Dnm1p Gtpase-Mediated Mitochondrial Fission Is a Multi-Step Process Requiring the Novel Integral Membrane Component Fis1p (original) (raw)
Abstract
Yeast Dnm1p is a soluble, dynamin-related GTPase that assembles on the outer mitochondrial membrane at sites where organelle division occurs. Although these Dnm1p-containing complexes are thought to trigger constriction and fission, little is known about their composition and assembly, and molecules required for their membrane recruitment have not been isolated. Using a genetic approach, we identified two new genes in the fission pathway, FIS1 and FIS2. FIS1 encodes a novel, outer mitochondrial membrane protein with its amino terminus exposed to the cytoplasm. Fis1p is the first integral membrane protein shown to participate in a eukaryotic membrane fission event. In a related study (Tieu, Q., and J. Nunnari. 2000. J. Cell Biol. 151:353–365), it was shown that the FIS2 gene product (called Mdv1p) colocalizes with Dnm1p on mitochondria. Genetic and morphological evidence indicate that Fis1p, but not Mdv1p, function is required for the proper assembly and distribution of Dnm1p-containing fission complexes on mitochondrial tubules. We propose that mitochondrial fission in yeast is a multi-step process, and that membrane-bound Fis1p is required for the proper assembly, membrane distribution, and function of Dnm1p-containing complexes during fission.
Keywords: Fis1p, Dnm1p GTPase, mitochondria, fission, fusion
Introduction
Maintenance of the tubular mitochondrial reticulum in budding yeast requires opposing fission and fusion events that regulate organelle copy number and morphology (Hoffman and Avers 1973; Stevens 1977; Nunnari et al. 1997). Time-lapse studies showed that these mitochondrial fusion and fission events occur at equal rates, with one event taking place on average every 2 min (Nunnari et al. 1997). The fusion reaction always occurs between the tips of two mitochondrial tubules or between a tip and the side of another mitochondrial tubule. In contrast, mitochondrial fission appears to initiate within a tubule, creating new mitochondrial tips.
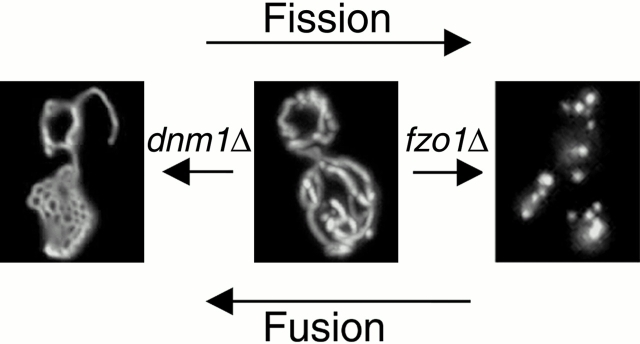
In Saccharomyces cerevisiae, two evolutionarily conserved GTPases act on the outer mitochondrial membrane to regulate opposing fission and fusion reactions (Fig. 1) (Hales and Fuller 1997; Hermann and Shaw 1998; Otsuga et al. 1998; Bleazard et al. 1999; Labrousse et al. 1999; Sesaki and Jensen 1999). Fission is regulated by the dynamin-related GTPase Dnm1p, which assembles on mitochondrial tubules at sites of future division (Otsuga et al. 1998; Bleazard et al. 1999; Sesaki and Jensen 1999). dnm1 mutations block mitochondrial fission and lead to formation of interconnected nets due to ongoing tip fusion (Fig. 1, left). This net formation does not affect mitochondrial function, mitochondrial DNA (mtDNA) maintenance, or mitochondrial transport into buds during division (Otsuga et al. 1998; Bleazard et al. 1999). Conversely, fusion is regulated by the Fzo1p transmembrane GTPase (Hermann et al. 1998; Rapaport et al. 1998). In fzo1 mutant strains, mitochondrial fusion is blocked and tubules fragment due to unopposed mitochondrial fission (Fig. 1, right). As a secondary consequence of this fragmentation, mtDNA is lost and the resulting respiratory-deficient fzo1 cells fail to grow on the nonfermentable carbon source glycerol (Hermann et al. 1998).
Figure 1.
Dnm1p and Fzo1p act in opposing fission and fusion pathways to maintain the yeast mitochondrial network. Ongoing fission and fusion events maintain a tubular mitochondrial network in wild-type yeast cells (middle). Loss of the Dnm1p GTPase (_dnm1_Δ) leads to net formation due to unopposed mitochondrial tip fusion (left); loss of the Fzo1p GTPase (_fzo1_Δ) leads to fragmentation due to unopposed mitochondrial fission (right).
In a previous study (Bleazard et al. 1999), we showed that dnm1 mutations prevent mitochondrial fission, fragmentation, and mtDNA loss at 37°C in a conditional fzo1-1 mutant. To identify new genes required for fission, we selected for mutations that, like dnm1, blocked mitochondrial fragmentation and restored glycerol growth in fzo1-1. Using this approach, we identified two new genes in the fission pathway, called FIS1 and FIS2 (FIS, fission). Here we show that FIS1 encodes an outer mitochondrial membrane protein required for the proper assembly and function of Dnm1p-containing fission complexes. In a related study, it was reported that the FIS2 gene product (named Mdv1p for mitochondrial division) forms a complex with Dnm1p on the outer mitochondrial membrane (Tieu and Nunnari 2000). We propose that mitochondrial fission in yeast is a multi-step process, and that membrane-bound Fis1p is directly involved in the assembly, membrane distribution, and function of Dnm1p-containing complexes during fission. Fis1p defines a novel protein family in eukaryotes and is the first integral membrane protein known to play a role in a membrane fission event regulated by a dynamin-like GTPase.
Materials and Methods
Strain and Plasmid Construction
Table lists the strains used in this study. All strains are derivatives of the FY10 strain (Winston et al. 1995). Standard genetic methods were used to grow, transform, and manipulate yeast (Sherman et al. 1986; Guthrie and Fink 1991) and bacterial (Maniatis et al. 1982) strains. All mutations, disruptions, tag insertions, and replacements were confirmed by PCR, DNA sequencing, and, where appropriate, Western blotting. The sequences of primers used in this study are available upon request.
Table 1.
Yeast Strains Used in this Study
| Strain | Genotype |
|---|---|
| ADM33 | _MATa ura3-52 leu2_Δ_1 trp_1Δ63 his3_Δ_200 DNM1 fzo1::HIS3 FIS1 fis2-5 with pRS416 + FZO1 |
| ADM176 | _MAT_α _ura_3_-52 LEU2 trp_1Δ63 his3_Δ_200 DNM1 fzo1-1 FIS1 fis2-5 |
| ADM179 | MATa ura3-52 LEU2 trp1_Δ_63 his3_Δ_200 DNM1 fzo1-1 FIS1 fis2-5 |
| ADM203 | MATa ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 DNM1 fzo1-1 fis1-21 FIS2 |
| ADM378 | MATa ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 dnm1::HIS3 fzo1-1 FIS1 FIS2 |
| ADM379 | _MAT_α ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 dnm1::HIS3 FZO1 FIS1 FIS2 |
| ADM415 | MATa ura3-52 TRP1 his3_Δ_200 dnm1::HIS3 FZO1 FIS1 FIS2 |
| ADM416 | _MAT_α ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 dnm1::HIS3 fzo1-1 FIS1 FIS2 |
| ADM547 | _MAT_α ura_3_-52 leu2 Δ 1 TRP1 his3 Δ 200 DNM1 fzo1-1 FIS1 FIS2 |
| ADM548 | MATa ura3-52 LEU2 trp1 Δ 63 his3 Δ 200 DNM1 FZO1 FIS1 FIS2 |
| ADM549 | _MAT_α ura3-52 leu2 Δ 1 TRP1 his3 Δ 200 DNM1 FZO1 fis1::HIS3 FIS2 |
| ADM550 | MATa ura3-52 LEU2 trp1 Δ 63 his3 Δ 200 DNM1 fzo1-1 fis1::HIS3 FIS2 |
| ADM551 | MATa ura3-52 leu2_Δ_1 trp1_Δ_63 his3_Δ_200 DNM1 FZO1 FIS1 FIS2 |
| ADM552 | MATa ura3-52 leu2_Δ_1 trp1_Δ_63 his3_Δ_200 DNM1 FZO1 fis1::HIS3 FIS2 |
| ADM554 | MATα ura3-52 LEU2 TRP1 his3_Δ_200 DNM1 fzo1-1 fis1::HIS3 FIS2 |
| ADM561 | MATα ura3-52 LEU2 trp1_Δ_63 his3_Δ_200 DNM1 FZO1 FIS1 FIS2 |
| ADM562 | MATa ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 DNM1 fzo1-1 FIS1 FIS2 |
| ADM574 | MATα ura3-52 leu2_Δ_1 trp1_Δ_63 his3_Δ_200 lys2_Δ_-202 DNM1 fzo1::HIS3 fis1::HIS3 FIS2 MGM1 |
| ADM575 | MATa ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 LYS2 DNM1 fzo1::HIS3 FIS1 FIS2 MGM1 |
| ADM742 | MATa ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 DNM1 FZO1 fis1::HIS3 fis2-5 |
| ADM750 | MATα ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 DNM1 fzo1-1 FIS1 fis2-5 |
| ADM752 | MATα ura3-52 LEU2 TRP1 his3_Δ_200 DNM1 FZO1 FIS1 fis2-5 |
| ADM763 | MATα ura3-52 leu2_Δ_1 TRP1 his3_Δ_200 DNM1 FZO1 fis1::HIS3 FIS2 mgm1::HIS3 |
| ADM764 | MATa ura3-52 leu2_Δ_1 trp1_Δ_63 his3_Δ_200 DNM1 FZO1 FIS1 FIS2 mgm1::HIS3 |
