Multiple Distinct Sets of Stereotyped Antigen Receptors Indicate a Role for Antigen in Promoting Chronic Lymphocytic Leukemia (original) (raw)
Abstract
Previous studies suggest that the diversity of the expressed variable (V) region repertoire of the immunoglobulin (Ig)H chain of B-CLL cells is restricted. Although limited examples of marked constraint in the primary structure of the H and L chain V regions exist, the possibility that this level of restriction is a general principle in this disease has not been accepted. This report describes five sets of patients, mostly with unmutated or minimally mutated IgV genes, with strikingly similar B cell antigen receptors (BCRs) arising from the use of common H and L chain V region gene segments that share CDR3 structural features such as length, amino acid composition, and unique amino acid residues at recombination junctions. Thus, a much more striking degree of structural restriction of the entire BCR and a much higher frequency of receptor sharing exists among patients than appreciated previously. The data imply that either a significant fraction of B-CLL cells was selected by a limited set of antigenic epitopes at some point in their development and/or that they derive from a distinct B cell subpopulation with limited Ig V region diversity. These shared, stereotyped Ig molecules may be valuable probes for antigen identification and important targets for cross-reactive idiotypic therapy.
Keywords: Ig variable region genes, human, B lymphocytes, antibodies, autoantibodies
Introduction
The B lymphocyte clone expanded in chronic lymphocytic leukemia (B-CLL) expresses low levels of surface membrane Ig, the B cell antigen receptor (BCR). The genetics of this Ig have clinical relevance, as patients with a clone whose Ig variable (V) region has no or few mutations have a significantly worse outcome than those with significant numbers of Ig V mutations (1, 2). The biology underlying this association is unclear.
Several lines of evidence support a role for the BCR in the evolution of B-CLL (for review see reference 3). The distribution of individual IgV H in B-CLL clones differs from that found in normal cells (4), with an increased frequency of V H1-69, V H4-34, and V H3-07 (4–6). In addition, the distribution of mutations among B-CLL cases using these specific VH genes is selectively biased (4, 5, 7).
Recently, two subgroups of B-CLL cases with remarkable similarity of the entire BCR (V regions of the H and L chains) were identified (8, 9). Although these findings are provocative, they have been considered rare and potentially anomalous because, in one instance, the clones expressed IgG (9) and in the other geography and ethnicity may be relevant (10). This report describes another five groups of B-CLL patients that express BCRs of strikingly similar primary structure defined by highly similar Ig V regions in the H and L chains, and, in particular, distinct H and L CDR3 configurations. Thus, a significant fraction of B-CLL clones derive from B lymphocytes with constrained antigen-binding sites that could recognize individual, discrete antigens or classes of structurally similar epitopes.
Materials and Methods
IgV Gene Sequencing.
VHDJH and VLJL sequences were determined by previously described methods (4, 9).
Database Searches.
B-CLL Ig H chain V sequences from our collection (n = 255) and the public databases (n = 197) were subjected to BLAST searches of both nucleotide and protein databases to identify similar sequences. The criteria used to define “sets” of similar rearranged VHDJH were as follows: (a) use of the same VH, D, and JH germline genes; (b) use of the same D segment reading frame and position relative to the VH, plus or minus one codon; and (c) an amino acid similarity within the HCDR3 of ≥60% identity. In addition, all B-CLL Ig H protein sequences were aligned and clustered using the ClustalW alignment algorithm. Sequences clustering tightly were visually inspected for similarity. All of these searches used the complete VHDJH and as such were weighted toward sequences that used the same VH gene. To identify sequences with similar HCDR3 but different VH genes, CDR3 motifs from the various sets were used to search the public databases with the ProteinInfo search engine (http://prowl.rockefeller.edu). The criteria for the members of set V were altered to permit the use of different IgV H genes that were members of the same IgV H clan, while retaining the criteria for the rearranged VLJL. Use of the same specific IgV L gene and ≥85% LCDR3 identity was required for the inclusion of a companion rearranged VLJL in a set.
538 V H sequences from CD5+ and CD5− peripheral B lymphocytes (10, 11) were downloaded from the public database. These 538 sequences were compared independently with the translated databases using tblastn on the BlastMachine at the AMDeC Bioinformatics Core Facility at the Columbia Genome Center at Columbia University.
