c-Jun NH2-Terminal Kinase (JNK)1 and JNK2 Have Distinct Roles in CD8+ T Cell Activation (original) (raw)
Abstract
The c-Jun NH2-terminal kinase (JNK) signaling pathway is induced by cytokines and stress stimuli and is implicated in cell death and differentiation, but the specific function of this pathway depends on the cell type. Here we examined the role of JNK1 and JNK2 in CD8+ T cells. Unlike CD4+ T cells, the absence of JNK2 causes increased interleukin (IL)-2 production and proliferation of CD8+ T cells. In contrast, JNK1-deficient CD8+ T cells are unable to undergo antigen-stimulated expansion in vitro, even in the presence of exogenous IL-2. The hypoproliferation of these cells is associated with impaired IL-2 receptor α chain (CD25) gene and cell surface expression. The reduced level of nuclear activating protein 1 (AP-1) complexes in activated JNK1-deficient CD8+ T cells can account for the impaired IL-2 receptor α chain gene expression. Thus, JNK1 and JNK2 play different roles during CD8+ T cell activation and these roles differ from those in CD4+ T cells.
Keywords: T lymphocytes, MAP kinases, IL-2Rα, AP-1, c-Jun
Introduction
CD4+ and CD8+ T cells are two subsets of peripheral T cells that play important yet distinct roles during an immune response. After antigen stimulation, CD4+ T cells differentiate into effector Th1 or Th2 cells that secrete cytokines to help modulate the type of immune response that develops. Th1 cells promote cell-mediated immunity against intracellular microbial pathogens whereas Th2 cells promote humoral immunity against parasites and extracellular pathogens. CD8+ T cells differentiate into cytotoxic T (Tc)* cells to help defend the host during the cell-mediated immune response. Tc cells secrete high levels of cytokines, mainly IFN-γ, and have cytotoxic activities that are mediated by secreted proteins such as perforin and granzyme (1). Thus, in addition to the balance between Th1 and Th2 differentiation, impaired or enhanced CD8+ T cell activation and differentiation play crucial roles in the susceptibility or resistance of the host to intracellular pathogens.
During the activation and differentiation of CD4+ and CD8+ T cells, a substantial reprogramming of gene expression occurs ultimately giving these cells the ability to perform their effector functions. To date, many studies have identified and characterized transcription factors and signal transduction pathways that regulate CD4+ T cell activation and differentiation. Little is known about molecular mechanisms and signaling pathways that control these events in CD8+ T cells. The divergent functions of CD4+ and CD8+ T cells suggest that distinct signaling requirements and molecular mechanisms exist for each subset.
Signal transduction via mitogen-activated protein (MAP) kinases is involved in a variety of cellular responses, including growth factor–induced proliferation, differentiation, and cell death. Several parallel MAP kinase signal transduction pathways have been defined in mammalian cells and implicated in the control of the immune response (2–4). These pathways include the extracellular signal–regulated kinases (ERKs) (5, 6), c-Jun NH2-terminal kinases (JNKs, also known as SAPKs) (7, 8), and p38 MAP kinases (9–11).
JNK, like the other MAP kinases, requires dual phosphorylation of threonine and tyrosine residues within the protein kinase subdomain VIII by MAP kinase kinases (MKKs). MKK4 (SEK1) and MKK7 phosphorylate JNK in response to environmental stress and mitogenic factors (12–17). While MKK7 specifically regulates JNK, MKK4 has been shown to also activate p38 MAP kinase (12, 18). Mice deficient for either MKK4 or MKK7 are embryonic lethal (19–22). Interestingly, studies of Mkk4 − / −, Mkk7 − / −, or Mkk4 − / − Mkk7 − / − murine embryonic fibroblasts have shown that MKK4 and MKK7 differentially regulate the JNK signal transduction pathway (17). In addition, studies of chimeric mice containing either MKK4 or MKK7-deficient CD4+ T cells have shown that both of these kinases are required for the normal functioning of peripheral T cells (19–21). Recently, it has been shown that Mkk7 −/− chimeric mice also have dramatic B cell and mast cell abnormalities (22). Together these studies support the critical role of the JNK signaling pathway in the immune response.
Within the JNK group of MAP kinases, three genes have been identified, Jnk1, Jnk2, and Jnk3 (2, 4). In addition, 10 isoforms generated by alternative splicing have been identified. Each isoform differs in its interaction with specific substrates, such as c-Jun, Elk-1, and ATF2 (23). JNK has also been shown to phosphorylate and regulate the transcription activities of JunB (24) and nuclear factor of activated T cells (NFAT) (25, 26). Jnk3 is selectively expressed in heart, brain, and testis, whereas Jnk1 and Jnk2 are constitutively expressed in many tissues, except spleen and lymph nodes (27, 28). Recent studies have shown that Jnk1 and Jnk2 gene expression is induced in peripheral CD4+ T cells after in vitro and in vivo activation (28). In T cells a costimulatory signal mediated by CD28 is required to induce JNK activity (28, 29). Activation of JNK in thymocytes appears to be required for negative selection (30–33). Studies using mice deficient of Jnk2 or Jnk1 have shown that JNK2 is required for IFN-γ production by peripheral CD4+ T cells and subsequent Th1 differentiation in vitro (34) whereas JNK1 appears to be a negative regulator of Th2 differentiation in vitro (35). In correlation, mice deficient for Jnk1 are unable to resolve Leishmania major infections in vivo due to predominant Th2 responses (36). Mice deficient for both Jnk1 and Jnk2 are embyronic lethal due to increased cell death in the developing forebrain (37, 38). Recent studies of chimeric mice containing Jnk1 −/− Jnk2 − / − compound mutant CD4+ T cells confirm that JNK is required for effector function but not for CD4+ T cell activation (21). In contrast, other groups using total T cells from JNK2 (31) or JNK1-deficient mice (32) have shown that the JNK signaling pathway is required for T cell activation and IL-2 production.
In this study, we have examined the role of the JNK signaling pathway in CD8+ T cell function. Activated CD8+ T cells that lack JNK2 hyperproliferate due to increased IL-2 production whereas activated CD8+ T cells that lack JNK1 hypoproliferate due to reduced expression of the α chain of the IL-2 receptor. These results therefore indicate that JNK1 and JNK2 have distinct roles in CD8+ T cell activation.
Materials and Methods
Mice.
Jnk1 − / − and Jnk2 − / − mice have been described previously (34, 35). These mice are on the C57BL/6 background (The Jackson Laboratory). The animal infection procedures used in this study were approved by the Wellington School of Medicine Animal Ethics Committee and performed in accordance with the guidelines of the University of Otago, New Zealand. All other procedures performed on the animals used in this study are in accordance with the institutional guidelines of the Animal Care Facility at the University of Vermont.
Virus Infection.
The A/HKx31 (H3N2) influenza A virus is a laboratory-generated recombinant with the external surface components of A/Aichi/2/68 (H3H2) and the internal components of A/PR8/8/34 (H1N1) (39). Virus stocks were grown and viral titers were determined as described previously (40). Mice were anesthetized with an intraperitoneal injection of a mixture of ketamine and xylazine (Phoenix) and 12 hemagglutination units of virus were administered by intranasal inoculation.
Virus titers in the lung tissue of infected mice were determined using the Madin-Darby kidney cell assay as described previously (40). Virus titers were graphed by interpolation of the last dilution that showed hemagglutination.
Antibody production in infected mice was measured by ELISA using purified Influenza HKx31 antigen (10 μg/ml; SPAFAS) as described previously (41).
Cell Preparation and Surface Staining.
Total CD8+ T cells were isolated from spleen and lymph nodes by negative selection using anti-NK 1.1 (BD PharMingen), anti-CD4 (GK 1.5), anti-Mac1 (BD PharMingen), and anti-MHC class II mAbs followed by depletion with magnetic beads (PerSeptive Biosystems) as described previously (42–44). Total CD4+ T cells were similarly isolated from spleen and lymph nodes with anti-NK1.1, anti-CD8, anti-Mac1, and anti-MHC class II mAbs. CD8+ or CD4+ T cells (106 cells/ml) were activated with plastic-immobilized anti-CD3 (2C11; 5 μg/ml) in the presence or absence of soluble anti-CD28 mAb (1 μg/ml; BD PharMingen), IL-2 (50 U/ml; R&D Systems), and anti-IL-2 mAbs (10 μg/ml; BD PharMingen).
Expression of IL-2Rα (CD25) was determined by cell surface staining and flow cytometry (EPICS; Coulter), using a cychrome-conjugated anti-CD8 mAb and a PE-conjugated anti-CD25 mAb (BD PharMingen).
Proliferation and Measurement of Cytokine Production.
The proliferative response of purified CD8+ T cells was determined by measurement of [3H]thymidine (Amersham Pharmacia Biotech) incorporation after 3 d. IC50 values were calculated by nonlinear regression.
ELISAs were performed using purified anti–IFN-γ or anti–IL-2 mAb (2 μg/ml), biotinylated anti-IFN-γ or anti–IL-2 mAb, horseradish peroxidase–conjugated avidin D (2.5 μg/ml; Vector Laboratories), peroxidase substrate and reaction stop solutions (Kirkegaard and Perry Laboratories, Inc.) following the recommended protocol (BD PharMingen). Recombinant mouse IFN-γ (GIBCO BRL) and mouse IL-2 (R&D Systems) were used as standards.
Cell Viability and Terminal Deoxynucleotidyltransferase-mediated dUTP Nick End-labeling Staining.
Apoptosis of total CD8+ T cells was determined by the terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling (TUNEL) assay (BD PharMingen) following the manufacturer's recommended protocol.
Western Blot Analysis.
c-Jun expression was measured by immunoblot analysis using a polyclonal antibody raised against or c-Jun (Cell Signaling Technologies) following the manufacturer's protocol. JNK protein levels were measured by immunoblot analysis using an anti–human JNK mAb (BD PharMingen) (28). Blots were then stripped and reprobed with an anti-actin mAb (AC40; Sigma-Aldrich). Immune complexes were detected by enhanced chemiluminescence (Kirkegaard & Perry Laboratories, Inc.).
Protein Kinase Assays.
Protein kinase assays were performed as described (7). Cell lysates were incubated in Triton lysis buffer (20 mM Tris pH 7.4, 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 2 mM pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin) with GST-c-Jun immobilized on GSH-agarose beads. After 12 h at 4°C, the beads were washed extensively in lysis buffer followed by kinase assay buffer (25 mM HEPES pH 7.4, 25 mM β-glycerophosphate, 25 mM MgCl2, 0.1 mM sodium orthovanadate, 0.5 mM DTT) and the activity of the bound JNK was detected by the addition of [γ-32P]ATP for 30 min at 30°C. The reaction products were resolved by SDS-PAGE and the incorporation of [32P]phosphate was quantitated by PhosphorImager analysis (Molecular Dynamics).
