Ku70 Is Required for DNA Repair but Not for T Cell Antigen Receptor Gene Recombination In Vivo (original) (raw)
Abstract
Ku is a complex of two proteins, Ku70 and Ku80, and functions as a heterodimer to bind DNA double-strand breaks (DSB) and activate DNA-dependent protein kinase. The role of the Ku70 subunit in DNA DSB repair, hypersensitivity to ionizing radiation, and V(D)J recombination was examined in mice that lack Ku70 (Ku70 −/−). Like Ku80 −/− mice, Ku70 −/− mice showed a profound deficiency in DNA DSB repair and were proportional dwarfs. Surprisingly, in contrast to Ku80−/− mice in which both T and B lymphocyte development were arrested at an early stage, lack of Ku70 was compatible with T cell receptor gene recombination and the development of mature CD4+CD8− and CD4−CD8+ T cells. Our data shows, for the first time, that Ku70 plays an essential role in DNA DSB repair, but is not required for TCR V(D)J recombination. These results suggest that distinct but overlapping repair pathways may mediate DNA DSB repair and V(D)J recombination.
Two distinct processes involving DNA double-strand breaks (DSB)1 have been identified in mammalian cells: the repair of DNA damage induced by ionizing radiation and V(D)J recombination during T and B cell development. So far, all mammalian cell mutants defective in DNA DSB repair share the common phenotype of hypersensitivity to radiation and impaired ability to undergo V(D)J recombination (1–6). Cell fusion studies using DSB repair mutants of human–rodent somatic hybrids have defined four ionizing radiation (IR) complementation groups: IR4, IR5, IR6, and IR7. Genetic and biochemical analyses have revealed that cells of IR5 (e.g., xrs-6) and IR7 (e.g., scid) are defective in components of the DNA-dependent protein kinase (DNA-PK) (2, 7–9). DNA-PK is a serine/ threonine kinase comprised of a large catalytic subunit and a DNA-targeting component termed Ku, which itself is a heterodimer of a 70- (Ku70) and a 86-(Ku80) kD polypeptide (10–12). Recently, the DNA-PK catalytic subunit has been shown to be the gene responsible for the murine scid defect (13–15), and Ku80 has been identified to be XRCC5 (16–18), the x-ray repair cross-complementing gene for IR5. Ku80 knockout mice were found to exhibit scid, defective processing of V(D)J recombination intermediates, and growth retardation (19, 20).
Although Ku70 has been designated as XRCC6 (7, 8) and is an important component of the DNA-PK complex, the function of Ku70 in vivo is hitherto unknown. To define the role of Ku70 in DNA repair and V(D)J recombination, we targeted the Ku70 gene in mice. Ku70 homozygotes exhibit proportional dwarfism, a phenotype of Ku80− /−, but not of scid mice. Absence of Ku70 confers hypersensitivity to ionizing radiation and deficiency in DNA DSB repair, which are characteristics of both Ku80− /− and scid mice. Surprisingly, in contrast to Ku80− /− and scid mice in which both T and B lymphocyte development are arrested at early stage, lack of Ku70 is compatible with T cell receptor gene recombination and the development of mature CD4+CD8− and CD4−CD8+ T cells. Our data, for the first time, provide direct evidence supporting that Ku70 plays an essential role in DNA DSB repair, but is not required for TCR gene recombination. These results suggest that distinct but overlapping repair pathways may mediate DSB repair and V(D)J rejoining; furthermore, it suggests the presence of a Ku70-independent rescue pathway in TCR V(D)J recombination. The distinct phenotype of Ku70− /− mice should make them valuable tools for unraveling the mechanism(s) of DNA repair and recombination.
Materials and Methods
Target Disruption of Ku70 and Generation of Ku70−/− Mice.
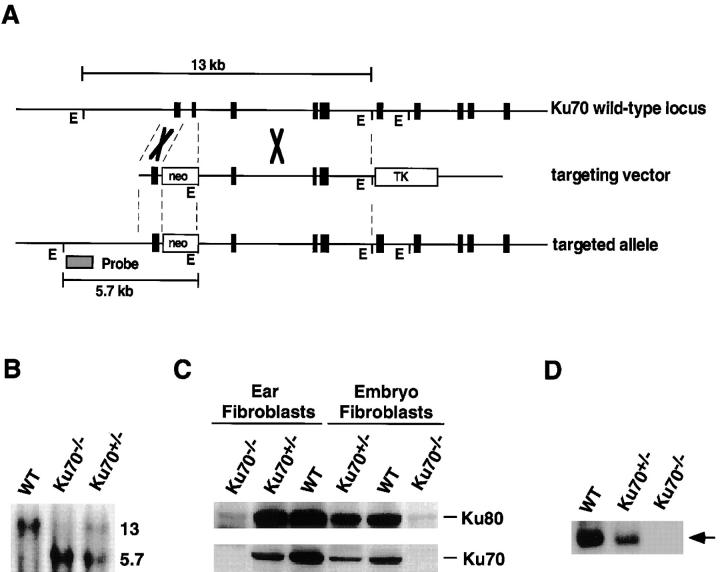
Mouse genomic Ku70 gene was isolated from a sCos-I cosmid library constructed from a mouse strain 129 embryonic stem (ES) cell lines (21). The replacement vector was constructed using a 1.5 kb 5′-fragment that contains the promoter locus with four GC boxes and exon 1, and an 8-kb EcoRV–EcoRI fragment extending from intron 2 to intron 5 as indicated in Fig. 1 A. Homologous replacement results in a deletion of 336-bp exon 2 including the translational initiation codon.
Figure 1.
Inactivation of Ku70 by homologous recombination. (A) Diagrammatic representation of the Ku70 locus (top), the targeting construct (middle), and the targeted allele and hybridization probe (bottom). EcoRI (E) restriction sites used to detect the targeted gene are indicated (21). (B) Southern blot of EcoRI-digested tail DNA from control wild-type (WT), heterozygous (+/−), and homozygous (−/−) _Ku70_-targeted mice. The wild-type and mutant fragments are 13 and 5.7 kb, respectively. (C) Western blot analysis showing that Ku70 protein is not expressed in Ku70− /− cells. Whole cell lysates prepared from mouse ear fibroblasts (50 μg) and mouse embryo fibroblasts (100 μg) were separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with polyclonal antibodies against full-length rodent Ku80 (top) and Ku70 (bottom), respectively. (D) Gel mobility shift assay (22) showing the lack of DNA-end–binding activity in Ku70− /− cells. Ku-DNA–binding complex is indicated by arrow on the right.
The targeting vector was linearized with NotI and transfected into CJ7 ES cells by electroporation using a gene pulser (Bio Rad Labs., Hercules, CA). 300 ES cell clones were screened, and five clones carrying the mutation in Ku70 were identified by Southern blotting. Positive ES clones were injected separately into C57BL/6 blastocysts to generate chimeric mice. One clone was successfully transmitted through the germline after chimeras were crossed with C57BL/6 females. Homozygous Ku70− /− mice were generated by crossing Ku70 +/− heterozygotes.