Online Supplemental Material.
Figs. S1–S7 depict detailed nucleotide and amino acid sequence alignments of the junctional regions and complete protein sequence alignments of the sequences described here. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20040544/DC1.
Results and Discussion
Identification of Subgroups of B-CLL Patients with Highly Restricted VHDJH Segments and Shared HCDR3 Configurations.
Each B-CLL–derived VHDJH sequence in our database was compared with every B-CLL sequence in our collection (n = 255) as well as with those in the public Ig V gene databases (n = 197) using nucleotide and protein sequence BLAST. In addition, all available B-CLL H chain V region sequences were phylogenetically grouped using the ClustalW method; sequences that clustered together were further analyzed for HCDR3 sequence similarity. These screening methods identified sets of sequences (Table I, sets I-Vb and Ve) consisting of the same IgV H with highly similar HCDR3 resulting from identical D (when identifiable) and J H segment use, D segment reading frame, similar D segment position relative to IgV H, and HCDR3 length, and significant (≥60%) amino acid sequence identity.
Table I.
Sets of B-CLL Cases That Share Ig V Region Genes
| No. of sequences identified/source | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B-CLL | Most similar germline gene | Median % mutation | |||||||||
| Set | Internal | Public | Non–B-CLL | Isotype | VH | D; reading frame | JH | VL | JL | VH | VL |
| I | 3 | 2 | 1a | IgMκ | 1-69 | 3-16; philic | 3 | κA27 | κ1/κ4/κ5 | 0.0 | 0.0 |
| II | 3 | 0 | 0 | IgMκ | 1-69 | 2-2; phobic | 6 | κL6 | κ3 | 0.0 | 0.0 |
| III | 4 | 3 | 0 | IgMλ | 1-69 | 3-10; phobic | 6 | λ1-16 | λ1/λ3 | 0.0 | 0.2 |
| IV | 3 | 2 | 0 | IgGκ | 4-34 | 5-5; phobic | 6 | κA17 | κ1/κ2 | 3.0 | 2.3 |
| Va | 4 | 2 | 1b | IgMκ | 1-02 | 6-19; stop | 4 | κ O12/2 | κ1/κ2 | 0.0 | 0.0 |
| Vb | 2 | 3 | 0 | IgMκ | 1-03 | 6-19; stop | 4 | κ O12/2 | κ1/κ2 | 0.8 | 0.0 |
| Vc | 1 | 0 | 0 | IgMκ | 1-18 | 6-19; stop | 4 | κ O12/2 | κ1 | 1.4 | 0.0 |
| Vd | 0 | 2 | 0 | IgM− | 1-46 | 6-19; stop | 4 | – | – | 0.0 | – |
| Ve | 2 | 5 | 0 | IgMκ | 5-51 | 6-19; stop | 4 | κ O12/2 | κ2 | 0.0 | 0.0 |
Three subsets of set V (Va, Vb, and Ve) contained sequences that used different IgV H genes, but used the same D and J H segments, the same _V_κ, and had highly similar HCDR3 configurations. Therefore, we used the HCDR3 motif common to these three subsets to search public databases for additional sequences with the same HCDR3 configuration potentially associated with a different IgV H segment. This search was not restricted to B-CLL sequences. The approach confirmed the previously identified subsets and identified two additional subsets of set V (Vc and Vd).
The public database searches identified 21 VHDJH sequences, belonging to one of the five individual sets, bringing the total number of sequences among these sets to 43. Interestingly, only 2 of the 21 sequences culled from the public databases were not derived from B-CLL cells. These two were from an anticardiolipin antibody-producing B cell (set I) and from a splenic marginal zone lymphoma (set Va). This distribution of similar sequences is particularly striking because, at the time of this search, the public databases contained only 197 Ig H chain V region sequences from B-CLL patients (excluding those from our laboratories) out of a total of >8,500 H chain V region sequences (search of Entrez with terms “human immunoglobulin heavy chain variable” produced 8,874 hits in the nucleotide database and >6,183 hits in the protein database on 12/16/03).