Northern Blot Analysis and Ribonuclease Protection Assays.
Total RNA from CD8+ T cells was isolated using the Ultraspec RNA isolation reagent (Biotecx Laboratory) as recommended by the manufacturer. 7 μg of RNA was examined by Northern blot as described previously (45). Specific cDNA probes for Jnk1 and Jnk2 (28) were labeled with [32P]dCTP using the Random Primer Kit (Stratagene).
Total RNA was extracted as above. 2–3 μg of total RNA was examined by ribonuclease protection assay (RPA) using [32P]UTP-labeled RNA probes transcribed in vitro from mCK-1 and mCR-3 DNA templates provided in the RPA kit following the manufacturer's recommended protocol (BD PharMingen).
Nuclear Extracts and Electrophoretic Mobility Shift Assays.
Nuclear extracts from CD8+ T cells or CD4+ T cells (2–3 × 106) were made as described previously (46, 47). Binding reactions for the electrophoretic mobility shift assays (EMSAs) were performed using 2 μg of nuclear protein and a [32P]dCTP-end-labeled double strand oligonucleotide as described previously (43). The oligonucleotides used in this study are as follows: activating protein 1 (AP-1) (5′-GTCGACGTGAGTCAGCGCGC-3′) (48, 49); IL-4-NFAT (5′-GTAATAAAATTTTCCAATGTAAA-3′) (50); cyclic AMP-responsive element (CRE) (5′-GATCTCTCTGACGTCAGCCAAGGAGGAGGCC-3′) (51); and IL-2Rα/AP-1(5′-GCTTTGTTGAGTCTTCTGG-3′) (52). 2 μl/reaction of anti-Jun family rabbit antiserum was used per reaction in competition studies.
Luciferase Activity.
Luciferase activity in cell extracts was measured as described previously (53) using the Luciferase Assay kit (Promega).
Results
Normal Viral Immune Response in JNK2-deficient Mice.
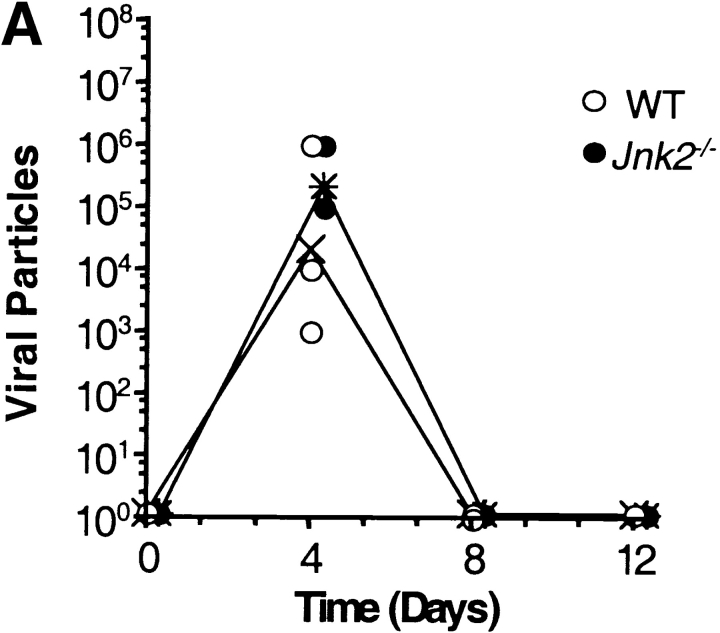
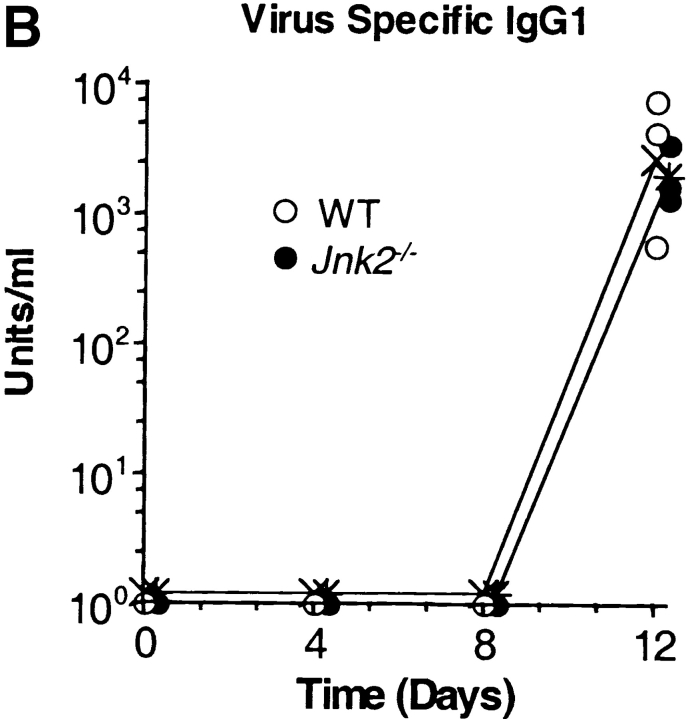
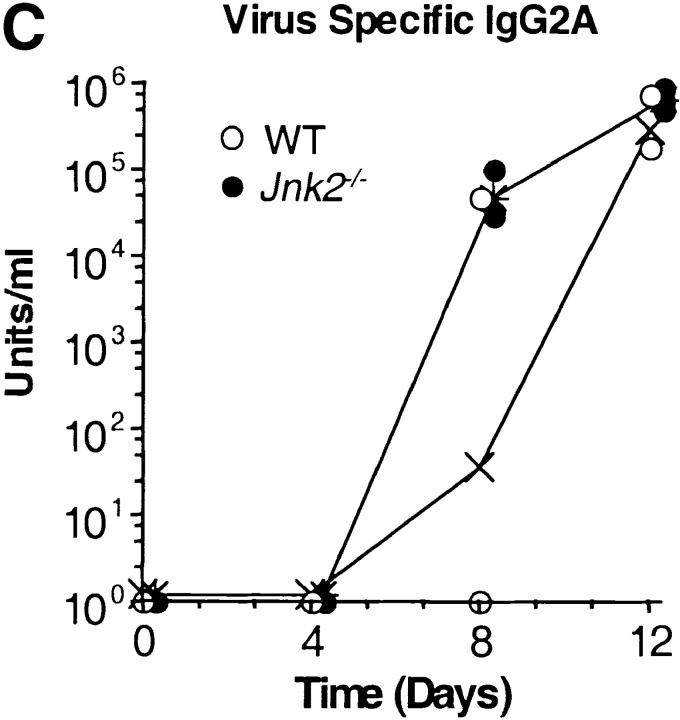
We have previously shown that CD4+ T cells from JNK2-deficient (Jnk2 − / −) mice had reduced IFN-γ production and impaired Th1 differentiation in vitro whereas Th2 differentiation was unaffected (34). To determine whether JNK2 was required for a Th1 immune response in vivo, wild-type and Jnk2 − / − mice were intranasally inoculated with the HKx31 strain of the human influenza virus. Both wild-type and Jnk2 − / − mice developed nonfatal pneumonia and displayed clinical signs of infection characterized by anorecticism, increased respiratory rates, ruffled fur, and grayish atelectatic areas on the lungs (data not shown). Furthermore, the size of the mediastinal lymph node increased in both wild-type and Jnk2 − / − mice after infection (data not shown). To examine virus titers, lung tissue was harvested from wild-type and Jnk2 − / − mice at different time points following the intranasal inoculation with HKx31. The virus titers in the lung tissue of Jnk2 − / − mice were similar to those observed in wild-type mice at the peak of infection (day 4; Fig. 1 A). Moreover, similar levels of virus-specific IgG1 and IgG2A antibodies were detected in the serum of infected Jnk2 − / − and wild-type mice (Fig. 1, B and C). Thus, despite the impaired Th1 differentiation in vitro, the absence of JNK2 did not interfere with the in vivo antiviral immune response.
Figure 1.
In vivo viral immune response in the JNK2-deficient mice. (A) Virus titers in the lung tissue of infected wild-type (WT) and Jnk2 − / − mice were determined at different time points during influenza infection. Open circles represent individual infected wild-type mice; filled circles represent individual infected Jnk2 − / − mice; (X) represents the geometric mean for wild-type mice; (*) represents the geometric mean for Jnk2 − / − mice. (B and C) Serum was collected from infected wild-type and Jnk2 − / − mice at different time points during influenza infection. Virus-specific IgG1 (B) and virus-specific IgG2A (C) Ab levels were determined by ELISA.
Regulation of the JNK2 During CD8+ T Cell Activation.