For pRS415-MET25 + 9xMYC-FIS1 (Myc-Fis1p), a FIS1 PCR fragment flanked by HindIII/XhoI sites was cloned into pRS415-MET25 +9xMYC (J. Thatcher and J.M. Shaw, manuscript in preparation). For pRS415-MET25 + rsGFP-FIS1aa1-155 (GFP-Fis1p), an XbaI/HindIII digest was first used to remove 9xMYC from the pRS415-MET25 + 9xMYC-FIS1 plasmid described above. The 9xMyc was then replaced by a red-shifted green fluorescent protein (GFP) variant (rsGFP) PCR amplified from pQBI25-fc1 (Quantum Biotechnologies Inc.) and flanked by XbaI/HindIII sites. An ATG initiation codon was engineered into the 5′ primer used to amplify rsGFP. For pRS416 + FIS1, a FIS1 PCR fragment flanked by XhoI sites was cloned into pRS416. For pGEX-4T-3 + FIS1 aa-1-127, a PCR fragment encoding amino acids 1–127 of FIS1 and flanked by BamHI/XhoI sites was cloned into pGEX-4T-3 (Amersham Pharmacia Biotech) to generate an in-frame GST-Fis1p fusion. For pRS414 + _DNM1_-rsGFP and pRS416 + _DNM1_-rsGFP, first a _DNM1_-containing PCR fragment flanked by SacII/SpeI sites was cloned into the SacII/NheI sites of pQBI25 (Quantum Biotechnologies Inc.), creating pQBI25 + _DNM1_-rsGFP. Second, an XhoI/BamHI fragment containing the _DNM1_-rsGFP sequence was cloned into pRS414 and pRS416. For pRS415-MET25 + rsGFP-FIS1aa1-127, a HindIII/XhoI digest was used to remove FIS1aa1-155 from pRS415-MET25 + rsGFP-FIS1aa1-155, which was then replaced by a HindIII/XhoI fragment containing FIS1aa1-127. For pRS416-GAL1 + _PrF0ATP9_-RFP (mito-RFP), a BamHI/XhoI fragment containing RFP (the red fluorescent protein from the pDsRed vector; CLONTECH Laboratories, Inc.) was cloned into pRS416-GAL1 + PrF0ATP9 (J. Thatcher and J.M. Shaw, manuscript in preparation). For pRS424-ADH1 + _PrF0ATP9_-RFP (mito-RFP), a SpeI/XhoI fragment containing _PrF0ATP9_-RFP from pRS416-GAL1 + _PrF0ATP9_-RFP was cloned into pRS424-ADH1.
GFP-Fis1p and Myc-Fis1p were expressed from CEN plasmids under control of the MET25 promoter. In the presence of methionine, this promoter is leaky and GFP-Fis1p and Myc-Fis1p are expressed at levels comparable with that of endogenous Fis1p in wild-type cells (verified by quantitative Western blotting). Under these conditions, GFP-Fis1p and Myc-Fis1p rescued the mitochondrial morphology defect in the _fis1_Δ mutant (83 and 55% wild type, respectively).
Identification of fis1 and fis2 and Cloning and Disruption of FIS1
14 spontaneous, second-site suppressors of the fzo1-1 temperature-sensitive (37°C) glycerol growth defect were isolated in JSY2788 (_MATa ura3-52 his3_Δ200 leu2Δ1 trp1Δ63 fzo1::HIS3 + pRS414-fzo1-1). Standard genetic methods were used to show that suppression in each case was due to a single recessive mutation. Complementation and meiotic segregation analyses revealed that the 14 fzo1-1 suppressors defined three linkage groups. Seven suppressors comprising one linkage group failed to complement the _dnm1_Δ mutation in diploid cells and a _DNM1_-containing plasmid reversed the fzo1-1 suppression phenotype of the mutants in this group. Sequence analysis indicated that these suppressors contained mutant dnm1 alleles. The remaining seven suppressor strains fell into two linkage groups (later named fis1 and fis2).
FIS1 was cloned by complementation of a fis1 allele (restoration of the fzo1-1 no-growth-on-glycerol-at-37°C phenotype) using a yeast genomic library in YEP213 (Lagosky et al. 1987). Standard procedures were used to identify the rescuing open reading frame as YIL065C in the Saccharomyces Genome Database, which we named FIS1. The predicted Fis1 protein sequence was compared with all available databases using the BLAST and Prosite programs.
A fis1::HIS3 disruption that precisely replaced the FIS1 coding region was generated by gene replacement in a diploid strain as described (Baudin et al. 1993). After sporulation and dissection to obtain a haploid fis1::HIS3 strain, integrative mapping studies confirmed that the original fis1 mutations were linked to _fis1_Δ::HIS3. Sequence analysis indicated that the fis1 suppressors contained mutations in FIS1.
Quantification of Mitochondrial Morphology and Fusion Phenotypes
Mitochondrial morphology was scored in wild-type and mutant cells expressing a mitochondrial-targeted form of GFP (mito-GFP) either from the plasmid pVT100UGFP (provided by B. Westermann and W. Neupert, Universitaet Muenchen, Muenchen, Germany) or the plasmid pRS416 + preCox4-GFP (Otsuga et al. 1998). In some cases, mitochondria were labeled with a matrix-targeted form of the red fluorescent protein, mito-RFP (A.D. Mozdy and J.M. Shaw) or MitoTracker Red CMXRos (Molecular Probes, Inc.). Growth conditions were essentially as described (Bleazard et al. 1999). DAPI (4′,6-diamidino-2-phenylindole) staining was used to assess the presence/absence of mtDNA (Pringle et al. 1991). Mitochondrial fusion was examined essentially as described previously (Nunnari et al. 1997), except that mito-RFP was used in place of the vital dye MitoTracker Red CMXRos (Molecular Probes, Inc.). Digital microscopic images of cells were acquired using a Axioplan microscope or a Confocal microscope (Carl Zeiss, Inc.), as described previously (Hermann et al. 1998; Otsuga et al. 1998).
Subcellular Localization of Fis1p and Dnm1p-GFP
To generate polyclonal serum specific for Fis1p, the soluble GST-Fis1paa1-127 fusion protein was expressed in Escherichia coli BL21-(DE3) cells from pGEX-4T-3 + FIS1 aa-1-127 and batch purified on Glutathione Sepharose 4B beads (Amersham Pharmacia Biotech). After separation by SDS-PAGE, the fusion protein was excised and used to immunize rabbits (Covance Research Products, Inc.).
For the protease protection experiments, a _fis1_Δ strain (ADM549) expressing GFP-Fis1p was grown in S-galactose medium lacking leucine to select for the pRS415-MET25 + rsGFP-FIS1 plasmid. Differential sedimentation experiments and protease protection experiments were performed using either wild-type cells (ADM548) or the ADM549 strain, as described previously (Hermann et al. 1998).
To localize Dnm1p in wild-type and mutant strains, Dnm1p-rsGFP expressed from either pRS414 + _DNM1_-rsGFP, pRS416 + _DNM1_-rsGFP, or pHS20 (Sesaki and Jensen 1999) was localized in strains expressing mito-RFP from pRS416-GAL1 + _PrF0ATP9_-RFP to visualize the mitochondrial membranes.
Immunoelectron microscopy was performed as described by Rieder et al. 1996 and Bleazard et al. 1999.
Results
Mutations in FIS1 and FIS2 Suppress mtDNA Loss and Glycerol Growth Defects in fzo1-1
To identify yeast genes required for mitochondrial fission, we selected spontaneous, second-site suppressor mutations that restored glycerol growth at 37°C to fzo1-1 cells. The 14 suppressor strains isolated were recessive in heterozygous diploids and segregated 2:2 in backcrosses with the fzo1-1 mutant. Complementation analysis revealed that seven of these strains represented new alleles of the previously characterized DNM1 gene (Gammie et al. 1995; Otsuga et al. 1998; Bleazard et al. 1999). The remaining suppressors defined two new genes, initially designated SFZ1 (four sfz1 alleles) and SFZ2 (three sfz2 alleles), for suppressor of fzo1-1. As described below, several lines of evidence indicated that SFZ1 and SFZ2 act together with DNM1 to regulate mitochondrial fission in yeast. Accordingly, we named them FIS1 and FIS2, respectively.
Like dnm1 mutations, fis1 and fis2 mutations rescued the fzo1-1 temperature-sensitive mtDNA loss and glycerol growth defects. Mitochondrial networks in wild-type cells retained their mtDNA nucleoids (DAPI staining, not shown) and these cells grew well on medium containing the nonfermentable carbon source glycerol at both 25° and 37°C (Fig. 2 A, WT). In contrast, the fzo1-1 strain failed to grow on glycerol at 37°C where the mitochondrial reticulum fragments and mtDNA is lost (Fig. 2 A, fzo1-1) (Hermann et al. 1998). As reported previously, this mtDNA loss was prevented when mitochondrial fission and fragmentation were blocked by introducing a _dnm1_Δ mutation into fzo1-1 (Fig. 2 A, _dnm1_Δ fzo1-1) or _fzo1_Δ cells (Bleazard et al. 1999). Although the single fis1 and fis2 mutations did not affect glycerol growth in an otherwise wild-type strain (Fig. 2 A, _fis1_Δ, _fis2_-5, both mutations suppressed the temperature-sensitive glycerol growth defect (Fig. 2 A, _fis1_Δ fzo1-1, fis2-5 fzo1-1) and mtDNA loss defect (not shown) of fzo1-1. (Note that fis2-5 is truncated after 191 of 714 amino acids and behaves like a null allele.)
