MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1 (original) (raw)
Abstract
Adhesion molecules expressed by activated endothelial cells play a key role in regulating leukocyte trafficking to sites of inflammation. Resting endothelial cells normally do not express adhesion molecules, but cytokines activate endothelial cells to express adhesion molecules such as vascular cell adhesion molecule 1 (VCAM-1), which mediate leukocyte adherence to endothelial cells. We now show that endothelial cells express microRNA 126 (miR-126), which inhibits VCAM-1 expression. Transfection of endothelial cells with an oligonucleotide that decreases miR-126 permits an increase in TNF-α-stimulated VCAM-1 expression. Conversely, overexpression of the precursor to miR-126 increases miR-126 levels and decreases VCAM-1 expression. Additionally, decreasing endogenous miR-126 levels increases leukocyte adherence to endothelial cells. These data suggest that microRNA can regulate adhesion molecule expression and may provide additional control of vascular inflammation.
Keywords: inflammation, nitric oxide, leukocyte, atherosclerosis
Adhesion molecules play a critical role in leukocyte trafficking. Selectins and their glycoprotein ligands mediate leukocyte rolling along endothelial cells, the first step in leukocyte trafficking (1–4). Subsequently, cytokines and chemokines activate leukocytes and endothelial cells to express intercellular adhesion molecules and their integrin ligands, which in turn mediate leukocyte adhesion to endothelial cells (5, 6). Leukocytes then transmigrate across the endothelial cell barrier and migrate through tissue to the site of injury.
Vascular cell adhesion molecule 1 (VCAM-1) is an intercellular adhesion molecule expressed by endothelial cells (7). VCAM-1 on the endothelial surface mediates leukocyte adhesion by interacting with its α4β1 integrin ligand, very late antigen 4 (VLA-4; CD49d/CD29), which is expressed on most leukocytes and on stimulated neutrophils (8). Although resting endothelial cells do not express VCAM-1, treatment with a variety of cytokines or bacterial products induce endothelial cells to express VCAM-1 within 4–12 h. VCAM-1 expression is thought to be regulated primarily at the level of transcription (9–11). Elegant cellular studies have shown that a set of transcription factors interact with each other and with the 5′ flanking region of the VCAM-1 gene to regulate VCAM-1 expression (9). These transcription factors include NF-κB, IFN regulatory factor-1 (IRF-1), Sp1, and scaffold factors such as high mobility group I(Y) [HMG I(Y)] (10, 11). Although VCAM-1 expression can be regulated at the posttranscriptional level, the mechanisms by which this occurs are not completely defined (12, 13).
We hypothesized that an endogenous microRNA (miRNA) molecule regulates VCAM-1 expression and vascular inflammation. miRNA are small noncoding RNA that regulate target genes posttranscriptionally. These endogenously expressed miRNA molecules are transcribed in the nucleus as long primary transcripts (pri-miRNA) that are further processed by the nuclear enzyme Drosha into shorter precursor species (premiRNA), which can then be exported from the nucleus by the Ran-GTP-dependent nuclear export factor, exportin-5 (14–16). Once in the cytoplasm, the premiRNA is processed by the cytosolic enzyme Dicer into the mature 20- to 24-nt miRNA species, which forms a complex with the RNA-induced silencing complex (RISC) and represses the expression of target transcripts via binding to complementary sequences in the 3′-UTR (17). miRNA regulate expression of genes by two distinct mechanisms: if miRNA precisely match their target sequence, then the mRNA is degraded by Ago2-RISC; however, if the miRNA contains mismatches to its target, then the mRNA translation is blocked by Ago1-RISC (18). Accordingly, we searched for miRNA expressed by endothelial cells that might regulate adhesion molecules.
We found that a particular miRNA, miRNA 126 (miR-126), is selectively expressed in endothelial cells. In silico analysis shows that VCAM-1 is a potential target of miR-126. We now show that endogenous miR-126 suppresses VCAM-1 expression, thereby decreasing leukocyte interactions with endothelial cells. Our data suggest that miRNA can regulate vascular inflammation.
Results
We measured the expression of miRNA in endothelial cells by using a microarray (see Materials and Methods). In brief, total RNA from human umbilical vein endothelial cells (HUVEC) was size fractionated, labeled with a fluorescent dye, and hybridized to a microarray chip (n = 3). Of the 500 miRNA probes on the microarray chip, the 26 miRNA with the highest level of expression in HUVEC were identified (Table 1). The most frequently expressed miRNA is miR-126.
