysis
Restriction endonuclease sites were introduced at 5′ and 3′ ends of _MDV1_and DNM1 by PCR with Vent polymerase (New England Biolabs, Inc.). MDV1 and DNM1 were separately subcloned into EcoRI and BamHI sites of pGAD-C1::LEU2 (AD vector for activation domain) and pGBDU-C1::URA3 (BD vector for binding domain) to generate the in frame fusions in the plasmids, pGAD-DNM1 and pGBDU-MDV1 (James et al. 1996). Pairwise combinations of AD and BD vectors were cotransformed into the yeast strain PJ69-4A (James et al. 1996). Interactions between AD and BD vectors were assessed by growth of transformants on SD media lacking leucine, uracil, and adenine as described (James et al. 1996).
Cytological Analyses
Mitochondrial morphology was analyzed and quantified as described using mito-GFP (kindly provided by B. Westermann, Ludwig Maximilians Universitaet, Muenchen, Germany; Wong et al. 2000). To assess mitochondrial fusion, haploid strains of opposite mating type were labeled with either mito-GFP or MitoTracker CMXR (Molecular Probes), mated at room temperature or 37°C, and analyzed as described (Nunnari et al. 1997). All samples were imaged using a Leica confocal microscope with a 100× 1.4 NA objective. Images were analyzed using Image Space software and figures were prepared using Adobe Photoshop.
Results
Identification of MDV Genes Required for Mitochondrial Fission
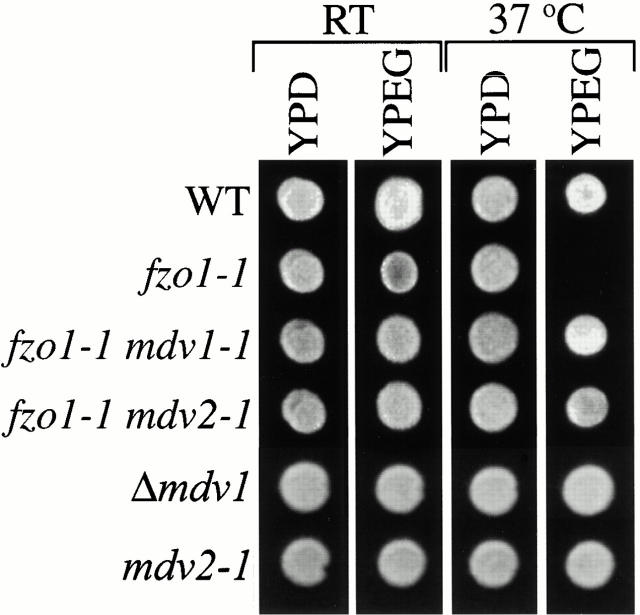
The conditional fzo1-1 mutation abolishes mitochondrial fusion in cells at the nonpermissive temperature of 37°C, causing mitochondrial membranes to fragment rapidly (Hermann et al. 1998). As a secondary consequence of this fragmentation, fzo1-1 cells lose mtDNA and are unable to grow on the nonfermentable carbon source glycerol (Fig. 1; Hermann et al. 1998). We isolated spontaneous suppressor mutants that rescued the fzo1-1 glycerol growth defect at 37°C (Fig. 1 A, see fzo1-1 mdv1-1 and fzo1-1 mdv2-1). These spontaneous suppressor mutations also rescued the glycerol growth defect of a Δ_fzo1_ strain, indicating that they suppress mtDNA loss by bypassing FZO1 function (not shown). Complementation analysis revealed that the fzo1-1 extragenic suppressor mutants represent three groups/genes. Not surprisingly, mutations in one group were allelic to DNM1, which encodes a dynamin-related GTPase required for mitochondrial fission. Previously, we have shown that both mitochondrial fragmentation and mtDNA loss in fzo1-1 cells are suppressed by deleting DNM1 and thus abolishing the opposing mitochondrial fission reaction (Bleazard et al. 1999). The remaining complementation groups represent two novel genes. Like the phenotype of dnm1 cells, mutations in the MDV genes caused almost no growth defects in cells on YPD or YPG media (Fig. 1 A and not shown). Based on our phenotypic analyses, we believe that these genes function in mitochondrial fission.
Figure 1.
Mutations in MDV genes suppress the loss of mtDNA in fzo1-1 cells. Indicated strains were plated onto YPD and YPG and grown at 25 and 37°C.
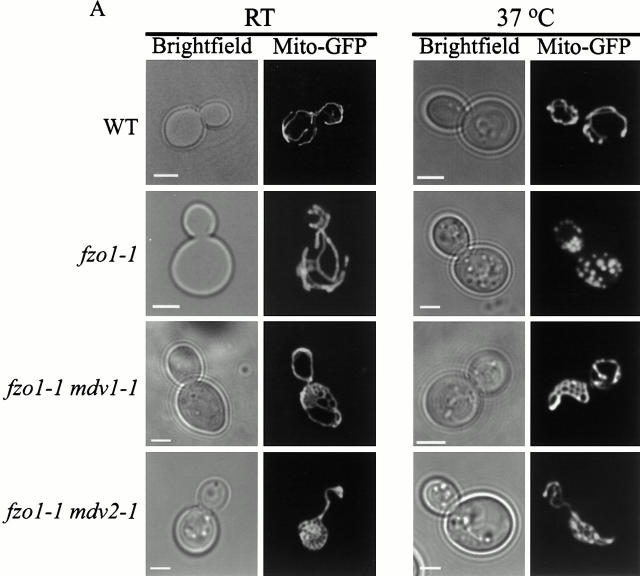
Several observations indicate that MDV1 and MDV2 function in mitochondrial fission. First, by examining mitochondrial morphology in mdv fzo1-1 cells, we determined that mutations in both MDV1 and MDV2 block mitochondrial fragmentation (Fig. 2 A and 3). Specifically, mitochondrial net-like structures are observed in mdv fzo1-1 cells grown at either permissive or nonpermissive temperature (Fig. 2 A and 3). These structures are also observed in mdv1-1 and mdv2-1 single mutant cells, consistent with the fact that these mutations suppress fzo1 phenotypes by bypassing FZO1 function (Fig. 2 B and 3). Mitochondrial net structures have been previously reported to occur in Δ_dnm1_ cells (Bleazard et al. 1999). These structures likely arise when mitochondrial ends fuse with tubule sides and, because fission is blocked, new tubule ends cannot be generated. Mitochondrial net structures also are observed in Δ_dnm1 fzo1-1_ cells under conditions where a block in mitochondrial fission precedes a block in fusion (Bleazard et al. 1999). Interestingly, mitochondrial nets are observed in double Δ_dnm1 mdv_ mutant cells, suggesting that these genes act in the same or in parallel pathways (not shown). Thus, the phenotypic characteristics of mdv cells are identical to those observed in Δ_dnm1_ cells, suggesting that the MDV genes act together with DNM1 to mediate mitochondrial fission.
Figure 2.