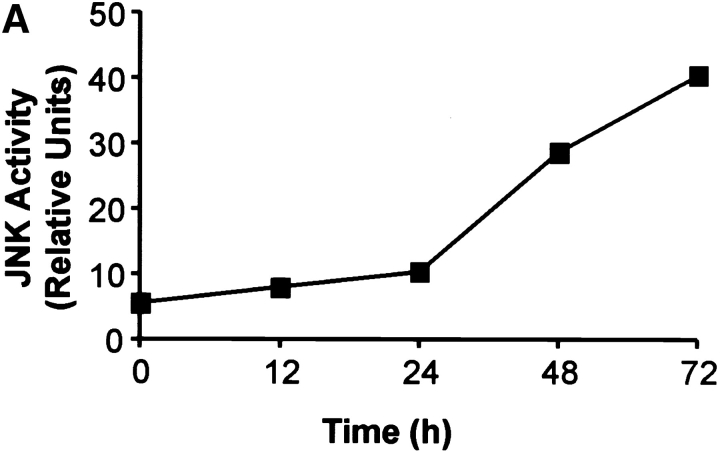
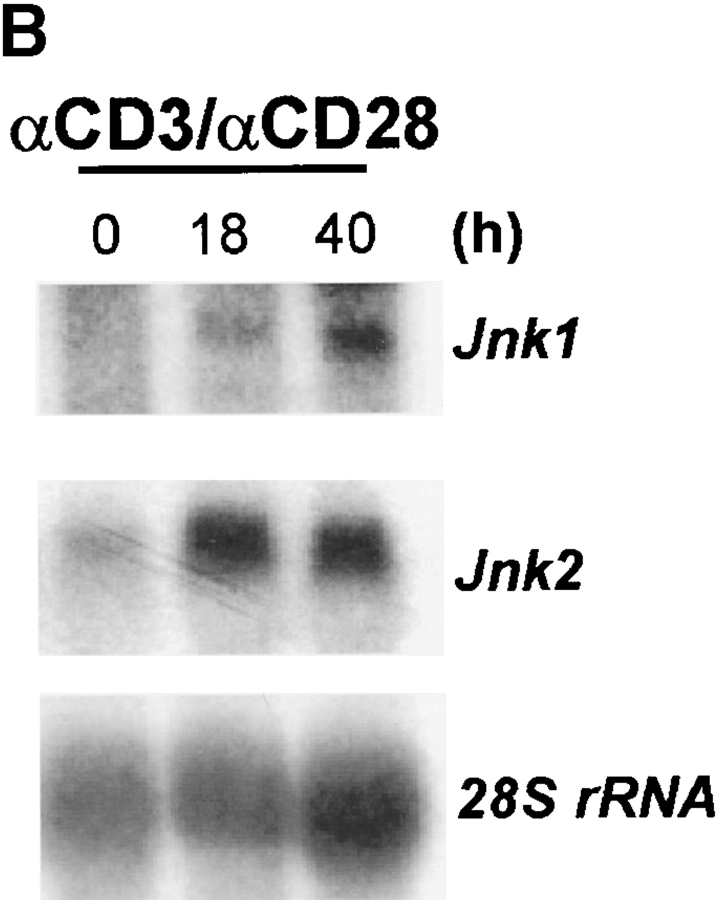
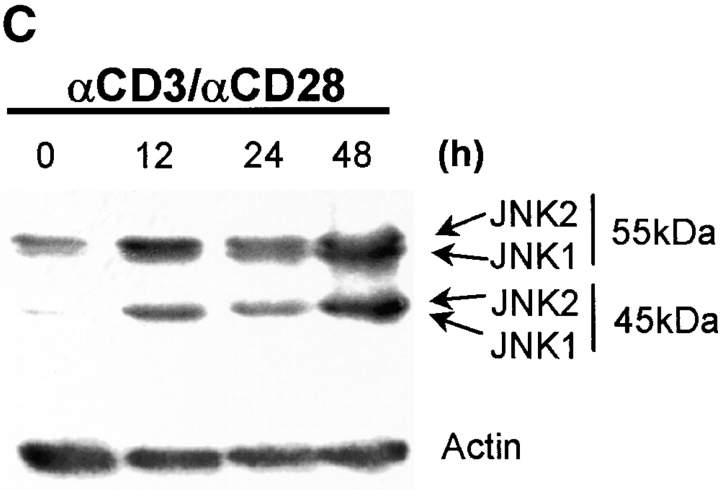
CD4+ T cells play an important role in mediating the antiviral immune response. However, during the influenza infection in mice, CD8+ T cells play a critical role in viral clearance from the host during the influenza infection. As the function of JNK2 in CD8+ T cells remained unknown, it was possible that the presence of functional CD8+ T cells in the Jnk2 − / − mice compensated for their impaired Th1 response. To investigate the role of JNK2 in CD8+ T cells, we examined JNK activity in CD8+ T cells isolated from wild-type mice in response to stimulation with anti-CD3 and anti-CD28 mAbs. Similar to the delayed kinetics of JNK activation in CD4+ T cells (28), activation of JNK in CD8+ T cells required long periods of stimulation (∼24 h; Fig. 2 A). We have previously shown that the delayed kinetics of JNK activation in CD4+ T cells was due to the low level of JNK1 and JNK2 proteins in freshly isolated cells and upregulation of Jnk1 and Jnk2 gene expression after stimulation (28). To determine whether the increase in JNK activity in activated CD8+ T cells correlated with increased Jnk1 and Jnk2 gene expression, we examined Jnk1 and Jnk2 mRNA levels in CD8+ T cells after stimulation with anti-CD3 and anti-CD28 mAbs by Northern blot analysis. Low levels of Jnk1 and Jnk2 mRNA were present in freshly isolated CD8+ T cells while high levels of Jnk1 and Jnk2 mRNA were found in activated CD8+ T cells (Fig. 2 B). In correlation, high levels of JNK1 and JNK2 protein were present in activated CD8+ T cells (Fig. 2 C).
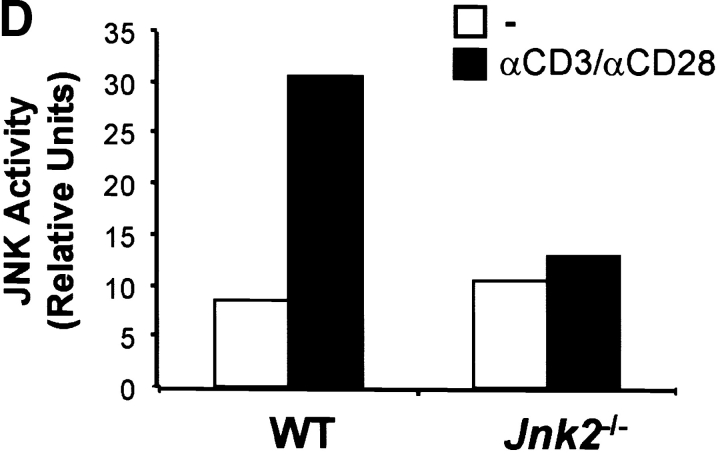
Figure 2.
Activation of the JNK signaling pathway in CD8+ T cells. (A) Total CD8+ T cells (106 cells/ml) from wild-type mice were stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs. Cells were harvested at the indicated time points, lysed, and cell extracts were assayed for JNK activity. Phosphorylated c-Jun was detected after SDS-PAGE by autoradiography. (B) Jnk expression was examined by Northern blot in CD8+ T cells stimulated as in A for the indicated time periods. Expression of 28S ribosomal RNA was used as a control. (C) JNK expression was examined by Western blot analysis in CD8+ T cells stimulated as in A for the indicated time periods. Actin expression was used as a control. (D) JNK activity was determined using cell extracts from freshly isolated (-) wild-type (WT) and Jnk2 − / − CD8+ T cells and wild-type and Jnk2 − / − CD8+ T cells stimulated as in A for 48 h.
To determine whether JNK2 contributed to the JNK activity observed in CD8+ T cells, we examined total JNK activity in CD8+ T cells isolated from wild-type and Jnk2 − / − mice activated with anti-CD3 and anti-CD28 mAbs. Reduced levels of JNK activity were observed in activated CD8+ T cells from Jnk2 − / − mice compared with activated wild-type CD8+ T cells (Fig. 2 D). These data indicated that JNK2 contributed to the total JNK activity in CD8+ T cells and that the presence of JNK1 did not completely compensate for the absence of JNK2.
JNK2 Is a Negative Regulator of CD8+ T Cell Activation.
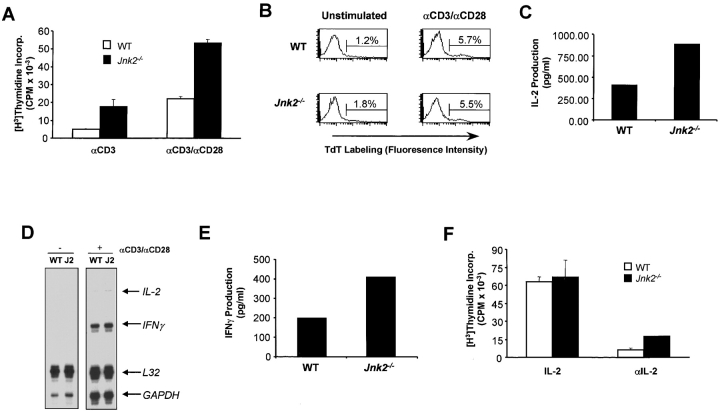
To determine whether the lack of JNK2 could affect CD8+ T cell activation, we examined the proliferative response of Jnk2 − / − CD8+ T cells. Increased proliferation of Jnk2 − / −CD8+ T cells was observed compared with wild-type CD8+ T cells after activation with anti-CD3 and anti-CD28 mAbs (Fig. 3 A). Despite the high rate of proliferation, no differences in the surface expression of activation markers (CD25 [IL-2Rα], CD69, CD44) were observed between activated wild-type and Jnk2 − / − CD8+ T cells (data not shown). Increased proliferation was also observed when Jnk2 − / −CD8+ T cells were activated with Concavalin A and antigen-presenting cells (data not shown). In correlation with our in vitro results, increased expansion of CD8+ T cells in Jnk2 −/− mice has also been observed during the lymphocytic choriomeningitis virus (LCMV) infection in vivo (Arbour et al. in this issue [53a]).
Figure 3.
JNK2 is a negative regulator of CD8+ T cells activation. (A) Total CD8+ T cells (2.5 × 105 cells/ml) from wild-type (WT) and Jnk2 − / − mice were stimulated with immobilized anti-CD3 (5 μg/ml) alone or in combination with soluble anti-CD28 (1 μg/ml) mAbs. The proliferative response was examined by [3H]thymidine incorporation. (B) Cell death determined by the TUNEL assay in freshly isolated (unstimulated) wild-type and Jnk2 − / − CD8+ T cells and wild-type and Jnk2 − / − CD8+ T cells (106 cells/ml) stimulated with anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 24 h. Numbers represent the percentage of TUNEL-positive cells. TdT, terminal deoxynucleotidyltransferase. (C) Wild-type and Jnk2 − / − CD8+ T cells were stimulated as described in B for 48 h IL-2 production was determined by ELISA. (D) Cytokine gene expression was determined by RPA using RNA extracted from freshly isolated (−) Wild-type and Jnk2 − / − (J2) CD8+ T cells and wild-type and J2 CD8+ T cells stimulated (+) as described in B for 24 h (E) Wild-type and Jnk2 − / − CD8+ T cells were stimulated as in B for 48 h IFN-γ production was determined by ELISA. (F) CD8+ T cells from wild-type and Jnk2 − / − mice (2.5 × 105 cells/ml) were stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAb in the presence of exogenous IL-2 (50 U/ml) or a neutralizing anti-IL-2 (10 μg/ml) mAb. The proliferative response was analyzed by [3H]thymidine incorporation.
Previous studies have demonstrated that the activation of JNK signaling pathway is implicated in cell death (for a review, see reference 4). We examined whether the increased proliferation of CD8+ T cells from Jnk2 − / − mice was due to increased resistance of these cells to activation-induced death. CD8+ T cells from wild-type and Jnk2 − / − mice were activated with anti-CD3 and anti-CD28 mAbs and cell death was determined by the TUNEL assay. Similar percentages of apoptotic cells were observed in freshly isolated and activated wild-type and Jnk2 − / − CD8+ T cell populations (Fig. 3 B). These data indicated that the absence of JNK2 did not affect the ability of these cells to undergo cell death.