Pairing Restricted VL JL Rearrangements with VHDJH Segments in Sets.
VLJL sequences corresponding to the shared VHDJH of the five sets were available for most of our B-CLL cases and for a few of those identified in the public databases. Remarkably, the available IgV L were highly conserved within the sets, and the corresponding J L were very restricted (Table I and Table S1, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1). Four out of the five sets with available L chains expressed the κ isotype.
IgV Gene Mutation Status and Isotype Restrictions of Individual Sets.
Most of the IgV H sequences in each set differed by <2.0% from the most similar germline gene, with the exception of set IV in which the median level of mutation was 3.0%. Notably, the deduced protein structures in those sequences that were considered “mutated” using the typical 2% threshold differed from the germline by relatively low levels. Only one sequence, from set IV (Table S1, CLL ID47), differed by >5% from its germline counterpart. The corresponding IgV L in each set exhibited low levels of mutation; in some cases, V L displayed <2.0% difference, whereas V H had ≥2% difference from the germline sequence (Table I and Table S1).
The H chain isotype was the same among members of a set. All sets expressed IgM, except for set IV that consisted of IgG+ cases, similar to a patient group reported previously (9).
H and L CDR3 Characteristics of the Individual Sets.
We identified trends in the chemical, structural, or functional nature of the residues that comprise the H and L CDR3s, and in particular their VH-D and D-JH junctions (Fig. 1). For example, the D segments in the HCDR3s of these sets were read in the hydrophobic and stop reading frames more often than in normal (12) and B-CLL (13) cells. For all cases in set V, the D6-19 segment is read in a nonproductive reading frame (Table I). However, the germline stop codon, located in the region of overlap with the terminal IgV H sequence, was trimmed, allowing productive rearrangements with the J H4 segment (Fig. S7, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1).
Figure 1.
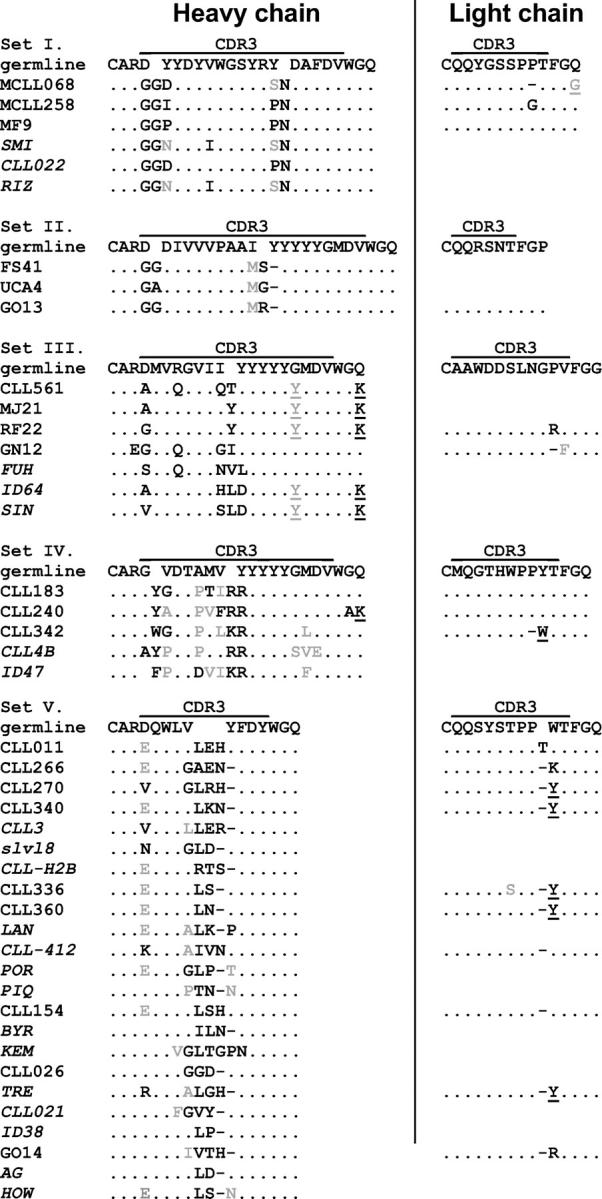
Sets of highly similar H and L CDRs among groups of B-CLL patients. Translated CDR3 regions are shown for all sequences within a set. Homology to the germline sequence is indicated by dots, and functionally similar replacement amino acids are indicated in gray. Amino acid replacements that may be attributed to known allelic differences are underlined.