Table 1.
Expression of microRNA in human umbilical vein endothelial cells
| MicroRNA name | Relative expression | SD |
|---|---|---|
| miR-126 | 1.3 | 0.1 |
| miR-23a | 0.8 | 0.1 |
| miR-23b | 0.8 | 0.0 |
| miR-22 | 0.6 | 0.1 |
| miR-221 | 0.6 | 0.2 |
| miR-16 | 0.5 | 0.1 |
| miR-103 | 0.5 | 0.1 |
| let-7d | 0.4 | 0.1 |
| miR-21 | 0.4 | 0.2 |
| miR-221 | 0.4 | 0.1 |
| miR-107 | 0.4 | 0.1 |
| miR-100 | 0.4 | 0.0 |
| let-7a | 0.4 | 0.1 |
| miR-27a | 0.4 | 0.0 |
| miR-26a | 0.3 | 0.0 |
| miR-24 | 0.3 | 0.0 |
| miR-20a | 0.3 | 0.0 |
| miR-17-5 | 0.3 | 0.1 |
| miR-181 | 0.2 | 0.1 |
| miR-106a | 0.2 | 0.1 |
| miR-29a | 0.2 | 0.1 |
| miR-130a | 0.2 | 0.0 |
To explore the biological role of miR-126, we first defined its expression in endothelial cells. We harvested RNA from a variety of mouse organs and analyzed it for miR-126 expression by Northern analysis. miR-126 is highly expressed in murine lung and heart tissues and also expressed at lower levels in the brain, liver, and kidney (Fig. 1A). Studying miR-126 expression in primary human endothelial cells and other cell lines, we found that endothelial cells from veins, arteries, skin, and brain all express miR-126 (Fig. 1 B and C). However, other cell lines such as vascular smooth muscle cells or leukocyte cell lines do not.
Fig. 1.
Endothelial cells express miR-126. (A) (Upper) Total RNA was harvested from various mouse tissues and analyzed by Northern blotting for miR-126 RNA. (Lower) Ethidium bromide staining of total RNA. (B) (Upper) Total RNA was harvested from various cell types and analyzed by Northern blotting for miR-126 RNA. (Lower) Ethidium bromide staining of total RNA. (C) Total RNA was harvested from various cell types and analyzed by quantitative RT-PCR for miR-126 RNA. (D) HUVEC were transfected with AS oligonucleotides or premiR-126 oligonucleotides and then cultured for 3 days. TNF-α at 1 ng/ml was added for 16 h. Total RNA was harvested and analyzed by Northern blotting for miR-126 RNA levels (Upper) and by ethidium bromide staining (Lower).
We next attempted to alter levels of endogenous miR-126 in endothelial cells. To decrease endothelial miR-126, we transfected HUVEC with an antisense miR-126 oligonucleotide (AS-miR-126) and measured miR-126 levels by Northern blotting. AS-miR-126 decreases endogenous miR-126 RNA (Fig. 1D Left). To increase endothelial miR-126, we transfected HUVEC with an RNA precursor to miR-126 (premiR-126). premiR-126 dramatically increases miR-126 RNA (Fig. 1D Right).
Computer analysis suggested that miR-126 may be a negative regulator of VCAM-1 expression (see Materials and Methods). To identify potential mRNA transcripts that are regulated by miR-126, we searched through a computer database (Human miRNA Targets, Computational Biology Center, Memorial Sloan–Kettering Cancer Center). In silico analysis revealed that VCAM-1 is a potential target of miR-126. miR-126 has sequence similarity to a region within the 3′-UTR of the transcript for human VCAM-1, extending between 604 and 625 (Fig. 2A). In particular, the first 6 nt of miR-126 can form base pairs that are complementary to an identical sense sequence of the VCAM-1 mRNA transcript, extending from 619 to 625 within the VCAM-1 3′-UTR. Furthermore, another 10 nt of miR-126 can base-pair within 604 and 617 of the VCAM-1 transcript.