IL-2 is the major growth factor for CD8+ T cells. IL-2 is also produced by CD8+ T cells but to a much lesser extent than CD4+ T cells. It was therefore possible that the increased proliferation of Jnk2 − / −CD8+ T cells resulted from increased production of IL-2. We therefore examined the IL-2 production by CD8+ T cells isolated from wild-type and Jnk2 − / − mice. Activated CD8+ T cells from Jnk2 − / − mice produced increased levels of IL-2 compared with activated CD8+ T cells from wild-type mice (Fig. 3 C). To determine whether the increased amount of IL-2 detected in the culture supernatant was due to increased production of IL-2 rather than reduced consumption, we examined IL-2 gene expression by an RPA. IL-2 mRNA was not detected in freshly isolated wild-type and Jnk2 − / −CD8+ T cells despite similar mRNA levels of the housekeeping genes L32 and GAPDH (Fig. 3 D). However, increased levels of IL-2 mRNA were detected in activated CD8+ T cells from Jnk2 − / − mice compared with activated wild-type CD8+ T cells (Fig. 3 D) while the levels of L32 and GAPDH mRNA were similar (Fig. 3 D). A slight increase in the expression of IFN-γ mRNA was also detected in activated Jnk2 − / −CD8+ T cells compared with activated wild-type CD8+ T cells (Fig. 3 D). In correlation, higher amounts of IFN-γ were detected in the culture supernatant of activated Jnk2 − / −CD8+ T cells (Fig. 3 E).
To determine whether the higher rate of proliferation observed in Jnk2 − / −CD8+ T cells compared with wild-type CD8+ T cells was due to the amount of IL-2 produced during activation, we examined the proliferative response of wild-type and Jnk2 − / −CD8+ T cells activated in the presence of exogenous IL-2. Similar amounts of proliferation were observed in wild-type and Jnk2 − / −CD8+ T cells (Fig. 3 F) in these conditions. Moreover, the presence of a neutralizing anti-IL-2 mAb during the activation of these cells abolished the hyperproliferation of Jnk2 − / −CD8+ T cells (Fig. 3 F). Together, these data indicated that JNK2 deficiency caused increased production of IL-2 and subsequent hyperproliferation of activated CD8+ T cells.
Impaired Proliferation of Jnk1−/−CD8+ T Cells.
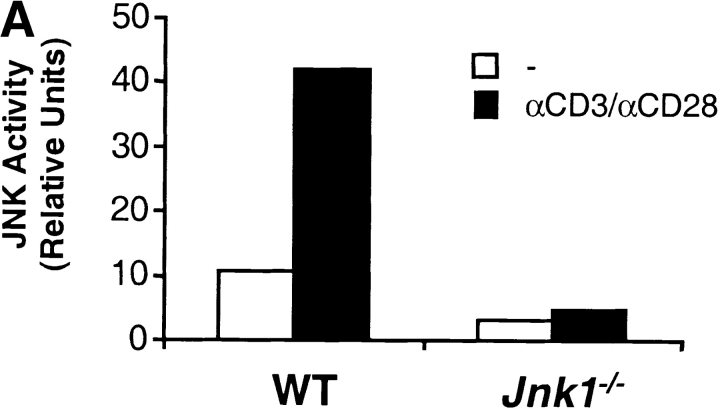
Both JNK1 and JNK2 are expressed at low levels in resting CD4+ T cells and high levels in activated CD4+ (28). While JNK2 is required for IFN-γ production by CD4+ T cells (34), JNK1 negatively regulates IL-4 production by CD4+ T cells (35). These studies indicate that these two isoforms control different aspects of CD4+ T cell effector function. To determine whether these two kinases also have distinct functions in CD8+ T cells, we examined JNK1-deficient CD8+ T cells. We first examined JNK activity in activated CD8+ T cells from wild-type and Jnk1 − / − mice stimulated with anti-CD3 and anti-CD28 mAbs. Unlike wild-type CD8+ T cells, very low levels of JNK activity were detected in Jnk1 − / −CD8+ T cells (Fig. 4 A). Interestingly, the reduction in JNK activity in activated Jnk1 − / −CD8+ T cells compared with wild-type CD8+ T cells (Fig. 4 A) was greater than the reduction observed in activated Jnk2 − / −CD8+ T cells (Fig. 2 D). These data suggested that JNK1 was the predominant contributor to the total JNK activity in CD8+ T cells.
Figure 4.
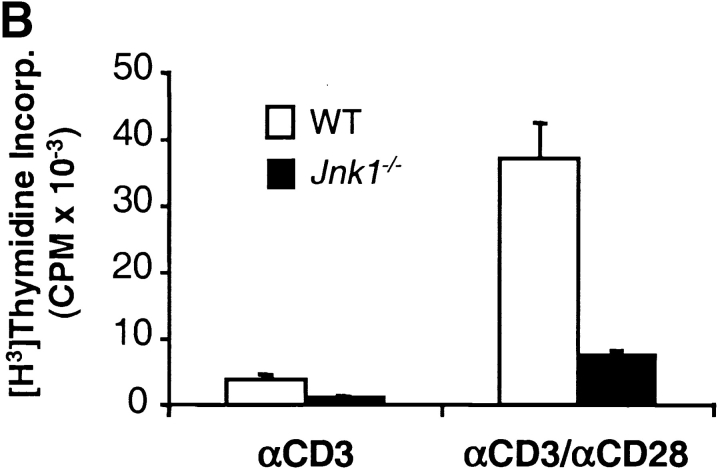
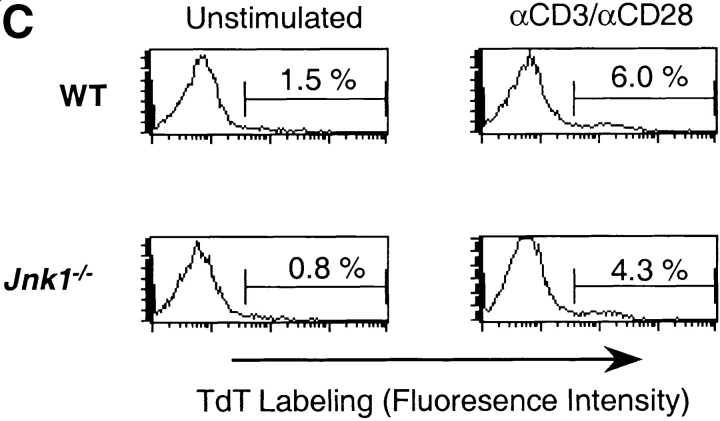
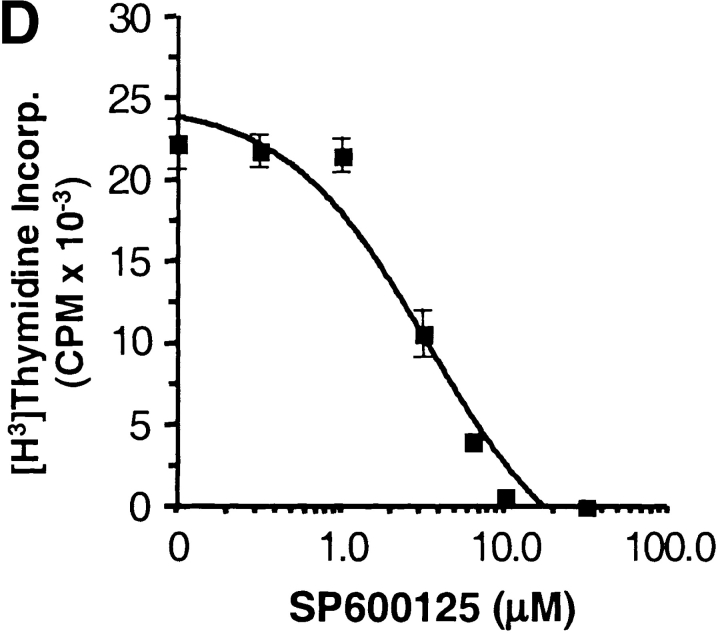
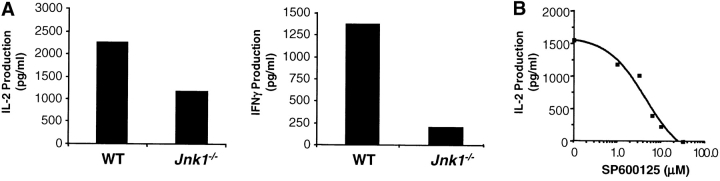
JNK1 is required for CD8+ T cell activation. (A) JNK activity was determined using cell extracts from freshly isolated (−) wild-type (WT) and Jnk1 − / − CD8+ T cells and wild-type and Jnk1 − / − CD8+ T cells (106 cells/ml) stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 48 h (B) CD8+ T cells (2.5 × 105 cells/ml) from wild-type and Jnk1 − / − mice were stimulated with anti-CD3 alone or in combination with soluble anti-CD28 mAbs. The proliferative response was examined by [3H]thymidine incorporation. (C) Cell death determined by the TUNEL assay in freshly isolated (unstimulated) wild-type and Jnk1 − / − CD8+ T cells and wild-type and Jnk1 − / − CD8+ T cells (106 cells/ml) stimulated as in A for 24 h. Numbers represent the percentages of TUNEL-positive cells. TdT, terminal deoxynucleotidyltransferase. (D) Wild-type CD8+ T cells were stimulated as in B in the presence of different concentration of the JNK inhibitor (SP600125). The proliferative response was examined by [3H]thymidine incorporation. An IC50 was calculated by nonlinear regression (IC50 = 3.4 μM).
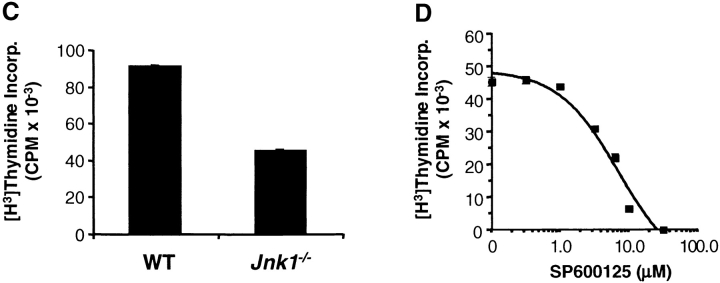
To determine whether the effects of JNK1-deficiency on CD8+ T cell proliferation were similar to JNK2-deficiency, we compared the proliferation of wild-type and Jnk1 − / −CD8+ T cells. In contrast to Jnk2 − / −CD8+ T cells, the proliferative response of Jnk1 − / −CD8+ T cells was substantially impaired in response to either anti-CD3 mAb alone or anti-CD3 plus anti-CD28 mAbs (Fig. 4 B). Similar percentages of apoptotic CD8+ T cells were observed after the activation of wild-type and Jnk1 − / −CD8+ T cells with anti-CD3 and anti-CD28 mAbs (Fig. 4 C), indicating that the hypoproliferation of these cells was not due to increased cell death. Together these data show that JNK1 was required for CD8+ T cell expansion in vitro. Arbour et. al. have also shown that JNK1 is required for the expansion of virus specific CD8+ T cells in vivo (53a).