Also of note was the repeated occurrence of certain nongermline encoded amino acids within D segments in some of the sets. For example, in all members of set II, a change to M is found at the 3′ end of the D segment (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1), a position that is not known to be polymorphic. Three out of seven sequences in set III had an R to Q change within the D3-10 segment that is also not listed as polymorphic (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1). In four out of five cases in set IV, P replaced A in the portion of HCDR3 encoded by the canonical D5-5 segment. Although this is most likely a polymorphism of the D segment rather than a common mutation, the last of the five sequences in this set (CLL ID47) also deviates from the canonical D5-5 sequence at this codon, substituting a D (Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1). Thus, even if these amino acid changes represent polymorphisms, their relative consistency within each set suggests a selection for these residues.
Members of several sets have common junctional residues that were not templated by any known germline gene segments and, therefore, presumably arose from trimming and/or addition during recombinational assembly. The sequences in set I all contain a pair of Gs at the VH-D junction and an N at the D-JH junction (Fig. 1 and Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20040544/DC1). A very similar VH-D junctional finding exists in set II (Fig. 1 and Fig. S4). All sequences in set IV contain an aromatic residue at the VH-D and a pair of basic residues (R or K) at the D-JH junction (Fig. 1 and Fig. S6).
Other trends in the composition of the H and L CDR3s are found in the other sets. These and the fine details of the nucleotide and amino acid sequences of the VHDJH and VL JL junctions for each set are shown and discussed in the online supplemental data (Figs. S3–S7).
Structural Similarities of the BCR among Members of the Sets.
The deduced VHDJH and VLJL protein sequences for each member of the stereotyped sets are presented in Figs. S1 and S2 (available at http://www.jem.org/cgi/content/full/jem.20040544/DC1). Because most members of the sets use the same IgV H, primarily in an unmutated form, associated with the same D and J H segments and because these rearrangements are virtually always paired with an identical IgV L that is restricted in its linked J L, the primary structural features of the entire BCR of each set are likely remarkably similar. Furthermore, the amino acid sequences of HCDR1, HCDR2, LCDR1, and LCDR2 of members of the individual sets are extremely similar, if not identical (e.g., sets I–III, and the set V subsets). In set IV, some amino acid differences exist in these regions due to somatic mutation.
These data indicate a much more marked constraint on the primary structure of the BCR in B-CLL than appreciated previously. They also indicate that this principle occurs in a sizeable number of patients. Collectively, ∼12% (31 out of 255: 22 from this work, 5 from our previous paper [9], and 4 that match another described set [8, 10]) of all of sequences in our internal laboratory B-CLL database and ∼20% (27 out of 131) of those with unmutated IgV belong to one out of the five stereotyped sets described here or one out of the two aforementioned patient groups (8–10). Approximately the same overall frequency (∼12%) was encountered among the sequences from the public databases (21 out of 197), although the proportion of the public B-CLL sequences that are unmutated was not determined. Most of the rearrangements in these sets lack or have few somatic mutations, and even those whose VH surpass the 2% threshold commonly used as the criterion to define significant IgV gene mutations (4, 5) are only slightly above that level. This suggests that restricted BCR structure is primarily a feature of those patients with the worse clinical course and outcome (1, 2). It appears that one out of five B-CLL cases with unmutated BCRs fit into one of these defined sets. Additional sets will likely be uncovered as more Ig V region sequences are defined in B-CLL, and all unmutated cases may be similar to one of a discrete number of archetypal sets. Although sets I–III use unmutated 1-69, they differ from previously described _1-69_–expressing B-CLL cases that have restrictions in specific D and J H segments associations (4, 6). These differences include JH (set I, J H3 vs. J H6), D (set II, D2 vs. D3 family and V_κ_L6 with an extremely short LCDR3), and L chain (set III, λ vs. κ) gene use.