Fig. 2.
miR-126 suppresses gene expression by targeting a miR-126-binding site. (A) miR-126 is partially complementary to a region in the VCAM-1 3′-UTR. (Upper) Schematic of VCAM-1 mRNA and miR-126. (Lower) The sequence of miR-126 and its potential matching site in the VCAM-1 3′-UTR. (B) The complementary miR-126-binding site was inserted downstream of a luciferase reporter on the pMIR-Report plasmid and transfected into HEK293 cells with varying concentrations of premiR-126 (n = 3 ± SD; *, P < 0.01 for 0 vs. 20 nM premiR-126). (C) The partially complementary miR-126-binding site found in the VCAM-1 3′-UTR (or a mutated binding site) was inserted downstream of a luciferase reporter on the pMIR-Report plasmid and transfected into HEK293 cells with varying concentrations of premiR-126 (n = 3 ± SD; *, P < 0.001 for 0 vs. 30 nM premiR-126).
To explore the effect of miR-126 on expression of mRNA, we first constructed a reporter vector containing a consensus miR-126-binding site: we synthesized a plasmid encoding the cDNA for luciferase, which included the antisense sequence for miR-126 within the 3′-UTR, transfected this reporter plasmid into HEK293 cells with or without premiR-126, and measured luciferase activity in the cell lysates. HEK293 cells transfected with this construct express luciferase (Fig. 2B). Transfection of premiR-126 into HEK293 cells decreases luciferase expression (Fig. 2B). These data suggest that miR-126 can regulate expression of transcripts containing an exact miR-126-binding site.
To explore the ability of miR-126 to regulate VCAM-1 mRNA translation, we created additional reporter vectors. These plasmids contained either the endogenous 21-nt miR-126-binding site found in the VCAM-1 3′-UTR or a corresponding 21-nt sequence with a scrambled seed sequence. [The miR-126-binding site in the VCAM-1 3′-UTR is only partially complementary to miR-126 and is not an exact match (Fig. 2A).] HEK293 cells transfected with these constructs express luciferase (Fig. 2C). Transfection of premiR-126 into HEK293 cells decreases the expression of the wild-type but not the mutant construct (Fig. 2C). Taken together, these data suggest that miR-126 regulates VCAM-1 expression through a miR-126-binding site in the VCAM-1 3′-UTR.
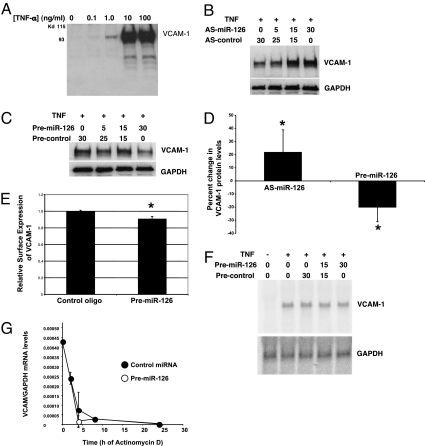
We next sought to determine the effect of miR-126 on VCAM-1 expression. To knockdown miR-126 levels, we transfected HUVEC with AS-miR-126 oligonucleotides for 3 days, then stimulated the transfected cells with TNF-α for 16 h, and then measured VCAM-1 expression by immunoblotting. TNF-α induces VCAM-1 expression as expected (Fig. 3A). Transfection of AS-miR-126 oligonucleotide leads to a dose-dependent increase in VCAM-1 expression without affecting other adhesion molecules such as intercellular adhesion molecule 1 (ICAM-1) [Fig. 3B and supporting information (SI) Fig. 5]. Conversely, overexpression of the precursor to miR-126 leads to a decrease in VCAM-1 expression (Fig. 3C). Quantification of immunoblots showed that AS-miR-126 increases VCAM-1 expression by ≈20%, and premiR-126 decreases it by ≈20% (Fig. 3D). Furthermore, premiR-126 decreases cell surface VCAM-1 expression by ≈10% (Fig. 3E).
Fig. 3.