Figure 2.
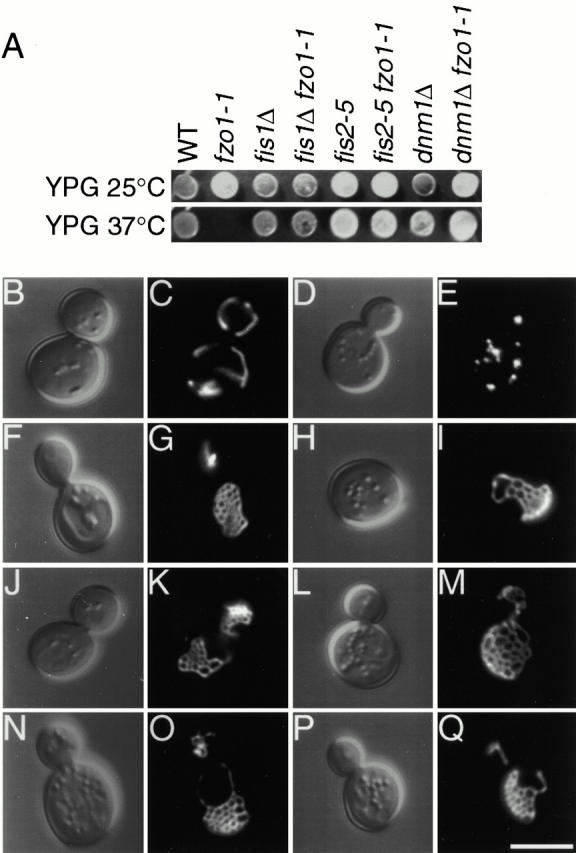
fis1 and fis2 mutations cause mitochondrial net formation and suppress glycerol growth defects and mitochondrial fragmentation in fzo1-1. (A) Wild-type (ADM548), fzo1-1 (ADM547), _fis1_Δ (ADM549), _fis1_Δ fzo1-1 (ADM550), _fis2_-5 (ADM752), _fis2_-5 fzo1-1 (ADM176), _dnm1_Δ (ADM379), and _dnm1_Δ fzo1-1 (ADM378) cells were spotted on YPGlycerol and grown at 25° or 37°C for 5 d. The presence/absence of mtDNA nucleoids in each strain was evaluated by DAPI staining (not shown). (B–Q) Morphology of mito-GFP–labeled (pVT100UGFP) mitochondria in (C) wild-type, (E) fzo1-1, (G) _fis1_Δ, (I) _fis1_Δ fzo1-1, (K) fis2-5, (M) fis2-5 fzo1-1, (O) _dnm1_Δ, and (Q) _dnm1_Δ fzo1-1 cells grown at 37°C. The corresponding differential interference contrast images are shown in B, D, F, H, J, L, N, and P. Bar, 5 μm.
Mitochondrial Membranes Form Nets in fis1 and fis2 Mutant Strains
The morphology of mitochondrial membranes in fis1 and fis2 mutant strains was consistent with a role for these genes in fission. As shown previously (Bleazard et al. 1999), the wild-type mitochondrial reticulum (Fig. 2 C; Table , 100% branched tubules) is converted into an interconnected “net” in dnm1 mutant cells due to unopposed tip fusion (Fig. 2 O; Table , 67% mitochondrial nets). Analysis of GFP-labeled mitochondria revealed that 85% of fis1 (Fig. 2 G; Table ) and 77% of fis2 (Fig. 2 K; Table ) mutant cells contained mitochondrial nets strikingly like those in _dnm1_Δ. These nets appeared identical when visualized with matrix-targeted, and inner and outer membrane-targeted forms of the red and green fluorescent proteins, indicating that fission of both the inner and outer membranes was abolished in these mutants (not shown). Mitochondrial nets were also observed in all combinations of dnm1, fis1, and fis2 double and triple mutant strains (Table and not shown), suggesting that FIS1 and FIS2 act in the same pathway as DNM1 to regulate mitochondrial fission.
Table 2.
Mitochondrial-Membrane Morphology in fis1 and fis2 Mutant Cells
| Strain | °C | No. cells scored | Branched tubules | Frayed/clumpy tubules | Collapsed tubules | Mitochondrial nets | Fragmented mitochondria |
|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | ||
| WT | 30 | 300 | 100 | ||||
| _fis1_Δ ‡ | 30 | 500 | 15 | 85 | |||
| fis2-5 ‡ | 30 | 100 | 1 | 22 | 77 | ||
| _dnm1_Δ ‡ | 30 | 200 | 33 | 67 | |||
| _fis1_Δ fis2-5 | 30 | 100 | 26 | 74 | |||
| _fis1_Δ _dnm1_Δ | 30 | 200 | 20 | 80 | |||
| _fis2-5_Δ _dnm1_Δ | 30 | 100 | 6 | 94 | |||
| _fis1_Δ _fis2-5 dnm1_Δ | 30 | 100 | 33 | 67 | |||
| WT | 25 | 100 | 96 | 4 | |||
| WT | 37 | 100 | 100 | ||||
| fzo1-1 | 25 | 100 | 51 | 46 | 3 | ||
| fzo1-1_§_ | 37 | 100 | 24 | 76 | |||
| _fis1_Δ fzo1-1 | 25 | 100 | 20 | 80 | |||
| _fis1_Δ fzo1-1 § | 37 | 100 | 14 | 86 | |||
| fis2-5 fzo1-1 | 25 | 100 | 15 | 41 | 44 | ||
| fis2-5 fzo1-1_§_ | 37 | 100 | 9 | 47 | 41 | 3 | |
| _dnm1_Δ fzo1-1 | 25 | 100 | 7 | 29 | 64 | ||
| _dnm1_Δ fzo1-1 § | 37 | 100 | 5 | 29 | 66 | ||
| _fzo1_Δ | 30 | 200 | 3 | 97 | |||
| _fis1_Δ _fzo1_Δ | 30 | 300 | 24 | 62 | 9 | 3 | 2 |
| _mgm1_Δ | 30 | 100 | 1 | 99 | |||
| _fis1_Δ _mgm1_Δ | 30 | 100 | 46 | 15 | 38 |
fis1 and fis2 Mutations Block Mitochondrial Fission and Fragmentation in fzo1-1
We previously reported (Bleazard et al. 1999) that dnm1 mutations blocked mitochondrial fission, fragmentation, and mtDNA loss in fzo1-1 cells at 37°C (compare fzo1-1 in Fig. 2 E and _dnm1_Δ fzo1-1 in Q; Table ). To determine whether fis1 and fis2 mutations also prevented mitochondrial fragmentation in fzo1-1, we examined mitochondrial morphology in _fis1_Δ fzo1-1 and _fis2_-5 fzo1-1 strains after a shift to the nonpermissive temperature. At 25°C, 80% of _fis1_Δ fzo1-1 and 44% of _fis2_-5 fzo1-1 cells contain mitochondrial nets similar to those observed in the single _fis1_Δ and fis2-5 mutants at 25°, 30°, and 37°C (Table ). Upon shifting _fis1_Δ fzo1-1 (Fig. 2 I; Table ) and fis2-5 fzo1-1 (Fig. 2 M; Table ) cells to 37°C, mitochondrial fragmentation was blocked despite the absence of FZO1 function and mitochondrial membranes remained net-like. These results were similar to those observed for the fzo1-1 dnm1 double mutant (Bleazard et al. 1999), and indicated that loss of Fis1p or Fis2p function in the _fis1_Δ fzo1-1 and fis2-5 fzo1-1 double mutants blocked mitochondrial fission and fragmentation in fzo1-1 cells at the nonpermissive temperature.
Because both _fis1_Δ and fis2-5 are null alleles, suppression of the fzo1-1 mitochondrial fragmentation, mtDNA loss and glycerol growth phenotypes cannot occur via direct interactions between Fzo1-1p and the FIS1 and FIS2 gene products. Indeed, the genetic interactions described above did not require expression of the Fzo1-1 protein, ruling out the possibility that fis1 and fis2 restore an Fzo1p-dependent fusion pathway. In _fzo1_Δ _fis1_Δ (Fig. 3A, second row, and E) and _fzo1_Δ fis2-5 (not shown), double mutant strains, the _fzo1_Δ glycerol growth defect was suppressed and mitochondrial membranes remained tubular, although the tubules sometimes appeared ragged or frayed (Table ). Interestingly, mitochondrial nets were largely absent when double mutants contained the _fzo1_Δ allele (Table ). This observation indicates that formation of mitochondrial nets in fis1 and fis2 mutant strains requires _FZO1_-mediated fusion.
Figure 3.