Mice with compound deficiencies of JNK1 and JNK2 die in utero (37, 38). As an alternative approach to address how the absence of JNK activity affects the activation of CD8+ T cells, we examined the effect of a newly described pharmacological compound (SP600125), which inhibits both JNK1 and JNK2 (54). Activation of wild-type CD8+ T cells in the presence of different concentrations of the JNK inhibitor caused a dose-dependent inhibition of proliferation (IC50 = 3.4 μM; Fig. 4 D). Complete inhibition of proliferation with 10 μM, however, could be due to nonspecific effects of the inhibitor on other signaling pathways. CD8+ T cells activated in the presence of the JNK inhibitor behaved like the Jnk1 − / −CD8+ T cells, indicating that the function of JNK1 predominates over the function of JNK2. Thus, while JNK2 appears to be a negative regulator for CD8+ T cells, JNK1 is required for CD8+ T cell activation.
To determine whether JNK-deficiency could affect the effector function of CD8+ T cells, we examined the amount of IL-2 and IFN-γ produced by CD8+ T cells isolated from wild-type and Jnk1 − / − mice. In correlation with their inability to undergo activation-induced expansion, Jnk1 − / −CD8+ T cells produced lower amounts of IL-2 and IFN-γ compared with activated wild-type CD8+ T cells (Fig. 5 A). Activation of wild-type CD8+ T cells in the presence of different concentrations of the JNK inhibitor caused a dose-dependent inhibition of IL-2 production (Fig. 5 B).
Figure 5.
IL-2 does not restore impaired proliferation of Jnk1 − / − CD8+ T cells. (A) Wild-type (WT) and Jnk1 − / − CD8+ T cells (106 cells/ml) were stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 48 h IL-2 (left panel) and IFN-γ (right panel) production was determined by ELISA. (B) CD8+ T cells (106 cells/ml) from wild-type mice were stimulated as in A for 48 h in the presence of different concentration of the JNK inhibitor (SP600125). IL-2 production was determined by ELISA. (C) CD8+ T cells from wild-type and Jnk1 − / − mice (2.5 × 105 cells/ml) were stimulated as in A in the presence of exogenous IL-2 (50 U/ml). The proliferative response was examined by [3H]thymidine incorporation. (D) Total CD8+ T cells (2.5 × 105 cells/ml) from wild-type mice were stimulated as in A in the presence of different concentrations of the JNK inhibitor and exogenous IL-2. The proliferative response was examined by [3H]thymidine incorporation. An IC50 was calculated by nonlinear regression (IC50 = 7.4 μM).
It was possible that the lower amounts of IL-2 produced by activated Jnk1 −/−CD8+ T cells accounted for their impaired proliferation. To determine whether IL-2 could rescue the low levels of proliferation of these cells, we examined the proliferative response of Jnk1 − / −CD8+ T cells activated in the presence of exogenous IL-2. Despite saturating levels of IL-2, the proliferation of activated Jnk1 − / − CD8+ T cells remained reduced compared with activated wild-type CD8+ T cells (Fig. 5 C). In correlation, the presence of exogenous IL-2 did not prevent the inhibition of CD8+ T cell proliferation caused by the JNK inhibitor (IC50 = 7.4 μM; Fig. 5 D). Thus the impaired proliferation of Jnk1 −/−CD8+ T cells was not due to low levels of IL-2 produced by these cells.
JNK1 Is Required for IL-2 Receptor α Chain Expression.
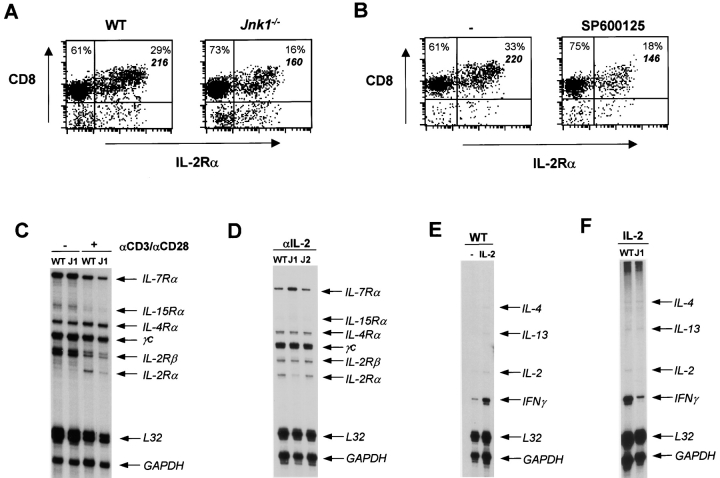
The inability of exogenous IL-2 to restore the reduced proliferation of Jnk1 −/−CD8+ T cells suggested that JNK1 could be required for IL-2 receptor expression. We examined the cell surface expression of the α chain of the IL-2 receptor (IL-2Rα/CD25) by flow cytometry on wild-type and Jnk1 −/−CD8+ T cells activated with anti-CD3 and anti-CD28 mAbs. The percentage of activated Jnk1 − / − CD8+ T cells that expressed IL-2Rα was significantly reduced compared with activated wild-type CD8+ T cells (Fig. 6 A). Moreover, the mean fluorescence intensity of IL-2Rα on Jnk1 − / −CD8+ T cells was substantially lower than the mean fluorescence intensity on wild-type CD8+ T cells (Fig. 6 A). Reduced expression of IL-2Rα in Jnk1 − / − CD8+ T cells was also observed even in the presence of exogenous IL-2 (data not shown). In correlation, IL-2Rα cell surface expression on wild-type CD8+ T cells activated in the presence of the JNK inhibitor was also substantially reduced (Fig. 6 B).
Figure 6.
Impaired IL-2Rα gene expression in Jnk1 − / − CD8+ T cells. (A) CD8+ T cells from wild-type (WT) and Jnk1 − / − mice (106 cells/ml) were stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 48 h and stained with anti-IL-2Rα (CD25) mAbs. Cell surface expression of IL-2Rα was determined by flow cytometry. Numbers represent the percentage of cells in each quadrant. The mean fluorescence intensity for the upper right quadrant of each plot is indicated in bold italics. (B) Cell surface expression of IL-2Rα was determined on wild-type CD8+ T cells (106 cells/ml) stimulated as in A in the presence of the JNK inhibitor (SP600125; 5.6 μM). (C) Cytokine receptor gene expression was determined by RPA using RNA extracted from freshly isolated (−) wild-type and Jnk1 − / − (J1) CD8+ T cells and wild-type and J1 CD8+ T cells stimulated (+) as in A for 24 h. (D) Cytokine receptor gene expression was determined by RPA using RNA extracted from wild-type, J1, and Jnk2 − / − (J2) CD8+ T cells stimulated as in A for 36 h in the presence of a neutralizing anti-IL-2 (10 μg/ml) mAb. (E) Cytokine gene expression was determined by RPA using RNA extracted from wild-type CD8+ T cells stimulated as in A in the absence (−) or presence of exogenous IL-2 (50 U/ml) for 24 h. (F) Cytokine gene expression was determined by RPA using RNA extracted from wild-type and J1 CD8+ T cells stimulated as in A for 24 h in the presence of exogenous IL-2.
To determine whether the reduced cell surface expression of IL-2Rα on activated Jnk1 − / −CD8+ T cells was due to impaired IL-2Rα gene expression, we examined IL-2Rα chain mRNA levels by RPA using RNA extracted from activated wild-type and Jnk1 − / −CD8+ T cells. Consistent with the reduced levels of IL-2Rα on the surface of Jnk1 − / −CD8+ T cells, the levels of IL-2Rα mRNA in activated Jnk1 − / −CD8+ T cells were also substantially lower than the levels detected in wild-type CD8+ T cells (Fig. 6 C). No significant differences in IL-2Rβ or the γc chain mRNA were observed between activated wild-type and Jnk1 − / −CD8+ T cells (Fig. 6 C), indicating the JNK1 was specifically required for the expression of the IL-2Rα chain in CD8+ T cells.
Unstimulated T cells do not express IL-2Rα gene. After TcR ligation, IL-2Rα gene expression is induced and subsequently upregulated in response to IL-2. To determine whether JNK1 was required for TcR-mediated upregulation of the IL-2Rα gene, we examined IL-2Rα gene expression in wild-type and Jnk1 −/−CD8+ T cells activated in the presence of a neutralizing anti–IL-2 mAb to prevent IL-2–induced upregulation of the IL-2Rα. Even in the presence of the anti–IL-2 mAb the levels of expression of IL-2Rα gene were significantly reduced in activated Jnk1 − / − CD8+ T cells compared with activated wild-type CD8+ T cells (Fig. 6 D). In contrast, normal levels of IL-2Rα mRNA were found in activated Jnk2 − / −CD8+ T cells (Fig. 6 D). Thus, JNK1 was required for TcR-induced IL-2Rα gene expression.
The impaired expression of IL-2Rα in Jnk1 − / −CD8+ T cells could explain the hypoproliferation of these cells. Could the impaired expression of IL-2Rα also explain the inability of Jnk1 − / −CD8+ T cells to produce IL-2 and IFN-γ? We first examined the effect of exogenous IL-2 on the expression of these genes during the activation of wild-type CD8+ T cells. The presence of exogenous IL-2 caused a dramatic increase of IFN-γ mRNA levels and a moderate increase in IL-2 mRNA levels (Fig. 6 E). To determine whether the low levels of IL-2Rα on the surface of Jnk1 − / −CD8+ T cells could prevent the upregulation of IL-2 and IFN-γ gene expression by IL-2, we examined cytokine mRNA levels in Jnk1 − / −CD8+ T cells activated in the presence of exogenous IL-2. Unlike activated wild-type CD8+ T cells, Jnk1 − / −CD8+ T cells contained low levels of IFN-γ and nearly undetectable levels of IL-2 mRNA (Fig. 6 F). Together these results support the impaired expression of IL-2Rα on Jnk1 − / −CD8+ T cells as the primary cause of the hypoproliferation and reduced production of effector cytokines by these cells.