Initial studies that considered only IgV H or VHDJH (4–6, 14) pointed toward limited structural diversity in the antigen-binding sites of B-CLL. However, our current results are much more striking because of the remarkable similarity of the sequences within a set and the virtual mathematic impossibility that this similarity arose by chance. If gene segment use in B-CLL was random, the probability of finding the same combination of VHDJH and VL JL segments in independent leukemic (or normal) B cells would be >10−6. Therefore, one would not expect to identify two B-CLL patients with BCRs comprised of the same VHDJH/VL JL until >106 cases were analyzed. This calculation is conservative because it does not account for diversity at the VH-D, D-JH, and VL-JL junctions that can be quite extensive (potentially exceeding 10−9 and reaching 10−12), although receptor editing and revision could limit these possibilities somewhat. Nevertheless, the level and frequency of BCR structural restriction in clusters of patients reported here is extraordinary and appears to be higher than any other B or T cell lymphoproliferative disorder reported to date.
Finding similar Ig H chain V region sequences by homology searches of the public databases is not, in itself, completely surprising because some IgV H are expressed in a biased fashion and ∼6,600 different VH-D-JH combinations can occur. Because the databases contain more than that number of Ig H chain V region sequences, identifying the same recombined gene segments is not improbable. When we analyzed 538 CD5+ and CD5− B cell–derived H chain V region sequences, we identified many pairs of similar sequences and some groups of similar sequences. However, these groups derived from B cells of diverse sources, as would be expected if the similarities were the product of random chance. In contrast, the similarity to a given B-CLL–derived sequence detected in our database comparisons arose almost exclusively from other B-CLL sequences (19/21) or other lymphoproliferative disorders (1/21), even though the entire database was searched. Only one identified sequence was from a non–B-CLL clone, one that coded an autoantibody (Table I and Table S1). Although the proper normal B cell repertoire against which B-CLL clones should be compared remains an open question (3), these results demonstrate that sequence sets of restricted cellular origin are not a generalized phenomenon in the public database.
Therefore, the development of B-CLL must involve B cell clones with restricted IgV and/or BCR structure. Although it seems unlikely that the expression of particular BCR gene combinations could be the sole promoting factor for leukemogenesis, a strong inherent bias in gene segment association and VHDJH/VL JL pairing in the B cell population that gives rise to B-CLL cannot be formally excluded, especially because the cell of origin for B-CLL is still uncertain (3). Although evidence exists in mice for biases in the recombination of particular Ig V gene segments before antigen experience (15), the extent of restriction imposed by recombination biases at both the H and L chain V gene loci in those instances, especially at the V-(D)-J junctions, are not as severe as in the sets described here. To our knowledge, there is no known subpopulation of human B cells in which the frequency of similar rearrangements, independent of antigen selection, is as great as among these B-CLL cases.
Therefore, antigen selection probably has a major restrictive influence on the transformation of a normal B lymphocyte to a B-CLL cell. A simple model would postulate that the transforming event is coupled with antigen specificity; i.e., an individual B lymphocyte from a highly diverse population could bind and internalize a transforming agent (e.g., virus) via its BCR. Although this seems unlikely, such a mechanism has been implied for B-CLL (16).
Alternatively, antigen could be a promoting factor for transformation, selecting specific clones for expansion from an initially diverse population of B lymphocytes and fostering their development to and in the transformed state (3). This would be the case if the B-CLL–susceptible cell population were preselected for antigen reactivity and, therefore, BCR structure, by exposure to distinct antigens or classes of antigens during their development. These clones could differ among patients, especially if the selecting antigens were foreign or autologous and possibly polymorphic. From within these clonal expansions, one member could develop an initial transforming lesion that would promulgate the leukemogenic cascade independent of antigen.
Finally, the initial transforming events could occur at random within a diverse B cell population or a previously antigen-selected population, and the subsequent nurturing of the transformed clone to clinical B-CLL could require ongoing BCR engagement by antigen (3). Recently, clonal expansions of B cells with phenotypic characteristics of B-CLL were found in normal elderly individuals (17, 18). The clinical relevance of these clones is not established. However, they may represent clones that have some of the genetic lesions of B-CLL, but lack BCR specificities that would result in sufficient ongoing stimulus to mature them into clinical B-CLL.