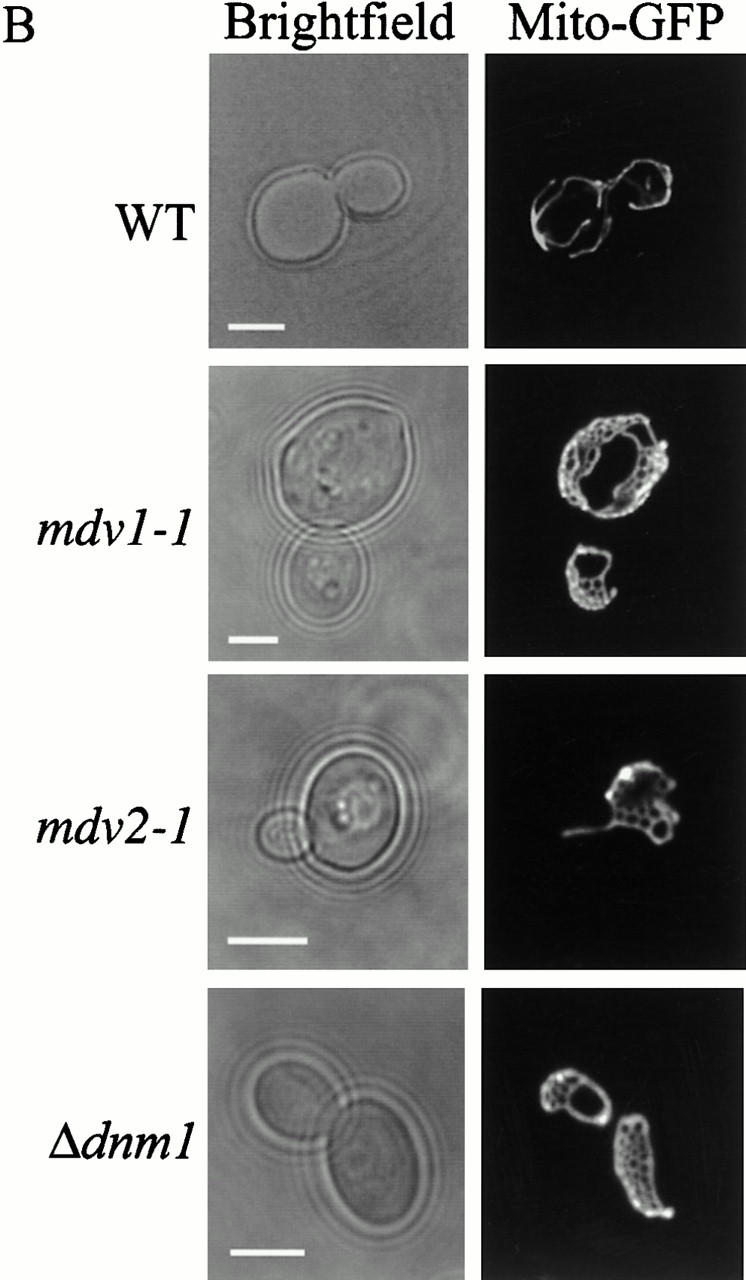
Mutations in MDV genes suppress mitochondrial fragmentation in fzo1-1 cells and cause mitochondrial membranes to form net-like structures. Mito-GFP was used to visualize mitochondrial morphology. All strains were grown at 25°C to log phase and imaged at 25°C, or after shifting to 37°C for 1 h. A, Mitochondrial morphology in fzo1-1 mdv double mutants. B, Mitochondrial morphology in mdv single mutants. Bars, 2 μm.
Mutations in DNM1, MDV1, and MDV2 also suppress mitochondrial fragmentation and genome loss in mgm1 cells (Wong et al. 2000 and not shown). MGM1 encodes a dynamin-related GTPase required for mitochondrial morphology and mtDNA maintenance (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999). Recently, we have shown that Mgm1p is localized to the mitochondrial intermembrane space and associated with the inner membrane (Wong et al. 2000). Our data indicate that loss of Mgm1p function results in mitochondrial fragmentation, similar to that observed in fzo1-1 cells (Wong et al. 2000). However, despite the similarities of mgm1 and fzo1 phenotypes, our analyses indicate that Mgm1p does not directly function in fusion (Wong et al. 2000). Rather, the observed mgm1 phenotypes and Mgm1p's localization suggest that Mgm1p functions in inner membrane remodeling events and that inner and outer membrane fission events are coupled. Thus, loss of MGM1 function may stimulate DNM1, MDV1, and _MDV2_-dependent outer membrane fission, explaining why mutations in these genes suppress mitochondrial fragmentation in mgm1 cells (Wong et al. 2000).
We also examined the possibility that mutations in MDV genes block mitochondrial fragmentation in fzo1-1 cells and cause mitochondrial nets to form by activating a _FZO1_-independent fusion pathway. To test this, we assayed mitochondrial fusion by labeling mitochondria in mdv fzo1-1 haploid cells of opposite mating type with either mito-GFP or a covalent vital probe, MitoTracker, and examined the distribution of these probes in zygotes formed at both permissive and nonpermissive temperature (Nunnari et al. 1997). We observed that mitochondrial fusion was blocked in mdv fzo1-1 double mutant zygotes formed at nonpermissive temperature, but not in mdv fzo1-1 double mutant zygotes formed at permissive temperature or in mdv single mutant zygotes formed under any condition (Table ). Thus, mutations in MDV genes block mitochondrial fragmentation in fzo1-1 cells by abolishing the opposing fission reaction and not by activating an _FZO1_-independent fusion pathway.
Table 2.
Quantification of Mitochondrial Fusion in Large Budded Zygotes during Mating
| Homozygous zygote | Temperature | Fused |
|---|---|---|
| (n) | % | |
| FZO1 MDV1 MDV2 | 25°C (52) | 98 |
| 37°C (53) | 98 | |
| fzo1-1 MDV1 MDV2 | 25°C (57) | 93 |
| 37°C (53) | 0 | |
| fzo1-1 mdv1-1 MDV2 | 25°C (49) | 92 |
| 37°C (57) | 7 | |
| FZO1 mdv1-1 MDV2 | 25°C (5) | 95 |
| 37°C (60) | 92 | |
| fzo1-1 MDV1 mdv2-1 | 25°C (56) | 90 |
| 37°C (60) | 2 | |
| FZO1 MDV1 mdv2-1 | 25°C (46) | 100 |
| 37°C (55) | 96 |
Interestingly, in certain allelic combinations of DNM1/dnm1; MDV1/mdv1 diploid cells, examination of mitochondrial morphology revealed that mitochondrial net-like structures persist, despite the presence of one wild-type copy of each gene (not shown). This genetic phenomenon, known as unlinked noncomplementation, suggests an interaction between the gene products of MDV1 and DNM1. Thus, MDV1 displays a genetic interaction with DNM1, the only characterized mitochondrial fission component, further indicating that it plays an essential role in this process.
Structural Features of the MDV1 Gene Product
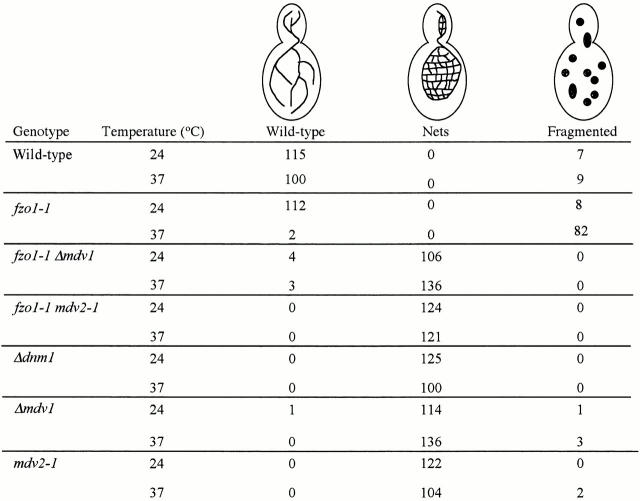
Given its genetic interaction with DNM1, we focused initially on the identification of the MDV1 gene. MDV1 was cloned by screening for yeast genomic library plasmids that would restore temperature-sensitive growth on glycerol to mdv1 fzo1-1 double mutant cells (i.e., fzo1-1 phenotype). We confirmed that our complementing gene, YJL112W, was MDV1 by replacing this ORF in the yeast chromosome with S. pombe his5 + and examining the linkage between histidine prototrophy and mdv phenotypes in tetrads from ΔYJL112W_mdv1_ diploids (4:0, n = 24 spores). Deletion of MDV1 caused mitochondria to form net-like structures in cells and blocked mitochondrial fragmentation in fzo1-1 cells, phenotypes identical to those observed in mdv1-1 cells (Fig. 3).
