Mechanism for elimination of a tumor suppressor: Aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma (original) (raw)
Abstract
Loss of tumor suppressors that restrain important oncoproteins such as c-Myc may contribute to malignant progression. Bin1 is an adapter protein with features of a tumor suppressor that was identified through its interaction with and inhibition of the oncogenic properties of c-Myc. In this study, we analyzed the patterns of Bin1 expression in normal melanocytes and melanoma cells at different stages of tumor progression. Evidence is provided that Bin1 function is abrogated in melanoma cells by a mechanism based on aberrant splicing of a tissue-specific exon. Specifically, most melanoma cells inappropriately expressed exon 12A, which is spliced alternately into Bin1 isoforms found in brain but not into isoforms found in melanocytes and many other nonneuronal cells. Exon 12A sequences abolished the ability of Bin1 to inhibit malignant transformation by c-Myc or adenovirus E1A. Similarly, these sequences abolished the ability of Bin1 to induce programmed cell death in melanoma cells that endogenously expressed exon 12A. Our findings suggest that aberrant splicing of Bin1 may contribute to melanoma progression, and they define a mechanism by which the activity of a tumor suppressor can be eliminated in cells.
Keywords: programmed cell death, Myc, oncogene, malignant transformation
Tumor development is driven by genetic events that fall into two general categories: activation of protooncogenes and elimination of tumor suppressor functions. The latter are caused by a variety of mechanisms, including loss of protein expression, expression of nonfunctional proteins, or expression of dominant inhibitory proteins. These events are achieved in tumors by genetic or epigenetic means and have been illustrated in studies of classical tumor suppressor proteins such as pRb, p53, and p16. Here we describe a mechanism for inactivation of the putative tumor suppressor protein Bin1 in human melanoma cells. Instead of alterations in the coding sequence, we found aberrant splicing of Bin1 transcripts that generated a protein that lacks growth-inhibitory activity and that has properties suggestive of a dominant inhibitor of wild-type Bin1.
Bin1 is a nucleocytosolic adapter protein that was identified initially through its ability to interact with Myc box regions at the N terminus of the Myc oncoprotein (1, 2). Although its actions as an adapter are complex, several lines of evidence support a role for Bin1 in the negative regulation of cell proliferation and malignancy. First, Bin1 inhibits the oncogenic activity of c-Myc, adenovirus E1A, and mutant p53 by domain-dependent mechanisms (1, 2). Second, Bin1 is structurally related to Rvs167, a negative regulator of the cell cycle in yeast that is needed to maintain viability after nutrient deprivation (1). Third, in cells where c-Myc is properly down-regulated after cytokine deprival, Bin1 promotes cell cycle exit and differentiation (3, 4). In contrast, if Myc is oncogenically activated and cell cycle exit is blocked, then cytokine deprival will cause programmed cell death, and Bin1 appears to be important for that process (5). Fourth, although normally ubiquitous, Bin1 is reduced or undetectable in some carcinomas, and its reintroduction blocks tumor cell proliferation (1). These results support the hypothesis that Bin1 is a tumor suppressor that may act in part by countering the transforming action of c-Myc. In this study, we present evidence that Bin1 is functionally altered by aberrant splicing in melanoma such that it loses its tumor suppressor activity. We propose that this event promotes tumor progression by allowing cells to overexpress c-Myc without apoptotic penalty.
MATERIALS AND METHODS
Cell Culture.
All cells were maintained in DMEM supplemented with 10% neonatal bovine serum (Life Technologies) and 50 units/ml penicillin and streptomycin (Fisher). Rat embryo fibroblast transformation assays were performed as described (1, 6) by using vectors for activated H-ras (pT22), human c-Myc vector (LTR Hm), or adenovirus E1A vector (p1A/neo), plus 10 μg of pcDNA3 vector (Invitrogen) or its derivatives CMV-99fE (Bin1), CMV-Bin1+12A, or CMV-Bin1–10+12A (7). COS cells were transfected with 10 μg of the vector indicated as described previously (1). For adenovirus infections, cells were seeded at 4 × 105 per well in a six-well tissue culture plate and allowed to attach overnight. A lacZ adenovirus was used to define the multiplicity of infection (moi) sufficient to cause infection of >90% of the culture, which was 100 moi for all cell lines except WM793, where 50 moi was sufficient and used in all experiments. Virus combinations were added at 100 moi, with each virus in a volume of 1 ml of DMEM supplemented with 4 μg/ml Polybrene (Sigma). After incubation for 3 hr at 37°C, cells were washed once with unsupplemented DMEM and fed with DMEM with 10% FBS. Cells were photographed or harvested for various purposes 24–48 hr later, as indicated.
Recombinant Adenoviruses.