miR-126 suppresses VCAM-1 expression. (A) TNF-α induction of VCAM-1 expression in HUVEC. HUVEC were stimulated for 16 h with increasing doses of TNF-α, and cell lysates were immunoblotted with antibody to VCAM-1. (B) Endogenous miR-126 decreases VCAM-1 protein levels, knocking down miR-126. HUVEC were transfected with a control oligonucleotide (AS-control) or with an oligonucleotide antisense to miR-126 (AS-miR-126) to knock down endogenous miR-126 and then cultured for 3 days. Total protein was harvested and analyzed by immunoblotting for VCAM-1. (C) Endogenous miR-126 decreases VCAM-1 protein levels, increasing miR-126. HUVEC were transfected with a premiR-126 oligonucleotide to increase miR-126 and then cultured for 3 days. TNF-α at 1 ng/ml was added for 16 h. Total protein was harvested and analyzed by immunoblotting for VCAM-1. (D) HUVEC were transfected with 30 nM AS-miR-126 or premiR-126 or a control oligonucleotide and stimulated with TNF-α, and immunoblots were performed as above. The intensity of the signal was quantified and compared with the control transfected cells (n = 6 ± SD; *, P < 0.03 compared with control). (E) HUVEC were treated with a control oligonucleotide or premiR-126 and stimulated with TNF-α as above, and the expression of VCAM-1 was measured by FACS (n = 4 ± SD; *, P < 0.01). (F) miR-126 does not affect steady-state VCAM-1 mRNA levels. HUVEC were transfected with premiR-126 and treated with TNF-α for 3 h, and total RNA was analyzed by Northern blotting for VCAM-1. (G) miR-126 does not affect the stability of VCAM-1 mRNA. HUVEC were transfected with premiR-126, treated with TNF-α for 3 h, and then exposed to actinomycin D. RNA was isolated at various times after actinomycin D and analyzed by quantitative RT-PCR for VCAM-1 and GAPDH mRNA levels.
We predicted that miR-126 regulates VCAM-1 expression at the level of translation, because VCAM-1 does not contain an exact match to miR-126 in its 3′-UTR. To confirm this prediction, we transfected HUVEC with premiR-126 for 2 days, then stimulated cells with TNF-α for 3 h, and measured mRNA expression by Northern blot. The precursor miR-126 does not affect VCAM-1 mRNA expression (Fig. 3F). Furthermore, experiments with actinomycin D revealed that miR-126 does not affect the half-life of VCAM-1 mRNA (Fig. 3G and SI Fig. 6). Taken together, these data show that endogenous miR-126 inhibits VCAM-1 protein expression but not mRNA levels in endothelial cells.
To explore the functional relevance of miR-126, we measured the influence of miR-126 on leukocyte adhesion to endothelial cells. We transfected HUVEC with premiR-126, treated the cells with TNF-α, added HRP- or 2′,7′-bis-carboxyethyl-5(6)carboxyfluorescein, acetoxymethyl ester (BCECF-AM)-labeled HL-60 leukocytes, washed the mixture, and measured leukocyte binding to the endothelial cells. TNF-α increases leukocyte adhesion to endothelial cells (Fig. 4). Overexpression of premiR-126 decreased leukocyte binding (Fig. 4 A–C). Thus, exogenous expression of miR-126 decreases leukocyte adherence.
Fig. 4.
miR-126 suppresses leukocyte adhesion ex vivo. (A) HUVEC were transfected with premiR-126 or a control oligonucleotide for 2 days and then treated with TNF-α or control for 6 h. BCECF-AM-labeled leukocytes were added to the HUVEC, incubated at 37°C for 45 min, and then washed. Adherent cells were photographed with a fluorescent camera. (B) HUVEC were transfected with premiR-126 or a control oligonucleotide for 2 days and then treated with TNF-α or control for 6 h. HRP-labeled leukocytes were added to HUVEC, incubated at 37°C for 45 min, and then washed. Adherent cells were lysed, and HRP was assayed by a spectrophotometer at _A_450, and then calibrated to a standard curve (n = 2 ± SD; P = 0.1 for 0 vs. 30 nM premiR-126). (C) HUVEC were transfected with premiR-126 or a control oligonucleotide for 2 days and then treated with TNF-α or control for 6 h. Cell lysates were immunoblotted with antibody to VCAM-1. (D) HUVEC were transfected with AS-miR-126 or an AS-control oligonucleotide for 2 days and then treated with TNF-α or control for 6 h. HRP-labeled leukocytes were added to the HUVEC, incubated at 37°C for 45 min, and then washed. Adherent cells were lysed, and HRP was assayed by a spectrophotometer at _A_450 and calibrated to a standard curve (n = 3 ± SD; *, P = 0.02 for 0 vs. 30 nM AS-miR-126). (E) HUVEC were transfected with AS-miR-126 or a control oligonucleotide for 2 days and then treated with TNF-α or control for 6 h. Cell lysates were immunoblotted with antibody to VCAM-1. (F) HUVEC were treated with TNF-α or control for 6 h. HUVEC were incubated with antibody to VCAM-1, ICAM-1, or control IgG for 30 min at 37°C. HRP-labeled leukocytes were then added to the HUVEC, incubated at 37°C for 45 min, and washed. Adherent cells were lysed, and HRP was assayed by a spectrophotometer at _A_450 and calibrated to a standard curve (n = 3 ± SD; *, P < 0.02).