The genotype of the mice was first determined by tail PCR analysis which distinguishes endogenous from the targeted Ku70 allele, and subsequently confirmed by Southern blot analysis. The PCR reaction contained 1 μg genomic DNA; 0.6 μM (each) of primers HO-2: GGGCCAGCTCATTCCTCCACTCATG, HO-3: CCTACAGTGTACCCGGACCTATGCC, and HO-4: CGGAACAGGACTGGTGGTTGAGCC; 0.2 mM (each) deoxynucleoside triphosphate; 1.5 mM MgCl2, and 2.5 U of Taq polymerase. Cycling conditions were 94°C for 1 min, 64°C for 1 min, 72°C for 1 min (30 cycles), followed by an extension at 72°C for 10 min. Primers HO-2 and HO-4 give a product of the targeted allele that is ∼380 bp; primers HO-3 and HO-4 yield a wild-type product of 407 bp.
Western Blot Analysis and Gel Mobility Shift Assay.
To confirm that the disruption of Ku70 produces a null mutation, Ku70 protein expression was measured by Western blotting using polyclonal antisera against intact mouse Ku70. The lack of Ku70 was also verified by a Ku–DNA-end–binding assay (gel mobility shift analysis). Cell extracts were prepared and gel mobility shift assays were performed as described (22). Equal amounts of cellular protein (50 μg) from Ku70 +/+ (wild type), Ku70 +/−, and Ku70− /− mouse embryo fibroblasts were incubated with a 32P-labeled double-stranded oligonucleotide, 5′-GGGCCAAGAATCTTCCAGCAGTTTCGGG-3′. The protein-bound and free oligonucleotides were electrophoretically separated on a 4.5% native polyacrylamide gel. Gel slabs are dried and autoradiographed with X-Omat film (Kodak, Rochester, NY).
Immunohistochemistry.
To determine the pathological changes, histological sections of various organs of Ku70− /−, Ku80− /−, and wild-type littermate mice were prepared and examined as previously described (23). Lymph nodes, spleens, and thymuses from 4–5-wk-old mice were fixed in 10% buffered formalin and embedded in paraffin, or embedded in (optimal cutting temperature) compound (Sakura Finetek, USA, Incorp., Torrance, CA) and frozen in liquid nitrogen at −70°C. Sections (5 μm) were stained with hematoxylin and eosin, and representative samples were selected for immunohistochemical analysis. Immunophenotyping was performed using an avidin-biotin immunoperoxidase technique (24). Primary antibodies included anti-CD3 (purified rabbit serum, 1:1,000; Dako Corp., Carpinteria, CA), anti-B220 (rat monoclonal, 1:1,000; PharMingen, San Diego, CA), and anti-CD19 (rat monoclonal, 1:1,000; PharMingen), and were incubated overnight at 4°C. Samples were subsequently incubated with biotinylated secondary antibodies (Vector Labs., Burlingame, CA) for 30 min (goat anti–rabbit, 1:100; rabbit anti–rat, 1:100), and then with avidin-biotin peroxidase (1:25 dilution; Vector Labs.) for 30 min. Diaminobenzadine was used as the chromogen and hematoxylin as the counter stain. Wild-type lymphoid organs including thymus, spleen, and lymph nodes from different mice were used for titration of the antibodies and positive controls. Anti-CD3 and anti-CD19 antibodies were tested in both frozen and paraffin sections of wild-type lymphoid organs and showed the expected specific patterns of staining (data not shown). For negative controls, primary antibodies were substituted with class-matched but unrelated antibodies at the same final working dilutions.
Cell Preparation and Flow Cytometric Analysis.
For flow cytometry, single cell suspensions from lymphoid organs of 4–6-wk-old mutant and littermate control mice were prepared for staining as described previously (19) and analyzed on a FACScan® with Cell Quest software (Becton Dickinson, San Jose, CA). Cells were stained with combinations of PE-labeled anti-CD4 and FITC-labeled anti-CD8, or PE-labeled anti-B220 and FITC-labeled anti-CD43, or FITC–anti-IgM and PE–anti-B220 (PharMingen), as needed. Bone marrow cells were harvested from femurs by syringe lavage, and cells from thymus and spleen were prepared by homogenization. Cells were collected and washed in PBS plus 5% FCS and counted using a hemacytometer. Samples from individual mice were analyzed separately. Dead cells were gated out by forward and side scatter properties. Experiments were performed at least three times and yielded consistent results.
DNA Preparation and Analysis of V(D)J Recombination Products.
To determine whether a null mutation in Ku70 affects the recombination of antigen-receptor genes in T and B lymphocytes in vivo, we measured the immunoglobulin and T cell antigen receptor (TCR) rearrangements by PCR. DNA from bone marrow was amplified with primers specific to immunoglobulin D-JH and V-DJH rearrangements, and DNA from thymus was amplified with primers that detect V-DJβ and Dδ-Jδ rearrangement (20, 25–28).
Oligonucleotides for probes and PCR primers specific to TCR Vβ-Jβ rearrangements and immunoglobulin D-JH and V-DJH rearrangements are as follows. For TCR-β Vβ8-Jβ2 rearrangements (28): Vβ8.1:5′-GAGGAAAGGTGACATTGAGC-3′, Jβ2.6: 5′-GCCTGGTGCCGGGACCGAAGTA-3′, Vβ8 probe: 5′-GGGCTGAGGCTGATCCATTA-3′. For Dδ2-Jδ1 rearrangements (20, 27): DR6: 5′-TGGCTTGACATGCAGAAAACACCTG-3′, DR53: 5′-TGAATTCCACAGTCACTTGGCTTC-3′, and DR2 probe: 5′-GACACGTGATACAAAGCCCAGGGAA-3′. For immunoglobulin D-JH and V-DJH rearrangements (26): 5′D: 5′-GTCAAGGGATCTACTACTGTG-3′, V7183: 5′-GAGAGAATTCAGAGACAATCCCAAGAACA C C C T G- 3′ , VJ558L: 5′-GAGAGAATTCTCCTCCAGCACAGCCTACATG-3′, J2: 5′-GAGAGAATTCGGCTCCCAATGACCCTTTCTG-3′, 5′ intervening sequence (IVS): 5′-GTAAGAATGGCCTCTCCAGGT-3′, 3′-IVS: 5′-GACTCAATCACTAA-GACAGCT-3′, and probe: a 6-kb EcoRI fragment covering the J region of mouse IgM.
Cell Survival Determination.
8–10-wk-old Ku70− /− and Ku80− /− mice and wild-type littermates were used for our studies. Bone marrow cell suspensions were prepared by flushing the femur with MEM supplemented with 15% FCS. The cell suspension was then counted using a hemacytometer and centrifuged at 1,000 rpm for 12 min. The resulting pellet was resuspended and diluted to ∼106 cells/ml in MEM plus 15% FCS for further experiments.