Figure 3.
Quantification of mitochondrial membrane morphology. Mitochondrial membrane was visualized with Mito-GFP.
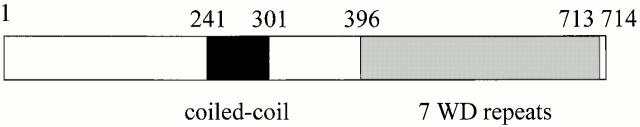
The MDV1 gene encodes a predicted ∼80-kD soluble cytosolic protein containing at least three distinct regions: a novel NH2-terminal region; a middle region predicted to form a coiled-coil structure; and a COOH-terminal region that contains seven WD repeats, predicted to form a circular seven-bladed propeller structure (Fig. 4; Lupas et al. 1991; Wall et al. 1995; Lambright et al. 1996; Sondek et al. 1996; Smith et al. 1999). The presence of the predicted coiled-coil and WD repeat domains indicates that Mdv1p functions in fission by mediating protein–protein interactions. Database searching identified proteins with similarity and identity to the coiled-coil and WD repeat regions of Mdv1p, but failed to identify any convincing Mdv1p structural homologues in higher eucaryotes outside of these regions (Altschul et al. 1997). Given that the mechanism of mitochondrial fission is conserved, it is likely that if no structural homologues exist, functional Mdv1p homologues will be present in other organisms.
Figure 4.
Mdv1p contains a novel NH2-terminal domain, a central coiled-coil domain, and a COOH-terminal WD repeat domain. The coiled-coil domain (amino acids 241–303) was identified as described (Lupas et al. 1991). The amino acid regions of the WD repeats are predicted to be the following: WD1 396–427, WD2 439–469, WD3 500–529, WD4 564–592, WD5 604–632, WD6 643–672, WD7 685–713.
Mdv1p Is Peripherally Associated with the Mitochondrial Outer Membrane
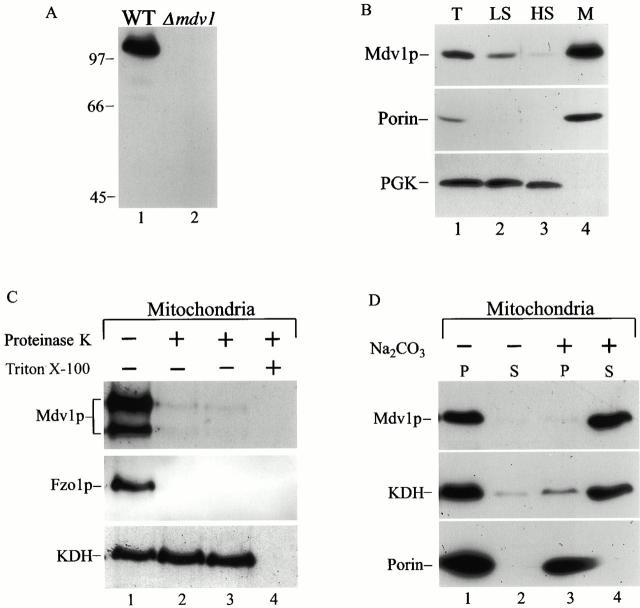
To detect Mdv1p and examine its subcellular localization, we raised polyclonal anti-Mdv1p antibodies. Western blot analysis of extracts made from wild-type cells with anti-Mdv1p antibodies detected a polypeptide migrating at ∼100 kD, an estimated molecular mass greater than that predicted from the primary amino acid sequence (Fig. 5 A, lane 1). In contrast, no species were detected by Western blotting in Δ_mdv1_ cells, indicating that anti-Mdv1p antibodies specifically recognize Mdv1p (Fig. 5 A, lane 2).
Figure 5.
Mdv1p is peripherally associated with the mitochondrial outer membrane. All samples were prepared and analyzed by SDS-PAGE and Western blot as described in Materials and Methods. A, Specificity of anti-Mdv1p polyclonal antibodies. Total extract was prepared from JSY1826 (wild-type, lane 1) and JNY552 (Δ_mdv1_, lane 2) cells and analyzed as described in Materials and Methods. B, Fractionation of total JSY1826 (wild-type) cell extract by differential centrifugation. T, Total extract (lane 1); LS, low-speed supernatant (lane 2); HS, high speed supernatant (lane 3); and M, high speed, mitochondrial-enriched pellet (lane 4). C, Topology of Mdv1p. Intact mitochondria were either mock-treated (lane 1), or treated with 50 μg/ml Proteinase-K (lane 2), 100 μg/ml Proteinase-K (lane 3), or 100 μg/ml Proteinase-K with 1% Triton-X 100 (lane 4) for 30 min on ice, and processed as described in Materials and Methods. D, Carbonate extraction of high speed mitochondrial-enriched pellet. Intact mitochondria were either mock-treated (lanes 1 and 2), or treated with 100 mM Na2CO3, pH 10.5 (lanes 3 and 4), subjected to ultracentrifugation at 100,000 g for 20 min, and fractionated into supernatant (S) and pellet (P) fractions. Multiple Mdv1p products observed by Western blotting were variable and likely the result of an in vitro proteolysis artifact.
To examine Mdv1p's subcellular localization, wild-type cell extracts were fractionated by differential centrifugation and analyzed by SDS-PAGE and Western blotting. Consistent with a predominantly mitochondrial localization, the majority of Mdv1p cofractionated with porin, the mitochondrial marker, in the mitochondrial-enriched pellet fraction, and not with the cytosolic marker, PGK (Fig. 5 B, lanes 3 and 4). We examined the submitochondrial localization and topology of Mdv1p by treating isolated mitochondria with exogenous proteases. In intact mitochondria, Mdv1p was accessible to Proteinase K, similar to the outer mitochondrial membrane protein, Fzo1p (Fig. 5 C, lanes 1–3). In contrast, the matrix marker protein, KDH, was protected from proteolysis, confirming that the mitochondrial outer and inner membranes were intact (Fig. 5 C, lanes 1–3). KDH became susceptible to proteolysis only after these membranes were solublized with detergent (Fig. 5 C, lane 4). To determine whether Mdv1p is an integral or peripheral outer membrane protein, we examined its sensitivity to sodium carbonate extraction. Mitochondria were extracted with 0.1 M Na2CO3 (pH 10.5) or mock-treated and fractionated into supernatant and pellet fractions by centrifugation (Fig. 5 D, lanes 1–4). As expected, the integral outer membrane protein, porin, was resistant to sodium carbonate extraction and was recovered in the pellet fraction (Fig. 5 D, compare lanes 3 and 4). In contrast, the majority of Mdv1p and the soluble matrix protein, KDH, were released into the supernatant upon sodium carbonate treatment, but not in control samples treated with buffer (Fig. 5 D, lanes 1–4). Based on this analysis, we conclude that the majority of Mdv1p is peripherally associated with the mitochondrial outer membrane in cells. Thus, Mdv1p, like Dnm1p, is a peripheral outer membrane component required for mitochondrial fission.
Mdv1p Localizes to Punctate Structures on Mitochondria
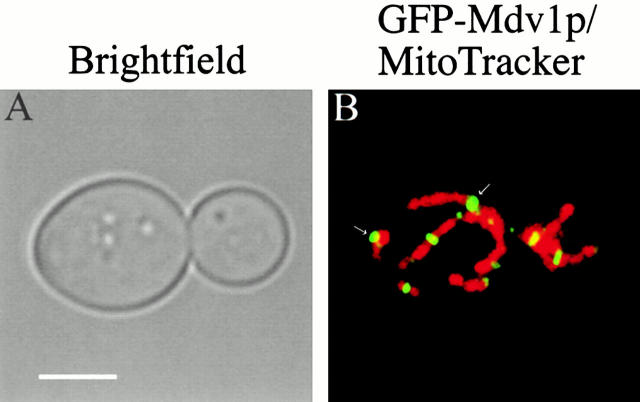
To localize Mdv1p in live yeast cells, we analyzed cells expressing an NH2-terminal GFP–Mdv1p fusion protein by fluorescence microscopy. We created a strain where the wild-type MDV1 locus was replaced with GFP–Mdv1p, whose expression is under the control of the regulated GAL1 promotor (JNY556). When JNY556 cells were grown under conditions where GFP–Mdv1p was not expressed (SRaf media), mitochondrial net-like structures were observed in cells, consistent with previous observations of Δ_mdv1_ cells (99%, n = 100). In contrast, when JNY556 cells expressed GFP–Mdv1p (SGal media), mitochondrial morphology was reticular and indistinguishable from wild-type cells (82%, n = 123), indicating that GFP–Mdv1p is functional. Interestingly, upon shifting JNY556 cells from SRaf to SGal media, mitochondrial net structures were transformed rapidly into wild-type reticular structures (average time, 30 min). This observation indicates that the block in fission in Δ_mdv1_ cells is reversible and suggests that a productive fission intermediate is present in these cells.
GFP–Mdv1p was observed in punctate structures within JNY556 cells (Fig. 6 B, in yellow). Visualization of mitochondria in these cells using the vital mitochondrial fluorescent probe, Mitotracker, indicated that these punctate structures were associated with mitochondrial membranes, consistent with biochemical analyses of Mdv1p's localization (Fig. 6 B, in red). Interestingly, the observed punctate labeling pattern for Mdv1p is similar to that reported for Dnm1p, suggesting that Mdv1p and Dnm1p may interact and colocalize in these structures in vivo.
Figure 6.
Mdv1p localizes to punctate structures on mitochondria. JNY556 (_GAL1_GFP-MDV1) cells were grown in SRaf to log phase and GFP–Mdv1p was expressed by shifting cells into SGal media and incubating at 30°C for 1 h. After expression of GFP–Mdv1p (in green/yellow, see arrows), mitochondria in JNY556 (_GAL1_GFP-MDV1) cells were labeled with MitoTracker (in red). Cells were visualized directly by confocal microscopy as described in Materials and Methods. Bar, 2 μm.
Mdv1p and Dnm1p Interact and Colocalize to Mitochondrially Associated Punctate Structures In Vivo
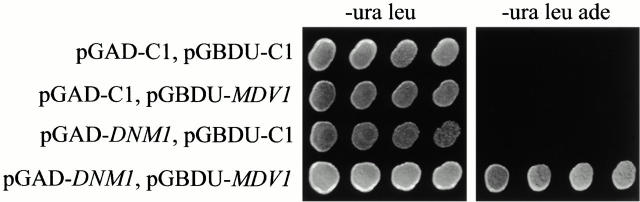
To test whether Mdv1p and Dnm1p interact, we used the established two-hybrid assay for protein–protein interactions (James et al. 1996). Interactions between AD fusion proteins and BD fusion proteins were assessed by monitoring the expression of the stringent GAL2-ADE2 reporter gene. Only cells expressing both AD-DNM1 and BD-MDV1 displayed robust growth on media lacking adenine, indicating that Mdv1p and Dnm1p strongly interact (Fig. 7). An interaction between Mdv1p and Dnm1p using a similar two-hybrid assay system also was reported in a high through-put two-hybrid screen of yeast gene products, confirming our observations (Uetz et al. 2000).
Figure 7.
MDV1 interacts with DNM1 by two-hybrid analysis. Interactions between activating domain (AD) and binding domain (BD) two-hybrid vectors were assessed by growth of indicated transformants on SD media lacking leucine (leu), uracil (ura), and adenine (ade), as described in Materials and Methods.
We also tested whether Mdv1p and Dnm1p interact in solubilized mitochondrial extracts by coimmunoprecipitation with both anti-Mdv1p and anti-Dnm1p antibodies. After extensive experimentation, we have failed to detect a Mdv1p/Dnm1p complex, suggesting that in vitro the interaction of Mdv1p with Dnm1p is labile (not shown).
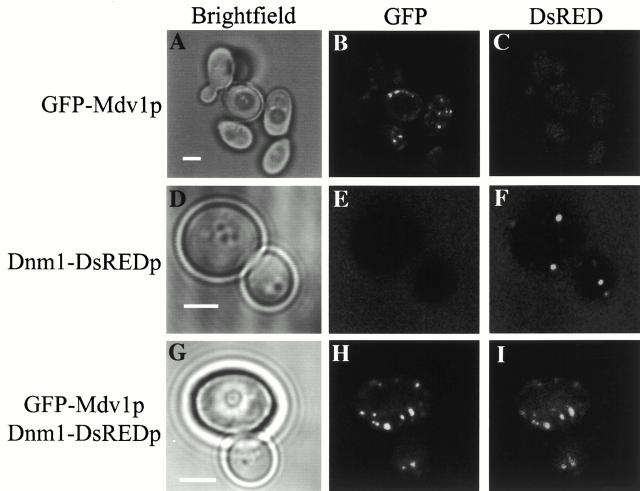
To test directly whether Mdv1p and Dnm1p colocalize in mitochondrially associated punctate structures, we examined localization patterns in cells simultaneously expressing differentially tagged versions of these proteins. To accomplish this, we constructed a yeast expression vector containing DsRed fused in frame at the COOH terminus to Dnm1p (Dnm1–DsRedp). DsRed is a fluorescent protein isolated from the IndoPacific sea anemone Discosoma that upon excitation emits light in the red spectral region (Matz et al. 1999). As expected, in cells expressing either GFP–Mdv1p or Dnm1–DsRedp, punctate structures were observed exclusively in images obtained from green or red emissions, respectively, indicating that DsRed and GFP spectra are resolvable by fluorescence microscopy under our experimental conditions (Fig. 8, A–C and D–F). In cells expressing both GFP–Mdv1p and Dnm1–DsRedp, both red and green fluorescence were observed in every punctate structure examined (Fig. 8, G–I). This observation indicates that Mdv1p and Dnm1p are both steady state components of these punctate structures and, together with data from genetic and two-hybrid analyses, further suggests that Mdv1p and Dnm1p interact directly in these structures. In addition, this observation indicates that the formation of these Mdv1p/Dnm1p structures is not a rate-limiting step in the process of fission.
Figure 8.
Mdv1p and Dnm1p colocalize in punctate structures in vivo. GFP–Mdv1p was expressed in JNY556 (_GAL1_GFP-MDV1) and JNY558 (_GAL1_GFP-MDV1, pRS315-_DNM1_-GFP) cells as described in Fig. 6. Cells expressing GFP–Mdv1p (A–C), Dnm1–DsREDp (D–F), or GFP–Mdv1p and Dnm1–DsREDp (G–I) were visualized directly by fluorescence confocal microscopy. Bars, 2 μm.
The Dnm1p Localization Pattern Is Unaffected in Δmdv1 Cells
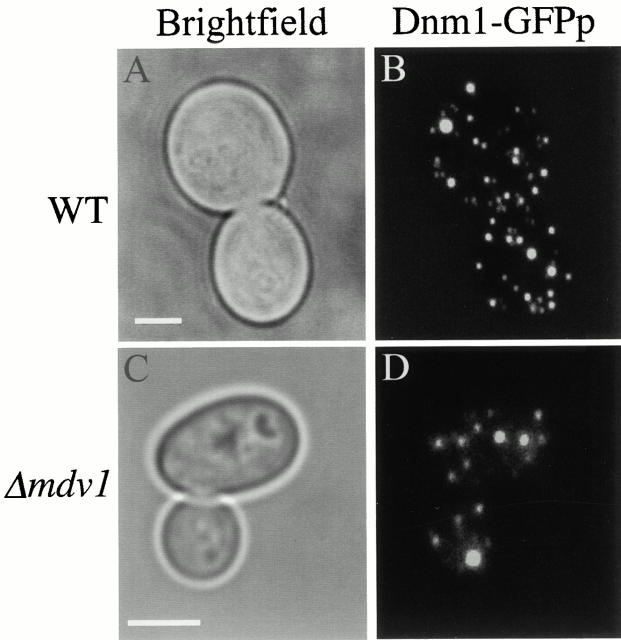
Given that Mdv1p and Dnm1p interact, we tested whether Mdv1p mediates the mitochondrial attachment and/or distribution of Dnm1p by comparing the localization pattern of Dnm1p in wild-type and Δ_mdv1_ cells. As expected, in wild-type cells, Dnm1–GFPp localizes to punctate structures on the mitochondrial membrane (Fig. 9 B; Otsuga et al. 1998; Sesaki and Jensen 1999). Interestingly, in Δ_mdv1_ cells, the pattern of Dnm1–GFPp localization remained punctate. Furthermore, the distribution and apparent size of these Dnm1p-containing structures on the mitochondria were indistinguishable from Dnm1p-containing punctate structures observed in wild-type cells (Fig. 9 compare B and D, 100%, n = 100). Thus, Dnm1p-containing punctate structures with cytologically wild-type characteristics can form in the absence of Mdv1p, but these structures are not able to complete the division of mitochondrial membranes. This observation suggests that Mdv1p is a relatively late recruit to Dnm1p-containing punctate structures, and thus may function during the process of fission to regulate a rate-limiting step such as membrane constriction and/or division via an interaction with another component.
Figure 9.
The assembly of Dnm1p into punctate structures does not required MDV1 function. JNY566 (wild-type, pRS315-_DNM1_-GFP; A and B) and JNY560 (Δ mdv1, pRS315-_DNM1_-GFP; C and D) cells were grown in YPG to log phase and Dnm1–GFPp was visualized directly by fluorescence confocal microscopy. Bars, 2 μm.
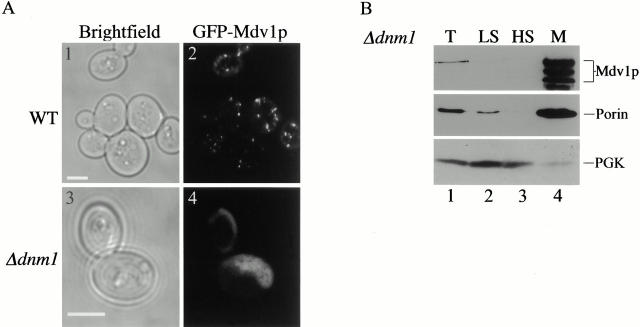
Mdv1p's Localization Pattern Is Altered in Δdnm1 Cells
We also examined whether Mdv1p's punctate localization pattern was dependent on Dnm1p by comparing the pattern of GFP–Mdv1p in wild-type and Δ_dnm1_ cells. In contrast to wild-type cells (Fig. 6 and Fig. 10 A, 2), GFP–Mdv1p in Δ_dnm1_ cells was dispersed uniformly on the mitochondrial membrane (Fig. 10 A, 4, 97%, n = 100). The mitochondrial localization of GFP–Mdv1p in Δ_dnm1_ cells in vivo was confirmed by double label experiments with MitoTracker (100%, n = 87). Consistent with this cytological observation, biochemical analysis indicates that the majority of Mdv1p cofractionated with the mitochondrial-enriched fraction derived from Δ_dnm1_ cell extracts by differential centrifugation (compare Fig. 5 B and 10 B, lane 4). These observations indicate that the localization of Mdv1p to punctate structures requires Dnm1p, consistent with our proposal that Mdv1p and Dnm1p interact in these structures to mediate the fission reaction. In addition, the ability of Mdv1p to localize to mitochondria in the absence of Dnm1p indicates that Mdv1p possesses a Dnm1p-independent mitochondrial targeting signal. This suggests that Mdv1p's localization to mitochondria is also dependent on one or more additional components/receptors on the mitochondrial membrane.
Figure 10.
Mdv1p localization pattern is altered in Δ_dnm1_ cells. A, GFP–Mdv1p was expressed in JNY556 (GAL1_GFP-MDV1) and JNY570 (GAL1_GFP-MDV1 Δ_dnm1) cells and visualized directly by fluorescence confocal microscopy as described in Fig. 6. B, Cell extract from JSY1371(Δ_dnm1; lanes 1–4) was fractionated and analyzed as described in Fig. 5. Bars, 2 μm.
MDV2 Regulates the Assembly of Dnm1p-containing Punctate Structures and Is Required for Mdv1p's Association with Mitochondrial Membranes in the Absence of Dnm1p
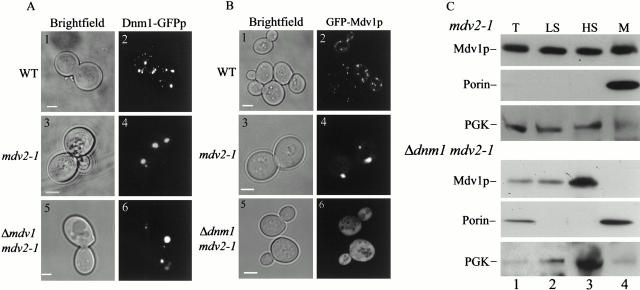
Given that MDV2 is also required for mitochondrial fission, we examined its role in Dnm1p's localization. Interestingly, the pattern of Dnm1–GFP labeling in mdv2-1 cells was altered as compared with the pattern observed in wild-type cells (Fig. 11 A, 4, 99% n = 173). Specifically, although Dnm1–GFP was still present in punctate structures within mdv2-1 cells, there were fewer (average of 2 foci, n = 126 in mdv2-1, compared with 22 foci in wild-type cells, n = 41) and the fluorescence intensity of a subset of these structures was significantly increased, suggesting that they contain a greater amount of Dnm1p and are either larger or that the organization of Dnm1p within these structures is altered.
Figure 11.
MDV2 regulates the assembly of Dnm1p-containing punctate structures and is required for Mdv1p's association with mitochondrial membranes in the absence of Dnm1p. A, Dnm1–GFPp in JSY1863 (wild-type, pRS315-_DNM1_-GFP; panel 2), JNY561 (mdv2-1, pRS315-DNM1_-GFP; panel 4), and JNY562 (Δ_mdv1 mdv2-1, pRS315-_DNM1_-GFP; panel 6) cells. B, GFP–Mdv1p in JNY 556 (_GAL1_GFP-MDV1; panel 2), JNY571 (GAL1_GFP-MDV1;mdv2-1; panel 4), and JNY572 (GAL1_GFP-MDV1;mdv2-1, Δ_dnm1; panel 6) cells. C, JNY567 (mdv2-1; top, lanes 1–5) and JNY569 (Δ_dnm1; bottom, lanes 1–4) cell extracts were fractionated and analyzed as described in Fig. 5.
The pattern of GFP–Mdv1p labeling in mdv2-1 cells also was altered as compared with wild-type cells (100%, n = 87). In mdv2-1 cells, GFP–Mdv1p was observed in punctate structures with similar characteristics to Dnm1–GFP-labeled structures and also was localized diffusely throughout mdv2-1 cells, indicating that a fraction of Mdv1p is present in the cytosol. Double label experiments in cells expressing both Dnm1–DsRedp and GFP–Mdv1p indicate that these proteins colocalized in the abnormal punctate structures in mdv2 cells, indicating that MDV2 is not required for the interaction between Dnm1p and Mdv1p within these structures (93%, n = 63). Thus, MDV2 is required for the normal assembly and/or distribution of Dnm1p/Mdv1p-containing punctate structures.
Analysis of cells expressing either Dnm1–GFP or GFP–Mdv1p in mdv2-1 cells in the presence of the mitochondrial membrane marker, MitoTracker, indicates that while the majority of abnormal Dnm1p/Mdv1p-containing structures were associated with mitochondrial membranes in vivo, a small fraction of these structures was not (9%, n = 54). In addition, a fraction of the abnormal Mdv1p/Dnm1p-containing structures associated with mitochondria in mdv2-1 cells possessed a brownian-type motion, in contrast to the Mdv1p/Dnm1p-containing punctate structures observed in wild-type cells, where this motion was not observed. These data indicate that the interaction of Mdv1p/Dnm1p structures with mitochondrial membranes is altered in mdv2-1 cells.
The altered nature of Mdv1p's interaction with mitochondrial membranes in mdv2-1 cells was further revealed by biochemical analyses. In contrast to wild-type or Δ_dnm1_ cell extracts, where the majority of Mdv1p was present in the mitochondrial-enriched fraction, Mdv1p in mdv2-1 cell extracts was observed in both the mitochondrial-enriched pellet and the postmitochondrial supernatant fraction (Fig. 11 B, lane 3). The postmitochondrial supernatant fraction contains the cytosol as indicated by the cofractionation of the cytosolic marker, PGK. These biochemical data are in agreement with our cytological data and suggest that a fraction of Mdv1p in mdv2 cells behaves as a soluble cytosolic protein. Taken together, these data suggest that MDV2 functions to target Mdv1p to the mitochondrial membrane, but that in the absence of MDV2 function, Mdv1p can still assemble with Dnm1p and associate with mitochondria, but with a lower efficiency.
Interestingly, in mdv1-1 mdv2-1 double mutant cells, abnormal Dnm1p-containing structures persisted and possessed the same characteristics as those observed for mdv2-1 single mutant cells. This observation indicates that the Dnm1p assembly defect observed in mdv2-1 cells is epistatic to the apparently wild-type Dnm1p localization pattern observed in mdv1-1 cells (99%, n = 100), and suggests that MDV2 functions upstream of MDV1 in the pathway of mitochondrial fission to mediate the correct assembly of fission-mediating Dnm1p/Mdv1p-containing punctate structures.
To test whether the dispersive mitochondrial localization of Mdv1p observed in the absence of Dnm1p requires Mdv2p, we examined the localization of GFP–Mdv1p in mdv2-1 Δ_dnm1_ double mutant cells. Interestingly, no GFP–Mdv1p-labeled punctate structures were observed (Fig. 11 A, 6). Instead, GFP–Mdv1p was diffusely distributed throughout these cells, indicating that Mdv1p is exclusively cytosolic in mdv2-1 Δ_dnm1_ cells (Fig. 11 A, 6, 100%, n = 198). Consistent with this cytological observation, differential centrifugation of mdv2-1 Δ_dnm1_ cell extracts indicates that the majority of Mdv1p cofractionated with the cytosol in the postmitochondrial supernatant (Fig. 11 B, lane 3). The apparent cytosolic localization of Mdv1p in mdv2-1 Δ_dnm1_ cells is in marked contrast to the dispersive mitochondrial localization of Mdv1p in Δ_dnm1_ cells and the abnormal punctate localization pattern of Mdv1p in mdv2-1 cells. These observations, taken together, indicate that Mdv1p interacts with mitochondria via two mechanisms: one dependent on its interaction with Dnm1p and one dependent on the function of the MDV2 gene product.
Based on our analyses of mdv2-1 cells, we postulate that Mdv2p plays multiple roles in the process of mitochondrial fission. Our data suggest that MDV2 functions early in the fission pathway to either directly or indirectly regulate the assembly of wild-type Dnm1p/Mdv1p containing punctate structures. Our data also suggest that Mdv2p functions to facilitate Mdv1p's association with the mitochondrial membrane in the absence of Dnm1p.
Discussion
Dnm1p-mediated Mitochondrial Fission Is a Multi-step Process Regulated by the Mdv1 and Mdv2 Genes
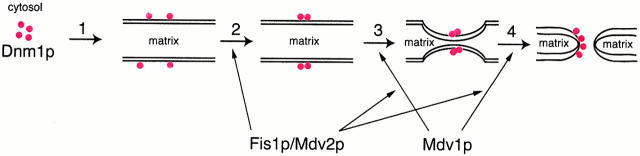
We have identified two genes, MDV1 and MDV2, that function with DNM1 to mediate mitochondrial fission. Our analysis of mdv1 and mdv2 cells, together with biochemical and ultrastructural analyses of the subcellular localization of Dnm1p, indicate that mitochondrial fission is a multi-step pathway, regulated by Mdv1p and Mdv2p, and similar in mechanism to dynamin-mediated endocytosis (Fig. 12; Otsuga et al. 1998; Schmid et al. 1998; Bleazard et al. 1999; Labrousse et al. 1999; Sesaki and Jensen 1999; Sever et al. 2000). These data support a model where Dnm1p is initially recruited from the cytosol to the mitochondrial outer membrane, where it subsequently assembles into punctate structures that ultimately mediate the division of mitochondrial membranes (Fig. 12, steps 1 and 2). Immunoelectron microscopy indicates that Dnm1p puncta are found at sites where the inner and outer mitochondrial membranes are coordinately constricted, and also on tubule tips that are presumably the end products of the division reaction (Bleazard et al. 1999). Because only a fraction of Dnm1p-containing structures are associated with constriction sites at steady state, it is probable that Dnm1p-mediated mitochondrial membrane constriction and/or mitochondrial membrane division are the rate-limiting steps in this pathway (Fig. 12, steps 3 and 4).
Figure 12.
Schematic model of the mitochondrial fission pathway. Mitochondrial tubules are diagramed in cross-section and red circles represent Dnm1p. Mitochondrial fission is initiated by the recruitment of Dnm1p from the cytosol to the mitochondrial membrane by an unknown regulatory mechanism (1). In a Fis1p-dependent regulated manner, mitochondrial-associated Dnm1p subsequently assembles into higher order structures that possibly form rings around the surface of the mitochondrial outer membrane (2). Via a direct interaction with Dnm1p, Mdv1p either coassembles with Dnm1p during the formation of these higher order structures or associates with Dnm1p in these structures after their formation. Mdv1p functions to recruit either Fis1p or a Fis1p-dependent component to the Mdv1p/Dnm1p structure, thereby catalyzing the potentially rate-limiting steps of membrane constriction (3) and membrane division (4).
Mdv1p Functions at a Late Step in the Fission Pathway by Interacting with Dnm1p
We have presented four independent lines of experimental evidence which suggest that Mdv1p regulates mitochondrial fission by directly interacting with Dnm1p. First, we have observed noncomplementation between mdv1 and dnm1 alleles. Second, two-hybrid analysis indicates that Mdv1p and Dnm1p strongly interact. Third, Mdv1p and Dnm1p colocalize in fission-mediating punctate structures associated with mitochondria. Finally, the localization of Mdv1p to these punctate structures requires Dnm1p function.
The specific role of the Mdv1p–Dnm1p interaction in mitochondrial fission can be inferred from the observations summarized above and from the analysis of mdv1 cells. In mdv1 cells, fission is blocked, but data indicate that a productive fission intermediate is assembled. This conclusion is supported by the observation that mitochondrial net structures in mdv1 cells, which form because of a fission block, are transformed rapidly into wild-type reticular networks when MDV1 is expressed from the regulated GAL1 promoter. Dnm1p in mdv1 cells is localized to punctate structures on mitochondrial membranes with characteristics similar to Dnm1p-containing structures in wild-type cells, however, these structures are not able to complete the fission reaction. These observations indicate that Mdv1p is not required for the recruitment of Dnm1p to the outer mitochondrial membrane or for its assembly into punctate structures (Fig. 12, steps 1 and 2). Rather, our data indicate that Mdv1p functions later in the fission pathway, by assembling with Dnm1p into punctate structures, and stimulating the rate-limiting steps of mitochondrial membrane constriction and/or mitochondrial division (Fig. 12, steps 3 and 4).
Mdv2p Possesses Two Separate Functions
During the course of our studies, we learned that Mozdy et al. 2000(this issue) also identified and cloned the MDV2 gene, which they term FIS1, for fission. FIS1 encodes an 18-kD protein that contains a single transmembrane spanning region at its COOH terminus (Mozdy et al. 2000). Fis1p is integrated into and localized uniformly on the mitochondrial outer membrane (Mozdy et al. 2000). Neither the targeting nor the integration of Fis1p to the outer membrane is dependent on Dnm1p or Mdv1p function (Mozdy et al. 2000). Biochemical studies of the topology of Fis1p in the outer membrane indicate that the NH2-terminal ∼13-kD region is cytoplasmic. The eight amino acids that are COOH-terminal to the transmembrane domain are predicted to be localized in the mitochondrial intermembrane space (Mozdy et al. 2000).
Based on our analysis, we have identified two separate functions for Mdv2p/Fis1p (referred to Fis1p hereafter). First, our data indicate that Fis1p functions to regulate the assembly of Dnm1p into punctate structures. Specifically, in the absence of Fis1p, Dnm1p can assemble into punctate structures, but these structures are abnormal and unable to mediate mitochondrial fission. Their characteristics indicate that they are larger, cells contain fewer of them, and their interaction with mitochondria is altered. However, because Dnm1p and Mdv1p associate within these structures, Fis1p is not required for the Dnm1p–Mdv1p interaction. In addition, the role of Fis1p in the assembly of puncta is not dependent on Mdv1p function, as Dnm1p-containing structures in mdv1 fis1 double mutants are morphologically identical to those observed in fis1 cells, indicating that this fis1 phenotype is epistatic to mdv1. This epistasis suggests that Fis1p's function in the assembly of Dnm1p puncta might occur before Mdv1p's function in the fission pathway (Fig. 12, step 2). Thus, one function of Fis1p in fission is to regulate the assembly and distribution of Dnm1p-containing punctate structures on mitochondria.
Secondly, we have shown that Fis1p is required for the interaction of Mdv1p with mitochondria in Δ_dnm1_ cells. In cells lacking Dnm1p, Mdv1p localizes uniformly on mitochondria. However, in cells lacking both Dnm1p and Fis1p, Mdv1p is found exclusively in the cytosol. These observations suggest that Fis1p also functions relatively early in the fission process, as a Mdv1p receptor to target Mdv1p to the mitochondrial membrane so that it can efficiently interact and assemble with Dnm1p (Fig. 12). This idea is consistent with our observations of cells lacking Fis1p, where Mdv1p's interaction with Dnm1p and mitochondria appears weakened. However, an alternative possibility is that the Fis1p-dependent association of Mdv1p with mitochondria in Δ_dnm1_ cells reflects a transient interaction that occurs between Fis1p and Mdv1p later in the fission pathway, within the context of Dnm1p-containing punctate structures. From this perspective, Fis1p might function relatively late in fission via a direct Mdv1p–Fis1p interaction or indirectly by facilitating an interaction between Mdv1p and another unidentified component to regulate membrane division (Fig. 12, steps 3 and 4). In either case, it is interesting to note that the uniform mitochondrial localization pattern observed for Fis1p is identical to that observed for Mdv1p in Δ_dnm1_ cells, consistent with a Fis1p (or Fis1p-containing complex)/Mdv1p interaction.
The two functions we observe for Fis1p might occur sequentially in the mitochondrial fission pathway. In this case, Fis1p (or a Fis1p-containing complex) might facilitate and/or stabilize the assembly and distribution of Dnm1p complexes and subsequently function as a switch that regulates the transition of assembled Dnm1p complexes to those that can be activated in an Mdv1p-dependent manner to complete division.
Proposed Molecular Interactions during Mitochondrial Fission
Using endocytosis as a paradigm, our data suggest that mitochondrial fission will be controlled by Dnm1p's GTPase cycle that in turn will be regulated by protein–protein interactions, specifically involving Mdv1p and Fis1p (Schmid et al. 1998). Although the exact nature of the molecular interactions between Dnm1p, Mdv1p, and Fis1p is unknown, any molecular model of mitochondrial fission must include our observations indicating two separate functions for Fis1p. Thus, if Fis1p is sufficient to mediate both the assembly of Dnm1p-containing structures and the targeting of Mdv1p to mitochondria, separate Fis1p functional domains might exist that interact directly or indirectly with both Mdv1p and Dnm1p. However, it is likely that additional components will be required for Dnm1p-mediated mitochondrial fission. Indeed, we have recently identified a fourth gene required for mitochondrial fission, MDV3. Future studies will determine the exact function of Mdv3p in the pathway of mitochondrial fission (Tieu, Q., S. Meeusen, and J. Nunnari, unpublished data).
Our proposed molecular role of Mdv1p in mitochondrial fission is based not only on our experimental data, but also on Mdv1p's structure. Mdv1p contains sequence motifs predicted to form two protein interaction domains, a middle coiled-coil domain and a COOH-terminal WD repeat domain, and a 27-kD NH2-terminal extension of unknown structure. The WD domain is predicted to form a seven-bladed β-propeller structure that alone has several surfaces capable of protein interaction (Wall et al. 1995; Lambright et al. 1996; Sondek et al. 1996; Smith et al. 1999). Thus, Mdv1p potentially can form reversible complexes with several proteins and coordinate sequential and/or simultaneous interactions among these proteins. We have identified Dnm1p as one Mdv1p-interacting protein. Thus, we propose that Mdv1p coassembles with Dnm1p into ring-like structures (visualized by fluorescence microscopy as puncta), similar to the dynamin-containing spirals observed on tubules associated with clathrin-coated pits (Takei et al. 1995). Within the context of these structures, our data suggest that Mdv1p functions late in the fission pathway as a molecular matchmaker that targets additional components to the fission site to catalyze the completion of membrane division. Our observations indicate that one of these components Mdv1p targets to the Dnm1p ring complex is Fis1p or Fis1p-dependent. Our preliminary data indicate that Mdv1p's novel NH2-terminal extension mediates its interaction with Dnm1p (Tieu, Q., and J. Nunnari, unpublished observations). Thus, consistent with our model of Mdv1p, the coiled-coil and WD repeat domains likely recruit additional components, such as Fis1p, to the fission site to catalyze the completion membrane division.
Although our data suggest that Mdv1p is required to complete membrane division during mitochondrial fission, the exact mechanism of the Dnm1p-mediated membrane division event is unknown. It is possible that Dnm1p acts as a mechanochemical enzyme, using the energy derived from GTP hydrolysis within an assembled Dnm1p ring structure to pinch mitochondrial membranes (Hinshaw and Schmid 1995; Sweitzer and Hinshaw 1998). Recent data, however, suggests that dynamin functions during endocytosis as a regulatory GTPase, where the GTP-bound active form recruits additional components that help catalyze the membrane scission event (Sever et al. 1999, Sever et al. 2000). In this context, Dnm1p in its GTP-bound form might also recruit components, perhaps via Mdv1p, that are responsible for membrane division. Recruited components may possess lipid modification activities, such as lysophosphatidic acid acyl transferase activity, shown to be associated with synaptic vesicle and Golgi fission events (Schmidt et al. 1999; Weigert et al. 1999). The development of in vitro assays for mitochondrial fission and the identification and characterization of additional components will ultimately shed light on the exact molecular mechanism of mitochondrial division.
Finally, coordinating the fission of two membranes in mitochondria and chloroplasts is essential for the preservation of the double-membraned nature of these organelles and presents an added topological complexity. In this report, we have shown that mitochondrial fission requires the activity of outer membrane associated components, namely, Dnm1p, Mdv1p, and Fis1p. However, fission in chloroplasts requires the action of two homologues of FtsZ, the bacterial cell division GTPase, termed FtsZ1 and FtsZ2, that are localized to outer membrane and inner membrane, respectively (Osteryoung and Vierling 1995; Osteryoung et al. 1998). Localizing fission machinery to each membrane may serve as a mechanism to coordinate the fission of those membranes. These observations raise the question of whether components on the mitochondrial inner membrane also are required for or regulate mitochondrial division. Indeed, another yeast dynamin-related GTPase, Mgm1p, has been shown to play a role in the maintenance of mitochondrial structure in cells (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999). We have recently shown that Mgm1p is localized to the intermembrane space and is associated with the mitochondrial inner membrane (Wong et al. 2000). Based on this localization and topology, and our phenotypic analysis of mgm1 cells, we postulate that Mgm1p functions in inner membrane remodeling and fission events (Wong et al. 2000). Our characterizations of outer membrane components required for mitochondrial fission leads us to believe that additional inner membrane components, similar in function to Mdv1p and Fis1p, will also be required for Mgm1p-mediated inner membrane remodeling events. Ultimately, the identification and characterization of these components will allow us to understand how the inner and outer mitochondrial membrane fission machineries are coordinated.
Acknowledgments
We are grateful to A. Mozdy (University of Utah), J.M. McCaffrey (Johns Hopkins University), and J. Shaw (University of Utah) for communicating their results before publication. We are also grateful to Voytek Okreglak, Edith Wong, and Jenny Wagner for their help in affinity purification of anti-Mdv1p antibodies, two-hybrid analysis, and quantification of mdv phenotypes. We thank Dave Wilson (UC Davis) for sharing his knowledge of WD repeat proteins, C. Koehler (UCLA) for antibodies, H. Sesaki and R. Jensen (Johns Hopkins University) for the DNM1-GFP plasmid, and the Nunnari lab for their helpful comments on the manuscript and for discussions.
This work was supported by the National Science Foundation Grant (MCB-9724143) to J. Nunnari.
Footnotes
Abbreviations used in this paper: AD, activation domain; BD, binding domain; MBP, maltose-binding protein; ORF, open reading frame.
References
- Altschul S.F., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.W., Baum P.R., Gesteland R.F. Processing of adenovirus-2 induced proteins. J. Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. The dynamin-related GTPases, Dnm1, regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziepluch C., Kordes E., Pujol A., Jauniax J.-C. Sequencing analysis of a 40.2 kb fragment of yeast chromosome X reveals 19 open reading frames including URA2 (5′ end), TRK1, PBS2, SPT10, GCD14, RPE1, PH086, NCA3, ASF1, CCT7, CZF3, two tRNA genes, three remnant delta elements and a Ty4 transposon. Yeast. 1996;12:1471–1474. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1471::AID-YEA30%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Transforming yeast with DNA. Methods Mol. Cell. Biol. 1994;5:255–269. [Google Scholar]
- Guan K., Farh L., Marshall T.K., Deschenes R.J. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genetics. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. Guide to Yeast Genetics and Molecular Biology. Academic Press; San Diego, CA: 1991. [Google Scholar]
- Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual 1998. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: pp. 285-316 [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–374. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E., Schmid S.L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hoffman H.-P., Avers C.J. Mitochondrion of yeastultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.E. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.V., Walsh M.L., Chen L.B. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.A., Fangman W.L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lambright D.G., Sondek J., Bohm A., Skiba N.P., Hamm H.E., Sigler P.B. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–325. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Longtine M., McKenzie A., Demarini D., Shah N., Wach A., Brachat A., Philippsen P., Pringle J. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotech. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., Yates J.R., Nunnari J. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Fox T.D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2005. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W., Straight A., Murray A., Sedat J.W., Walter P. Mitochondrial transmission during mating in S. cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mtDNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–476. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thantcher J.W., Hermann G.J., Bleazard W., Shaw J. The dynamin GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae . J. Biol.Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Broach J.R. Cloning genes by complementation in yeast. In: Guthrie C., Fink G.R., editors. Methods in Enzymology. Vol. 194. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc; San Diego, CA: 1991. pp. 195–230. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Novick P., Thomas J.H., Botstein D., Fink G.R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–244. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Schmid S.L., McNiven M.A., De Camilli P. Dynamin and its partnersa progress report. Curr. Opin. Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A.V., Witke W., Huttner W.B., Soeling H.-D. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 1999;401:133–149. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. Division versus fusionDnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S., Muhlberg A.B., Schmid S.L. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Sever S., Damke H., Schmid S.L. Garrotes, springs, ratchets, and whipsputting dynamin models to the test. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- Shepard K., Yaffe M. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–719. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Shurland D., Ryazantsev S., Van Der Bliek A. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. The WD repeata common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm A., Lambright D.G., Hamm H.E., Sigler P.B. Crystal structure of a G-A protein beta-gamma dimer at 2.1Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Stevens B.J., White J.G. Computer reconstruction of mitochondria from yeast. Methods Enzymol. 1979;56:718–728. doi: 10.1016/0076-6879(79)56064-9. [DOI] [PubMed] [Google Scholar]
- Strathern J.N., Higgins D.R. Recovery of plasmids from yeast into Escherichia coli shuttle vectors. In: Guthrie C., Fink G.R., editors. Methods in Enzymology, Vol. 194. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc; San Diego, CA: 1991. pp. 319–329. [DOI] [PubMed] [Google Scholar]
- Sweitzer S.M., Hinshaw J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K., McPherson P.S., Schmid S.L., Camilli P.D. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma-S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T., Judson R., Knight J., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:601–613. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wall M.A., Coleman D.E., Lee E., Iniquez-Lluhi J.A., Posner B.A., Gilman A.G., Sprang S.R. The structure of the G protein heterotrimer Gi-alpha-1-beta-1-gamma-2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Weigert R., Silletta M.G., Spano S., Turacchio G., Cericola C., Colanzi A., Senatore S., Mancini R., Polishchuk E.V., Salmona M. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Gorsich S.W., McCaffery J.M., Shaw J.M., Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]