We next tested whether endogenous miR-126 regulates leukocyte adherence to endothelial cells. We transfected HUVEC with antisense oligonucleotide to miR-126, and then measured leukocyte adherence to TNF-α-stimulated cells as above. Increasing antisense to miR-126 increases leukocyte adherence (Fig. 4 A, D, and E). We confirmed that leukocyte binding in this assay depends on VCAM-1 by using an antibody that blocks VCAM-1 (Fig. 4F). Collectively, these data suggest that endogenous miR-126 inhibits leukocyte adherence through the regulation of VCAM-1.
Discussion
Summary.
The major finding of our study is that miR-126 inhibits VCAM-1 expression and limits leukocyte adherence to endothelial cells. Our data suggest that miRNA can regulate expression of adhesion molecules and regulate vascular inflammation.
miRNA Regulation of VCAM-1.
We have discovered a pathway that controls adhesion molecule expression. Resting endothelial cells do not transcribe mRNA of VCAM-1, but proinflammatory cytokines such as TNF-α activate the transcription factors NF-κB and IRF-1, which induce VCAM-1 transcription (9). VCAM-1 expression can also be modulated by unknown factors that control its mRNA stability (12, 13). Our studies define a mechanism for the regulation of VCAM-1 expression: miRNA can inhibit expression of VCAM-1. Because resting endothelial cells express miR-126, perhaps miR-126 normally functions to suppress inflammation in resting cells.
We show that a miR-126-binding site mediates miR-126 suppression of VCAM-1 expression. Exogenous miR-126 suppresses expression of luciferase when a miR-126-binding site is fused to the luciferase 3′-UTR (Fig. 2B). Furthermore, the magnitude of suppression is relatively large, because it results from an exact match between miR-126 and miR-126-binding site sequences. Exogenous miR-126 also suppresses expression of luciferase when its sequence is fused to the miR-126-binding site from VCAM-1 3′-UTR. However, because this endogenous miR-126-binding site is only partially complementary to miR-126, the magnitude of suppression is relatively less (Fig. 2C). This difference in magnitude of miR-126 suppression is explained by studies that demonstrate that miRNA regulate gene expression through two distinct pathways (18). Precise matches between miRNA and their targets form a complex with RISC-Ago2, which degrades mRNA. Imprecise matches lead to a complex of RISC-Ago1, which suppresses mRNA translation.
Regulation of miR-126 Expression.
The regulation of miR-126 expression is unknown. In particular, the 5′ flanking region that controls miR-126 expression has not been characterized. The 5′ flanking region of miR-126 may contain response elements that limit its expression to a subset of cells, including endothelial cells. Although our data suggest that miR-126 regulates an inflammatory pathway, our studies show that TNF has no effect on miR-126 expression (data not shown), implying that the 5′ flanking region of miR-126 does not contain response elements for inflammatory transcription factors.
Endothelial Cell miRNA.
Recent studies have identified specific miRNA expressed in endothelial cells. Sessa et al. (19) used microarray to identify miRNA expression in HUVEC. Of the 25 miRNA that we found most highly expressed in endothelial cells, 10 were also identified by Sessa et al. (19). The miRNA identified by both us and Sessa et al. (19) include miR-222, miR-221, miR-23a, miR-181a, miR-107, miR-31, miR-103-1,2, miR-320, miR-106, and miR-22. Another study identified 12 miRNA highly expressed in HUVEC that we also found, including miR-126 (20). Thus, these three studies establish a set of miRNA that are expressed in endothelial cells.
Many of these endothelial miRNA are individually expressed in other cell types. However, it is possible that this set of miRNA could be a combination unique to endothelial cells. A recent miRNA atlas presented miRNA expression profiles for multiple cells and tissues (21). In this study, most miRNA were expressed in a variety of tissues. But one-third of all miRNA were expressed with a greater degree of specificity in particular tissues. miR-126 was highly specific for cardiovascular tissue. Our data confirm these studies: we found that miR-126 was highly expressed in the heart and lungs (Fig. 1). In addition, the miRNA atlas showed that miR-126 was expressed at lower levels elsewhere, including hematopoietic, endocrine, and reproductive tissues (21). This prior report of miR-126 in various tissues might simply reflect the vascularity of these organs. We found that miR-126 is expressed in endothelial cells, but not in vascular smooth muscle cells nor in lymphocytic cells (Fig. 1).