To measure the survival of granulocyte–macrophage progenitors, the method of Van Zant et al. (29) was used with minor modifications (30). In brief, α-MEM contained 30% heat-inactivated FCS and 1% bovine serum albumin; in addition, 0.5 ng/ml GM-CSF (R & D Sys. Inc., Minneapolis, MN) was used as a source of colony-stimulating factor. 1 d before each experiment, 2.0 ml of the above media containing 0.5% noble agar (Difco Labs., Detroit, MI) was added to individual 60-mm petri dishes. Immediately after radiation exposure, cells were diluted in 2 ml of the above media with 0.3% noble agar and poured over the prepared dishes with 0.5% noble agar underlayer. The cells were then incubated at 37°C with 5% CO2 and 95–98% humidity. The colonies were counted on day 8 with a dissecting microscope. Macrophage and granulocyte colonies were counted separately and then summed together for survival calculations of granulocyte–macrophage progenitors (granulocyte/macrophage CFUs, CFU-GM). Only colonies containing ⩾50 cells were scored. The colony forming efficiency of CFU-GMs was 60–100/105 nucleated cells for untreated controls. Surviving fraction was defined as the cloning efficiency of irradiated marrow cells relative to that of untreated controls. All experiments were performed at least twice and yielded consistent results.
Asymmetric Field Inversion Gel Electrophoresis.
To determine the rate and extent of DNA DSB repair in Ku-deficient cells after exposure to ionizing radiation, primary embryo fibroblasts derived from Ku70− /−, Ku80− /− and wild-type littermate mice were used. Mouse embryo fibroblasts from day 13.5 embryos growing in replicate cultures for 3 d in the presence of 0.01 μCi/ml [l4C]thymidine (New England Nuclear, Boston, MA) and 2.5 μM cold thymidine were exposed to 40 Gray (Gy) of x-rays and returned to 37°C. At various times thereafter, one dish was removed and trypsinized on ice; single cell suspensions were made and embedded in an agarose plug at a final concentration of 3 × 106 cells/ml. Asymmetric field inversion gel electrophoresis (AFIGE) was carried out in 0.5% Seakem agarose (FMC Bioproducts, Rockland, ME; cast in the presence of 0.5 μg/ml ethidium bromide) in 0.5 × TBE (45 mM Tris, pH 8.2, 45 mM boric acid, 1 mM EDTA) at 10°C for 40 h by applying cycles of 1.25 V/cm for 900 s in the direction of DNA migration, and 5.0 V/cm for 75 s in the reverse direction as described (31).
Quantification and analysis for DNA DSB present were carried out in a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Levels of DNA DSB were quantified by calculating the fraction of activity released from the well into the lane in irradiated and unirradiated samples, which equals the ratio of the radioactivity signal in the lane versus that of the entire sample (well plus lane).
Results
Targeted Disruption of Ku70 Gene.
To study the role of Ku70 in vivo, we generated mice containing a germline disruption of the Ku70 gene. Murine genomic Ku70 gene was isolated and a targeting vector was constructed (Fig. 1 A). Homologous replacement results in a deletion of 336-bp exon 2, including the translational initiation codon. Two targeted ES clones carrying the mutation in Ku70 were injected into C57BL/6 blastocysts to generate chimeric mice. One clone was successfully transmitted through the germline after chimeras were crossed with C57BL/6 females. No obvious defects were observed in Ku70 +/− heterozygotes, and these Ku70 +/− mice were subsequently used to generate Ku70 −/− mice (Fig. 1 B). 25% of the offspring born from Ku70 +/− × Ku70 +/− crosses were Ku70 −/−. Adult Ku70 −/− mice are fertile, but give reduced litter size (two to four pups) as compared to the Ku70 +/− or Ku70 +/+ mice (about eight pups).
To confirm that the disruption produced a null mutation, Ku70 protein expression was analyzed by both Western blotting (Fig. 1 C) and a DNA end binding assay (Fig. 1 D). Ku70 immunoreactivity was undetectable (Fig. 1 C), and there was no Ku–DNA-end–binding activity in Ku70− /− fibroblasts (Fig. 1 D). The Ku80 subunit of the Ku heterodimer was found, but at much reduced levels (Fig. 1 C), suggesting that the stability of Ku80 is compromised by the absence of Ku70. These observations are consistent with the finding that the level of Ku70 was significantly reduced in Ku80− /− fibroblasts and Ku80− /− ES cells (19). Taken together, these data suggest that the stability of either component of Ku is compromised by the absence of the other.
Newborn Ku70− /− mice were 40–60% smaller than their Ku70 +/− and Ku70 +/+ littermates. During a 5-mo observation period, Ku70− /− mice grew and maintained body weight at 40–60% of controls. Thus, Ku70− /− mice, like Ku80− /− mice, are proportional dwarfs (19).
Development of B Lymphocytes, but Not T Lymphocytes, Is Blocked at Early Stage in Ku70−/− Mice.
Examination of various organs from Ku70 −/− mice showed abnormalities only in the lymphoid system (Fig. 2 A). Spleen and lymph nodes were disproportionately smaller by 5–10-fold relative to controls. In particular, splenic white pulp nodules were significantly reduced. Immunohistochemistry on deparaffinized tissue sections revealed that the splenic white pulp contained cells that stained with anti-CD3 (i.e., CD3-positive T cells), but there were no CD19-positive B cells (Fig. 2 A, k and n). The Ku70 −/− thymus was also disproportionately smaller and contained 50–100-fold fewer lymphocytes than Ku70 +/+ littermates (3 × 106 in the former versus 2 × 108 in the latter; measured in three mice of each genotype). In contrast to the Ku80 −/− mice, the Ku70 −/− thymus displayed normal appearing cortical-medullary junctions (Fig. 2 A, g and j). Overall, the lymphoid tissues and organs of Ku70 −/− mice are somewhat disorganized and much smaller than Ku70 +/+ mice (Table 1); yet, they are relatively more developed and slightly larger than in Ku80 −/− mice.
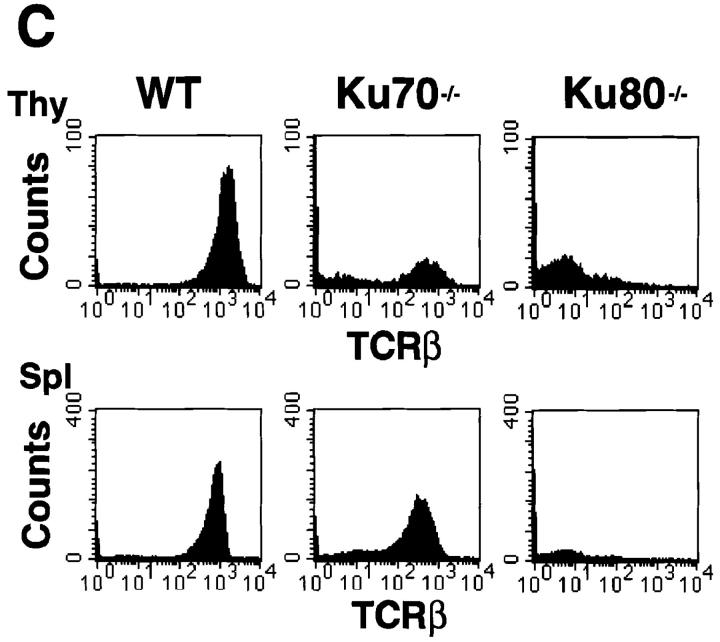
Figure 2.