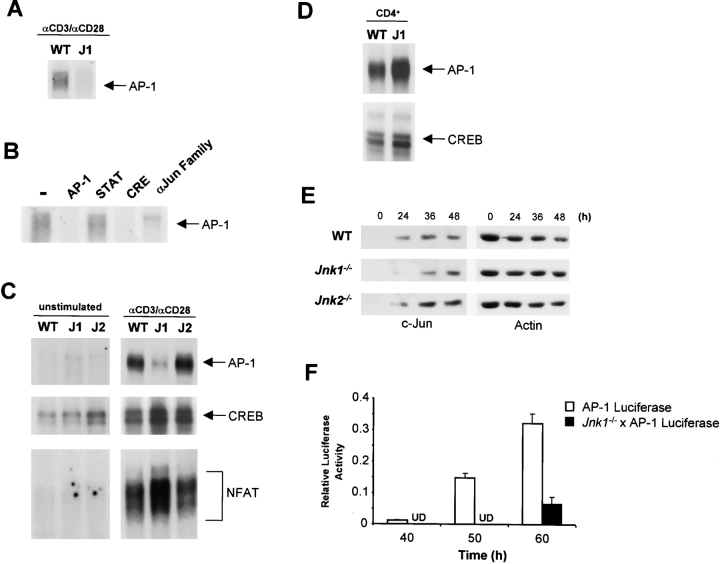
JNK1 Is Required for AP-1 DNA Binding to the IL-2Rα Promoter in CD8+ T Cells.
Several substrates of JNK have been identified. Specifically, JNK has been shown to regulate the Jun family of proteins and, therefore, the activity of the transcription factor AP-1 (2). Two AP-1–like sites have been identified in the −415–+1 promoter region of the IL-2Rα gene (52) but they have not been well characterized. To determine whether AP-1 complexes were able to bind to these AP-1-like sites, we performed EMSAs using specific oligonucleotides that spanned the AP-1–like sites from the IL-2Rα promoter and nuclear extracts from activated wild-type CD8+ T cells. We could not detect nuclear complexes bound to the –336 to −321 AP-1–like consensus sequence (GGACTCA) (data not shown). However, a –365 to –347 oligonucleotide containing the AP-1–like sequence (TGAGTCT) (IL-2Rα/AP-1) was able to bind nuclear complexes (Fig. 7 A). Interestingly, the level of binding to this IL-2Rα/AP-1 element was reduced in activated JNK1-deficient CD8+ T cells compared with activated wild-type CD8+ T cells (Fig. 7 A). To verify that the complexes bound to the IL-2Rα/AP-1 element were indeed AP-1, we performed competition studies using a cold oligonucleotide containing an AP-1 consensus sequence from the human collagenase promoter (53). The presence of the cold AP-1 oligonucleotide completely abrogated DNA binding to the IL-2Rα/AP-1 element (Fig. 7 B). In contrast, cold oligonucleotides containing unrelated consensus sequences such as those for the transcription factors signal transducer activator of transcription (STAT; Fig. 7 B) or nuclear factor (NF)κB (data not shown) did not affect the DNA binding to the IL-2Rα/AP-1 element. A cold oligonucleotide containing the consensus CRE, which has homology with the AP-1 consensus sequence (51), also competed with the IL-2Rα/AP-1 DNA binding (Fig. 7 B). Competition with anti-Jun family antisera confirmed the presence of Jun family members in the IL-2Rα/AP-1 complex (Fig. 7 B). In addition, competition with antisera raised against CRE binding protein (CREB) also showed the presence of CREB in the IL-2Rα/AP-1 complex (data not shown).
Figure 7.
Reduced binding of AP-1 DNA binding in Jnk1 − / −CD8+ T cells. (A) DNA binding of transcription factors to the AP-1–like site in the IL-2Rα promoter (AP-1/IL-2Rα) was determined using nuclear extracts prepared from wild-type (WT), and Jnk1 − / − (J1) CD8+ T cells stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 36 h and a [32P]-end-labeled double stranded oligonucleotide spanning –365 to –347 of the IL-2Rα promoter. (B) Composition of the AP-1 complex detected in activated CD8+ T cells. Nuclear extracts were prepared from wild-type CD8+ T cells stimulated with immobilized anti-CD3 and soluble anti-CD28 mAbs as in A. DNA binding to the AP-1/IL-2Rα element was performed in the absence (−) or presence of cold AP-1, STAT, or CRE oligonucleotides. DNA binding to the AP-1/IL-2Rα element was also examined in the presence of anti-Jun family (αJun Family) antiserum. (C) AP-1, CREB, and NFAT DNA binding was determined using nuclear extracts from unstimulated wild-type, J1, and Jnk2 −/− (J2) CD8+ T cells and wild-type, J1, and J2 CD8+ T cells stimulated as in A using [32P]-end-labeled double stranded oligonucleotides containing either an AP-1, CREB, or NFAT consensus binding site. (D) AP-1 and CREB DNA binding was determined using nuclear extracts from wild-type and J1 CD4+ T cells stimulated with immobilized anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs for 36 h using [32P]-end-labeled double stranded oligonucleotides containing either an AP-1 or CRE consensus sequence. (E) Western blot of c-Jun and actin in wild-type, Jnk1 − / −, and Jnk2 − / −CD8+ T cells stimulated as in A for the indicated time periods. (F) CD8+ T cells were isolated from AP-1-luciferase and Jnk1 −/− × AP-1-luciferase reporter transgenic mice, stimulated with immobilized anti-CD3 and soluble anti-CD28 as in A for different time periods, harvested, and assayed from luciferase activity. UD, undetectable.
To determine whether the reduced IL-2Rα/AP-1 DNA binding found in activated Jnk1 − / −CD8+ T cells was due to reduced amounts of AP-1 or CREB we examined the DNA binding to consensus AP-1 and CRE DNA regulatory sequences. A substantial reduction in the amount of AP-1 DNA binding was observed in activated Jnk1 −/− CD8+ T cells compared with that in activated wild-type CD8+ T cells (Fig. 7 C). In contrast, the levels of CREB DNA binding in activated Jnk1 − / −CD8+ T cells was not reduced (Fig. 7 C). Unlike Jnk1 −/−CD8+ T cells, activated Jnk2 −/−CD8+ T cells had levels of AP-1 and CREB DNA binding that were similar to those in activated wild-type CD8+ T cells (Fig. 7 C). These results indicated that the reduced levels of IL-2Rα/AP-1 DNA binding in Jnk1 −/− CD8+ T cells was due primarily to the low levels of nuclear AP-1 complexes in these cells. We also examined NFAT DNA binding and found that NFAT nuclear complexes were increased in activated Jnk1 −/−CD8+ T cells (Fig. 7 C). These results correlated with the previously described negative regulation of NFAT by JNK (25, 55). In contrast to the reduced levels of nuclear AP-1 observed in activated Jnk1 − / −CD8+ T cells, the levels of nuclear AP-1 DNA complexes in activated Jnk1 −/−CD4+ T cells were normal or slightly increased (Fig. 7 D). CREB DNA binding was also examined as a control for protein loading (Fig. 7 D). These results indicate that the targets of JNK1 differ in CD4+ and CD8+ T cells.
Phosphorylation of Jun family members by JNK has been shown to regulate AP-1 transcriptional activity but does not appear to be required for AP-1 DNA binding (7). Thus, the reduction of AP-1 DNA binding in Jnk1 −/− CD8+ T cells could not be directly explained by the lack of JNK1-mediated phosphorylation. As c-Jun expression is upregulated by AP-1 (3), it was possible that the reduced levels of nuclear AP-1 complexes in activated Jnk1 −/− CD8+ T cells could be due to decreased levels of c-Jun. We examined the levels of c-Jun in activated wild-type and Jnk1 −/−CD8+ T cells by Western blot analysis. c-Jun expression was induced in activated Jnk1 −/−CD8+ T cells but delayed compared with the kinetics of c-Jun expression in activated wild-type CD8+ T cells (Fig. 7 E). The levels of JunD, however, were comparable in wild-type and Jnk1 −/− CD8+ T cells (data not shown). The kinetics of c-Jun expression in activated Jnk2 −/−CD8+ T cells were comparable to those in activated wild-type CD8+ T cells (Fig. 7 E). Together, these results suggested that the reduced expression of IL-2Rα in activated Jnk1 −/−CD8+ T cells was most likely due to reduced levels of c-Jun in these cells.
To confirm that the reduced AP-1 DNA binding in Jnk1 −/−CD8+ T cells resulted in impaired AP-1 transcriptional activity, Jnk1 −/− mice were backcrossed with AP-1 luciferase reporter transgenic mice (53) and AP-1 transcriptional activity was measured by the luciferase assay. AP-1 transcriptional activity in activated wild-type CD8+ T cells was detected at 40 h and upregulated in a time-dependent fashion (Fig. 7 F). In contrast, AP-1 transcriptional activity in activated Jnk1 −/−CD8+ T cells was undetectable until 60 h and dramatically reduced compared with the activity detected in activated wild-type CD8+ T cells (Fig. 7 F). Thus, JNK1 was required for AP-1 mediated transcription in CD8+ T cells.
Discussion
MAP kinase signaling pathways are implicated in several biological processes, including growth, differentiation, and cell death. The ERK signaling pathway has been primarily associated with cell growth and differentiation whereas the JNK and p38 MAP kinase signaling pathways have been associated with cell death and differentiation. The specific role of each of these pathways, however, ultimately depends on the cell type. The immune system is a complex organization of cells, each with a specialized function. In this study we show that the regulatory roles of JNK1 and JNK2 in CD8+ T cells are distinct from the previously described function of these two kinases in CD4+ T cells.
Three Jnk genes (Jnk1, Jnk2, and Jnk3) that encode for at least 10 different isoforms have been identified (2, 4). Although these isoforms may have different affinities for specific substrates, no clear specificity has been found. Are these kinases redundant within the same cell type? Jnk3 is predominantly expressed in heart, brain, and testis whereas Jnk1 and Jnk2 are widely expressed with the exception of spleen and lymph nodes (27, 28). Disruption of Jnk3 in mice causes the failure of hippocampal neurons to undergo apoptosis in response to excitotoxic stress (27). In this case, the presence of JNK1 and JNK2 does not compensate for the lack of JNK3. Similarly, the absence of both JNK1 and JNK2 during neuronal tube formation cannot be compensated for by the presence of JNK3 (37, 38). Here, we show that the absence of JNK1 is not completely compensated for by JNK2 in CD8+ T cells. Likewise, the absence of JNK2 is not completely compensated for by JNK1. Similar results have been found in CD4+ T cells (34, 35). Thus, if there is some redundancy of these kinases, it is limited and depends on the specific tissue or cell type.