The remarkable protein similarity of the entire BCR among members of each set (Figs. S1 and S2) suggests that they could recognize the same or similar antigens. Although the nature of the antigens cannot be directly deduced from the Ig sequences presented here, there are several reasons to suspect that they are autoantigens or carbohydrates possibly derived from bacterial or viral coats, or a combination of the two.
V H1-69 (sets I–III) and V H3-21 (previously described set [8, 10]) are enriched among rheumatoid factors (19, 20). V H4-34 (set IV) is used in every case of monoclonal cold agglutinin disease (21) and in autoimmune conditions. Indeed, the inherent autoreactivity of this VH segment elicits a major inhibitory process by the immune system that keeps 4-34 + B cells from diversifying into high affinity, isotype-switched B cells (22). The anticardiolipin antibody identified as a member of set I implies that the other members of that set may be specific for cardiolipin or DNA because some antibodies to the former react with the latter (23). In addition, restricted VHDJH and/or VL JL gene segments are features of B cells that produce anticarbohydrate mAb in human (24) and mouse (25).
Characteristic junctional residues are also a feature of anticarbohydrate mAb and autoantibodies and basic junctional residues, as seen in sets I, IV, and Ve (Fig. 1 and Fig. S1), often indicate reactivity with acidic targets such as DNA (26). The synthesis of autoreactive Ig/BCR molecules by many B-CLL clones (27, 28) supports a link between the unique BCR structural features of these sets and autoantibodies.
The non–B-CLL Ig sequences that matched these B-CLL stereotypes may give insight into the identity of the B-CLL progenitor cells. One of those two was derived from a splenic marginal zone lymphoma (Table S1, set Va), and the other was derived from an autoantibody-producing B cell (Table S1, set I). Interestingly, normal MZ B cells produce mAb that can recognize thymus-independent type II antigens and autoantigens (29). In addition, the Ig V region repertoire of murine MZ B cells is very restricted in gene segment use and structure that requires intact BCR signal transduction to develop (30). MZ B cells appear to be progenitors for gastric MALT lymphoma (31) and have been proposed as precursors of B-CLL cells (3). If one infers common antigenic reactivity based on the similar sequences within a set, a significant fraction of B-CLL cases, and in particular those with unmutated IgV genes, could produce mAb that recognize one of a limited, discrete array of antigens or epitopes. With such an interpretation, some B-CLL cases may resemble gastric MALT lymphoma regarding the role of antigenic drive (in that instance, Helicobacter pylori) in the promotion of malignancy. The stereotyped Ig molecules reported here might be valuable probes to identify antigens that drive the leukemogenic process in B-CLL.
Finally, these sets of stereotyped Ig molecules may serve as therapeutic targets on B-CLL cells. A conceptual drawback to targeting the BCR as a tumor-specific antigen has been the apparent need to create an individualized reagent for each patient. However, because our data indicate that there is potentially extensive overlap in BCR structure and specificities among groups of B-CLL cases, this approach may be far less daunting. Indeed, because ∼20% of the cases with unmutated IgV H genes fall into one of these sets, such targeting might be most effective in those cases that have the worst prognosis, are least responsive to therapy, and have the most aggressive clinical courses (1, 2).
Acknowledgments
This work was supported in part by RO1 grants (nos. CA 81554 and CA 87956) from the National Cancer Institute, an M01 General Clinical Research Center Grant (no. RR018535) from the National Center for Research Resources, the Associazione Italiana Ricerca sul Cancro, and MIUR. The Peter J. Sharp Foundation, The Marks Family Foundation, the Jean Walton Fund for Lymphoma and Myeloma Research, the Joseph Eletto Leukemia Research Fund, the Laurie Strauss Leukemia Foundation, and the S.L.E. Foundation, Inc. also provided support for this work.
The authors have no conflicting financial interests.
B.T. Messmer and E. Albesiano contributed equally to this paper.