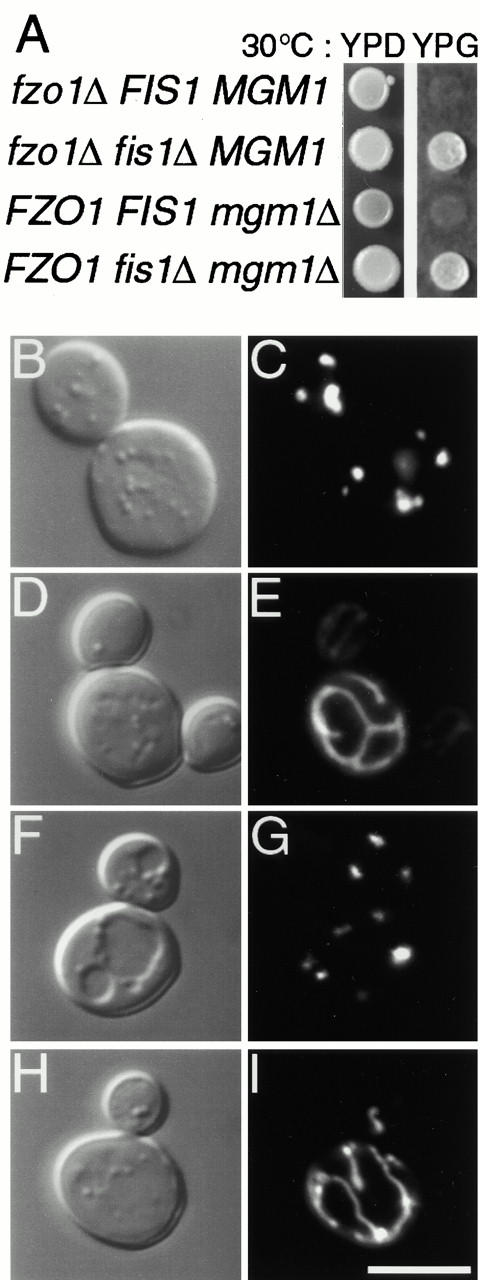
_fis1_Δ acts as a bypass suppressor of mitochondrial fragmentation defects. (A) _fzo1_Δ FIS1 MGM1 (ADM575), _fzo1_Δ fis1_Δ MGM1 (ADM574), FZO1 FIS1 mgm1_Δ (ADM764), and _FZO1 fis1_Δ _mgm1_Δ (ADM763) strains were spotted on YPDextrose or YPGlycerol and grown at 30°C for 5 d, respectively. Morphology of mito-GFP-labeled (pVT100UGFP) mitochondria in (C) _fzo1_Δ FIS1 MGM1, (E) _fzo1_Δ _fis1_Δ MGM1, (G) _FZO1 FIS1 mgm1_Δ, and (I) _FZO1 fis1_Δ _mgm1_Δ cells grown at 30°C. The corresponding differential interference contrast images are shown in B, D, F, and H. Bar, 5 μm.
dnm1, fis1 and fis2 Mutations Are General Suppressors of Mitochondrial Fragmentation Defects
To determine whether the dnm1, fis1, and fis2 mutations were specific suppressors of the fzo1 fusion defect or were general suppressors of mitochondrial fragmentation defects, we examined the effect of dnm1, fis1, and fis2 mutations in an _mgm1_Δ mutant (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999). Cells with mutations in MGM1 have fragmented mitochondrial membranes (Fig. 3 G), although the mgm1 mutant does not block _FZO1_-mediated mitochondrial fusion (Wong et al. 2000). The results of these studies with the _fis1_Δ mutant are shown in Fig. 3 and Table . As observed for the fis1 Δ fzo1 Δ double mutant, _fis1_Δ is able to suppress the _mgm1_Δ mtDNA loss (not shown), glycerol growth defect (Fig. 3 A, bottom two rows) and mitochondrial fragmentation defect (Fig. 3 I; Table ) in the _fis1_Δ _mgm1_Δ double mutant strain. Similar results were observed for the _mgm1_Δ fis2-5 double mutant strains (not shown) and _mgm1_Δ _dnm1_Δ (Gorsich, S., and J.M. Shaw, unpublished data), consistent with the idea that dnm1, fis1, and fis2 mutations prevent fragmentation of mitochondrial membranes by blocking the fission pathway.
Mutations in fis1 and fis2 Do Not Restore Mitochondrial Fusion in fzo1-1 Cells
The observation that fis1 and fis2 mutations block mitochondrial fragmentation in fzo1-1 cells is consistent with a role for Fis1p and Fis2p in mitochondrial fission. However, similar results could be obtained if mutations in fis1 and fis2 activated an _FZO1_-independent fusion pathway. To determine whether fis1 and fis2 mutations restored fusion in fzo1-1, we assayed mitochondrial fusion in zygotes formed from either _fis1_Δ fzo1-1 or fis2-5 fzo1-1 double mutants. Fusion was assayed essentially as described by Nunnari et al. 1997 by labeling mitochondria in haploid cells of opposite mating type with either mito-GFP or -RFP (instead of MitoTracker Red CMXRos), and then examining the distribution of the two fluorescent markers in zygotes formed at the permissive and nonpermissive temperatures.
Both mito-GFP and -RFP colocalized in 100% of wild-type zygotes formed at 25° and 37°C, indicating that mitochondria from each haploid parent had fused and their contents had mixed (Table ) (Nunnari et al. 1997). Mitochondrial content mixing also occurred efficiently in fis1 X fis1, fis2 X fis2, and dnm1 X dnm1 zygotes formed at both temperatures, indicating that these three genes are not required for fusion (Table ). As reported previously, mitochondrial fusion occurred in fzo1-1 X fzo1-1 zygotes at 25°C, but fragmented and failed to fuse at 37°C, consistent with Fzo1p's role in fusion (Table ) (Hermann et al. 1998). Although disruption of FIS1 or FIS2 prevented mitochondrial fragmentation in fzo1-1 zygotes at the nonpermissive temperature, fusion was not restored (Table ). These findings are similar to those reported for DNM1 (Table ) (Bleazard et al. 1999) and indicate that the maintenance of mitochondrial tubules in _fis1_Δ fzo1-1 and fis2-5 fzo1-1 cells results from a block in mitochondrial fission rather than the activation of an _FZO1_-independent fusion pathway. Together, the results presented here indicate that FIS1 and FIS2 act in the same pathway as DNM1 to regulate mitochondrial fission.
Table 3.
Mitochondrial Fusion in Budded Zygotes
| Strains crossed | °C | No. Fused | Mitochondria |
|---|---|---|---|
| % | |||
| FIS1 FIS2 DNM1 FZO1 X | 25 | 35/35 | 100 |
| FIS1 FIS2 DNM1 FZO1 | 37 | 76/76 | 100 |
| FIS1 FIS2 DNM1 fzo1-1 X | 25 | 24/30 | 80 |
| FIS1 FIS2 DNM1 fzo1-1 | 37 | 0/55 | 0 |
| _fis1_Δ FIS2 DNM1 FZO1 X | 25 | 50/50 | 100 |
| _fis1_Δ FIS2 DNM1 FZO1 | 37 | 75/75 | 100 |
| _fis1_Δ FIS2 DNM1 fzo1-1 X | 25 | 40/40 | 100 |
| fis1_Δ_ FIS2 DNM1 fzo1-1 | 37 | 0/77 | 0 |
| FIS1 fis2-5 DNM1 FZO1 X | 25 | 29/30 | 97 |
| FIS1 fis2-5 DNM1 fzo1 X with pRS416 + FZO1 | 37 | 29/30 | 97 |
| FIS1 fis2-5 DNM1 fzo1-1 X | 25 | 48/50 | 96 |
| FIS1 fis2-5 DNM1 fzo1-1 | 37 | 0/50 | 0 |
| FIS1 FIS2 dnm1 Δ FZO1 X | 25 | 40/40 | 100 |
| FIS1 FIS2 dnm1 Δ FZO1 | 37 | 50/50 | 100 |
| FIS1 FIS2 dnm1 Δ fzo1-1 X | 25 | 30/30 | 100 |
| FIS1 FIS2 dnm1 Δ fzo1-1 | 37 | 0/35 | 0 |
FIS1 Defines a Novel Gene Family
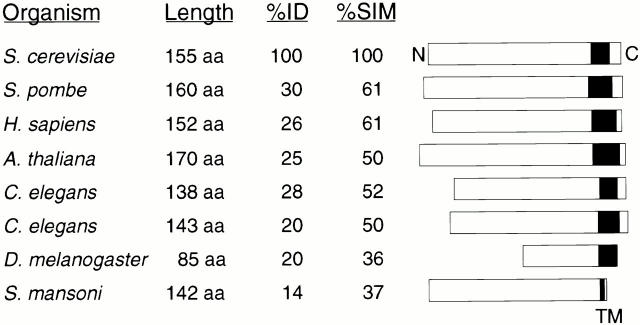
The FIS1 gene (YIL065C in the Saccharomyces Genome Database) was cloned by library complementation of the glycerol growth suppression phenotype in fis1 fzo1-1 strains. A fragment of the rescuing plasmid containing FIS1/YIL065C also rescued the mitochondrial morphology defects in a fis1 mutant strain. Integrative mapping studies and DNA sequencing were used to show that the original fis1 mutations were allelic to the YIL065C cloned open reading frame. FIS1 encodes a novel protein of 155 amino acids with an estimated molecular mass of 17,000 D (Fig. 4). The only identifiable protein motif in the open reading frame is a single, predicted, COOH-terminal transmembrane domain (Fig. 4, TM). A database search identified Fis1p orthologs in fission yeast, humans, plants, worms, and flies, suggesting that Fis1p function has been conserved during evolution (Fig. 4). Whether these orthologs participate in membrane division events mediated by Dnm1p-like GTPases remains to be seen.