The adenoviral cre-loxP inducible gene expression system used in this study was provided kindly by F. L. Graham, MacMaster Univ., Hamilton, ON (8). Briefly, the luciferase cDNA in the vector pMA19 (8) was replaced with human Bin1 cDNA 99fE (1), which represents the nonneuronal +10+13 splice isoform, or with the aberrant Bin1 −10+12A splice isoform cDNA (7). The resulting plasmids were introduced into 293 cells with _Cla_I-digested dl7001 adenovirus DNA, and the Ad-MABin1 or Ad-MABin1−10+12A viruses were obtained by standard methods (9). Ad-vect is an analogous control virus provided by J. M. Wilson (Univ. of Pennsylvania, Philadelphia) that lacks a transgene (9).
Apoptosis Assays.
Cells were harvested 48 hr after infection by trypsinization and fixed with 1% paraformaldehyde followed by 80% ethanol. For terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assay, samples were incubated in terminal deoxynucleotidyltransferase reaction buffer (BMB, Nutley, NJ) for 1 hr at 37°C and then stained with fluoresceinated streptavidin (DuPont) for 30 min at room temperature followed by propidium iodide for another 30 min at room temperature. Alternatively, cells were fixed in 70% ethanol and permeabilized in PBS/0.2% Tween 20 followed by propidium iodide staining and flow cytometry.
Reverse Transcription–PCR (RT-PCR).
For RT, 1 μg of total cytoplasmic RNA in 5 μl of water was denatured for 5 min at room temperature in 2 μl of 0.1 M methylmercury hydroxide, followed by the addition of 2.5 μl of 0.7 M 2-mercaptoethanol and 0.5 μg of random hexanucleotides in 1 μl of aqueous solution. After a 2-min incubation at 70°C, RNAs were incubated on wet ice and 2.0 μl of 5 mM dNTPs, 0.5 μl of RNase inhibitor (Promega), 4 μl of 5× RT buffer, 2 μl of 0.1 M DTT, and 1.0 μl Moloney murine leukemia virus reverse transcriptase (Life Technologies, Grand Island, NY) were added, incubated for 60 min at 37°C, and stopped for 5 min at 95°C. The product was used as template in three separate PCRs that amplified the 5′, middle, and 3′ ends of the coding region of Bin1. For the 5′ region (exons 1–7), the primers were 5′-TTTTGGATCCTTGCCACCGGGCGAGGCCTGCGC (Bam 5′UT Bin1) and 5′-TTTTTCTAGATCACACATTCATCTCCTCAAACACC (PTX7α/Stop/Xba). For the middle region (exons 7–11), the primers were 5′-TTTTGGATCCAGCCATGGATGAAGCCAAAATTGCCAAGGC (Bam/dT3ext/kozak) and 5′-TTTTTCTAGATCATGCTGGCTGAGATGGGGACTTG (5ATG99/Stop/Xba). For the 3′ region (exons 11–16), the primers were 5′-TTTTGGATCCAGCCATGGCGCCTGCAAAAGGGAACAAGAGC (Bam/99Fsp/kozak) and 5′-TTTTTCTAGATCATCGGGAGGAGGTGTTCTTCACAC (3UT99fe/Stop/Xba). Reactions were performed in 100 μl containing 200 ng of template, 50 pmol of each primer, 0.1 mM each dNTP, 1× Taq PCR buffer (BMB), and 2.5 units of Taq polymerase (BMB). PCR was performed by 2-min incubation at 96°C and then 35 cycles of 30 sec at 96°C/45 sec at 58°C/45 sec at 72°C followed by a final extension of 10 min at 72°C. Products were purified from 1.4% TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) agarose gels and subjected to direct DNA sequencing. A second set of PCRs using a different primer set was performed to confirm evidence of the presence of exon 12A in melanoma messages (results shown in Fig. 2 were derived from this set). The primer sequences were 5′-GCCTGCAAAAGGGAACAAGAGC and 5′-GGTTCTGGAAGGGGATCACC. The reaction conditions were identical to before except that the cycling program started with a 30-sec incubation at 96°C followed by 30 cycles of 30 sec at 96°C/30 sec at 59°C/45 sec at 72°C.
Figure 2.
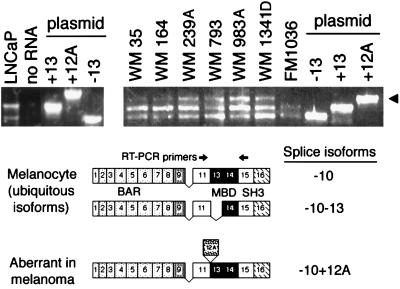
Nonphysiological splicing of brain-specific Bin1 exon 12A in melanoma. Splice isoforms expressed in fetal melanocytes and melanoma cell lines were identified by RT-PCR. Results from DNA sequencing of the RT-PCR products is presented in Table 1. (Upper) RT-PCR products derived from the 3′ region of Bin1 RNA in which splice variations occur were fractionated on agarose gels and stained with ethidium bromide. LNCaP is a control that identifies the two ubiquitous isoforms found all in nonneuronal cells (−10+13 and −10−13 isoforms), which also are expressed in fetal melanocytes (FM1036). Plasmid cDNA clones of different splice isoforms also were used as template controls (7). The arrowhead identifies 12A isoforms expressed in melanoma cell lines. (Lower) Cartoons illustrate splice isoforms identified (exons numbered), with arrows indicating the position of the primers used for the RT-PCR (Upper).
Northern Blot Analysis.
Twenty micrograms of total cytoplasmic RNA per lane was prepared and analyzed essentially as described (10). Blots were hybridized with human c-Myc or Bin1 (99fE) cDNAs (1, 6) washed at a final stringency of 0.2× SSC at 50°C and subjected to autoradiography at −80°C for 1–3 days. Loading controls were provided by ethidium bromide staining of ribosomal RNA (gel) or by hybridization of stripped blots with a β-actin cDNA probe.
mAb Generation.
A mAb was raised to a glutathione _S_-transferase (GST) fusion protein that included sequences encoded by exon 12A (7) by using methods described previously (11). Briefly, BALB/c mice were immunized with a GST-12A polypeptide. Hybridomas were generated by using the nonsecreting murine myeloma T3X63Ag8.Sp2/0, and the supernatant of hybridoma clones was screened by ELISA as described (12). One hybridoma that was specific for GST-12A immunogen and judged to be specific by Western analysis and immunohistochemistry (anti-12A) was used in this study.
Western Blot Analysis.
Cell lysates were prepared in Nonidet P-40 lysis buffer, and 50 μg/lane of each sample was analyzed by SDS/PAGE and Western blotting (13). Blots were probed with hybridoma supernatant 99D (1:20, vol/vol) or with anti-12A (1:10, vol/vol). Blots were developed by using a horseradish peroxidase-conjugated goat anti-mouse IgG (1:5,000) and a chemiluminescence kit, using the conditions recommended by the vendors (BMB; Pierce).
RESULTS
Frequent Aberrant Splicing of a Brain-Specific Exon in Bin1 in Melanoma.
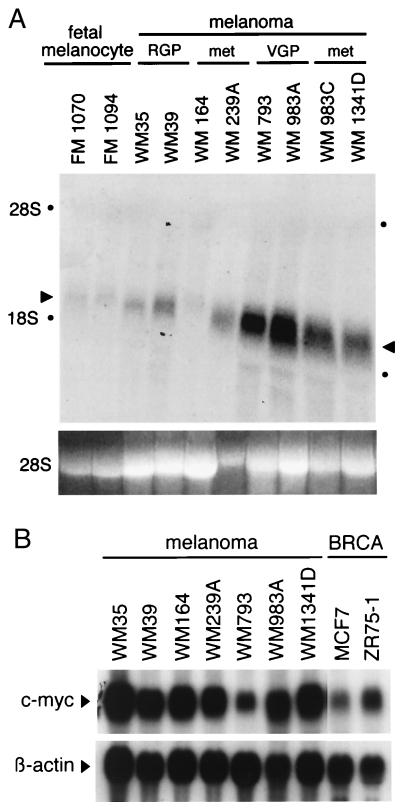
Northern blot analysis was performed to compare the level of Bin1 expression in normal neonatal melanocytes with that in cell lines derived from different types of sporadic melanoma, including two preparations of normal human neonatal melanocytes (FM1070 and FM1094), two cell lines derived from nonmalignant radial growth phase (RGP) melanoma (WM35 and WM39), two cell lines derived from malignant vertical growth phase (VGP) melanoma (WM793, WM983A), and four cell lines derived from metastatic melanoma [WM164, WM239A, WM983C (a metastatic variant of WM983A), and WM1341D]. Loss of message was anticipated given such events in carcinoma cells (1). However, although normal melanocytes and RGP tumor cells expressed basal levels similar to those found in other cell types (1, 7), both of the VGP lines and three of four of the metastatic lines actually exhibited elevated levels of Bin1 RNA (see Fig. 1A). This result was not an artifact of in vitro establishment of melanoma cells, because elevated protein levels were confirmed in six of eight metastatic melanomas examined by immunohistochemistry with anti-Bin1 antibody 99D (data not shown). c-Myc message levels were examined to assess the relationship with Bin1 (see Fig. 1B). c-Myc levels were at least as high as two breast carcinoma lines, MCF7 and ZR75–1, where c-Myc is overexpressed. Bin1 and c-Myc levels, therefore, were not coordinated because c-Myc was uniformly high whereas Bin1 levels varied with progression stage.
Figure 1.
Northern blot analysis of Bin1 and c-Myc in melanoma cell lines. Fifteen micrograms of total cytoplasmic RNA from each cell line was analyzed by Northern blotting and hybridization with 32P-labeled human Bin1 or c-Myc cDNA probes. (A) Bin1. Arrows indicate the Bin1 mRNAs, and dots indicate the position of 28S and 18S ribosomal RNAs. (Lower) Ethidium bromide-stained 28S RNA is shown as a loading control. (B) c-Myc. MCF7 and ZR75–1 are breast carcinoma cell lines that overexpress c-Myc and that are provided for comparison with the melanoma lines. (Lower) β-Actin hybridization to the same blot is shown as a loading control.
To determine whether Bin1 was overexpressed because of genetic alteration, we investigated Bin1 gene structure by Southern analysis, RT-PCR, and DNA sequencing. Southern analysis with cDNA probes revealed no gross aberrations, arguing against gene rearrangement or amplification (data not shown). RNA splice patterns were examined because Bin1 undergoes alternate splicing in a tissue-specific manner (7). Total cytoplasmic RNA isolated from primary melanocytes or melanoma cell lines was analyzed by RT-PCR as described (7), and the complete DNA sequence was determined by a direct sequencing method. This analysis, which spanned the entire coding region of Bin1, revealed two types of genetic alteration in melanoma cells (see Fig. 2 and Table 1). Neonatal melanocytes expressed the two major splice isoforms characteristic of nonneuronal tissues, whereas melanoma cell lines expressed, in addition to these isoforms, a larger splice isoform that included the brain-specific exon 12A (7). Primary melanomas also expressed this exon, ruling out the possibility that this event was an artifact of in vitro establishment (data not shown). Exon 12A is spliced in various combinations with exons 12B, 12C, and 12D (7, 14, 15), none of which are appreciably expressed outside the central nervous system. Exons 12B–D were not seen in melanoma, arguing that the splice event in melanoma was aberrant, because exon 12A is not alternately spliced by itself even in brain. DNA sequencing of the RT-PCR products revealed missense mutations in WM35, which had five alterations, in WM983A and its metastatic variant WM983C, and in WM1341D, the latter of which had one missense mutation each (see Table 1). In each case, the mutations mapped to the N-terminal BAR domain, which is crucial for tumor suppressor activity (2). We did not investigate these alterations in detail because subsequent investigations indicated that exon 12A splicing was sufficient by itself to cause loss of tumor suppressor activity (see below). Similarly, we did not explore the significance of other abnormal splice events detected by RT-PCR in WM164 and WM239A, because accumulation of the predicted truncated species was not seen by either Northern or Western blot analyses (data not shown).
Table 1.
Structure of Bin1 RNA in melanoma cells
| Cell line | Cell type | Sequence | Exon 12A splicing* | Level of 12A protein | Susceptibility to ectopic Bin1 |
|---|---|---|---|---|---|
| FM 1036 | Melanocyte | WT | − | − | − |
| WM 35 | RGP | Five muts | + | + | ++ |
| WM 39 | RGP | ND | ND | ND | ND |
| WM 164 | Met | WT*† | + | − | − |
| WM 239A | Met | WT*† | + | +++ | +++ |
| WM 793 | VGP | WT | + | ++ | +++ |
| WM 983A | VGP | E180K | + | ++ | ++ |
| WM 983C‡ | Met | E180K | + | ND | ND |
| WM 1341D | Met | D146E | + | +/− | − |
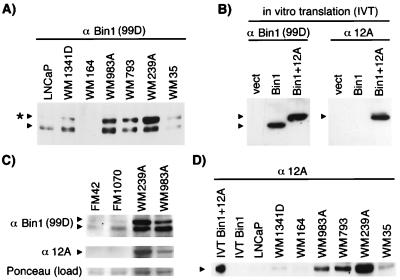
Western blot analyses with mAbs recognizing different epitopes within Bin1 were performed to confirm specific expression of 12A sequences in melanoma cells. The first antibody, 99D, which recognizes an exon 13-encoded epitope (11), detected a single band in extracts from human fetal melanocytes and control LNCaP prostate cells, which, as other nonmalignant cells of nonneuronal origin, do not express exon 12A at detectable levels. In contrast, 99D detected two bands in several melanoma cell lines (see Fig. 3 A and C). The size of the upper band matched an in vitro translated Bin1 isoform that included exon 12A-encoded residues (see Fig. 3B). The presence of 12A sequences in this band was confirmed with anti-12A, a mAb that was raised against exon 12A residues (see Fig. 3B). Western analysis of extracts with anti-12A revealed bands of the predicted size in melanoma cells that were not seen in fetal melanocytes or control LNCaP cells (see Fig. 3 C and D). These results corroborated the RT-PCR results and confirmed that 12A was expressed at the level of protein in melanoma cells. We concluded that exon 12A was spliced frequently and aberrantly in melanoma cells and that exon 12A-containing isoforms were overexpressed in tumor cells at the VGP and metastatic stages of progression.
Figure 3.
Western blot analysis of Bin1 in melanoma. Cell extracts were subjected to SDS/PAGE and Western analyses. Where indicated, extracts from control LNCaP cells were used to identify nonneuronal Bin1 isoforms. (A) 99D Western blot. The asterisk marks an additional polypeptide uncharacteristic of nonneuronal tissues. (B) Specificity of anti-12A. Vectors for Bin1, Bin1+12A, or no insert (vect) were subjected to in vitro transcription, and translation and products were analyzed by SDS/PAGE and Western blotting with 99D or anti-12A. (C) Bin1 expression in fetal melanocytes. Identical Western blots were probed with either 99D or anti-12A. (Bottom) A blot was stained with Ponceau S to provide a normalization control for protein loading. (D) Expression of 12A isoforms in melanoma cells. The blot was probed with anti-12A.
Aberrant Splicing of Exon 12A Leads to Loss of the Tumor Suppressor Activity of Bin1.
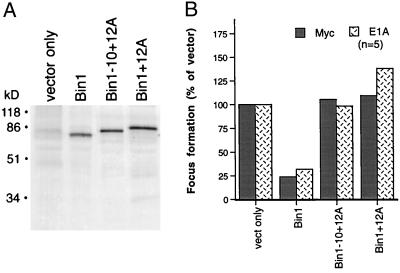
The effect of 12A sequences on the tumor suppressor activity of Bin1 was tested in primary rat embryo fibroblast transformation assays and in melanoma cells. The 12A isoforms tested accumulated to a similar degree in COS cells, so 12A sequences did not destabilize Bin1 (see Fig. 4A). In rat embryo fibroblast assays, the presence of 12A sequences relieved the ability of Bin1 to inhibit the focus-forming activity of c-Myc or adenovirus E1A when cotransfected with activated Ras (see Fig. 4B). Expression of transgenes was confirmed by Northern analysis of RNA from pooled foci (data not shown). Similarly, 12A sequences also relieved the ability of Bin1 to suppress proliferation of HepG2 cells (data not shown), a model used to measure tumor suppressor activity of Bin1 (1, 2).
Figure 4.
Exon 12A sequences abolish the antioncogenic activity of Bin1. (A) Protein expression. COS cells were transiently transfected with vectors for the splice isoforms indicated and metabolically labeled with [35S]methionine, and cell extracts were prepared and analyzed by immunoprecipitation and SDS/PAGE 48 hr later with 99D antibody. The Bin1-positive control used was the +10+13 splice isoform. (B) Transformation assay. Primary rat embryo fibroblasts were cotransfected with 5 μg each of vectors for activated H-Ras, c-Myc or adenovirus E1A, and 10 μg of the splice isoforms indicated, and the number of transformed foci were scored 2 weeks later. The Bin1-positive control was the +10+13 splice isoform. The data are depicted as the percentage of focus formation by c-Myc or E1A plus Ras and vector only. The mean of five trials of each experiment is shown.
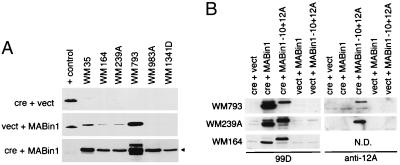
A cre-inducible adenovirus vector system was used to test the effect of Bin1 splice isoforms in human melanoma cells, the construction and characterization of which will be described in detail elsewhere (K. Elliott, K.G., and G.C.P., unpublished data). Briefly, two vectors expressing splice isoforms that lacked or included exon 12A were generated (Ad-MABin1 or Ad-MABin1−10+12A), the latter of which corresponded to the aberrant splice isoform from melanoma cells. Coinfection with a recombinant adenovirus-expressing Cre recombinase (Ad-cre) results in removal of an adjacent stuffer fragment, bringing the Bin1 cDNAs under the control of the cytomegalovirus early-region enhancer/promoter. An moi required to infect ≥90% of the target population for each of the melanoma cell lines was determined by using a β-galactosidase vector (Ad-lacZ). Several cell lines infected with 100 moi of Ad-MABin1 exhibited leaky transgene expression, the basis for which was unclear. Nevertheless, cells infected with Ad-MABin1 plus Ad-cre exhibited Bin1 induction in all cell lines (see Fig. 5A). Cells infected with Ad-MABin1−10+12A and Ad-cre showed similar induction of the 12A isoform in the subset of cell lines employed for biological analysis (see Fig. 5B).
Figure 5.
Bin1 expression from cre-inducible adenoviral vectors. (A) Bin1 expression. The melanoma cells indicated were infected with Ad-cre + Ad-vect (Top), Ad-vect + Ad-MABin1 (Middle), or Ad-cre + Ad-MABin1 (Bottom). Cell lysates were prepared 48 hr later and subjected to Western blotting with Bin1 antibody 99D. The positive control (+ control) for expression is an extract from murine C2C12 cells, which robustly express the −10+13 splice isoform (3). The exposure times were short to identify only exogenous levels of Bin1 expression (exposure in Bottom was shortest). (B) Bin1–10+12A expression. Cells indicated were infected and analyzed as above, except that Ad-MABin1−10+12A was used in addition to Ad-MABin1 and parallel blots were processed with 99D or anti-12A antibodies. N.D., not done.
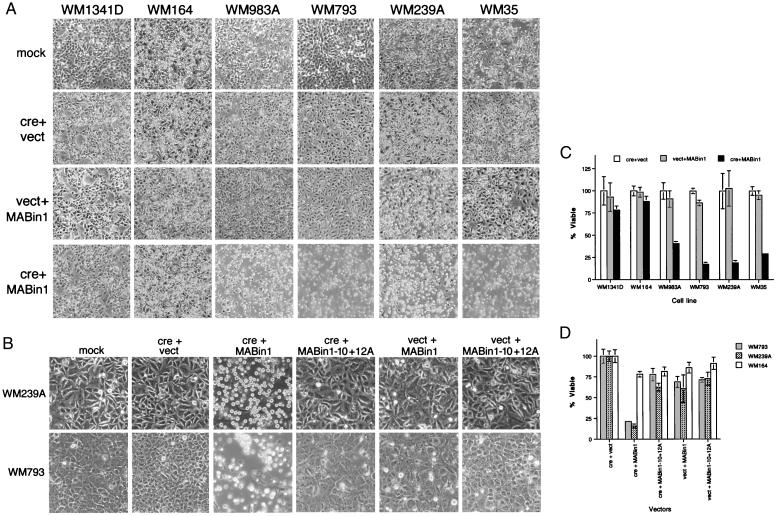
Ectopic Bin1 elicited substratum detachment and morphological features of programmed cell death in the melanoma cell lines that expressed 12A isoforms (WM35, WM239A, WM793, and WM983A). The extent of this phenomenon was correlated with loss of viability and with the relative ratio of endogenous 12A isoform to nonneuronal isoform in each cell line (see Fig. 6 A and C). For example, WM164 and WM1341D did not express much 12A species and Ad-MABin1 had little effect. WM35 expressed low levels of 12A species but also low levels of the wild-type species, and it responded to Bin1, as did all the remaining cell lines that robustly expressed 12A species. Experiments in the most susceptible cell lines (WM239A and WM793) showed that Ad-MABin1−10+12A caused neither cellular demise nor loss of viability (see Fig. 6 B and D). Death was not a nonspecific feature of Bin1 overexpression, because WM164 and WM1341D did not lose viability. Moreover, Bin1 did not cause death in fetal melanocytes or in IMR90 human diploid fibroblasts (data not shown). Loss of viability in cells was correlated with an accumulation of sub-G1 phase DNA and susceptibility to TUNEL reaction, consistent with the induction of programmed cell death (see Table 2 and Fig. 7). In summary, we concluded that aberrant splicing of exon 12A led to a loss of tumor suppressor activity of Bin1 in melanoma cells.
Figure 6.
Selective cell death elicited by ectopic Bin1 but not by a 12A-containing isoform. (A) Cell morphology. Cells were infected with the viruses indicated as in Fig. 5 and photographed 48 hr later. (B) Lack of effect of Bin1−10+12A. Cells were infected with the viruses indicated as in Fig. 5 and photographed 48 hr later. (C) Cell viability. Viable cell counts were determined by the trypan blue-exclusion method 42–46 hr after viral infection. (D) Bin1−10+12A does not affect cell viability. Viable cell counts were determined as before. The data presented in C and D represent the mean and SE of three separate determinations each.
Table 2.
Fraction of sub-G1 phase DNA detected by flow cytometry in cells infected with induced Bin1 adenovirus
| Cell line | cre only (Ad-cre + Ad-vect) | Uninduced Bin1 (Ad-vect + Ad-MABin1) | Induced Bin1 (Ad-cre + Ad-MABin1) | Level of 12A protein |
|---|---|---|---|---|
| WM 35* | 1.6 | 3.5* | 15.2 | + |
| WM 164* | 6.0 | 4.7* | 5.7 | − |
| WM 239A* | 6.3 | 12.8* | 20.9 | +++ |
| WM 793* | 6.1 | 10.7* | 58.8 | ++ |
| CM 983A | 3.5 | 3.7 | 10.3 | ++ |
| WM 1341D | 5.9 | 4.1 | 4.2 | +/− |
Figure 7.
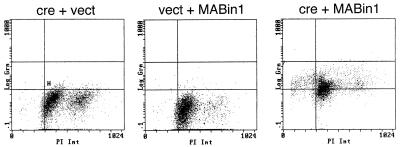
Loss of viability caused by ectopic Bin1 is due to programmed cell death. WM793 cells were mock-infected or infected with the viruses indicated. Cells were processed 48 hr later for TUNEL reaction, propidium iodide staining, and flow cytometry. For each data point, the x axis measures DNA content (propidium iodide binding) and the y axis measures relative TUNEL staining.
DISCUSSION
This study defines a loss-of-function alteration of the tumor suppressor Bin1 through a splicing mechanism. Bin1 is a nucleocytoplasmic adapter protein whose functions are regulated by alternate splicing. A nonneuronal species of Bin1 originally was identified through its ability to interact with c-Myc, and recent investigations have implicated it in the control of programmed cell death and differentiation (16). In contrast, brain-specific splice isoforms of Bin1, alternately termed amphiphysin II (14), apparently lack growth-inhibitory activity but, instead, influence endocytosis as amphiphysin (17). Notably, the determinants needed for interaction with the endocytosis machinery are encoded by the brain-specific exons (18). Consistent with this evidence, nonneuronal species that lack these exons do not affect appreciably receptor-mediated or fluid-phase endocytosis when overexpressed (unpublished observations). Thus, Bin1 isoforms that are specific to brain have a unique role in endocytosis that is conferred by tissue-specific splicing. Exon 12A is one of at least 4 brain-specific exons in the Bin1 gene (7, 14, 15). However, exon 12A was the only one that was spliced in melanoma, and this event was sufficient to eliminate the tumor suppressor activity of Bin1. We did not investigate the significance of several N-terminal mutations in certain melanoma lines, because 12A splicing was sufficient for loss of function, but these mutations might be germane at some level because they were clustered in the highly conserved and functionally important BAR region (2). Exon 12A is not spliced by itself in the absence of the other brain-specific exons 12B–D (7, 14, 15, 18, 19), in which 12B and 12C encode clathrin-binding determinants (18). Thus, although 12A splicing is sufficient to eliminate tumor suppressor properties, it is not sufficient to confer full endocytotic properties. This is, therefore, aberrant in the sense that it appears to be nonphysiological. 12A splicing has been documented in cell types other than melanoma cells (ref. 7 and unpublished observations), suggesting Bin1 may be inactivated by a similar mechanism more broadly. Other tumor suppressors have been shown to undergo loss of function because of aberrant splicing, such as DCC (20) and FHIT, in which exons are deleted at the level of genomic DNA (21). However, this is a case where tumor suppressor activity is abolished by splicing of a tissue-specific exon in an inappropriate tissue.
Exon 12A sequences abrogated the ability of Bin1 to suppress malignant transformation by c-Myc or adenovirus E1A in primary fibroblasts and to drive programmed cell death in melanoma cells. The mechanism for the loss of activity was undetermined. Cell localization may be important, because immunohistochemical experiments indicate that exon 12A-containing isoforms are preferentially located in the cytosol of neuronal cells and transiently transfected COS cells (refs. 14 and 15 and unpublished observations). Thus, one interpretation of our findings is that 12A isoforms may act in a dominant inhibitory manner to sequester factors in the cytosol that are required for the action of nuclear-localized Bin1 isoforms. If so, the level of 12A isoforms needed to block function would depend on the relative level of nuclear isoforms expressed. The ability of ectopic Bin1 to drive programmed cell death in this case would represent a successful competition for such nonproductively sequestered factors. Further investigation is required to validate this interpretation as well as to determine exactly how 12A sequences cause a loss of tumor suppressor activity.
12A splicing was detected in melanoma cell lines derived from several stages of progression, but overexpression of 12A isoforms was seen only in VGP and metastatic cells that have poor prognosis. These observations suggest that loss of function involves two steps, one involving aberrant splicing and a second involving overexpression of the aberrant isoform. Melanocytes are derived from the neural crest during development, so it is conceivable that dedifferentiation events characteristic of neoplastic progression may reintroduce patterns of neuronal splicing, and a second step causing overexpression would be consistent with selection for a dominant inhibitory capability. The tendency to overexpress 12A isoforms in VGP cells is interesting because progression to VGP status is equivalent to malignant conversion. Bin1 is one of an emerging network of proteins that interact with the Myc N terminus (16), and recent unpublished investigations in our laboratory suggest that Bin1 may have a specific role in mediating apoptotic signals from c-Myc (5). c-Myc activation occurs in as many as 50% of cases of sporadic melanoma (22, 23) and has been associated with poor prognosis (24, 25). Thus, inactivation of Bin1 may allow melanoma cells to activate c-Myc without apoptotic penalty. In support of this idea, insulin-like growth factor I (IGF-I) is a potent suppressor of c-Myc-mediated apoptosis (26), and whereas RGP cells require IGF-I for survival, VGP cells do not (27). In future work, it will be important to investigate the link between Bin1 inactivation and melanoma development in vivo and to examine other malignancies for similar splice alterations in Bin1.
Acknowledgments
We thank F. L. Graham for providing the adenovirus-mediated cre-loxP system, J. Wilson for a no-insert adenoviral vector (Ad-vect), and R. Buccafusca and P. Scott Donover for expert technical assistance. We gratefully acknowledge assistance from the Wistar Nucleic Acids, Histology, Hybridoma, and Flow Cytometry Core Facilities. This work was supported by National Institutes of Health Grants CA10851 to The Wistar Institute and P30 DK50306 to the Center for Digestive and Liver Diseases at the University of Pennsylvania and by grants to G.C.P. from the American Cancer Society (CN160). K.G. is the recipient of a fellowship from the Adler Foundation. G.C.P. is a Pew Scholar in the Biomedical Sciences.
ABBREVIATIONS
moi
multiplicity of infection
TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
RT-PCR
reverse transcription–PCR
RGP
radial growth phase melanoma
VGP
vertical growth phase melanoma
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sakamuro D, Elliott K, Wechsler-Reya R, Prendergast G C. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 2.Elliott K, Sakamuro D, Basu A, Du W, Wunner W, Staller P, Gaubatz S, Zhang H, Prochownik E, Eilers M, et al. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- 3.Wechsler-Reya R, Elliott K, Prendergast G C. Mol Cell Biol. 1998;18:566–575. doi: 10.1128/mcb.18.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao N C, Steingrimsson E, DuHadaway J, Ruiz J C, Wasserman W, Copeland N G, Jenkins N A, Prendergast G C. Genomics. 1999;56:51–58. doi: 10.1006/geno.1998.5709. [DOI] [PubMed] [Google Scholar]
- 5.Prendergast G C. Oncogene. 1999;18:2966–2986. [Google Scholar]
- 6.Prendergast G C, Hopewell R, Gorham B, Ziff E B. Genes Dev. 1992;6:2429–2439. doi: 10.1101/gad.6.12a.2429. [DOI] [PubMed] [Google Scholar]
- 7.Wechsler-Reya R, Sakamuro D, Zhang J, Duhadaway J, Prendergast G C. J Biol Chem. 1997;272:31453–31458. doi: 10.1074/jbc.272.50.31453. [DOI] [PubMed] [Google Scholar]
- 8.Anton M, Graham F L. J Virol. 1995;69:4600–4606. doi: 10.1128/jvi.69.8.4600-4606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis A R, Wilson J M. Adenoviral Vectors. New York: Wiley; 1996. pp. 12.4.1–12.4.18. [Google Scholar]
- 10.Prendergast G C, Cole M D. Mol Cell Biol. 1989;9:124–134. doi: 10.1128/mcb.9.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wechsler-Reya R, Elliott K, Herlyn M, Prendergast G C. Cancer Res. 1997;57:3258–3263. [PubMed] [Google Scholar]
- 12.Koprowski H, Steplewski Z, Mitchell K, Herlyn M, Herlyn D, Fuhrer P. Somatic Cell Genet. 1979;5:957–972. doi: 10.1007/BF01542654. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 14.Butler M H, David C, Ochoa G-C, Freyberg Z, Daniell L, Grabs D, Cremona O, De Camilli P. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramjaun A R, Micheva K D, Bouchelet I, McPherson P S. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 16.Sakamuro D, Prendergast G C. Oncogene. 1999;18:2942–2953. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 17.Wigge P, Kohler K, Vallis Y, Doyle C A, Owen D, Hunt S P, McMahon H T. Mol Biol Cell. 1997;8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramjaun A R, McPherson P S. J Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsutsui K, Maeda Y, Tsutsui K, Seki S, Tokunaga A. Biochem Biophys Res Commun. 1997;236:178–183. doi: 10.1006/bbrc.1997.6927. [DOI] [PubMed] [Google Scholar]
- 20.Ekstrand B C, Mansfield T A, Bigner S H, Fearon E R. Oncogene. 1995;11:2393–2402. [PubMed] [Google Scholar]
- 21.Huebner K, Garrison P N, Barnes L D, Croce C M. Annu Rev Genet. 1998;32:7–31. doi: 10.1146/annurev.genet.32.1.7. [DOI] [PubMed] [Google Scholar]
- 22.Chenevix-Trench G, Martin N G, Ellem K A. Oncogene. 1990;5:1187–1193. [PubMed] [Google Scholar]
- 23.Husain Z, FitzGerald G B, Wick M M. J Invest Dermatol. 1990;95:571–575. doi: 10.1111/1523-1747.ep12505549. [DOI] [PubMed] [Google Scholar]
- 24.Grover R, Ross D A, Richman P I, Robinson B, Wilson G D. Eur J Surg Oncol. 1996;22:342–346. doi: 10.1016/s0748-7983(96)90154-7. [DOI] [PubMed] [Google Scholar]
- 25.Ross D A, Wilson G D. Br J Surg. 1998;85:46–51. doi: 10.1046/j.1365-2168.1998.00528.x. [DOI] [PubMed] [Google Scholar]
- 26.Harrington E, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herlyn M, Kath R, Williams N, Valyi-Nagy I, Rodeck U. Adv Cancer Res. 1990;54:213–234. doi: 10.1016/s0065-230x(08)60812-x. [DOI] [PubMed] [Google Scholar]