Do JNKs regulate similar biological functions in specific cell types? It has been shown that a mixed population of lymphocytes containing both CD4+ and CD8+ T cells from either Jnk1 − / − or Jnk2 − / − mice hypoproliferate and produce lower levels of IL-2 in response to TcR engagement and suboptimal doses of costimulatory signals (31, 32). Together, these results suggest that JNK1 and JNK2 may have similar functions. However, analysis of CD4+ T cells in these mice has shown that the absence of JNK1 results in increased Th2 cytokine production (35) whereas the absence of JNK2 impairs Th1 differentiation (34). These results indicate that JNK1 and JNK2 have distinct functions in CD4+ T cell differentiation. In this study we show that Jnk2 − / − CD8+ T cells hyperproliferate and secrete increased amounts of IL-2, while Jnk1 − / − CD8+ T cells have impaired expression of IL-2Rα expression and thereby hypoproliferate following TcR ligation. Thus, JNK1 and JNK2 regulate different aspects of CD8+ T cell function. In correlation with these results, the cosubmitted study (53a) demonstrates that JNK1 and JNK2 also play distinct roles in vivo since they differentially regulate T cell expansion during the LCMV infection. Specifically, impaired expansion of LCMV-specific CD8 T+ cells was observed in JNK1-deficient mice compared with wild-type mice while increased expansion of LCMV-specific CD8+ T cells was observed in JNK2-deficient mice (Arbour et al. in this issue [53a]). Together our results, therefore, demonstrate that the JNK signaling pathway does indeed play different roles in CD4+ and CD8+ T cell activation and differentiation. Previous studies showing that other signaling pathways also differentially regulate CD4+ and CD8+ T cell activation confirm our findings (56–59).
Like CD4+ T cells, CD8+ T cells synthesize and secrete IL-2 upon activation, but the levels and kinetics of IL-2 production are lower and delayed, respectively. Although IL-2 production is reduced in the Jnk1 − / − CD8+ T cells, this is not the major cause of the hypoproliferation of these cells because exogenous IL-2 did not restore the proliferation of these cells to normal levels. A critical event required for the expansion of CD8+ T cells after activation is the upregulation cell surface expression of the IL-2 receptor (IL-2R) α chain and its association with β and γ chains to form the high affinity IL-2R (αβγ) (for a review, see reference 60). Here we show that IL-2Rα gene expression is severely compromised in activated Jnk1 − / −CD8+ T cells.
Several regulatory elements within the IL-2Rα promoter such as those for NFκB, Elf, HMG-I(Y), and NFAT have been described (52, 61–63). NFκB and Elf DNA binding were not affected by the lack of JNK1 in CD8+ T cells (data not shown). In correlation with the negative regulation of NFAT by JNK (55), we observed increased NFAT DNA binding in activated Jnk1 −/−CD8+ T cells. Two AP-1 like consensus sites have also been identified within the proximal IL-2Rα promoter (52). Here we show that AP-1 can bind to one of these elements (IL-2Rα/AP-1) and is reduced in JNK1-deficient CD8+ T cells. Based on our results, we propose that the reduced expression of IL-2Rα in Jnk1 −/− CD8+ T cells is due to reduced levels of c-Jun and AP-1–mediated transcription. Interestingly, this effect is also specific to CD8+ T cells because IL-2Rα expression (35) and AP-1 DNA binding was not affected in JNK1-deficient CD4+ T cells.
Considering the distinct roles of the different JNK family members in CD4+ and CD8+ T cell activation and differentiation, predicting the final outcome of an in vivo immune response in mice deficient of JNK1 or JNK2 could be difficult. Consistent with the predominant Th2 bias of Jnk1 − / −CD4+ T cells in vitro, JNK1-deficient mice fail to resolve the Leishmania major infection (36) which requires CD4+ Th1 effector cells for pathogen clearance (64). In contrast, despite impaired CD4+ Th1 differentiation observed in vitro (34), JNK2-deficient mice are able to resolve the influenza infection (Fig. 1 A) which requires both effector Th1 and CD8+ T cells for viral clearance. The increased expansion and production of IFN-γ by Jnk2 − / −CD8+ T cells shown here could compensate for the impaired Th1 differentiation in vivo and account for the normal viral clearance observed in these mice. Enhanced CD8+ T cell expansion in Jnk2 − / − while impaired expansion of CD8+ T cells in Jnk1 − / − mice during the LCMV infection have been described (53a). Together, these results are consistent with our in vitro studies showing that JNK1-deficient CD8+ T cells hypoproliferate after TcR ligation due to impaired IL-2Rα expression whereas JNK2-deficient CD8+ T cells hyperproliferate after TcR ligation. Our studies support the distinct roles for JNK1 and JNK2 in CD8+ T cell activation both in vitro and in vivo.
Acknowledgments
We graciously thank Julie Rodgers for her excellent technical assistance, the personnel of the Animal Facility of the Wellington School of Medicine for Animal Husbandry, Brydon Bennett and David Anderson for providing the JNK inhibitor (SP600125), and Collette Charland for flow cytometry analysis and helpful discussions.
This work was supported by grants from Arthritis Foundation (M. Rincon), and the Marsden Fund (G. Le Gros). R.A. Flavell and R.J. Davis are Investigators of the Howard Hughes Medical Institute.
Footnotes
*
Abbreviations used in this paper: AP-1, activator protein 1; CRE, cyclic AMP-responsive element; CREB, CRE binding protein; JNK, c-Jun NH2-terminal kinase; LCMV, lymphocytic choriomeningitis virus; MAP, mitogen-activated protein; MKK, MAP kinase kinase; NF, nuclear factor; NFAT, nuclear factor of activated T cells; RPA, ribonuclease protection assay; Tc, cytotoxic T; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling.
References
- 1.Stenger, S., J.-P. Rosat, B. Bloom, A. Krensky, and R. Modlin. 1999. Granulysin: a lethal weapon of cytolytic T cells. Immunol. Today. 20:390–394. [DOI] [PubMed] [Google Scholar]
- 2.Ip, Y.T., and R.J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK) - from inflammation to development. Curr. Opin. Cell Biol. 10:205–219. [DOI] [PubMed] [Google Scholar]
- 3.Whitmarsh, A.J., and R.J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinases signal transduction pathways. J. Mol. Med. 74:589–607. [DOI] [PubMed] [Google Scholar]
- 4.Davis, R.J. 2000. Signal transduction by the JNK group of MAP kinases. Cell. 103:239–252. [DOI] [PubMed] [Google Scholar]
- 5.Boulton, T.G., G.D. Yancopoulos, J.S. Gregory, C. Slaughter, C. Moomaw, J. Hsu, and M.H. Cobb. 1990. An insulin-stimulated protein kinase similar to yeast kinase involved in cell cycle control. Science. 249:64–67. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, T.G., S.H. Nye, D.J. Robbins, N.Y. Ip, E. Radziejewska, S.D. Morgembesser, A. DePinho, N. Panayotatos, M.H. Cobb, and G.D. Yancopoulos. 1991. ERKs: family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 65:663–675. [DOI] [PubMed] [Google Scholar]
- 7.Dérijard, B., M. Hibi, I.-H. Wu, T. Barret, B. Su, T. Deng, M. Karin, and R.J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 76:1025–1037. [DOI] [PubMed] [Google Scholar]
- 8.Kyriakis, J.M., P. Banerjee, E. Nikolakaki, T. Dai, E.A. Rubie, M.F. Ahmad, J. Avruch, and J.R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 369:156–160. [DOI] [PubMed] [Google Scholar]
- 9.Han, J., J.-D. Lee, L. Bibbs, and R.J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 265:808–811. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J.C., T. Laydon, P.C. McDonnell, T.F. Gallagher, S. Kumar, D. Gree, D. McNulty, M.J. Blumenthal, J.R. Heys, S.W. Landvatter, et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 372:739–746. [DOI] [PubMed] [Google Scholar]
- 11.Rouse, J., P. Cohen, S. Trigon, M. Morange, A. Alonso-Llamazares, D. Zamanillo, T. Hunt, and A.R. Nebreda. 1994. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 78:1027–1037. [DOI] [PubMed] [Google Scholar]
- 12.Dérijard, B., J. Rainjeaud, T. Barret, I.-H. Wu, J. Han, R.J. Ulevitch, and R.J. Davis. 1995. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 267:682–685. [DOI] [PubMed] [Google Scholar]
- 13.Nishina, H., K.D. Fischer, L. Radvanyl, A. Shahinian, R. Haken, E.A. Rubie, A. Bernstein, T.W. Mak, J.R. Woodgett, and J.M. Penninger. 1997. Stress-signalling kinase Sek 1 protects thymocytes from apoptosis mediated by CD95 and CD3. Nature. 385:350–353. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez, I., R.T. Hughes, B.J. Mayer, K. Yee, J.R. Woodgett, J. Avruch, J.M. Kyriakis, and L.I. Zon. 1994. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 372:794–798. [DOI] [PubMed] [Google Scholar]
- 15.Yang, D., C. Tournier, M. Wysk, H.-T. Lu, X. Jie, R.J. Davis, and R.A. Flavell. 1997. Targeted disruption of the MKK4 gene causes embryonic death inhibition of c-Jun NH2 terminal kinase activation, and defects in AP-1 transcriptional activity. Proc. Natl. Acad. Sci. USA. 94:3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tournier, C., A.J. Whitmarsh, J. Cavanagh, T. Barrett, and R.J. Davis. 1997. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc. Natl. Acad. Sci. USA. 94:7337–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tournier, C., C. Dong, T. Turner, S. Jones, R. Flavell, and R. Davis. 2001. MKK7 is an essential component of the JNK signal transduction pathway activated by pro-inflammatory cytokines. Genes Dev. 15:1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, A., A. Minden, H. Martinetto, F.-X. Claret, C. Lange-Carter, F. Mercurio, G.L. Johnson, and M. Karin. 1995. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 268:286–290. [DOI] [PubMed] [Google Scholar]
- 19.Nishina, H., M. Bachmann, A.J. Oliveira-dos-Santos, B. Odermatt, A. Wakeham, A. Shahinian, H. Takimoto, A. Bernstein, T.M. Mak, J.R. Woodgett, et al. 1997. Impaired CD28-mediated interleukin 2 production and proliferation in stress kinase SAPK/ERK1 Kinase (SEK1)/mitogen-activated protein kinase kinase 4 (MKK4-deficient T lymphocytes. J. Exp. Med. 186:941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swat, W., K. Fujikawa, S. Ganiatsas, D.D. Yang, R.J. Xavier, N.L. Harris, L. Davidson, R. Ferrini, R.J. Davis, M.A. Labow, et al. 1998. SEK1/MKK4 is required for maintenance of a normal peripheral lymphoid compartment but not for T lymphocyte development. Immunity. 8:625–634. [DOI] [PubMed] [Google Scholar]
- 21.Dong, C., D. Yang, C. Tournier, A. Whitmarsh, J. Xu, R. Davis, and R. Flavell. 2000. JNK is required for effector T-cell function but not for T-cell activation. Nature. 405:91–94. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki, T., T. Wada, H. Kishimoto, J. Irie-Sasaki, G. Matsumoto, T. Goto, Z. Yao, A. Wakeham, T.W. Mak, A. Suzuki, et al. 2001. The stress kinase mitogen-activated protein kinase kinase (MKK)7 is a negative regulator of antigen receptor and growth factor receptor-induced proliferation in hematopoietic cells. J. Exp. Med. 194:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, S., T. Barret, A.J. Whitmarsh, J. Cavanagh, H.K. Sluss, B. Dérijard, and R.J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 24.Li, B., C. Tournier, R.J. Davis, and R.A. Flavell. 1998. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow, C.-W., M. Rincón, J. Cavanagh, M. Dickens, and R.J. Davis. 1997. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 278:1638–1641. [DOI] [PubMed] [Google Scholar]
- 26.Chow, C.-W., M. Rincón, and R.J. Davis. 1999. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell. Biol. 19:2300–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, D.D., C.-Y. Kuan, A.J. Whitmarsh, M. Rincón, T.S. Zheng, R.J. Davis, P. Rakic, and R.A. Flavell. 1997. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 389:865–870. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, L., A. Whitmarsh, D. Yang, M. Rincon, R. Davis, and R. Flavell. 2000. Regulation of c-Jun NH2 terminal kinase (Jnk) gene expression during T cell activation. J. Exp. Med. 191:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su, B., E. Jacinto, M. Hibi, T. Kallunki, M. Karin, and Y. Ben-Neriah. 1994. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 77:727–736. [DOI] [PubMed] [Google Scholar]
- 30.Rincón, M., A. Whitmarsh, D.D. Yang, L. Weiss, B. Dérijard, P. Jayaraj, R.J. Davis, and R.A. Flavell. 1998. The JNK pathway regulates the in vivo deletion of immature CD4+ CD8+ thymocytes. J. Exp. Med. 188:1817–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabapathy, K., Y. Hu, T. Kallunki, M. Schreiber, J.P. David, W. Jochum, E.F. Wagner, and M. Karin. 1999. b. JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 9:116–125. [DOI] [PubMed] [Google Scholar]
- 32.Sabapathy, K., T. Kallunki, J.-P. David, I. Graet, M. Karin, and E. Wagner. 2001. c-Jun NH2-terminal Kinase (JNK)1 and JNK2 have similar and stage-dependent roles in regulating T cell apoptosis and proliferation. J. Exp. Med. 193:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behrens, A., K. Sabapathy, I. Graef, M. Cleary, G. Crabtree, and E. Wagner. 2001. Jun N-terminal kinase 2 modulates thymocyte apoptosis and T cell activation through c-Jun and nuclear factor of activated T cell (NFAT). Proc. Natl. Acad. Sci. USA. 98:1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, D., D. Conze, A.J. Whitmarsh, T. Barrett, R.J. Davis, M. Rincón, and R.A. Flavell. 1998. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity. 9:575–585. [DOI] [PubMed] [Google Scholar]
- 35.Dong, C., D. Yang, M. Wysk, A.J. Whitmarsh, R.J. Davis, and R.A. Flavell. 1998. Defective T cell differentiation in the absence of JNK1. Science. 282:2092–2095. [DOI] [PubMed] [Google Scholar]
- 36.Constant, C., C. Dong, D. Yang, M. Wysk, R. Davis, and R. Flavell. 2000. JNK1 is required for T cell-mediated immunity against Leishmania major infection. J. Immunol. 165:2671–2676. [DOI] [PubMed] [Google Scholar]
- 37.Kuan, C.Y., D.D. Yang, D.R. Samanta Roy, R.J. Davis, P. Rakic, and R.A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 22:667–676. [DOI] [PubMed] [Google Scholar]
- 38.Sabapathy, K., W. Jochum, K. Hochedlinger, L. Chang, M. Karin, and E.F. Wagner. 1999. a. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89:115–124. [DOI] [PubMed] [Google Scholar]
- 39.Kilbourne, E. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Organ. 41:643–645. [PMC free article] [PubMed] [Google Scholar]
- 40.Lumsden, J., J. Roberts, N. Harris, R. Peach, and F. Ronchese. 2000. Differential requirement for CD80 and CD80/86-dependent costimulation in the lung immune response to an influenza virus infection. J. Immunol. 164:79–85. [DOI] [PubMed] [Google Scholar]
- 41.Harris, N., C. Campbell, G.L. Gros, and F. Ronchese. 1997. Blockade of CD28/B7 co-stimulation by mCTLA4-Hγ1 inhibits antigen-induced lung eosinophilia but not Th2 cell development or recruitment in the lung. Eur. J. Immunol. 27:155–161. [DOI] [PubMed] [Google Scholar]
- 42.Rincón, M., J. Anguita, T. Nakamura, E. Fikrig, and R.A. Flavell. 1997. IL-6 directs the differentiation of IL-4–producing CD4+ T cells. J. Exp. Med. 185:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rincón, M., B. Dérijard, C.-W. Chow, R.J. Davis, and R.A. Flavell. 1997. Reprogramming the signaling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T cells to effector Th1 and Th2 cells. Genes Funct. 1:51–68. [DOI] [PubMed] [Google Scholar]
- 44.Kamogawa, Y., L.-A.E. Minasi, S. Carding, K. Bottomly, and R.A. Flavell. 1993. The relationship of IL-4 and IFNγ-producing T cells studied by lineage ablation of IL-4-producing cells. Cell. 75:985–995. [DOI] [PubMed] [Google Scholar]
- 45.Rincón, M., A. Tugores, A. López-Rivas, A. Silva, M. Alonso, M.O. de Lándazuri, and M. López-Botet. 1988. Prostaglandin E2 and the increase of intracellular cAMP inhibit the expression of interleukin 2 receptors in human T cells. Eur. J. Immunol. 18:1791–1796. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber, E., P. Matthias, M.M. Müller, and W. Shaffner. 1989. Rapid detection of octamer binding proteins with ‘mini’-extracts prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tugores, A., M.A. Alonso, F. Sánchez-Madrid, and M.O. de Landázuri. 1992. Human T cell activation through the activation-inducer molecule/CD69 enhances the activity of transcription factor AP-1. J. Immunol. 148:2300–2306. [PubMed] [Google Scholar]
- 48.Angel, P., M. Imagawa, R. Chiu, B. Stein, R.J. Imbra, H.J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 49:729–739. [DOI] [PubMed] [Google Scholar]
- 49.Lee, W., P. Mitchell, and R. Tjian. 1987. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 49:741–752. [DOI] [PubMed] [Google Scholar]
- 50.Rooney, J.W., M.R. Hodge, P.G. McCaffrey, A. Rao, and L.H. Glimcher. 1994. A common factor regulates both Th1- and Th2-specific cytokine gene expression. EMBO J. 13:625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montminy, M.R., and L.M. Bilezikijan. 1987. Binding of nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 328:175–178. [DOI] [PubMed] [Google Scholar]
- 52.Sperisen, P., S.M. Wang, E. Soldaini, M. Pla, C. Rusterholz, P. Bucher, P. Corthesy, P. Reichenbach, and M. Nabholz. 1995. Mouse interleukin-2 receptor alpha gene expression. Interleukin-1 and interleukin-2 control transcription via distinct cis-acting elements. J. Biol. Chem. 270:10743–10753. [DOI] [PubMed] [Google Scholar]
- 53.Rincón, M., and R.A. Flavell. 1994. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 13:4370–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Arbour, N. D. Naniche, D. Homann, R.J. Davis, R.A. Flavell, and M.B.A. Oldstone. 2002. c-Jun NH2-terminal kinase (JNK)1 and JNK2 signaling pathways have divergent roles in CD8+ T cell–mediated antiviral immunity. J. Exp. Med. 195:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett, B.L., D.T. Sasaki, B.W. Murray, E.C. O'Leary, S.T. Sakata, W. Xu, J.C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, et al. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA. 98:13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chow, C.W., and R.J. Davis. 2000. Integration of calcium and cyclic AMP signaling pathways by 14-3-3. Mol. Cell. Biol. 20:702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuford, W.W., K. Klussman, D.D. Tritchler, D.T. Loo, J. Chalupny, A.W. Siadak, T.J. Brown, J. Emswiler, H. Raecho, C.P. Larsen, et al. 1997. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J. Exp. Med. 186:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merritt, C., H. Enslen, N. Diehl, D. Conze, R. Davis, and M. Rincón. 2000. Activation of p38 mitogen-activated protein kinase in vivo selectively induces apoptosis of CD8+ but not CD4+ T cells. Mol. Cell. Biol. 20:936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carter, L., and K. Murphy. 1999. Lineage-specific requirement for signal transducer and activator of transcription (Stat)4 in interferon γ production from CD4+ versus CD8+ T cells. J. Exp. Med. 189:1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki, I., and P.J. Fink. 1998. Maximal proliferation of cytotoxic T lymphocytes requires reverse signaling through Fas ligand. J. Exp. Med. 187:123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson, B., and D. Willerford. 1998. Biology of the interleukin-2 receptor. Adv. Immunol. 70:1–81. [DOI] [PubMed] [Google Scholar]
- 61.John, S., R.B. Reeves, J.X. Lin, R. Child, J.M. Leiden, C.B. Thompson, and W.J. Leonard. 1995. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol. Cell. Biol. 15:1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freimuth, W.W., J.M. Depper, and G.J. Nabel. 1989. Regulation of the IL-2 receptor alpha-gene. Interaction of a kappa B binding protein with cell-specific transcription factors. J. Immunol. 143:3064–3068. [PubMed] [Google Scholar]
- 63.Serdobova, I., M. Pla, P. Reichenbach, P. Sperisen, J. Ghysdael, A. Wilson, J. Freeman, and M. Nabholz. 1997. Elf-1 contributes to the function of the complex interleukin (IL)-2-responsive enhancer in the mouse IL-2 receptor alpha gene. J. Exp. Med. 185:1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiner, S.L., and R.M. Locksley. 1995. The regulation of immunity to Leishmania major. Annu. Rev. Immunol. 13:151–177. [DOI] [PubMed] [Google Scholar]