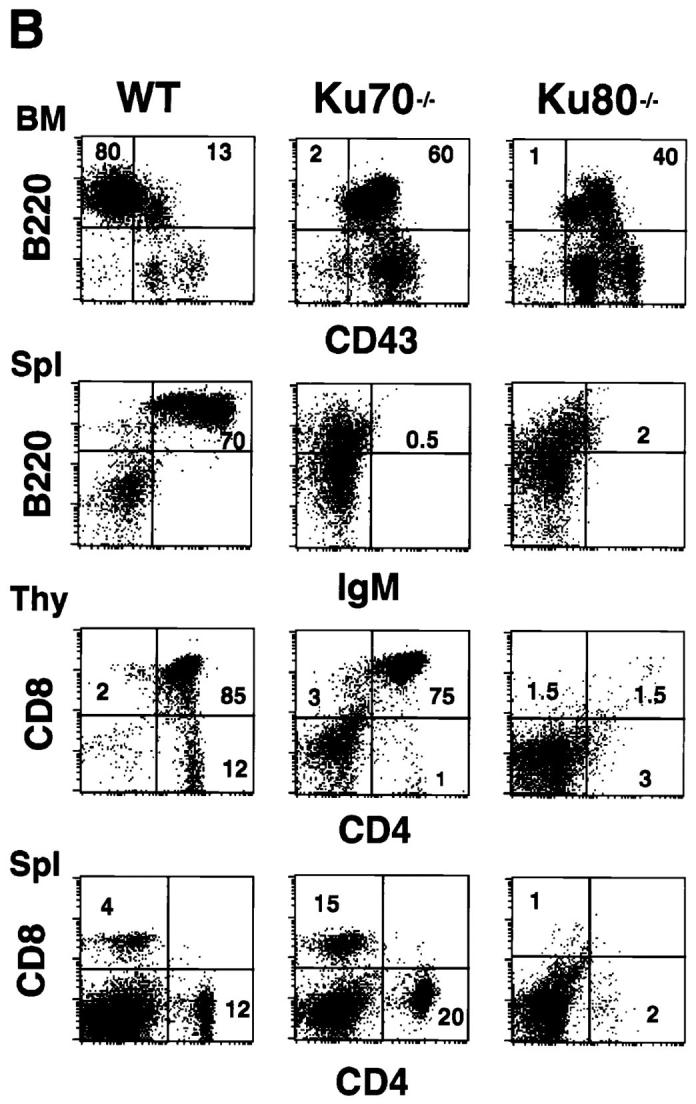
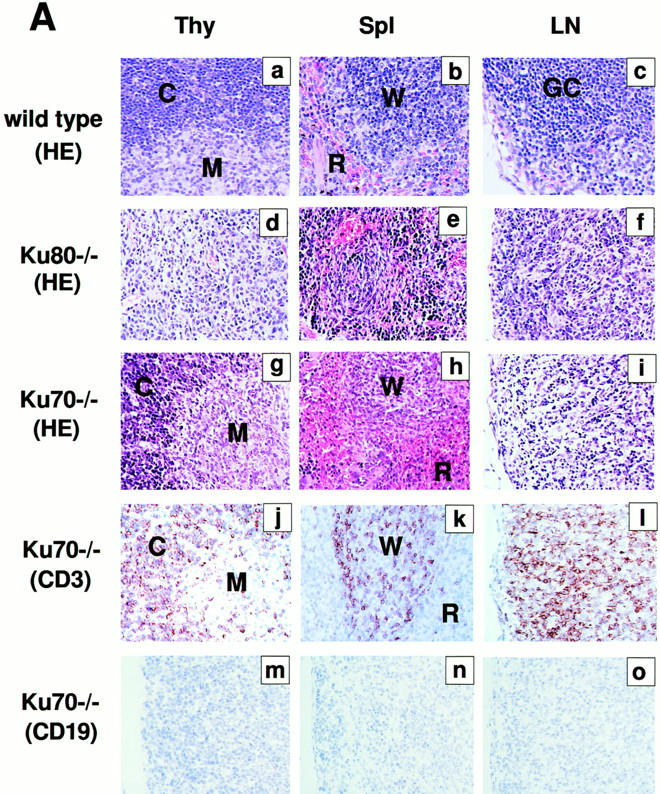
Development of B lymphocytes, but not T lymphocytes, is blocked at an early stage in Ku70− /− mice. (A) Histology of thymus (Thy), lymph nodes (LN) and spleens (Spl) from wild-type control mice, Ku70− /− mice, and Ku80− /− mice (23). Cortex (C) and medulla (M) are indicated. W, white pulp; R, red pulp; GC, germinal center. (a–i) Tissue sections were stained with haematoxylin and eosin (HE); (j– l) tissue sections were stained with anti-CD3 (CD3); and (m–o) tissues were stained with anti-CD19 (CD19). Anti-CD3 and anti-CD19 antibodies were tested in both frozen and paraffin sections of wild-type lymphoid organs and showed the expected specific patterns of staining (data not shown). (B) Flow cytometric analysis of thymocytes (Thy) bone marrow (BM) and spleen (Spl) cells from Ku70− /− mice, Ku70+ /+ littermates, and Ku80− /− mice. CD4, anti-CD4 monoclonal antibody; CD8, anti-CD8 monoclonal antibody; B220, anti-B220 monoclonal antibody; CD43, anti-CD43 monoclonal antibody; IgM, anti-Igμ heavy chain monoclonal antibody. The data were gated for live lymphoid cells based on forward and side scatter properties; 10,000–20,000 cells were analyzed per sample. (C) Analysis of TCR-β chain expression in Ku70− /− mice. Thymocytes and spleen cells were obtained from Ku70− /−, Ku80− /−, and wild-type littermates and analyzed for expression of CD4, CD8, and TCR-β by three-color flow cytometry. The TCR-β expression of both CD4+ and CD8+ SP T cells were shown.
Table 1.