Figure 4.
Domain structure of the Fis1p Family. Domain structure of eight Fis1p family members with the predicted carboxy-terminal transmembrane domain shown in black (TM). The length in amino acids, percent sequence identity, and percent sequence similarity between the Saccharomyces cerevisiae predicted protein (NCBI RefSeq NP012199) and the predicted Schizosaccharomyces pombe (PIR T39328), Caenorhabiditis elegans (T16053 and T16332), Homo sapiens (NP057152), Arabidopsis thaliana (CAB72179.1), Schistosoma mansoni (AAA99799), and Drosophila melanogaster (AAF45343) orthologs are indicated. The D. melanogaster and S. mansoni amino acid sequences may be incomplete. All sequences can be accessed through the NCBI database (http://www.ncbi.nlm.nih.gov). The Jotun Hein program (MegAlign, DNA*) was used to construct pairwise alignments between the S. cerevisiae protein sequence and each Fis1p ortholog.
FIS1 Encodes an Integral, Outer Mitochondrial Membrane Protein with Its NH2 Terminus Facing the Cytoplasm
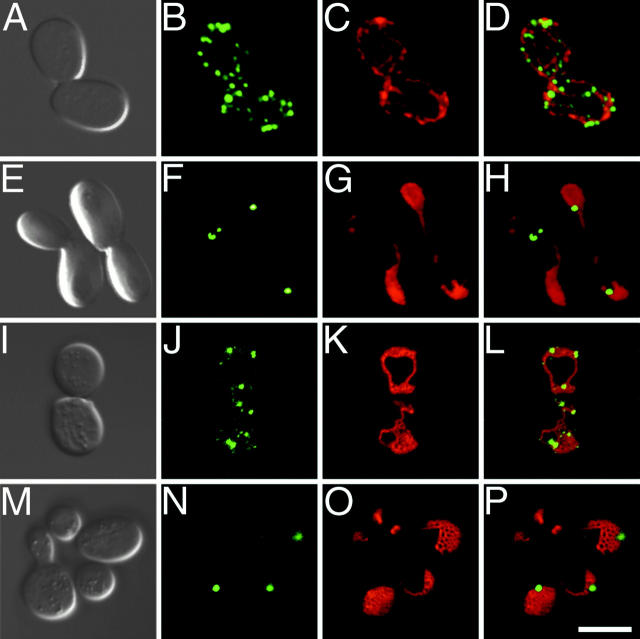
The subcellular localization of Fis1p was investigated by fluorescence microscopy and cryoimmunoelectron microscopy using functional, amino-terminal GFP-tagged (GFP-Fis1p) and Myc-tagged (Myc-Fis1p) forms of the protein (see Materials and Methods; GFP-Fis1p and Myc-Fis1p were expressed at levels comparable with that of endogenous Fis1p in these experiments). When expressed in yeast, GFP-Fis1p and Myc-Fis1p rescued the mitochondrial net morphology defect in _fis1_Δ and the glycerol growth suppression phenotype in the _fis1_Δ fzo1-1 double mutant.
In dividing yeast cells, GFP-Fis1p colocalized completely with the MitoTracker red CMXRos-labeled mitochondrial reticulum (Fig. 5, A–C), indicating that Fis1p is a mitochondrial protein. The identical Fis1p localization pattern was observed in _dnm1_Δ, fis2-5, and _fis2-5 dnm1_Δ mutants, suggesting that Fis1p's localization is not determined or affected by other known components of the fission machinery (not shown). In addition, the predicted COOH-terminal transmembrane domain (Fig. 4) was required for Fis1p's mitochondrial localization and function. A GFP-Fis1p fusion protein lacking the transmembrane domain (GFP-Fis1paa1-127) failed to colocalize with MitoTracker red CMXRos-labeled mitochondria (Fig. 5, D–F) and did not rescue fis1 mitochondrial morphology defects or the glycerol growth suppression phenotype in a _fis1_Δ fzo1-1 strain (not shown). Carbonate and detergent extraction studies confirmed that Fis1p was an integral mitochondrial membrane protein (not shown). Interestingly, overexpression of cytoplasmic GFP-Fis1paa1-127 from a _MET25-_inducible promoter did not cause dominant mitochondrial morphology defects in wild-type cells, suggesting that this form of Fis1p is not competent to titrate essential components of the fission reaction (data not shown). Cryo-immunoelectron microscopy revealed that, unlike Dnm1p, which was found in discrete spots associated with mitochondrial constriction sites (Bleazard et al. 1999), Myc-Fis1p appeared to be distributed evenly on the surface of the mitochondrial compartment (Fig. 5 G; see H for anti–Myc detection of Myc-Fis1p). These localization studies were consistent with biochemical studies (described below) demonstrating that Fis1p was an outer mitochondrial membrane protein.
Figure 5.
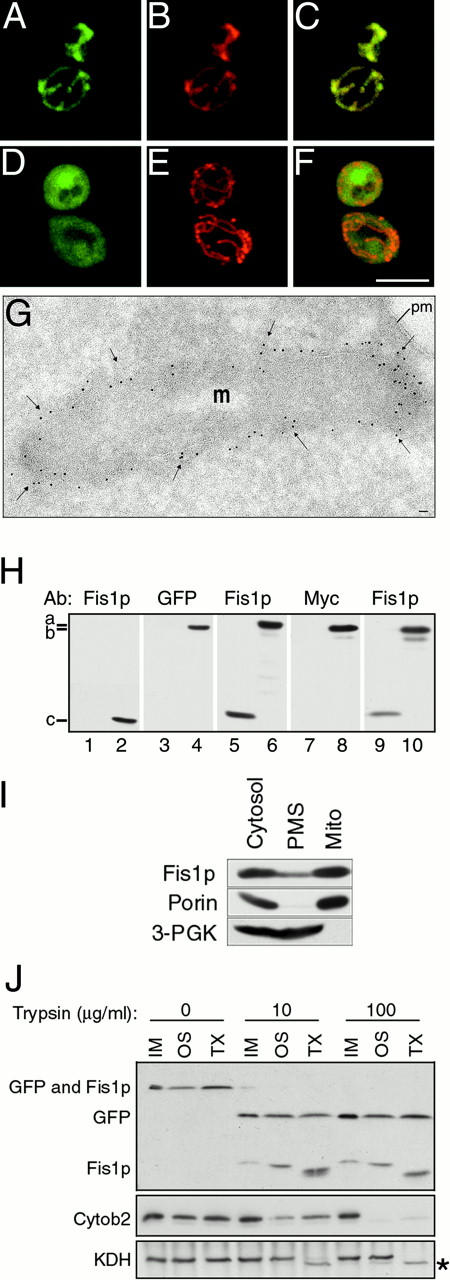
Fis1p is a mitochondrial membrane protein with its amino terminus exposed to the cytoplasm. (A–F) Wild-type (ADM551) cells expressing GFP-Fis1p (A–C; pRS415MET25 + rsGFP-FIS1 aa1-155) or GFP-Fis1p lacking the COOH-terminal transmembrane domain (D–F; pRS415MET25 + rsGFP-FIS1 aa1-127) were stained with MitoTracker red CMXRos (B, C, E, and F). Confocal microscopy was used to evaluate the distribution of the GFP fusion proteins (green) and mitochondrial compartments (red) in individual (A, B, D, and E) and merged (C and F) images. Bar, 5μm. (G) The distribution of Myc-Fis1p (black arrows) (ADM551; pRS415_MET25_ + 9xMYC-FIS1) was determined by immunogold labeling of ultrathin cryosections. 5-nm gold particles were found concentrated at the periphery of the mitochondrial compartment. Percentage distribution of 1,807 gold particles counted: mitochondrial tips 8.5%, mitochondrial sides (excluding constriction sites) 77.6%, mitochondrial constriction sites 12.5%, and cytoplasm 1.4%. m, mitochondria; pm, plasma membrane. Bar, 0.1 μm. (H) Total cell extracts from _fis1_Δ (lane 1, ADM549), wild-type (lane 2, ADM548), or _fis1_Δ cells expressing either Fis1p (lanes 3, 5, 7, and 9, ADM549; pRS416 + FIS1), GFP-Fis1p (lanes 4 and 6, ADM549; pRS415MET25 + rsGFP-FIS1) or Myc-Fis1p proteins (lanes 8 and 10, ADM549; pRS415MET25 + 9xMYC-FIS1) were analyzed by Western blotting with anti–Fis1p, –GFP, and –Myc serum as indicated. a, the position of GFP-Fis1p (47 kD); b, Myc-Fis1p (35 kD); and c, native Fis1p (17 kD). (I) Wild-type protein extract was fractionated by differential centrifugation, separated by SDS-PAGE, and analyzed by Western blotting with anti–Fis1p, –porin, and –3-PGK serum. Cytosol, postnuclear cytoplasmic extract; PMS, supernatant depleted of mitochondria; Mito, mitochondrial pellet. (J) Untreated (IM), osmotically shocked (OS), and Triton X-100 solubilized (TX) mitochondrial fractions were treated with 0, 10, or 100 μg/ml Trypsin and analyzed by SDS-PAGE and Western blotting with anti–GFP, –Fis1p, –cytochrome b2, and –KDH serum. *Protease-clipped form of KDH.
Subcellular fractionation experiments using anti–Fis1p and anti–GFP antibodies confirmed that Fis1p is a mitochondrial protein. Anti–Fis1p antibodies recognized a 17-kD protein in extracts prepared from FIS1 wild-type but not _fis1_Δ cells (Fig. 5 H, compare 1 and 2, band c). Similarly, anti–GFP antibodies detected a 47-kD fusion protein in extracts prepared from _fis1_Δ cells expressing GFP-Fis1p but not in extracts prepared from _fis1_Δ cells expressing untagged Fis1p (Fig. 5 H, compare 3 and 4, band a). As shown in Fig. 4 I, native Fis1p cofractionated predominantly with the mitochondrial pellet, along with the outer mitochondrial membrane protein porin, during differential centrifugation of a postnuclear cell extract. In extracts prepared from _fis1_Δ cells expressing GFP-Fis1p, GFP-Fis1p also cofractionated with porin, and not the cytoplasmic marker, 3-phosphoglycerate kinase (3-PGK; data not shown).
Protease protection experiments revealed that Fis1p is an outer mitochondrial membrane protein with its NH2 terminus exposed to the cytoplasm. Anti–GFP and anti–Fis1p antibodies detected the full-length GFP-Fis1p fusion protein in untreated mitochondrial fractions (Fig. 5 J, 0 μg/ml Trypsin). Treatment of intact mitochondria with 10 or 100 μg/ml trypsin resulted in the clipping and release of the NH2-terminal, 29-kD GFP tag from Fis1p (Fig. 5 J, 10 and 100 μg/ml trypsin, IM; the linker region between GFP and Fis1p contains a predicted trypsin cleavage site). In the presence of 100 μg/ml trypsin, the intermembrane space protein cytochrome b2 was not digested, indicating that the outer mitochondrial membrane remained intact (Fig. 5 J, 100 μg/ml trypsin, IM). When the outer membrane was disrupted by osmotic shock, however, cytochrome b2 was degraded by trypsin, although the matrix marker alpha-ketoglutarate dehydrogenase (KDH) was still protected (Fig. 5 J, 100 μg/ml trypsin, OS). Protease clipping of KDH (*) only occurred when mitochondria were solubilized with Triton X-100 in the presence of trypsin (Fig. 5 J, 100 μg/ml trypsin, TX). Interestingly, significant degradation of Fis1p was only observed after treatment of mitochondria with detergent, suggesting that Fis1p's small size and outer membrane association affects its protease susceptibility (Fig. 5 J, note shifted and smeared Fis1p bands in TX lanes treated with trypsin). The fact that (a) the GFP tag is at the NH2 terminus of Fis1p, (b) Fis1p has only one predicted transmembrane domain, and (c) there are only nine amino acids COOH-terminal of the transmembrane domain suggests that the COOH-terminal nine amino acids of Fis1p face the mitochondrial intermembrane space.
Proper Assembly and Distribution of Dnm1p on the Outer Mitochondrial Membrane Requires Fis1p, but Not Fis2p Function
Two-hybrid, coimmunoprecipitation (Fukushima, N.H., B.R. Keegan, and J.M. Shaw. 1999. Mol. Biol. Cell. 10:315a. [Abstr.]), and genetic studies (Mozdy and Shaw, unpublished observations) indicate that Dnm1p interacts with itself and probably functions as a multimer. Our previous work demonstrates that these Dnm1p multimer-containing complexes assemble on the cytoplasmic face of the outer mitochondrial membrane at sites of future constriction and fission (Otsuga et al. 1998; Bleazard et al. 1999). Both the fis1 mitochondrial fission defect and the topology of Fis1p on the outer mitochondrial membrane raised the possibility that Fis1p was required to localize or activate the Dnm1p GTPase. To test this idea, we examined the localization of a Dnm1p-GFP fusion protein in the presence and absence of Fis1p. We also labeled mitochondria in the same cells with mito-RFP to evaluate colocalization of Dnm1p-GFP with mitochondrial membranes.
In 97% of wild-type cells and 92% of _dnm1_Δ cells examined, Dnm1p-GFP was organized in eight or more punctate structures that colocalized completely with wild-type mito-RFP–labeled networks (Fig. 6, A–D; Table ). This pattern was consistent with the Dnm1p localization pattern determined previously by indirect immunofluorescence studies (Otsuga et al. 1998), immunogold labeling studies (Bleazard et al. 1999), and Dnm1p-GFP studies (Sesaki and Jensen 1999). In contrast, 81% of cells lacking the Fis1 protein contained only one to three of the Dnm1p-GFP structures and these structures often appeared larger than normal (Fig. 6, E–H, Table ). This change in Dnm1p-GFP localization is not a secondary consequence of net formation since Dnm1p-GFP distribution appears wild type in fis2 mutants, which also contain mitochondrial nets (see below). Interestingly, the Dnm1p-GFP spots in _fis1_Δ cells colocalized with mitochondrial membranes in this mutant. However, these structures were not able to catalyze fission, as evidenced by the mitochondrial nets formed in the _fis1_Δ strain (Table ).
Figure 6.
Dnm1-GFP localization requires FIS1 but not FIS2 function. (A, E, I, and M) differential interference contrast, (B, F, J, and N) Dnm1-GFP (pRS414-_DNM1_-rsGFP, PRS416-_DNM1_-rsGFP, or pHS20), (C, G, K, and O) mito-RFP (pRS416-GAL1 + _PrF0ATP9_-RFP), and (D, H, L, and P) merged Dnm1-GFP/mito-RFP images of (A–D) wild-type (ADM548), (E–H) _fis1_Δ (ADM552), (I–L) fis2-5 (ADM752), and (M–P) _fis1_Δ fis2-5 (ADM742) cells grown at 30°C. Bar, 5 μm.
Table 4.
Number of Dnm1p-GFP Complexes in Wild-Type and Mutant Strains
| Strain | No. of cells scored | >8 | 4–8 | 1–3 |
|---|---|---|---|---|
| % | % | % | ||
| WT | 100 | 97 | 3 | 0 |
| _fis1_Δ | 100 | 3 | 16 | 81 |
| fis2-5 | 100 | 86 | 14 | 0 |
| _fis1_Δ fis2-5 | 100 | 0 | 7 | 93 |
| _dnm1_Δ | 100 | 92 | 5 | 3 |
| fis1-21 fzo1-1 | 100 | 61 | 32 | 7 |
Although Dnm1p-GFP complexes colocalized with mitochondria in the absence of Fis1p, Fis1p was clearly required to properly distribute these complexes into punctate structures along mitochondrial tubules. Analysis of one FIS1 allele, fis1-21, suggested that Fis1p may also be required for the function of these punctate Dnm1p-containing structures. The fis1-21 allele blocked mitochondrial fission and fragmentation in a fis1-21 fzo1-1 double mutant at 37°C (mitochondrial membranes remained tubular in 97% of the cells). However, the number and distribution of punctate Dnm1p-GFP structures on fis1-21 fzo1-1 mitochondria appeared more wild type (Table ). These data suggested that the rate or extent of fission by properly distributed Dnm1p complexes was impaired in the fis1-21 fzo1-1 strain. Thus, while Fis1p plays a role in determining the punctate distribution of Dnm1p complexes on mitochondrial tubules, it is also required for Dnm1p complex function once that distribution is achieved.
Similar experiments demonstrated that wild-type Dnm1p-GFP distribution did not require FIS2 gene function. In the majority (86%) of fis2-5 cells, the number and distribution of Dnm1p-GFP structures was indistinguishable from wild type (Fig. 6, I–L; Table ). Once again, these structures colocalized with fis2-5 mitochondrial tubules and nets, suggesting that the Fis2 protein was required for the function but not the localization of Dnm1p-containing complexes (see Discussion for more information about Fis2p). Finally, the number and distribution of Dnm1p-GFP structures in a _fis1_Δ fis2-5 double mutant was similar to that in a _fis1_Δ single mutant, indicating that fis1 is epistatic to fis2 with respect to Dnm1p-GFP localization (Fig. 6, M–P). Thus, Fis1p apparently functions upstream of Fis2p to localize Dnm1p.
Immunogold labeling of ultrathin cryosections confirmed that Dnm1p localization was altered in the fis1 mutant. In wild-type cells expressing the tagged Dnm1p-HAc protein, 89.3% of the 5-nm gold particles were associated with the tips and sides of mitochondrial tubules and with constriction sites in these tubules (Table ). These results are similar to those reported previously for Dnm1p-HAc localization (Bleazard et al. 1999). In contrast, in _fis1_Δ cells, only 8% of gold particles were found on the tips and sides of mitochondrial tubules, while the majority (92%) were found in the cytoplasm (Table ). Quantitative Western blot analysis indicated that this difference was not due to reduced Dnm1p expression in _fis1_Δ relative to FIS1 wild type (not shown). Rather, in _fis1_Δ cells, the total amount of Dnm1p associated with mitochondria appeared to be reduced, and the distribution of this mitochondrial-associated Dnm1p appeared to be altered.
Table 5.
Distribution of Dnm1-HAc Protein in FIS1 and fis1_Δ_ Cells
| Compartment | Dnm1p-HA in wild-type cells | Dnm1p-HA in _fis_Δ cells |
|---|---|---|
| % total gold particles, n = 531 | % total gold particles, n = 121 | |
| Mitochondrial tips | 38.4 | 6.2 |
| Mitochondrial sides | 15.4 | 1.8 |
| Mitochondrial constriction sites | 35.5 | 0 |
| Mitochondrial other‡ | 1.1 | 0 |
| Mitochondrial total | 90.4 | 8.0 |
| Plasma membrane | 2.2 | 0 |
| Cytoplasm | 7.5 | 92.0 |
| Vacuole | 0 | 0 |
Discussion
Our previous studies established that the S. cerevisiae dynamin-related GTPase, Dnm1p, associates with the outer mitochondrial membrane at sites of future constriction and fission (Otsuga et al. 1998; Bleazard et al. 1999). Here we provide genetic and morphological evidence that FIS1 and FIS2 encode two additional components of the Dnm1p fission apparatus. Like dnm1 (Bleazard et al. 1999), fis1 and fis2 mutations abolish fission and suppress fragmentation in the fzo1-1 strain. This suppression occurs via a bypass mechanism since _fis1_Δ and fis2-5 (null) alleles prevent fragmentation in an _fzo1_Δ strain and in _mgm1_Δ cells (Fig. 3). Fis1p is a novel outer mitochondrial membrane protein essential for the proper distribution and function of the Dnm1p GTPase during the fission reaction. Fis1p is, to our knowledge, the first integral membrane protein known to function with a dynamin-related GTPase in a membrane fission event. In contrast, the FIS2 gene product is required for the function, but not the mitochondrial association and distribution, of Dnm1p.
The results described here raised the possibility that Fis1p functions early in the fission pathway to recruit Dnm1p or Dnm1p-containing complexes to the mitochondrial membrane. Although we do not exclude such a function for Fis1p, several lines of evidence argue against this model. First, we have been unable to detect interactions of Fis1p with Dnm1p using coimmunoprecipitation and yeast two-hybrid studies (not shown). It is possible that such interactions are simply too transient to detect by these methods. Alternatively, one or more additional molecules may serve as a bridge between Fis1p and its binding partner(s). It is also possible that the methods we used failed to mimic a guanine nucleotide-bound state of Dnm1p required for Fis1p binding interactions. Second, we found that one to three large structures containing Dnm1p-GFP continued to colocalize with mitochondria in the absence of Fis1p (Fig. 6). This result suggests that Dnm1p contains Fis1p-independent signals for protein or lipid binding that mediate its mitochondrial attachment. A number of previously identified Dnm1p structural domains might function in this manner, including the middle domain, insert B, or the alpha-helical domain (van der Bliek 1999). Third, one of our FIS1 alleles, fis1-21, interferes with fission and suppresses fragmentation in fzo1-1 but does not completely disrupt the mitochondrial distribution of Dnm1p-GFP (Table ). This last observation is consistent with the idea that Fis1p plays a role in the activation or function of Dnm1p complexes after they have assembled on the outer mitochondrial membrane. Interestingly, we recently detected Fis1p self-interactions using the two-hybrid assay. We are currently exploring the possibility that Fis1p self-interactions are required for Dnm1p mitochondrial distribution or function.
Even though punctate Dnm1p-GFP complexes are properly distributed in fis1-21 fzo1-1 cells (Table ), mitochondrial fragmentation is still blocked in this double mutant at 37°C (97% tubular mitochondria), suggesting that the ability of these complexes to catalyze fission is impaired. Thus, Fis1p may be required (a) to distribute Dnm1p complexes on mitochondrial tubules and (b) to activate the fission activity of Dnm1p complexes. One intriguing possibility is that Fis1p functions late in the fission pathway (after mitochondrial recruitment of Dnm1p complexes) as an effector molecule to regulate some aspect of the Dnm1p GTPase cycle. Preliminary studies of wild-type Dnm1p-GFP localization in the presence of a dominant dnm1-T62A allele (Otsuga et al. 1998) are consistent with this idea. The dnm1-T62A mutation is equivalent to a mutation in the ras GTPase that is predicted to abolish effector binding (Vojtek et al. 1993; Adari et al. 1988; Cales et al. 1988). In FIS1 wild-type cells, over-expressed Dnm1-T62A protein interacts with wild-type Dnm1p-GFP, causing the wild-type protein to redistribute into one to three large structures on the mitochondrial compartment (not shown). This altered Dnm1p-GFP localization pattern is similar to that observed in the _fis1_Δ mutant, suggesting that the _fis1_Δ and dnm1-T62A mutations affect the same step in Dnm1p assembly, distribution, or activation.
A related study reported the cloning and characterization of the FIS2 gene, which they named MDV1 for mitochondrial division (Tieu and Nunnari 2000). MDV1 encodes an 80-kD protein containing seven COOH-terminal WD-40 repeats. Together, these WD-40 repeats are predicted to form a tertiary structure called the β propeller that has the potential to interact with multiple protein-binding partners (Smith et al. 1999). Several lines of evidence suggest that Mdv1p and the Dnm1p GTPase interact (Tieu and Nunnari 2000). In genetic crosses, nonallelic noncomplementation is observed between specific MDV1 and DNM1 alleles. In addition, fluorescence microscopy studies reveal that Mdv1p and Dnm1p colocalize in punctate structures on the cytoplasmic face of mitochondrial tubules. Finally, Mdv1p and Dnm1p have been shown to interact in a yeast two-hybrid assay (Tieu and Nunnari 2000; Uetz et al. 2000). Although Mdv1p is not necessary for the proper localization of Dnm1p, Dnm1p is required for the proper mitochondrial distribution of Mdv1p (Tieu and Nunnari 2000). In cells lacking Dnm1p, Mdv1p is uniformly dispersed on the outer mitochondrial membrane (Tieu and Nunnari 2000), similar to the localization pattern we observe for Fis1p (Fig. 5 A). This observation suggests that Mdv1p contains signals that allow targeting to the mitochondrial compartment in the absence of Dnm1p. In cells lacking Fis1p, Mdv1p continues to colocalize with Dnm1p in one to three large structures associated with mitochondria (see Results, and Tieu and Nunnari, 2000). Thus, while Fis1p is required for proper mitochondrial distribution of Dnm1p, Mdv1p's association with Dnm1p is unaffected by Fis1p. In the absence of both Dnm1p and Fis1p, Mdv1p is cytoplasmic, indicating that Fis1p is also required for the mitochondrial recruitment of Mdv1p. These combined results suggest that Mdv1p and Dnm1p bind to the mitochondrial outer membrane as a complex, or that Dnm1p binds to mitochondria first and Mdv1p is a late recruit to Dnm1p-containing structures. In both cases, association of Mdv1p with Dnm1p appears to be required for the function of Dnm1p-containing complexes during fission.
The data presented here and in the accompanying study by Tieu and Nunnari 2000 suggest that mitochondrial fission is a multi-step process requiring functional Dnm1p GTPase, its putative binding partner Mdv1p, and the integral, outer membrane protein, Fis1p. The results of our studies indicate that Fis1p is involved in the assembly, spatial distribution, and function of Dnm1p-containing fission complexes on the outer mitochondrial membrane (Fig. 7, step 1). Based on the results of Tieu and Nunnari 2000, these Dnm1p-containing complexes also contain Mdv1p, although the stoichiometry of this interaction is not known. We are currently determining whether Dnm1p or Mdv1p bind directly or indirectly to Fis1p in the mitochondrial membrane. The Dnm1p-containing fission complexes may polymerize to form rings or collars surrounding the mitochondrial outer membrane (visualized as punctate structures on mitochondria; Fig. 7, step 1), similar to the collars formed at the base of clathrin-coated pits by the dynamin GTPase (Takei et al. 1995). Formation of such a structure could coincide with, or subsequently result in, membrane constriction and fission. Alternatively, Dnm1p might be required to recruit or activate additional components that mediate constriction and fission (Fig. 7, steps 2 and 3).
Figure 7.