References
- 1.Damle, R.N., T. Wasil, F. Fais, F. Ghiotto, A. Valetto, S.L. Allen, A. Buchbinder, D. Budman, K. Dittmar, J. Kolitz, et al. 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847. [PubMed] [Google Scholar]
- 2.Hamblin, T.J., Z. Davis, A. Gardiner, D.G. Oscier, and F.K. Stevenson. 1999. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 94:1848–1854. [PubMed] [Google Scholar]
- 3.Chiorazzi, N., and M. Ferrarini. 2003. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Ann. Rev. Immunol. 21:841–894. [DOI] [PubMed] [Google Scholar]
- 4.Fais, F., F. Ghiotto, S. Hashimoto, B. Sellars, A. Valetto, S.L. Allen, P. Schulman, V.P. Vinciguerra, K. Rai, L.Z. Rassenti, et al. 1998. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 102:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder, H.W.J., and G. Dighiero. 1994. The pathogenesis of chronic lymphocytic leukemia: analysis of the antibody repertoire. Immunol. Today. 15:288–294. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, T.A., L.Z. Rassenti, and T.J. Kipps. 1997. Ig VH1 genes expressed in B cell chronic lymphocytic leukemia exhibit distinctive molecular features. J. Immunol. 158:235–246. [PubMed] [Google Scholar]
- 7.Kipps, T.J., E. Tomhave, L.F. Pratt, S. Duffy, P.P. Chen, and D.A. Carson. 1989. Developmentally restricted immunoglobulin heavy chain variable region gene expressed at high frequency in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 86:5913–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobin, G., U. Thunberg, A. Johnson, I. Eriksson, O. Soderberg, K. Karlsson, M. Merup, G. Juliusson, J. Vilpo, G. Enblad, et al. 2003. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vlambda2-14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood. 101:4952–4957. [DOI] [PubMed] [Google Scholar]
- 9.Ghiotto, F., F. Fais, A. Valetto, E. Albesiano, S. Hashimoto, M. Dono, H. Ikematsu, S.L. Allen, K.R. Rai, M. Nardini, et al. 2004. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J. Clin. Invest. 113:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin, G., U. Thunberg, A. Johnson, I. Thorn, O. Soderberg, M. Hultdin, J. Botling, G. Enblad, J. Sallstrom, C. Sundstrom, et al. 2002. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 99:2262–2264. [DOI] [PubMed] [Google Scholar]
- 11.Geiger, K.D., U. Klein, A. Brauninger, S. Berger, K. Leder, K. Rajewsky, M.L. Hansmann, and R. Kuppers. 2000. CD5-positive B cells in healthy elderly humans are a polyclonal B cell population. Eur. J. Immunol. 30:2918–2923. [DOI] [PubMed] [Google Scholar]
- 12.Zemlin, M., M. Klinger, J. Link, C. Zemlin, K. Bauer, J.A. Engler, H.W. Schroeder Jr., and P.M. Kirkham. 2003. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 334:733–749. [DOI] [PubMed] [Google Scholar]
- 13.Fais, F., G. Gaidano, D. Capello, A. Gloghini, F. Ghiotto, S. Roncella, A. Carbone, N. Chiorazzi, and M. Ferrarini. 1999. Immunoglobulin V region gene use and structure suggest antigen selection in AIDS-related primary effusion lymphomas. Leukemia. 13:1093–1099. [DOI] [PubMed] [Google Scholar]
- 14.Chiorazzi, N., and M. Ferrarini. 2001. Immunoglobulin variable region gene characteristics and surface membrane phenotype define B-CLL subgroups with distinct clinical courses. Chronic Lymphoid Leukemias. B.D. Cheson, editor. Marcel Dekker, New York. 81–109.
- 15.Seidl, K.J., J.D. MacKenzie, D. Wang, A.B. Kantor, E.A. Kabat, and L.A. Herzenberg. 1997. Frequent occurrence of identical heavy and light chain Ig rearrangements. Int. Immunol. 9:689–702. [DOI] [PubMed] [Google Scholar]
- 16.Mann, D.L., P. DeSantis, G. Mark, A. Pfeifer, M. Newman, N. Gibbs, M. Popovic, M.G. Sarngadharan, R.C. Gallo, J. Clark, et al. 1987. HTLV-I–associated B-cell CLL: indirect role for retrovirus in leukemogenesis. Science. 236:1103–1106. [DOI] [PubMed] [Google Scholar]
- 17.Rawstron, A.C., M.J. Green, A. Kuzmicki, B. Kennedy, J.A. Fenton, P.A. Evans, S.J. O'Connor, S.J. Richards, G.J. Morgan, A.S. Jack, and P. Hillmen. 2002. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 100:635–639. [DOI] [PubMed] [Google Scholar]
- 18.Ghia, P., G. Prato, C. Scielzo, S. Stella, M. Geuna, G. Guida, and F. Caligaris-Cappio. 2004. Monoclonal CD5+ and CD5- B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 103:2337–2342. [DOI] [PubMed] [Google Scholar]
- 19.Silverman, G.J., R.D. Goldfien, P. Chen, R.A. Mageed, R. Jefferis, F. Goni, B. Frangione, S. Fong, and D.A. Carson. 1988. Idiotypic and subgroup analysis of human monoclonal rheumatoid factors. Implications for structural and genetic basis of autoantibodies in humans. J. Clin. Invest. 82:469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, X., J.J. Goronzy, W. Zhong, C. Xie, and C.M. Weyand. 1995. VH3-21 B cells escape from a state of tolerance in rheumatoid arthritis and secrete rheumatoid factor. Mol. Med. 1:768–780. [PMC free article] [PubMed] [Google Scholar]
- 21.Pascual, V., K. Victor, M. Spellerberg, T.J. Hamblin, F.K. Stevenson, and J.D. Capra. 1992. VH restriction among human cold agglutinins. The VH4-21 gene segment is required to encode anti-I and anti-i specificities. J. Immunol. 149:2337–2344. [PubMed] [Google Scholar]
- 22.Pugh-Bernard, A.E., G.J. Silverman, A.J. Cappione, M.E. Villano, D.H. Ryan, R.A. Insel, and I. Sanz. 2001. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J. Clin. Invest. 108:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., S. Nagl, J.K. Kalsi, C.T. Ravirajan, D. Athwal, D.S. Latchman, L.H. Pearl, and D.A. Isenberg. 2003. Anti-cardiolipin/beta-2 glycoprotein activities co-exist on human anti-DNA antibody light chains. Mol. Immunol. 40:517–530. [DOI] [PubMed] [Google Scholar]
- 24.Scott, M.G., D.L. Crimmins, D.W. McCourt, I. Zocher, R. Thiebe, H.G. Zachau, and M.H. Nahm. 1989. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. III. A single VKII gene and one of several JK genes are joined by an invariant arginine to form the most common L chain V region. J. Immunol. 143:4110–4116. [PubMed] [Google Scholar]
- 25.Potter, M. 1977. Antigen-binding myeloma proteins of mice. Adv. Immunol. 25:141–211. [PubMed] [Google Scholar]
- 26.Radic, M.Z., and M. Weigert. 1994. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu. Rev. Immunol. 12:487–520. [DOI] [PubMed] [Google Scholar]
- 27.Sthoeger, Z.M., M. Wakai, D.B. Tse, V.P. Vinciguerra, S.L. Allen, D.R. Budman, S.M. Lichtman, P. Schulman, L.R. Weiselberg, and N. Chiorazzi. 1989. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J. Exp. Med. 169:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borche, L., A. Lim, J.L. Binet, and G. Dighiero. 1990. Evidence that chronic lymphocytic leukemia B lymphocytes are frequently committed to production of natural autoantibodies. Blood. 76:562–569. [PubMed] [Google Scholar]
- 29.Bendelac, A., M. Bonneville, and J.F. Kearney. 2001. Autoreactivity by design: innate B and T lymphocytes. Nat. Rev. Immunol. 1:177–186. [DOI] [PubMed] [Google Scholar]
- 30.Martin, F., and J.F. Kearney. 2000. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a “natural immune memory”. Immunol. Rev. 175:70–79. [PubMed] [Google Scholar]
- 31.Isaacson, P.G. 1999. Gastric MALT lymphoma: from concept to cure. Ann. Oncol. 10:637–645. [DOI] [PubMed] [Google Scholar]