Lymphoid Cellularity of Ku70−/− Mice
| Cell content (x 106) | |||
|---|---|---|---|
| Tissue and genotype | Total | B220+ | CD4+CD8+ |
| Thymus | |||
| wild type (n = 4) | 155 ± 42 | – | 104 ± 28 |
| Ku70−/− (n = 3) | 2.98 ± 0.91 | – | 0.6 ± 0.2 |
| Ku80−/− (n = 2) | 1.0 ± 0.5 | – | – |
| Bone marrow | |||
| wild type (n = 4) | 11.9 ± 3.3 | 5.5 ± 1.5 | – |
| Ku70−/− (n = 3) | 7.2 ± 2.9 | 1.1 ± 0.4 | – |
| Ku80−/− (n = 2) | 9.0 ± 3.0 | – | – |
| Spleen | |||
| wild type (n = 4) | 53 ± 20 | 29 ± 11 | – |
| Ku70−/− (n = 3) | 6.5 ± 1.3 | 0.4 ± 0.2 | – |
| Ku80−/− (n = 2) | 1.2 ± 0.5 | – | – |
To further examine the immunological defect in Ku70− /− mice, cells from thymus, bone marrow, and spleen were analyzed using monoclonal antibodies specific for lymphocyte surface markers and flow cytometry (19). Consistent with the immunohistological data, there was a complete block in B cell development at the B220+CD43+ stage in the bone marrow, and there were no mature B cells in the spleen (Fig. 2 B). In contrast, thymocytes developed through the CD4+CD8+ double-positive stage and matured into CD4+CD8− and CD4−CD8+ single-positive (SP), TCR-β–positive cells (Fig. 2, B and C). In six 4-wk-old Ku70_−_ /− mice analyzed, the percentage of CD4−CD8− double-negative thymocytes ranged from 11 to 62%, and the CD4+CD8+ double-positive cells varied from 35 to 73%. CD4−CD8+ (1–11%) and CD4+CD8− (1–3%) SP cells were also detected in the thymus. Furthermore, CD4+ CD8− or CD4−CD8+ SP T cells were found in the spleen in 67% of the mice studied (Fig. 2 B), which expressed surface TCR-β (Fig. 2 C) and CD3 (data not shown). Thus, in contrast to the early arrest of both T and B cell development in Ku80− /− mice (Fig. 2 B), lack of Ku70 is compatible with the maturation of T cells.
T-Cell Receptor and Immunoglobulin Gene Rearrangement.
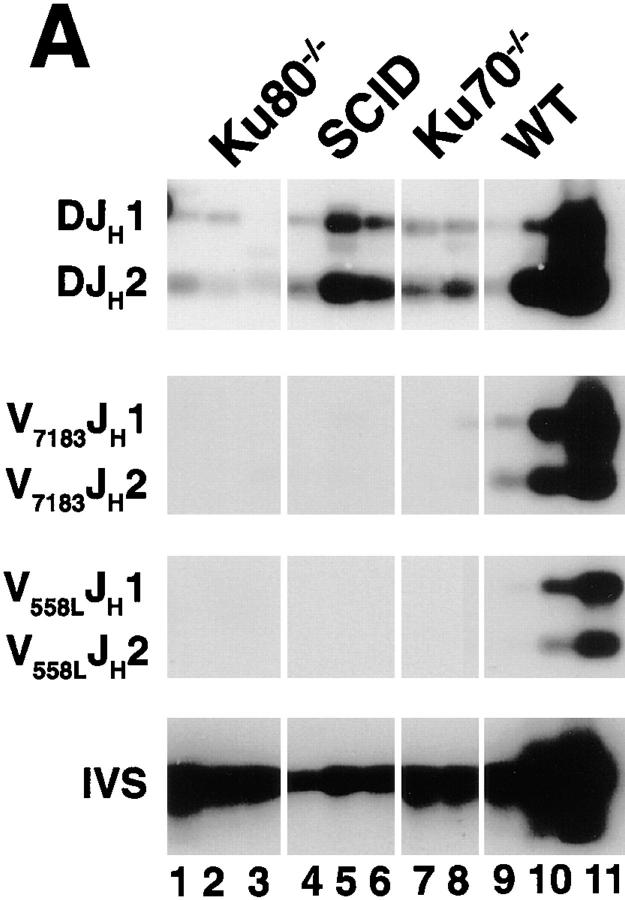
To determine whether a null mutation in Ku70 affects antigen-receptor gene recombination, DNA from bone marrow was amplified with primers specific to immunoglobulin D-JH and V-DJH rearrangements and DNA from thymus was amplified with primers that detected V-DJβ and Dδ-Jδ rearrangements (20, 25–28). Fig. 3 A shows that Ku70 −/− B cells do undergo D-JH recombination at a level which is similar to Ku80 −/− B cells, but is 2–3-fold lower than the level found in scid mice, and 10–50-fold lower than wild-type littermates. It is possible that some, but not all, of the decrease in D-JH rearrangement is due to a lower fraction of B lineage cells in the mutant sample, since the wild-type littermate mice have only approximately 5-fold more B220+ cells than the Ku70 −/− mice (see Table 1). V-DJH rearrangements were not detected in either Ku70 −/−, Ku80 −/−, or scid bone marrow samples, possibly accounting for the absence of mature B cells in these mutant mice (Fig. 3 A).
Figure 3.
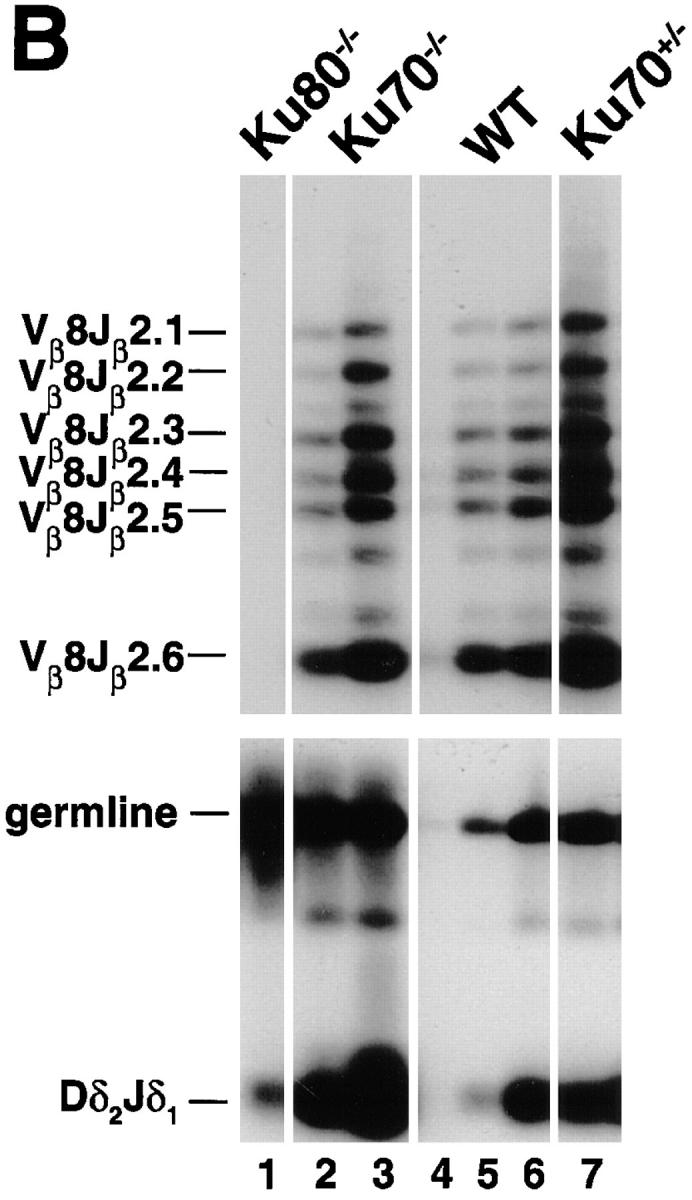
TCR and immunoglobulin gene rearrangement in Ku70− /− mice. (A) Recombination of V558L, V7183 to DJH, and DH to JH gene segments (26). 100 ng DNA was used for Ku70− /− (lanes 7 and 8), Ku80− /− (lanes 1–3), and scid mice (lanes 4–6), and 1, 10, and 100 ng for wild-type (WT) mice (lanes 9– 11). For IVS controls, DNA was diluted 100-fold before PCR. (B) PCR analysis of TCR gene rearrangements. Thymus DNA was assayed for recombination of Vβ8-Jβ2 and Dδ2 to Jδ1 rearrangements (20, 27, 28). 100 ng DNA was used for Ku70− /− (lanes 2 and 7), Ku80− /− (lane 1), and Ku70+ /− mice (lane 7), and 1, 10, and 100 ng for wild-type mice (lanes 4–6). Controls include a 1-kb germline interval amplified in the Dδ2 to Jδ1 intervening region (germline), and a nonrecombining segment of the Ig locus between JH and CH1 (not shown). The same thymus DNA samples were examined for Vβ8-Jβ2 and Dδ2 to Jδ1 recombination. Abbreviations: DJH, DH to JH rearrangements; V7183JH and V558LJH, V7183 and V558L to DJH rearrangements (26); Vβ8Jβ2.1 to Vβ8Jβ2.6, Vβ8 to DJβ2 rearrangements (28); germline, unrecombined DNA from the Dδ2 to Jδ1 interval; Dδ 2 Jδ 1, Dδ2 to Jδ1 rearrangements (20, 27); IVS, nonrecombining segment of the Ig locus between JH and CH1 (26). Multiple lanes underneath each genotype label (Ku70− /−, Ku80− /−, and SCID) represent different individual animals.
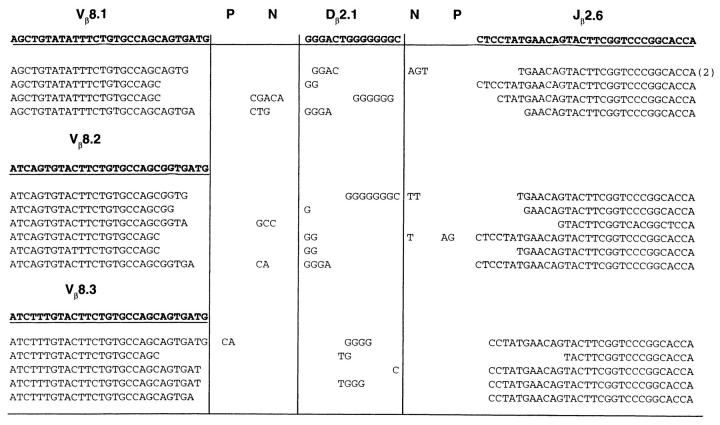
In contrast to the immunoglobulin heavy chain gene recombination, semiquantitative PCR analysis of thymocyte DNA for V-DJβ joints showed normal levels of TCR-β rearrangements on a per cell basis (Fig. 3 B). Similarly, Dδ2 and Jδ1 coding joints were found in Ku70− /− thymocytes at levels that resembled the wild type. To determine the molecular nature of the amplified coding joints, cloned Vβ8-DJβ2.6 joints were sequenced. We found normal numbers of N and P nucleotides, as well as normal levels of coding end deletions (Fig. 4). Thus, coding joints in Ku70− /− thymocytes differ from coding joints produced in xrs6 Ku80-deficient cells in that there were no large aberrant deletions (4, 18). We conclude that TCR V(D)J recombination in vivo does not require Ku70.
Figure 4.
Nucleotide sequences of Vβ8Dβ2.1Jβ2.6 junctions from the thymus of a 4-wk-old Ku70− /− mouse. Products corresponding to Vβ8.1, Vβ8.2, or Vβ8.3 rearrangement with Jβ2.6 were cloned and sequenced. TCR Vβ8-Jβ2 joints were amplified by PCR (20, 27, 28) as described (see Fig. 3 B). PCR cycling conditions were 94°C for 45 s, 58°C for 30 s, and 72°C for 30 h (30 cycles). The band corresponding to Vβ8-Jβ2.6 was purified, reamplified for 20 cycles, and then subcloned into the pCRII vector (Invitrogen, San Diego, CA). DNA was extracted from individual colonies and sequenced using the universal T7 and M13 reverse primers. Germline sequences are written in boldface; N and P, nucleotides not present in the germline sequences.
Absence of Ku70 Confers Radiation Hypersensitivity and Deficiency in DNA DSB Repair.
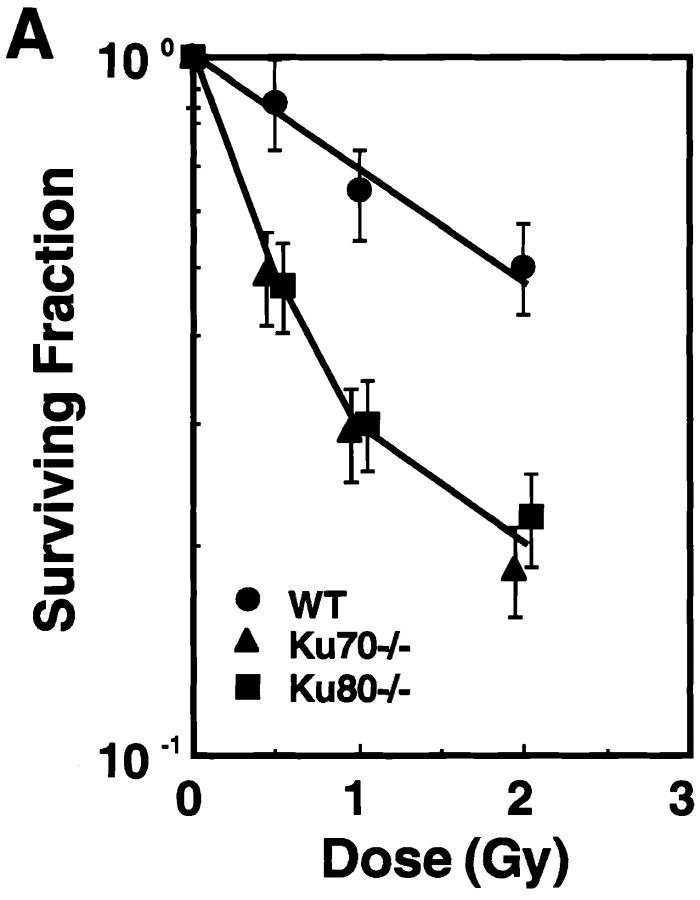
To assess radiation sensitivity in the absence of Ku70, cells from the bone marrow were exposed to ionizing radiation and were assayed for colony formation (30, 32). Fig. 5 A shows the survival curves of the CFU-GM from Ku70 −/−, Ku80 −/−, and wild-type control mice. CFU-GM from Ku70-deficient mice were more sensitive to ionizing radiation than those from Ku-proficient control mice (Fig. 5 A). Similar hypersensitivity to radiation was seen for Ku80−/− CFU-GM (Fig. 5 A).
Figure 5.
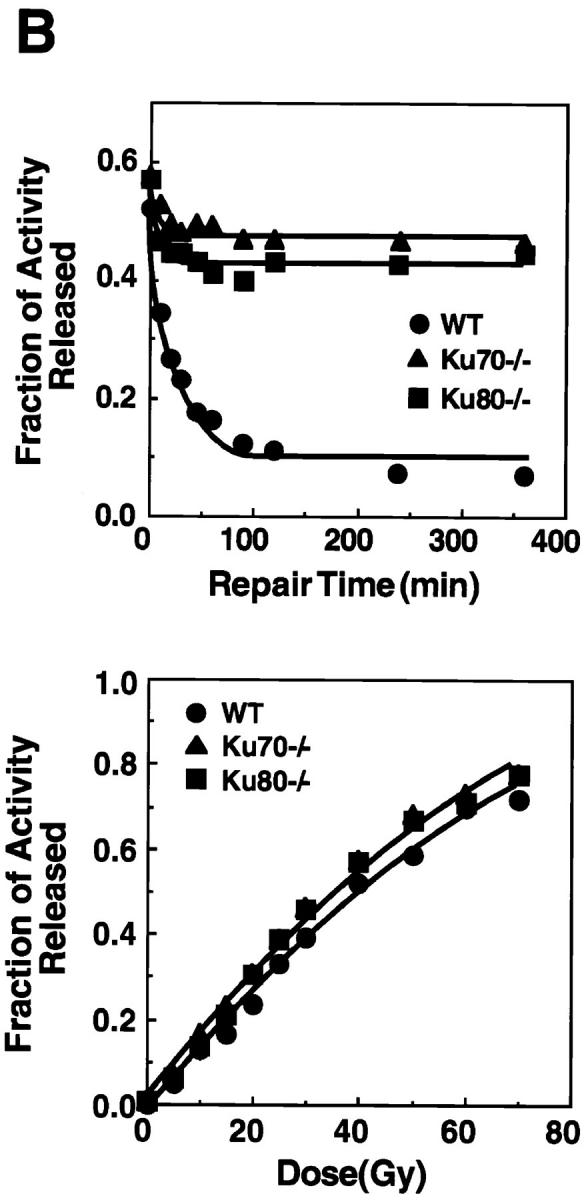
Disruption of Ku70 confers radiation hypersensitivity and a deficiency in DNA DSB repair. (A) Radiation survival curves for the CFU-GM in the bone marrow of wild type (WT), Ku70− /−, and Ku80− /− mice (30, 32). (B) Deficiency in the repair of radiation-induced DSB in Ku70− /− and Ku80− /− cells (31). (Top) Rejoining of DNA DSB produced by 40 Gy x-ray. (Bottom) Induction of DNA DSB as a function of the radiation dose in wild-type, Ku70− /−, and Ku80− /− cells. •_,_ wild type; ▴, Ku70− /− ; and ▪, Ku80− /− cells, respectively.
The rate and extent of rejoining of x-ray–induced DNA DSB in Ku70− /−, Ku80− /−, and Ku70 +/+ cells were measured using AFIGE (31). Fibroblasts derived from day 13.5 embryos were exposed to 40 Gy of x-rays and returned to 37°C for repair. At various times thereafter, cells were prepared for AFIGE to quantitate DNA DSB (Fig. 5 B, top). DNA DSB were nearly completely rejoined in wild-type cells within ∼2 h after radiation exposure. However, fibroblasts derived from Ku70− /− mice showed a drastically reduced ability to rejoin DNA DSB. A similar deficiency in DNA DSB rejoining was also observed in fibroblasts derived from Ku80− /− embryos. Despite the large differences observed in rejoining of DNA DSB between wild-type fibroblasts and fibroblasts derived from Ku70− /− or Ku80− /− mouse embryos, dose-response experiments showed that Ku70− /−, Ku80− /−, and wild-type fibroblasts were equally susceptible to x-ray–induced damage (Fig. 5 B, bottom). Thus, Ku deficiency primarily affects the ability of cells to rejoin radiation-induced DNA DSB without significantly affecting the induction of DNA damage.
Discussion
Absence of Ku70 results in radiation hypersensitivity and proportional dwarfism, as well as deficiencies in DNA DSB repair and V(D)J recombination. Thus, Ku70 −/− mice resemble Ku80 −/− mice in several respects, but the two mutations differ in their effects on T and B cell development. Lack of Ku70 was compatible with TCR gene rearrangement and development of mature CD4+CD8− and CD4−CD8+ T cells, whereas mature T cells were absent in Ku80 −/− mice. In contrast, B cells failed to complete antigen receptor gene rearrangement and did not mature in either Ku70 −/− or Ku80 −/− mice.
What could account for the differences we find in TCR and immunoglobulin gene rearrangements in the Ku70 −/− mice? One implication of our findings is that there are alternative Ku70-independent rescue pathways that are compatible with completion of V(D)J recombination in T cells. It is likely at the critical phase of T cell maturation, other DNA repair activity may be stimulated (33, 34) and can functionally complement the Ku70 gene in T cell–specific V(D)J recombination. Since Ku80 −/− mice are deficient in both T and B lymphocyte development, it is plausible that these yet to be identified alternative DNA repair pathways include Ku80. The much reduced level of Ku80 protein in Ku70 −/− cells may in part account for the hypocellularity of Ku70− /− thymuses.
Although the role of Ku in V(D)J recombination is not molecularly defined, Ku has been proposed to protect DNA ends from degradation (18, 35), to activate DNA-PK (10, 11), and to dissociate the recombination-activating protein RAG–DNA complex to facilitate the joining reaction (20). These functions are not mutually exclusive, and they are all dependent on the interaction of Ku with DNA. Thus, the finding that Ku70 is not required for TCR gene rearrangement is particularly unexpected because the Ku70 subunit is believed to be the DNA-binding subunit of the Ku complex (36), and DNA-end–binding activity was not detected in Ku70-deficient cells (Fig. 1 D).