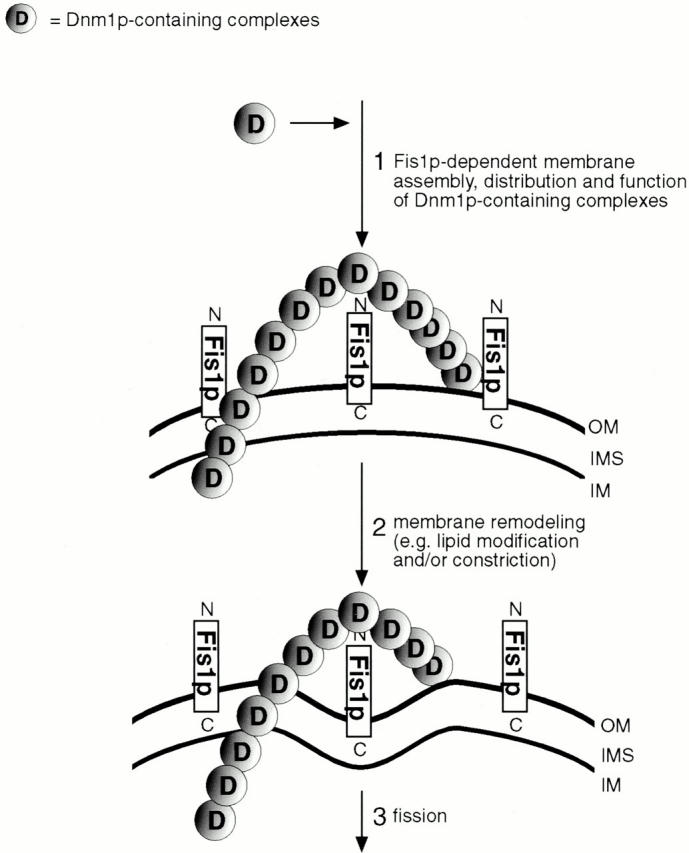
Model for mitochondrial constriction and fission mediated by Dnm1p-containing complexes. In response to an unknown signal(s), fission complexes containing the Dnm1p GTPase and probably Mdv1p are assembled in a Fis1p-dependent manner on the outer mitochondrial membrane (step 1). Dnm1p-containing fission complexes may assemble into higher-order structures, possibly forming rings or collars around the outer surface of mitochondrial tubules, similar to structures formed by dynamin during endocytosis (rings/collars are visualized as punctate structures in intact yeast cells). Dnm1p-containing complexes could play a role in the mechanical constriction and fission of mitochondrial tubules, or, alternatively, these complexes might recruit or activate additional components that mediate constriction and fission (steps 2 and 3). Although both the inner and outer mitochondrial membranes are shown in cross section, Dnm1p-containing fission complexes are predicted to contact only the outer membrane. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
Whether or not this model is correct, it seems likely that Dnm1p's GTPase cycle will play an important role in regulating one or more steps in the division process. For example, GTP binding or hydrolysis by Dnm1p might regulate: (a) self assembly into a complex with itself or other proteins, (b) its association with the outer mitochondrial membrane, (c) the assembly of Dnm1p complexes into higher order structures, or (d) the recruitment of additional components to the site of fission. A model in which GTP hydrolysis by Dnm1p provides mechanical energy for membrane constriction and fission cannot yet be excluded. It is likely that lipid modification activities are also required to remodel the outer mitochondrial membrane during constriction and fission, similar to endophilin's proposed activity during endocytosis (Ringstad et al. 1999; Schmidt et al. 1999). Future studies should identify any additional activities of Dnm1p, Fis1p, and Mdv1p as well as new proteins required for outer membrane division.
Although molecules required for outer mitochondrial membrane fission have been identified in a number of organisms (Otsuga et al. 1998; Smirnova et al. 1998; Bleazard et al. 1999; Labrousse et al. 1999; Sesaki and Jensen 1999), essentially nothing is known about molecules that mediate inner mitochondrial membrane fission. Recently, however, another dynamin-related GTPase in budding yeast, called Mgm1p, was shown to function in mitochondrial membrane dynamics (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999; Wong et al. 2000). The Mgm1p GTPase localizes to the mitochondrial intermembrane space and associates with the inner membrane, suggesting that it may play a role analogous to Dnm1p in inner membrane fission (Wong et al. 2000). If this model is correct, there may be structural or functional homologues of Mdv1p and Fis1p that work together with Mgm1p to regulate fission of the mitochondrial inner membrane. It seems likely that at least some of the molecules required for outer and inner membrane fission will also play a role in coordinating the behavior of both membranes during division. The integral outer membrane protein, Fis1p, is one potential candidate, since its COOH terminus probably extends into the intermembrane space and is exposed to the inner mitochondrial membrane. We are currently determining the submitochondrial topology of the Fis1p COOH terminus and its role in mitochondrial fission.
Fis1p and Mdv1p are new additions to the list of players that regulate division of prokaryotic cells and eukaryotic organelles of prokaryotic origin, including mitochondria and chloroplasts. E. coli cell division is mediated by the FtsZ GTPase that forms a ring on the cytoplasmic face of the inner membrane and constricts during cytokinesis (Rothfield et al. 1991; Bramhill 1997; Lutkenhaus and Addinall 1997; Nanninga 1998; Erickson 2000). In this case, an integral inner membrane protein called ZipA binds directly to FtsZ and is thought to be required for the function of the FtsZ ring (Hale and de Boer 1997). In the chloroplast, two different FtsZ homologues, FtsZ1 and FtsZ2, form rings at the organelle midpoint and coordinate division of the inner and outer membranes, respectively (Osteryoung and Vierling 1995; Osteryoung and Pyke 1998; Osteryoung et al. 1998). FtsZ homologues have not been identified in higher eukaryotes, although FtsZ homologues that localize to mitochondria have been identified in unicellular alga (Beech and Gilson 2000; Beech et al. 2000). During mitochondrial division in higher eukaryotes, the role of the FtsZ GTPase appears to have been replaced by dynamin-related GTPases like S. cerevisiae Dnm1p. Homologues of the Dnm1p GTPase have been identified in a wide range of organisms including humans (Smirnova et al. 1998) and worms (Labrousse et al. 1999), and a role for the Caenorhabditis elegans Dnm1p counterpart, DRP-1, in outer mitochondrial membrane fission has been established (Labrousse et al. 1999). Although homologues of Mdv1p have not yet been identified (Tieu and Nunnari 2000), the fact that S. cerevisiae Fis1p defines a novel family of outer mitochondrial membrane proteins conserved in humans, plants, worms, and flies (Fig. 4) strongly suggests that the molecular machinery regulating the Dnm1p-mediated type of mitochondrial fission has been conserved during eukaryotic evolution.
Acknowledgments
We are grateful to Q. Tieu and J. Nunnari (University of California, Davis, Davis, CA) for communicating their MDV1 results before publication. We also thank Steven Gorsich for providing the _mgm1_Δ and _mgm1_Δ _dnm1_Δ strains, C. Koehler (University of California, Los Angeles, Los Angeles, CA) for providing anti–KDH and anti-cytochrome b2 antibodies, H. Sesaki and R. Jensen (Johns Hopkins University, Baltimore, MD) for the pHS20 plasmid, and members of the Shaw laboratory for stimulating discussions and careful review of the manuscript.
This work was supported by grants from the American Cancer Society (CB-97) and the National Institutes of Health (GM53466) awarded to J.M. Shaw. The Utah Health Sciences Sequencing Facility is supported by a National Cancer Institute grant (5-P30CA42014).
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; KDH, alpha-ketoglutarate dehydrogenase; RFP, red fluorescent protein.
References
- Adari H., Lowy D.R., Willumsen B.M., Der C.J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech P.L., Gilson P.R. FtsZ and organelle division in Protists. Protist. 2000;151:11–16. doi: 10.1078/1434-4610-00003. [DOI] [PubMed] [Google Scholar]
- Beech P.L., Nheu T., Schultz T., Herbert S., Lithgow T., Gilson P.R., McFadden G.I. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- Cales C., Hancock J.F., Marshall C.J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988;332:548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. Dynamin and FtsZ. Missing links in mitochondrial and bacterial division. J. Cell Biol. 2000;148:1103–1105. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A.E., Kurihara L.J., Vallee R.B., Rose M.D. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K., Farh L., Marshall T.K., Deschenes R.J. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R. Guide to yeast genetics and molecular biology. In: Abelson J.N., Simon M.I., editors. Methods in Enzymology. Vol. 194. Academic Press, Inc; San Diego, CA: 1991. pp. 1–933. [PubMed] [Google Scholar]
- Hale C.A., de Boer P.A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Shaw J.M. Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H., Avers C.J. Mitochondrion of yeastultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Jones B.A., Fangman W.L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lagosky P.A., Taylor G.R., Haynes R.H. Molecular characterization of the Saccharomyces cerevisiae dihydrofolate reductase gene (DFR1) Nucleic Acids Res. 1987;15:10355–10371. doi: 10.1093/nar/15.24.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J., Addinall S.G. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular CloningA Laboratory Manual 1982. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: pp. 545 [Google Scholar]
- Nanninga N. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Marshall W.F., Straight A., Murray A., Sedat J.W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K., Pyke K. Plastid divisionevidence for a prokaryotically derived mechanism. Curr. Opin. Plant Biol. 1998;1:475–479. doi: 10.1016/s1369-5266(98)80038-1. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J.M. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R., Adams A.E.M., Drubin D.G., Haarer B.K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rieder S.E., Banta L.M., Köhrer K., McCaffery J.M., Emr S.E. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N., Gad H., Low P., Di Paolo G., Brodin L., Shupliakov O., De Camilli P. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Rothfield L.I., Cook W.R., de Boer P.A. Biogenesis of the Escherichia coli cell division system. Cold Spring Harbor Symp. Quant. Biol. 1991;56:751–756. doi: 10.1101/sqb.1991.056.01.084. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A.V., Witke W., Huttner W.B., Soling H.D. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. Division versus fusionDnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K.A., Yaffe M.P. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Methods in Yeast Genetics 1986. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: pp. 186 [Google Scholar]
- Smirnova E., Shurland D.-L., Ryazantsev S.M., van der Bliek A.M. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. The WD repeata common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Stevens B.J. Variation in number and volume of mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstruction. Biol. Cell. 1977;28:37–56. [Google Scholar]
- Takei K., McPherson P.S., Schmid S.L., De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. A novel WD protein, Mdv1p, interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial fission in yeast. J. Cell Biol. 2000;151:353–365. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van der Bliek A.M. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg S.M., Cooper J.A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S228C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Gorsich S., McCaffery J.M., Shaw J.M., Nunnari J. The dynamin-related GTPase, Mgm1p, is a mitochondrial intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]