The role of miRNA in endothelial biology is largely unknown, and many individual targets of miRNA within endothelial cells have not yet been identified. Sessa et al. (19) blocked global miRNA by knocking down Dicer and found that miRNA are necessary for endothelial cell proliferation and cord formation. Furthermore, miR-221/222 regulates endothelial NOS expression and c-Kit expression. Our data extend these studies by identifying a miRNA that regulates VCAM-1 expression. We also show that miRNA can target a different aspect of endothelial biology: miRNA can regulate vascular inflammatory pathways. Future studies may identify sets of targets and pathways regulated within endothelial cells by endothelial miRNA.
Materials and Methods
Reagents.
HUVEC and endothelial cell basal media (EBM-2) and growth factors were purchased from Lonza. RNA oligonucleotides were purchased from Integrated DNA Technologies or Ambion. The sequence of the antisense miR-126 (referred to as miR-126AS) from Integrated DNA Technologies is as follows: GCAUUAUUACUCACGGUACGA. All bases were modified with a 2′-OMe. HUVEC were transfected with siPort NeoFX reagent or Amine reagent (Ambion). VCAM-1 monoclonal antibody, ICAM-1 monoclonal antibody, and control IgG were purchased from Santa Cruz Biotechnology. Luciferase Reporter plasmid was purchased from Ambion. Microarrays were purchased from Combimatrix. Quantitative RT-PCR reagents were purchased from Applied Biosystems.
Endothelial miRNA Analysis.
Total RNA was harvested from HUVEC by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Small RNA species, ≈20–30 nt, were fractionated by size-exclusion column chromatography by using Microcon YM-100 (Amicon). Small RNA species were fluorescently labeled with Cy3 by using a Label IT miRNA labeling kit by Mirus and hybridized to a human miRNA 4 × 2K microarray by Combimatrix according to the manufacturer's instructions. Expression data were normalized to the level of the highest expressed miRNA (n = 3). Data were visualized by using the Axon GenePixTM 4200AL scanner and imaging software. We used a computer to search for potential mRNA targets of miR-126. (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl, Computational Biology Center, Memorial Sloan–Kettering Cancer Center).
Northern Blot Analysis.
Total RNA harvested as above were ran on a 15% TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3)–urea gel (Invitrogen) and transferred to a Nytran nylon transfer membrane (Schleicher and Schuell). A 32P-probe was synthesized from miRNA-126 antisense oligonucleotides (Integrated DNA Technologies) and hybridized by using UltraHyb reagents (Ambion) according to the manufacturer's instructions.
miRNA Real-Time PCR.
Total RNA was harvested from various mouse tissue by using RNA Later (Ambion). RNA was isolated from tissue by using the TRIzol reagent (Invitrogen). miR-126 expression was analyzed by TaqMan miRNA assays according to the manufacturer's protocol (Applied Biosystems).
Cell Culture and miRNA Transfection.
HUVEC were obtained from Cambrex and grown in EBM-2 media supplemented with essential growth factors. By using 1–3 μl siPort NeoFx (Ambion) reagent, 0–40 nM of the precursor miR-126 oligonucleotide (Ambion) or a 2′-OMe-modified antisense miR-126 oligonucleotide (Integrated DNA Technologies) were transfected into HUVEC for 3 days.
Western Blotting.
Western blotting was performed as described previously (22). In brief, HUVEC were lysed with Laemmli sample buffer (Bio-Rad), sonicated, boiled, fractionated on a 7.5% Tris·HCl gel (Bio-Rad), and transferred to a nitrocellulose membrane, which was hybridized with an antibody to VCAM-1, ICAM-1, or GAPDH (Santa Cruz Biotechnology).
Generation of Luciferase Reporter Construct.