In summary, our studies provide direct evidence supporting the involvement of Ku70 in the repair of DNA DSB and V(D)J recombination and the presence of a Ku70-independent rescue pathway(s) in TCR V(D)J rearrangement. The distinct phenotype of Ku70− /− mice should make them valuable tools for unraveling the mechanism(s) of DNA repair and recombination.
Acknowledgments
We thank D. Roth for PCR primers, D. Kim and L. Wu for Ku antiserum, T. Deloherey for FACS® analysis, P. Krechmer for word processing, A. Haimovitz-Friedman and Hatsumi Nagasawa for valuable suggestions, and C.C. Ling and Z. Fuks for advice and support.
Footnotes
The work was supported in part by National Institutes of Health grants CA-31397 and CA-56909 (to G.C. Li), CA-42026 (to G. Iliakis), CA-50519 (to D.J. Chen), and Department of Energy Office of Health and Environmental Research (to D.J. Chen). A. Nussenzweig is a research fellow supported by National Institutes of Health training grant CA61801 and M. Nussenzweig is an associate investigator in the Howard Hughes Medical Institute.
1
Abbreviations used in this paper: AFIGE, asymmetric field inversion gel electrophoresis; CFU-GM, granulocyte/macrophage CFUs; DNA-PK, DNA-dependent protein kinase; DSB, double-strand break, ES, embryonic stem; Gy, Gray; IR, ionizing radiation; IVS, intervening sequence; SP, single-positive.
References
- 1.Li Z, Otevrel T, Gao Y, Cheng H-L, Sneed B, Stamato T, Taccioli G, Alt FW. The XRCC4gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 2.Hendrickson EA, Qin X-Q, Bump EA, Schatz DG, Oettinger M, Weaver DT. A link between double-strand break-related repair and V(D)J recombination: the scidmutation. Proc Natl Acad Sci USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pergola F, Zdzienicka MZ, Lieber MR. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science (Wash DC) 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 5.Roth DB, Lindahl T, Gellert M. How to make ends meet. Curr Biol. 1995;5:496–499. doi: 10.1016/s0960-9822(95)00101-1. [DOI] [PubMed] [Google Scholar]
- 6.Bogue M, Roth DB. Mechanism of V(D)J recombination. Curr Opin Immunology. 1996;8:175–180. doi: 10.1016/s0952-7915(96)80055-0. [DOI] [PubMed] [Google Scholar]
- 7.Jeggo PA, Taccioli GA, Jackson SP. Menage a trois: double strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995;17:949–956. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 8.Weaver DT. What to do at an end: DNA double-strand-break repair. TIG (Trends Genet) 1995;11:388–392. doi: 10.1016/s0168-9525(00)89121-0. [DOI] [PubMed] [Google Scholar]
- 9.Biedermann KA, Sun J, Giaccia AJ, Tosto LM, Brown JM. scidmutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 12.Lees-Miller SP. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 13.Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, Oettinger MA, Brown JM. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science (Wash DC) 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 15.Blunt T, Finnie NJ, Taccioli GE, Smith GCM, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, Jackson SP. Defective DNA- dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 16.Boubnov NV, Hall KT, Wills Z, Lee SE, He DM, Benjamin DM, Pulaski CR, Band H, Reeves W, Hendrickson EA, Weaver DT. Complementation of the ionizing radiation sensitivity, DNA end binding, and V(D)J recombination defects of double-strand break repair mutants by the p86 Ku autoantigen. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smider V, Rathmell WK, Lieber MR, Chu G. Restoration of x-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science (Wash DC) 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 18.Taccioli GE, Gottlieb TM, Blunt T, Priestly A, Demengeot J, Mizuta R, Lehmann AR, Alt FA, Jackson SP, Jeggo PA. Ku80: product of the XRCC5gene and its role in DNA repair and V(D)J recombination. Science (Wash DC) 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 19.Nussenzweig A, Chen C, da Costa V, Soares, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature (Lond) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhu C, Bogue MA, Lim D-S, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 21.Takiguchi Y, Kurimasa A, Chen F, Pardington PE, Kuriyama T, Okinaka RT, Moyzis R, Chen DJ. Genomic structure and chromosomal assignment of the mouse Ku70 gene. Genomics. 1996;35:129–135. doi: 10.1006/geno.1996.0331. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Ouyang H, Yang S-H, Nussenzweig A, Burgman P, Li GC. A constitutive heat shock element– binding factor is immunologically identical to the Ku- autoantigen. J Biol Chem. 1995;270:15277–15284. doi: 10.1074/jbc.270.25.15277. [DOI] [PubMed] [Google Scholar]
- 23.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4ain tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 24.Cordon-Cardo C, Richon VM. Expression of the retinoblastoma protein is regulated in normal human tissue. Am J Pathol. 1994;144:500–510. [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1997. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- 26.Costa TEF, Suh H, Nussenzweig M. Chromosomal position of rearranging gene segments influences allelic exclusion in transgenic mice. Proc Natl Acad Sci USA. 1992;89:2205–2208. doi: 10.1073/pnas.89.6.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogue MA, Zhu C, Aguilar-Cordova E, Donehower LA, Roth DB. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- 29.Van Zant G, Flentje D, Flentje M. The effect of hyperthermia on hemopoietic progenitor cells of the mouse. Radiat Res. 1983;95:142–149. [PubMed] [Google Scholar]
- 30.Mivechi NF, Li GC. Thermotolerance and profile of protein synthesis in murine bone marrow cells after heat shock. Cancer Res. 1985;45:3843–3849. [PubMed] [Google Scholar]
- 31.Illiakis G, Metzger L, Denko N, Stamato TD. Detection of DNA double-strand breaks in synchronous cultures of CHO cells by means of asymmetric field inversion gel electrophoresis. Int J Radiat Biol. 1991;59:321–341. doi: 10.1080/09553009114550311. [DOI] [PubMed] [Google Scholar]
- 32.Fulop GM, Phillips RA. The scidmutation in mice causes a general defect in DNA repair. Nature (Lond) 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 33.Strasser A, Harris AW, Corcoran LM, Cory S. Bcl-2 expression promotes B- but not T-lymphoid development in scidmice. Nature (Lond) 1994;368:457–460. doi: 10.1038/368457a0. [DOI] [PubMed] [Google Scholar]
- 34.Danska JS, Pflumio F, Williams CJ, Huner O, Dick JE, Guidos CJ. Rescue of T cell–specific V(D)J recombination in SCID mice by DNA-damaging agents. Science (Wash DC) 1994;266:450–455. doi: 10.1126/science.7524150. [DOI] [PubMed] [Google Scholar]
- 35.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 36.Chou CH, Wang J, Knuth MW, Reeves WH. Role of a major autoepitope in forming the DNA binding site of the p70 (Ku) antigen. J Exp Med. 1992;175:1677–1684. doi: 10.1084/jem.175.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]