A miR-126 consensus response element, GCATTATTACTCACGGTACGA, VCAM-1 wild-type 3′-UTR response element, GTATAGTACTGGCATGGTACGG, and a VCAM-1 mutant 3′-UTR response element, GTATAGTACTGGCATGGCGATG, were inserted into the multiple cloning site of the miRNA expression vector pMIR-REPORT downstream of the cDNA for luciferase (Ambion). Each vector, along with varying doses of precursor miR-126 oligonucleotide (Ambion), was transfected into HEK293 cells by using the Amine reagent (Ambion) or Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cells were cultured for 2 days and assayed by using the Dual-Luciferase Reporter Assay System (Promega).
VCAM-1 Real-Time PCR.
HUVEC were cultured and transfected as above for 2 days and stimulated with 1 ng/ml TNF-α for 3 h. HUVEC were then treated with 5 μg/ml actinomycin D (Sigma) for 0, 2, 4, 8, and 24 h. RNA was isolated from cells by using the TRIzol reagent (Invitrogen). VCAM-1 and GAPDH mRNA expression were analyzed by the TaqMan gene expression assay according to the manufacturer's instructions (Applied Biosystems).
HL-60 Cell Binding to HUVEC.
The human promyelocytic cell line HL-60 was obtained from American Type Culture Collection. HUVEC were transfected for 2 days with varying doses of premiR-126 or antisense miR-126 oligonucleotides. Transfected HUVEC were treated with TNF-α for 6–8 h. HL-60 cells were labeled with HRP and incubated with transfected HUVEC for 45 min. The culture wells were washed several times with serum-free media and lysed with 1% Triton solution. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate was added, and the concentration of HRP was measured at _A_450. To quantitate the precise number of adherent HL-60 cells, a standard curve was constructed by using known amounts of HRP-labeled HL-60 cells.
Statistical Analyses.
Data are expressed as the mean ± SD. Statistical comparisons were made between two groups with the t test and between multiple groups by ANOVA. A value of P < 0.05 was considered significant.
Supplementary Material
Supporting Figures
ACKNOWLEDGMENTS.
We thank Dr. Craig Fletcher (The Johns Hopkins University School of Medicine) for assistance with the leukocyte adherence assay. This work was supported by National Institutes of Health Grants R01 HL63706-04, R01 HL074061, P01 HL65608, and P01 HL56091; American Heart Association Grant EIG 0140210N; The Ciccarone Center; The John and Cora H. Davis Foundation; The Clarence P. Doodeman Professorship (C.J.L.); National Institutes of Health Grant R01 CA120185 and the Rita Allen Foundation (J.T.M.); and by a training grant from the Cellular and Molecular Medicine Program (to T.A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bevilacqua MP. Annu Rev Immunol. 1993;11:767–804. doi: 10.1146/annurev.iy.11.040193.004003. [DOI] [PubMed] [Google Scholar]
- 2.Choi J, Enis DR, Koh KP, Shiao SL, Pober JS. Annu Rev Immunol. 2004;22:683–709. doi: 10.1146/annurev.immunol.22.012703.104639. [DOI] [PubMed] [Google Scholar]
- 3.Smalley DM, Ley K. J Cell Mol Med. 2005;9:255–266. doi: 10.1111/j.1582-4934.2005.tb00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo BH, Carman CV, Springer TA. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber C. J Mol Med. 2003;81:4–19. doi: 10.1007/s00109-002-0391-x. [DOI] [PubMed] [Google Scholar]
- 6.Salmi M, Jalkanen S. Nat Rev Immunol. 2005;5:760–771. doi: 10.1038/nri1705. [DOI] [PubMed] [Google Scholar]
- 7.Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, Lobb R. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- 8.Alon R, Kassner PD, Carr MW, Finger EB, Hemler ME, Springer TA. J Cell Biol. 1995;128:1243–1253. doi: 10.1083/jcb.128.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]
- 10.Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essani NA, Bajt ML, Farhood A, Vonderfecht SL, Jaeschke H. J Immunol. 1997;158:5941–5948. [PubMed] [Google Scholar]
- 12.Croft D, McIntyre P, Wibulswas A, Kramer I. Am J Pathol. 1999;154:1149–1158. doi: 10.1016/s0002-9440(10)65367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietersma A, Tilly BC, Gaestel M, de Jong N, Lee JC, Koster JF, Sluiter W. Biochem Biophys Res Commun. 1997;230:44–48. doi: 10.1006/bbrc.1996.5886. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 15.Yi R, Qin Y, Macara IG, Cullen BR. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohnsack MT, Czaplinski K, Gorlich D. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 20.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Tuschl T. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, et al. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures