Theoretical Design of a Gene Therapy To Prevent AIDS but Not Human Immunodeficiency Virus Type 1 Infection (original) (raw)
Abstract
Recent reports confirm that, due to the presence of long-lived, latently infected cell populations, eradication of human immunodeficiency virus type 1 (HIV-1) from infected patients by using antiretroviral drugs will be exceedingly difficult. An alternative to virus eradication may be to use gene therapy to induce a pseudo-latent state in virus-producing cells, thus transforming HIV-1 into a lifelong, but manageable, virus. Conditionally replicating HIV-1 (crHIV-1) gene therapy vectors provide an avenue for subduing HIV-1 expression in infected cells (by creating a parasite, crHIV-1, of the parasite HIV-1), potentially reducing the HIV-1 set point and delaying AIDS onset. Development of crHIV-1 vectors has proceeded in vitro, but the requirements for a crHIV-1 vector to proliferate and persist in vivo have not been explored. We expand a widely accepted mathematical model of HIV-1 in vivo dynamics to include a crHIV-1 gene therapy virus and derive a simple criterion for designing crHIV-1 viruses that will persist in vivo. The model introduces only two new parameters—HIV-1 inhibition and crHIV-1 production—and both can be experimentally engineered and controlled. Analysis demonstrates that crHIV-1 gene therapy can indefinitely reduce HIV-1 set point to levels comparable to those achieved with highly active antiretroviral therapy, provided crHIV-1 production is more efficient than HIV-1. Paradoxically, highly efficient therapeutic inhibition of HIV-1 was found to be disadvantageous. Thus, the field may benefit by shifting the search for more potent antiviral genes toward engineering optimized therapy viruses that package ultraefficiently while downregulating viral production moderately.
Although huge strides have been made in the treatment of human immunodeficiency virus type 1 (HIV-1) infection and AIDS during the past 20 years, an HIV-1 vaccine still appears many years away (20). Currently, the only effective HIV-1 drug therapy, highly active antiretroviral therapy (HAART), is highly toxic to patients, expensive, and quickly overcome by resistant mutants (for reviews, see references 1 and 25). Furthermore, due to the presence of long-lived, latently infected cell populations, HAART appears to be incapable of eradicating HIV-1 from patients (12, 50).
HIV-1 infection is characterized by a rapid increase in viremia, peaking several weeks after infection, followed by a drop in viremia to a quasi-steady-state level termed the HIV-1 set point. This set point is maintained for an average of 10 years, during which time infected individuals are largely asymptomatic (for a review, see reference 25). After ∼10 years the HIV-1 viral load begins to increase, and the CD4+-T-cell count begins to drop, eventually leading to the onset of AIDS. The HIV-1 set point level is inversely correlated with the time to AIDS onset and is the best clinical measure of disease progression (34). It has been established that HAART subdues HIV-1 replication, reduces the HIV-1 set point, and prolongs the time to AIDS onset (for reviews, see references 19 and 25). Importantly, recent evidence (11) suggests that reducing the HIV-1 set point below some absolute value may not be necessary; rather, the relative extent of HIV-1 set point reduction in a patient appears to be the significant predictor of patient health and immune recovery.
The development of HAART regimens has been significantly aided by mathematical modeling of HIV-1 dynamics in vivo. The basic three-equation model of HIV-1 in vivo dynamics helped quantify key parameters in the HIV-1 life cycle (39, 44) and has gained widespread acceptance due to its relative simplicity and excellent fidelity in fitting patient drug perturbation data. Nearly all of the parameters in this “Basic Model” can be measured experimentally or derived through analysis, allowing it to predict numerous features of HIV-1 viremia with impressive accuracy. For example, modeling, coupled with accurate experimental measurement, quantified the rapid production of virus in vivo (∼1010 virions produced per day per individual) (21, 51), the viral eclipse phase (∼1.5 days between initial infection of the cell and virus production) (35), and the short half-life of infected CD4+ T cells that are actively producing virus (∼1 day) (44).
Mathematical analysis of experimental data also suggested that, in addition to short-lived cells actively producing virus, HIV-1-infected individuals harbor long-lived latent reservoirs of infection. These reservoirs, the most problematic of which are the memory CD4+ T lymphocytes with integrated HIV-1 provirus, are not affected by HAART, and recent estimates predict that it may take many decades of continuous therapy in order to purge these reservoirs of infected cells (12, 50; for a review, see reference 6). However, such long-term HAART is highly problematic due to drug toxicity in patients and HIV-1's ability to develop viral escape mutants that render most triple-drug regimens ineffective in ∼1.5 years (41).
An alternative to purging latent reservoirs and eradicating virus from infected individuals could be to convert virus-producing cells into a pseudolatent state in which HIV-1 production from these cells is highly diminished. The HIV-1 set point could thereby be reduced, and the time to AIDS onset could be delayed or even forestalled. Gene therapy approaches, including the conditionally replicating HIV-1 (crHIV-1) gene therapy vectors first proposed by Dropulic et al. in 1996 (14) in particular, have the potential to “prod” or convert the population of cells actively producing virus into a “pseudolatent” state in which HIV-1 production is greatly diminished.
crHIV-1 vectors contain only the cis and none of the trans elements necessary for viral packaging and instead carry an antiviral gene that inhibits any of numerous wild-type HIV-1 functions (31). If a cell carrying an integrated copy of the crHIV-1 becomes infected with wild-type virus, the antiviral vector payload acts to limit the production of HIV-1. Also, a fraction of virions produced from these dually infected cells carry crHIV-1 genomes, leading to the further propagation of the therapy vector. crHIV-1 vectors having an HIV-1-induced promoter have the benefit of being expressed only in HIV-1-infected cells, thereby reducing vector toxicity concerns. Importantly, crHIV-1 vectors have the potential to encode for inhibitors of cellular factors necessary for HIV-1 replication. These inhibitors should be significantly less susceptible to mutational escape by HIV-1, since the virus would need to evolve away from its dependency on this cellular factor.
Although the successful development of crHIV-1 vectors has proceeded in vitro (13), the requirements for a crHIV-1 vector to persist and to reduce the HIV-1 set point in vivo have not yet been explored. Fortunately, as mentioned above, a widely accepted, experimentally justified differential equation model of HIV-1 in vivo dynamics exists (44). Here we expand the Basic Model to incorporate a crHIV-1 gene therapy virus and therapeutically infected cell populations. Extensive numerical simulations of the Expanded Model were performed to explore any sensitivity of the HIV-1 set point to specific parameters. We analyze the steady states with respect to two important parameters and derive a simple equation for designing crHIV-1 viruses that can persist in vivo. Our results show that HIV-1 set point can decrease to levels comparable with HAART when the intracellular crHIV-1 mRNA concentration is ∼100-fold greater than the full-length, genomic HIV-1 mRNA concentration. Paradoxically, highly efficient viral downregulation caused a rebound in HIV-1 steady state to pretreatment levels. Furthermore, our model suggests that it is important for the burst size of cells producing therapeutic virus to exceed the burst size of HIV-1-infected cells. We rely on reports that HIV-1-infected cells in vivo have the potential to produce much more virus than typically seen (23), implying that packaging resources within the infected cell (such as the HIV-1 Gag-Pol polyprotein that is cleaved into the HIV-1 capsid) are not limiting. (“Competition” for these increased packaging resources within infected cells is not ruled out and is the focus of ongoing research). The study presented here highlights the need to determine both the maximum possible viral output from HIV-1-infected cells and the potential of Gag-Pol to act as a limiting intracellular resource in vivo.
THEORY
Expansion of the Basic Model.
The Basic Model (reviewed in reference 40) is a set of three nonlinear ordinary differential equations that describes population densities of uninfected CD4+ T cells (T), productively infected T cells (I), and free virus (V) circulating in the body. To explore the dynamics of a crHIV-1 gene therapy virus in vivo, the Basic Model was expanded to incorporate populations of crHIV-1 therapy virus (V T), therapeutically infected cells (I T), and cells that were dually infected with both HIV-1 and crHIV-1 therapy virus (I D). However, to maintain the validity of the Basic Model, only two new parameters were introduced: the efficiency of crHIV-1 mediated viral inhibition (D) and the intracellular concentration of crHIV-1 mRNA relative to HIV-1 mRNA (P). Both parameters can be experimentally engineered or controlled, but a significant feature of the model is that D is an extracellularly measured parameter related to viral production, whereas P is an intracellular parameter related to mRNA concentration.
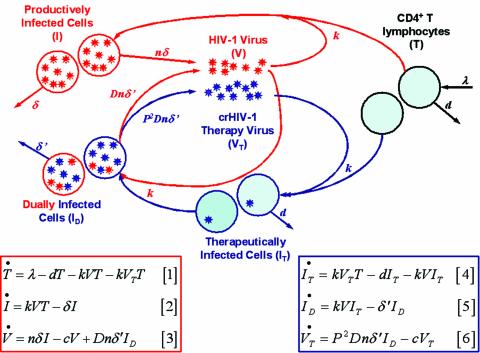
A schematic of the expanded gene therapy model, along with the equations, is presented in Fig. 1. Briefly, crHIV-1 gene therapy virus (V T), indistinguishable from HIV-1 virus (V) except by its RNA payload, infects target cells (T) at the same rate as HIV-1. HIV-1 infection converts T cells to productively infected cells (I), which produce HIV-1 virus, whereas crHIV-1 infection converts T cells to I T cells that do not produce any virus. HIV-1 can infect I T cells, which carry an integrated copy of crHIV-1 and are functionally equivalent to T cells, and the resulting dually infected (I D) cells produce both HIV-1 and crHIV-1. Within I D cells, crHIV-1 can inhibit HIV-1 production via any of the antiviral payload genes described in the literature (e.g., ribozymes, antisense, RNAi, intrabodies, or transdominant-negative mutants [see reference 31 for a review]) and can compete for packaging with HIV-1 mRNA transcripts via increased cytoplasmic mRNA abundance or half-life (32).
FIG. 1.
Schematic and equations for Expanded Model of HIV-1 in vivo dynamics. The schematic gives an overview of the processes described by model equations 1 to 6, with red and blue populations, arrows, and parameters describing HIV-1-mediated processes and crHIV-1-mediated processes, respectively. The T population has black arrows since its processes are independent of virus. The light blue background of the T and I T cells represents the assumed functional similarity between these cells; I T should be equivalent to “healthy” T cells. For simplicity, viral clearance has been omitted from the schematic. All parameters are described in Table 1. The red and blue boxes encasing the equations roughly correspond to red and blue color coding of the schematic. The red box encloses the (original) Basic Model, equations 1 to 3, with two added terms (see text), while the blue box encloses the three new equations, equations 4 to 6, added to describe crHIV-1 and associated populations.
The Basic Model makes the simplifying assumption that once a cell is infected by HIV-1, superinfection is unlikely due to CD4 downregulation by HIV-1 Nef (28) (the Nef protein, expressed shortly after HIV-1 infection, is known to cause internalization of the CD4 receptor from the cell surface [for a review, see reference 5]). Similarly, our expanded gene therapy model neglects superinfection of I D cells by HIV-1. However, a central point of our model is that HIV-1 can infect I T cells, since their integrated copy of crHIV-1 does not encode for HIV-1 Nef. Another outcome of the lack of CD4 downregulation in I T cells is that crHIV-1 can also superinfect these cells. We explored this possibility of crHIV-1 superinfection of I T cells and found that it modestly improves therapy (additional supporting information [SI] is available online [http://genomics.lbl.gov/suppinfo/JVI/index.html]). Thus, the model and results presented here (no superinfection) appear to be a lower limit of the efficacy of this therapy.
The three equations in the red box in Fig. 1 extend the Basic Model by the addition of two terms: a term for infection by therapy virus, −kV T T, added to the equation 1 and a term for HIV-1 production by dually infected cells, Dnδ′ID, added to the equation 3 (Fig. 1). The equations in the blue box in Fig. 1 represent the expansion to the Basic Model. I T cells are assumed to be infected at the same “per-contact” rate as I cells, since the crHIV-1 virus is phenotypically identical to wild-type HIV-1 and differs only in its genotypic, internal RNA payload. I T cells are also assumed to die at the same rate as T cells since the crHIV-1 is not produced in the absence of HIV-1 infection (an assumption that can be relaxed for some specific therapeutic payloads). The new parameter P is the fold increase in crHIV-1 “production” relative to HIV-1 production from I D cells (but mechanistically is the increase in cytoplasmic vector mRNA concentration) and is written as _P_2 in equation 6, since two mRNA transcripts are incorporated into each diploid lentivirus (see below). The new parameter D is the fraction by which the HIV-1 titer produced from I D cells is therapeutically downregulated (e.g., transcriptional and/or translational inhibition) by a crHIV-1-encoded antiviral gene. All other parameters, with the exception of δ′, are carried from the original model and have previously been measured or estimated (40, 43). δ′ is the death rate of I D cells, and we assumed that δ′ = D × δ since I D cells will likely die at an altered rate compared to I cells (which have a death rate δ) due to the decreased expression of cytotoxic HIV-1 proteins. We also relaxed this assumption and used in vivo data (16) to derive a dimensionless function, F(δ,n), that describes the death rate of I D cells based on the number of HIV-1 virions produced (see SI). Fortunately, all assumptions related to δ′ were “benign” in the sense that δ′ algebraically cancels out of the steady-state solutions for all equations except the I D equation (see SI). Parameter values used in the model are presented in Table 1.
TABLE 1.
Explanation of parameters and values used in the crHIV-1 gene therapy model
| Parametera | Biological interpretation | Value (units)b | Reference(s)c |
|---|---|---|---|
| λ | Birth rate constant of uninfected CD4+ T cells (T) | 31 (cells/[μl × day]) | 45 |
| d | Death rate of uninfected CD4+ T cells (T) | 0.02 (1/day) | 36 |
| k | Infection rate of activated CD4+ T cells per virion | 1.875 × 10−4 (μl/virions) | No data |
| δ | Death rate of HIV-1-infected cells (I) | 0.7 (1/day) | 21, 51 |
| δ′ | Death rate of dually infected cells (ID) | δ′ = D × δ (1/day) | †† |
| n | Burst size (no. of virions released from HIV-1-infected cell) (I) | 200 (†) | 17 |
| c | Clearance rate of HIV-1 (V) and crHIV-1 (VT) virions | 30 (1/day) | 21, 47 |
| P | Fold increase in crHIV-1 mRNA concentration compared to HIV-1 mRNA in cytoplasm of dually infected cells (ID) | 1-100 (†) | †† |
| D | Antiviral effect: fractional therapeutic downregulation that crHIV-1 achieves on HIV-1 virion production from ID cells | 0.0-1.0 (†) | †† |
| T0 | Initial concn of uninfected CD4+ T cells in plasma | 800 (cells/μl) | 40 |
| V0 | Initial HIV-1 set point (before crHIV-1 administration) | 105 (virions/ml) | 40 |
The Basic Model neglects viral loss due to infection (a term −kVT in the dV/dt equation) by assuming that its effect is minimal. We have followed this standard in our model, but we also explored the effect of relaxing this assumption. Inclusion of viral loss due to infection had no qualitative effect and minimal quantitative effect on the results (see SI).
Viral competition and recombination.
Retroviruses are diploid and incorporate two single strands of mRNA into each capsid. Thus, three species of diploid virions can be produced from an I D cell: homozygous virions containing two strands of HIV-1 mRNA, heterozygous virions containing one strand of crHIV-1 mRNA and one strand of HIV-1 mRNA, and homozygous virions containing two strands of crHIV-1 mRNA. By using a binomial distribution, the ratio of RNA pairs in the cytoplasm of an I D cell (homozygous HIV-1:heterozygous crHIV-1:homozygous crHIV-1) can be determined as 1:2_P_:_P_2, respectively (since, by definition, the ratio of HIV-1 mRNA to crHIV-1 mRNA in the cytoplasm of an I D cell is 1:P).
Since all of these RNA pairs have equal chance of being packaged into a capsid (explained further in the Discussion), this ratio of cytoplasmic RNA pairs is also the ratio of diploid virions released from an I D cell. However, not all of these virions go on to infect and integrate into CD4+ T cells. Specifically, An et al. (2) show that while heterozygous lentiviral virions may infect cells and be internalized, there is some preintegration (but postfusion) block in the retroviral life cycle.
Thus, the ratio of infectious virions released from an I D cell is just 1:_P_2 since heterozygous virions do not produce viable infections. HIV-1- and simian immunodeficiency virus (SIV)-infected cells have been observed to produce an average of 200 virions (the parameter n) in vivo (17). However, SIV-infected cells in rhesus macaques can be perturbed to produce 3 to 5 times more virions when CD8+ cells are depleted via an anti-CD8 antibody (23) or even up to 40,000 virions when env is unnaturally deleted from the SIV genome (Alan Perelson and David Ho, unpublished data). Thus, encapsidation resources within HIV-1-infected cells do not appear to not be the factor limiting production to ∼200 virions. Rather, immune recognition and killing of infected cells, as well as the pathogenicity of HIV-1 proteins (such as Nef), seem to be destroying the cell before it can produce its full potential of virus and thereby only allowing ∼200 virions to leave the cell. In our model, an I D cell will produce D × 200 HIV-1 virions and _P_2 × D × 200 crHIV-1 virions. If no therapeutic gene is encoded in the crHIV-1 and D = 1, then _P_2 × 200 crHIV-1 virions are still produced, but there is no reduction in the number of HIV-1 virions produced and the HIV-1 set point remains unchanged.
Recombination between HIV-1 and crHIV-1, i.e., heterozygous recombination, would occur after infection of a cell with a heterozygous virion. Retroviral recombination occurs intracellularly during the reverse transcription step when the reverse transcriptase enzyme jumps between the two mRNA strands (25). Apparently, the large size discrepancy between HIV-1 and a smaller HIV-1-based vector (10 versus ∼4 kb) does not permit reverse transcriptase to proceed through all of the steps required to make a viable convalently closed circular DNA needed for nuclear import and integration into host genomic DNA. An et al. (2) go on to show that virtually none of the these heterozygous integrations occur in vitro. Others report that these heterozygous recombinations are absent in mice (46).
Analysis. (i) dynamics of crHIV-1 and HIV-1 (numerical simulations).
To investigate how HIV-1 viremia evolved in time after the addition of the crHIV-1 therapy, numerical simulations on equations 1 to 6 were performed for a large spectrum of parameter values by using the Berkeley Madonna differential equation solving software (http://www.berkeleymadonna.com). The viral eclipse phase (an ∼1.5-day delay in the HIV-1 life cycle between the time a cell is initially infected by HIV-1 and the time that cell begins producing daughter virions (38) was included in the simulations, and equations 1 to 6 were altered accordingly (see SI). Neglecting the viral eclipse phase did not alter the qualitative behavior of the system.
Since the crHIV-1 gene therapy model is a system of stiff differential equations, a Rosenbrock method was used to solve the system of equations. The solutions were checked by using both a fourth order Runge-Kutta algorithm and an adaptive step size method.
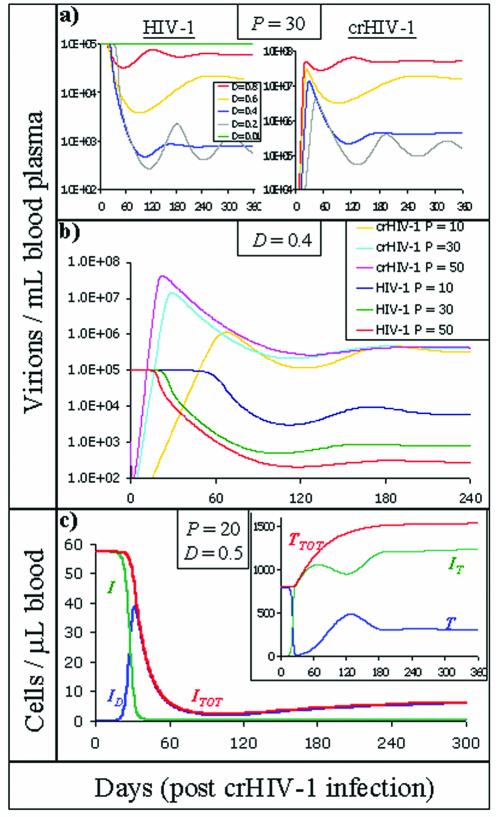
Representative simulations, shown in Fig. 2, demonstrate that the new reduced HIV-1 set point was typically reached ∼100 days after crHIV-1 administration but varied slightly based on the D values (Fig. 2a) and P values (Fig. 2b) used: higher P and D values reduced the time needed to establish the new steady state. crHIV-1 viral load reached high values (107 virions/ml), but HIV-1-infected cells counts dropped to very low levels (Fig. 2c) and total CD4+ cells (T + I T) leveled off at ∼1,500 cells/μl (Fig. 2c, inset), a finding consistent with healthy individuals not infected with HIV-1. Time to achieve steady state varied slightly with respect to initial crHIV-1 values (inoculation volume) used, in agreement with the numerical findings of Lund et al. (27), who analyzed the criteria for a successful nonreplicating gene therapy for HIV-1.
FIG. 2.
Representative numerical simulations. Equations 1 to 6 (with viral eclipse phase delay incorporated) were numerically solved for a large spectrum of parameter values (see Table 1) by using Berkeley Madonna. The system was initially at steady state for the Basic Model. At day 0 crHIV-1 virus was introduced (initial value was V T = 105 virions/ml of blood plasma; a value equivalent to an inoculation of ∼5 × 107 total crHIV-1 virions). (a) HIV-1 and crHIV-1 viral loads are shown for a range of different D values (P was fixed at 30; the legend in the left panel applies to the right panel as well). Increasing the therapeutic downregulation of HIV-1 expression (i.e., decreasing D values) up to a point (D ≈ 0.4) caused a reduction in the HIV-1 set point but also increased the time needed to attain the new HIV-1 set point. As D is further decreased (D = 0.1), oscillations arise (oscillations for D = 0.1 persist and are stable for many years [see SI]), and highly efficient downregulation (D = 0.01) causes the HIV-1 set point to rebound to pretherapy levels. (b) HIV-1 and crHIV-1 viral loads shown for a range of different P values (D was fixed at 0.4). Increasing the crHIV-1 mRNA cytoplasmic concentration (i.e., increasing P values) caused a reduction in HIV-1 set point and also decreased the time needed to attain the new HIV-1 set point. (c) Uninfected cell levels and infected cell levels (inset) after crHIV-1 infection (P and D were fixed at representative values of 20 and 0.5, respectively). I TOT refers to the total HIV-1 infected cell population and was obtained by summing all HIV-1-infected cell populations (I TOT = I + I D), whereas T TOT refers to all non-HIV-1-infected cells (i.e., “healthy” cells, T TOT = T + I T). All infected-cell levels dropped well below pretherapy levels. Although uninfected CD4+-T-cell levels (inset) drop into the AIDS regime (<200 cells/μl), most are converted into therapeutically infected I T cells. Thus, the total non-HIV-1-infected T-cell level (T + I T) climbs to levels seen in healthy, uninfected individuals.
Importantly, the simulation results are not qualitatively unique to equations 1 to 6. We have expanded various other Basic Model architectures, i.e., models incorporating an immune response (7), to include a crHIV-1 gene therapy virus and associated infected cells and found qualitatively equivalent results (see SI). None of these models qualitatively influence the effect of crHIV-1 on HIV-1 because crHIV-1 essentially creates a different reservoir of HIV-1-susceptible cells (I T). This I T cell reservoir becomes predominant, such that any circulating HIV-1 virion is more likely to encounter, and infect, a therapeutically infected (I T) cell and convert it to a dually infected cell rather than encounter an uninfected (T) cell and convert it to a productively infected cell (I). Therefore, the crHIV-1 effect on HIV-1 can be roughly mimicked by decreasing λ or increasing d (uninfected CD4 T-cell production and death, respectively) in the Basic Model. This perturbation to the Basic Model also decreases the pool of uninfected cells that can be infected and converted to HIV-1-producing cells and thus decreases the HIV-1 set point. Although the dynamics of crHIV-1 introduction and uninfected T-cell perturbation can have roughly the same effect on HIV-1 set point, it is important that crHIV-1 introduction does so without decreasing CD4+-T-cell counts (Fig. 2b).
(ii) steady states of the Expanded Model.
To relate the HIV-1 set point to the new experimentally controllable parameters P and D, steady-state solutions for equations 1 to 6 were algebraically determined. As reviewed by Nowak and May (40), the Basic Model of HIV-1 in vivo dynamics has two possible solutions: infection is either cleared or it persists and reaches a steady-state level determined by the parameter values used (the transition between clearance and persistence is described by R 0 [see below]). The expanded crHIV-1 gene therapy model has two additional solutions: one of these solutions is nonphysical (or biologically irrelevant, i.e., one or more populations have negative concentrations) and therefore ignored, and the other solution describes a new, reduced viral set point. Thus, there are two possible outcomes when crHIV-1 is introduced to an HIV-1-infected individual: either crHIV-1 therapy virus propagates and the system (equations 1 to 6) reaches a new steady state or crHIV-1 dies out (i.e., the V T, I T, and I D populations go to zero) and the system “reverts” to the original HIV-1 infection set point.
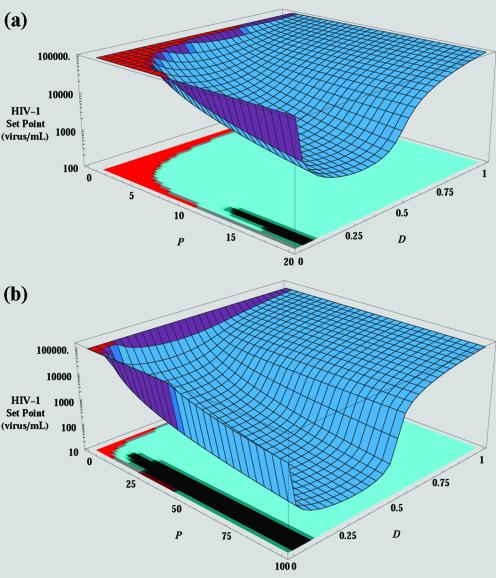
To predict the effect that a crHIV-1 gene therapy virus might have on disease progression in HIV-1-infected individuals, this new HIV-1 viral set point reached after crHIV-1 therapy virus administration was examined in greater detail. We began by calculating a steady state for the Basic Model of HIV-1 in vivo dynamics by using typical patient values for CD4+-T-cell and HIV-1 concentrations in blood plasma (see Table 1). Parameter values were taken from the literature (42), except that λ and k were calculated by steady-state analysis, as described earlier (23). The steady states for equations 1 to 6 were solved algebraically by using parameter values from Table 1. Figure 3 shows the steady-state value of V (the HIV-1 set point) as a function of parameters P and D, with Fig. 3a and b presenting steady-state values for 0 < P ≤ 20 and 0 < P ≤ 100, respectively. When P ≤ 1 (more HIV-1 than crHIV-1 was produced from I D cells), crHIV-1 virus dies out (the red area in Fig. 3a). As P is increased in value beyond ∼3 (i.e., a crHIV-1 hypothetically engineered to have threefold-higher cytoplasmic mRNA concentration), crHIV-1 virus reaches a steady state of coexistence with HIV-1, and the HIV-1 set point is reduced compared to the original pretherapy set point. Reduction in the HIV-1 set point is also influenced by the parameter D (therapeutic reduction in HIV-1 titer produced). As D decreases in value (reduced HIV-1 production from I D cells), the HIV-1 set point is also reduced (the blue area in Fig. 3a).
FIG. 3.
Reduced HIV-1 set point as a function of crHIV-1 packaging (P) and therapeutic downregulation (D). Steady-state solutions of equations 1 to 6 were algebraically determined (using values in Table 1) and plotted by using Mathematica (sensitivity analysis of parameters P and D). Steady states were also analyzed for stability (the sensitivity matrix has been plotted as a projection on the floor on the graphs). (a) The steady-state solution of equation 3 is plotted as a function of parameters P and D. The red area corresponds to parameter regimes where the original HIV-1 set point is stable and the crHIV-1 solution is nonphysical (or biologically irrelevant). As P is increased, a transcritical bifurcation occurs (red-to-blue transition) and the crHIV-1 solution (now the biologically relevant solution) becomes the stable solution. In the blue region, crHIV-1 persists and the HIV-1 set point is lowered. Low values of D (highly efficient HIV-1 downregulation) cause crHIV-1 infection to die out and allow HIV-1 levels to rebound to the original set point level. The projection on the floor of the graph shows the stability (i.e., where all eigenvalues of the Jacobian are negative) of the solutions: the solution of the Basic Model is stable in the red area of the projection, while the crHIV-1 solution is stable in the cyan area of the projection. (b) The P axis has been expanded out to 100 in order to show that the HIV-1 set point can be reduced to levels comparable to reduction caused by HAART when P ≈ 100 and D ≈ 0.4 (i.e., downregulation is ∼60% effective). Clinically, HAART failure occurs when the HIV-1 set point rebounds to >400 copies/ml (18). The transition from the cyan region to the black is referred to mathematically as a supercritical Hopf bifurcation; solutions above the black region oscillate about the value predicted. Stable oscillations arise for the same reason as in a classic predator-prey system: crHIV-1 inhibits HIV-1 to such an extent that crHIV-1 can no longer be packaged, which allows HIV-1 levels to rise, and subsequently permits crHIV-1 to again be produced (see SI for oscillation figure).
However, if HIV-1 downregulation is highly efficient (D ≤ 0.1), crHIV-1 propagation itself is constrained by the dearth of HIV-1 packaging proteins in the cell. In fact, as D is decreased to <0.1, viral production from I D cells becomes so efficiently inhibited that these cells stop producing virus altogether and crHIV-1 infection and propagation within the individual die out. Importantly, I cells still produce HIV-1 at the same rate, allowing the HIV-1 set point to rebound to the pretherapy level. We refer to this rebound effect as crHIV-1 “shooting itself in the molecular foot”; crHIV-1 inhibits HIV-1 so efficiently that its own production is inhibited.
Very importantly, if P and D are optimized, Fig. 3b shows that crHIV-1 gene therapy can reduce HIV-1 set point to levels comparable to HAART reduction. The optimal value of D is ∼0.2 for the parameter values used here. It should also be noted that, as mentioned above, the altered death rate δ′ algebraically cancels out of the steady-state solutions of all populations (except the I D population). This occurs because viral production and cell death are reciprocally linked: increasing cell death increases the viral production term by exactly the same amount that infected cell lifetime is decreased.
Overall, algebraic results agreed precisely with simulation results, in both analyses the same HIV-1 set point was achieved for a given P and D.
(iii) stability of the model and defining a new R 0.
We next investigated the stability of crHIV-1 propagation and maintenance to small perturbations in the system (by examining the eigenvalues of the Jacobian for equations 1 to 6). The results are shown in Fig. 3 as a shadow beneath the curves. crHIV-1 infection is stable when it is the valid, biologically relevant solution (blue region of figure), whereas the pretherapy HIV-1 steady state (red area of figure) is stable when it is the valid, biologically relevant solution. In addition, mathematically, there is an exchange of stability (or transcritical bifurcation) when crHIV-1 with a large enough P is introduced into a system at steady state for HIV-1. When P > 10 and D ≈ 0.1, the HIV-1 steady state becomes oscillatory (black area in Fig. 3) much like a classic predator-prey system: crHIV-1 inhibits HIV-1 to such an extent that crHIV-1 can no longer be packaged, which allows HIV-1 levels to rise and subsequently permits crHIV-1 to again be produced. Mathematically, this is referred to as a supercritical Hopf bifurcation (see SI for oscillation figure).
To characterize crHIV-1 persistence, a “basic reproductive ratio” (R 0) was calculated for the gene therapy model. R 0 is defined as the number of new HIV-1 infections produced from a single infected cell, assuming that virtually all cells are uninfected, and is commonly used to characterize disease spread in epidemiological models (3). In the Basic Model, R 0 = λ_kn_/_c_δ and characterizes the transition between the uninfected and infected steady states (40). When R 0 is less than 1 infection dies out, and when R 0 is greater than 1 infection persists—a stability exchange (transcritical bifurcation) occurs at R 0 = 1.
We have defined a new therapeutic reproductive ratio R0T as the number of new crHIV-1 infections produced from one I T cell, assuming that the system is at steady state for HIV-1 infection (i.e., virtually all cells are uninfected by crHIV-1). The result is surprisingly simple and depends only upon the new parameters P and D and the original R 0 of the system, with the following form (derived in the SI): R0T = P_2_D(1 − 1/R 0). Since an R 0 greater than 1 was assumed (by the Basic Model being at steady state) and 1 − 1/R 0 is therefore less than 1, crHIV-1 persistence in vivo is predicated upon choosing a large enough value of the product P_2_D such that R0T is greater than 1. We verified that an R0T greater than 1 is obtained in precisely the parameter regimes that the crHIV-1 steady-state solution dominates (the blue region in Fig. 3 [see SI]). Since R0T is an “invasion parameter” that describes the rate of infection spread (i.e., a higher R0T implies a faster crHIV-1 spread), we can now understand how lower D values increase the time needed to establish the new steady state in Fig. 2a and b.
RESULTS AND DISCUSSION
In this report we examined the effect of crHIV-1 gene therapy on the HIV-1 in vivo set point by constructing and analyzing a novel mathematical model founded upon the Basic Model of HIV-1 in vivo dynamics. The goal of this therapy is not to fully inhibit HIV-1 but to prevent disease progression by reducing the HIV-1 set point low enough for the patient to be asymptomatic. Importantly, some HIV-1 must be maintained so that crHIV-1 can persistently replicate along with HIV-1. In essence, the aim of this therapy is to create a parasite (crHIV-1) of a parasite (HIV-1). We derived a simple criterion for designing crHIV-1 gene therapy viruses that will persist in vivo: crHIV-1 persists when its basic reproductive ratio R0T = P2 D(1 − 1/R 0) is >1. Algebraic analysis showed that increasing the intracellular mRNA concentration of crHIV-1 relative to that of HIV-1 is the most important consideration for establishing a persistent crHIV-1 infection and reducing the HIV-1 set point. Paradoxically, therapeutic genes that are too potent (i.e., they downregulate HIV-1 expression with high efficiency) were found to be detrimental to crHIV-1 therapy virus persistence (Fig. 2) and HIV-1 set point stability (Fig. 3) and caused the HIV-1 set point to rebound. This occurs because crHIV-1 inhibits the production of HIV-1 packaging materials to such an extent that dually infected cells no longer produce any virus and only HIV-1-infected cells continue to produce HIV-1.
Numerous antiviral genes have been described in the literature, including antisense, RNAi, ribozymes, intrabodies, transdominant Rev mutants, and TAR RNA decoys (see references 31 and 32 for reviews). Depending on the specific gene utilized, the value of D will change, but our model still applies. For a given antiviral cargo, D can be experimentally measured simply by quantifying the HIV-1 titer in the supernatant of cell cultures in the absence or presence of the crHIV-1. We do not consider D to be an intracellular parameter here in order to retain generality for the model. If D is defined as reduction in full-length HIV-1 cytoplasmic mRNA, then each class of antiviral genes described in the literature requires a different model (L. Weinberger, D. Schaffer, and A. Arkin, unpublished data).
The model results highlight the critical importance of maximizing the ratio of crHIV-1 to HIV-1 virus generated from dually infected cells (described by the parameter P), and this ratio can be improved either by increasing the quantity of cytoplasmic crHIV-1 mRNA or increasing the efficiency of their incorporation into virion particles. Efforts to improve the HIV-1 packaging signal (Ψ) will likely fail since Ψ and other HIV-1 regulatory domains are believed to be optimally evolved (29). However, methods to increase the intracellular concentration of lentiviral vector mRNA relative to HIV-1 transcripts have been explored. These methods include the following: removing vector splice signals to increase the proportion of full-length genomic mRNA (10); abrogating vector dependence on HIV-1 Rev by including an mRNA nuclear export signal, such as the Mason-Pfizer monkey virus constitutive transport element (30); and including mRNA stabilizing elements. For example, the woodchuck hepatitis virus posttranscriptional regulatory element has been shown to increase mRNA half-life by up to 10-fold in vitro (52). When the effects of these and other approaches are combined, they may yield crHIV-1 vectors capable of packaging 100 times more efficiently than HIV-1 (Weinberger et al., unpublished). However, future work may yield further means to increase P.
One concern is the prudence of replacing a high HIV-1 viral load with a new, even higher, crHIV-1 viral burden. However, a high crHIV-1 viral burden may not necessarily be harmful since high lentiviral vector loads do not produce disease in rats (24). Furthermore, wild African green monkeys naturally infected with SIV have viral loads 3 logs higher than human AIDS patients, and the monkeys remain healthy (22). Thus, high viral titers are not necessarily harmful; pathogenicity depends on the identity and properties of the virus that constitutes this high viral load.
Despite their promise, potential concerns with crHIV-1 therapies include recombination and mutation. Any therapy that utilizes a competing virus strategy must broach the issue of recombination but, as mentioned above, it has been reported that heterozygous virions do not efficiently infect cells due to a block in reverse transcription (2). We have addressed the issue of recombination between crHIV-1 and HIV-1 above.
Mutation is of more concern, since HIV-1 has the potential to develop viral escape mutants that could abolish the therapeutic effect of crHIV-1, and crHIV-1 could also potentially mutate into a nontherapeutic virus (a possibility that is highly dependent upon the type and size of antiviral gene encoded within the crHIV-1). Dropulic appears to have addressed the former mutation issue by using a vector encoding an antisense env, which is currently entering phase I clinical trials (13). The preliminary results from a mouse model indicate that it is difficult for HIV-1 to mutate around this construct (46). Furthermore, mutations in HIV-1 tat (the transcriptional transactivator) and TAR (the RNA target of Tat) loci induce latency in cell lines (15), and viral escape mutants are believed to arise early in infection (48). Thus, crHIV-1 therapies that attack the tat and TAR loci may be less susceptible to HIV-1 escape since mutants that evade this attack would likely be latent. Models that examine the likelihood of crHIV-1 mutation into a nontherapeutic virus must be examined. We also believe that crHIV-1 mutation may be beneficial since crHIV-1 has the potential to reciprocally match any mutation that HIV-1 might make to escape crHIV-1 therapy.
Another important consideration is whether a complex biological system, such as the interaction between the immune system, HIV-1, and crHIV-1, will even reach steady state, a consideration that calls into question the relevance of mathematical steady-state analysis. Numerous experimental perturbations have been administered to HIV-1-infected patients, including HAART and plasma apheresis (47), and these perturbations have the effect of forcing the patient to new, reduced set points. In addition, SIV-infected rhesus macaques given anti-CD8 antibodies, which eliminate their CD8+ lymphocytes, show an increase in SIV to a new set point (23, 49). Similarly, HIV-1-infected hu-PBL-SCID mice infused with CD8+ lymphocytes reach new, reduced set points (33). Admittedly, these perturbations are caused by drug therapy or enhancement or depletion of immune cells. Perturbations that are more similar to crHIV-1 introduction include: patients coinfected with HIV-1 and either HIV-2 (4) (a completely different virus evolutionarily) or hepatitis C virus (47). In each of these cases, patients develop new, stable HIV-1 set points. Therefore, there is ample precedent for assuming that a perturbation, such as the introduction of a crHIV-1, will force HIV-1 levels to new a steady state rather than eliminate system stability.
As stated above, our model assumes that the I D cell burst size is greater than n, implying that intracellular packaging resources are not limited. The issue of Gag-Pol being a limiting resource is a potential concern for crHIV-1 therapy. While in vitro studies have demonstrated that encapsidation resources, most likely Gag-Pol, within a dually infected cell are a limited resource (8), we discussed above in vivo evidence (23) that SIV-infected rhesus macaques cells can produce many more virions than the ∼200 observed (17). This suggests that, rather than resource depletion, immune recognition and killing of infected cells, as well as the pathogenicity of HIV-1 proteins (such as Nef), seem to be destroying the cell before it can produce its full potential of virus. Our results emphasize the importance of quantifying both the maximum viral output from HIV-1-infected cells and determining conclusively whether HIV-1 encapsidation proteins are a limited intracellular resource in vivo.
Finally, there are different relative benefits and risks associated with attempting crHIV-1 gene therapy. Conditionally replicating gene therapy vectors, ones mobilized by wild-type HIV-1, have a distinct advantage over both autonomously replicating and nonreplicating vectors from the standpoint of long-term therapeutic intervention for HIV-1. Self-replicating or competing HIV-1 therapy viruses have been theoretically examined (37) but have the potential to outcompete HIV-1 and run the risk of establishing a new uncontrolled infection by the principle of competitive exclusion.
Nonreplicating gene therapy vectors, which have been mathematically examined by Lund et al. (27), have the disadvantage of a finite therapeutic lifetime. To achieve long-term therapeutic effects, nonreplicating vectors must integrate into hematopoietic progenitor or stem cells. However, it is currently difficult to efficiently transduce these cells even with ex vivo infection; furthermore, current therapeutic genes would not provide the progeny of these cells with any selective long-term survival advantage over other cells in the CD4+-T-cell population (9). Therefore, the clinically expensive process of harvesting, transducing, reimplanting blood cells would likely have to be repeated multiple times for nonreplicating vectors. In contrast, because they would have the ability to propagate in the presence of an HIV-1 infection, crHIV vectors have long-term therapeutic potential with a single administration, thereby reducing expense, eliminating the need for strong patient compliance, and significantly expanding the patient population this therapy could reach.
However, the most significant risk of the crHIV-1 approach is the potential for crHIV-1 to be transmitted between individuals. The propagation of this genetic material within the HIV-1-infected population poses significant concerns, but we propose that it may also be worthwhile to examine the accompanying benefit: can crHIV-1 spread through a population act to mitigate HIV-1 and AIDS spread through a population?
Acknowledgments
L.S.W. thanks T. Cooke, J. Lloyd-Smith, M. Mautino, and R. Ribeiro for many helpful discussions and acknowledges G. Funk for providing data.
D.V.S. was partially supported by an NSF CAREER Award. A.P.A. was supported in part by the Howard Hughes Medical Institute and Defense Advanced Projects Research Agency. L.S.W. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
REFERENCES
- 1.Allen, T. M., A. D. Kelleher, J. Zaunders, and B. D. Walker. 2002. STI and beyond: the prospects of boosting anti-HIV immune responses. Trends Immunol. 23**:**456-460. [DOI] [PubMed] [Google Scholar]
- 2.An, D. S., K. Morizono, Q. X. Li, S. H. Mao, S. Lu, and I. S. Chen. 1999. An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J. Virol. 73**:**7671-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. M., and R. M. May. 1991. Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford, England.
- 4.Andersson, S., H. Norrgren, Z. da Silva, A. Biague, S. Bamba, S. Kwok, C. Christopherson, G. Biberfeld, and J. Albert. 2000. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 160**:**3286-3293. [DOI] [PubMed] [Google Scholar]
- 5.Arora, V. K., B. L. Fredericksen, and J. V. Garcia. 2002. Nef: agent of cell subversion. Microbes Infect. 4**:**189-199. [DOI] [PubMed] [Google Scholar]
- 6.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53**:**557-593. [DOI] [PubMed] [Google Scholar]
- 7.Bonhoeffer, S., J. M. Coffin, and M. A. Nowak. 1997. Human immunodeficiency virus drug therapy and virus load. J. Virol. 71**:**3275-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukovsky, A. A., J. P. Song, and L. Naldini. 1999. Interaction of human immunodeficiency virus-derived vectors with wild-type virus in transduced cells. J. Virol. 73**:**7087-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazzana-Calvo, M., S. Hacein-Bey, G. de Saint Basile, F. Gross, E. Yvon, P. Nusbaum, F. Selz, C. Hue, S. Certain, J. L. Casanova, P. Bousso, F. L. Deist, and A. Fischer. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288**:**669-672. [DOI] [PubMed] [Google Scholar]
- 10.Cui, Y., T. Iwakuma, and L. J. Chang. 1999. Contributions of viral splice sites and _cis_-regulatory elements to lentivirus vector function. J. Virol. 73**:**6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeks, S. G., J. D. Barbour, R. M. Grant, and J. N. Martin. 2002. Duration and predictors of CD4 T-cell gains in patients who continue combination therapy despite detectable plasma viremia. AIDS 16**:**201-207. [DOI] [PubMed] [Google Scholar]
- 12.Di Mascio, M., G. Dornadula, H. Zhang, J. Sullivan, Y. Xu, J. Kulkosky, R. J. Pomerantz, and A. S. Perelson. 2003. In a subset of subjects on highly active antiretroviral therapy, human immunodeficiency virus type 1 RNA in plasma decays from 50 to <5 copies per milliliter, with a half-life of 6 months. J. Virol. 77**:**2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dropulic, B. 2001. Lentivirus in the clinic. Mol. Ther. 4**:**511-512. [DOI] [PubMed] [Google Scholar]
- 14.Dropulic, B., M. Hermankova, and P. M. Pitha. 1996. A conditionally replicating HIV-1 vector interferes with wild-type HIV-1 replication and spread. Proc. Natl. Acad. Sci. USA 93**:**11103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emiliani, S., W. Fischle, M. Ott, C. Van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72**:**1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funk, G. A. 2003. Models of human immunodeficiency virus type 1 infection: exploring viral fitness, cytopathicity and spatial structure. Ph.D. thesis. ETH (Swiss Federal Institute of Technology), Zurich, Switzerland.
- 17.Haase, A. T., K. Henry, M. Zupancic, G. Sedgewick, R. A. Faust, H. Melroe, W. Cavert, K. Gebhard, K. Staskus, Z. Q. Zhang, P. J. Dailey, H. H. Balfour, Jr., A. Erice, and A. S. Perelson. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274**:**985-989. [DOI] [PubMed] [Google Scholar]
- 18.Havlir, D. V. 2002. Structured intermittent treatment for HIV disease: necessary concession or premature compromise? Proc. Natl. Acad. Sci. USA 99**:**4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermans, P. 2001. Current review and clinical management of patients with primary HIV-1 infection: limits and perspectives. Biomed. Pharmacother. 55**:**301-307. [DOI] [PubMed] [Google Scholar]
- 20.Ho, D. D., and Y. Huang. 2002. The HIV-1 vaccine race. Cell 110**:**135-138. [DOI] [PubMed] [Google Scholar]
- 21.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373**:**123-126. [DOI] [PubMed] [Google Scholar]
- 22.Holzammer, S., E. Holznagel, A. Kaul, R. Kurth, and S. Norley. 2001. High virus loads in naturally and experimentally SIVagm-infected African green monkeys. Virology 283**:**324-331. [DOI] [PubMed] [Google Scholar]
- 23.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T-cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189**:**991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kafri, T., U. Blomer, D. A. Peterson, F. H. Gage, and I. M. Verma. 1997. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat. Genet. 17**:**314-317. [DOI] [PubMed] [Google Scholar]
- 25.Knipe, D. M., B. N. Fields, P. M. Howley, and D. E. Griffin. 2001. Fields virology, p. 1227-1279, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 26.Lisziewicz, J., G. Zeng, C. Gratas, J. N. Weinstein, and F. Lori. 2000. Combination gene therapy: synergistic inhibition of human immunodeficiency virus Tat and Rev functions by a single RNA molecule. Hum. Gene Ther. 11**:**807-815. [DOI] [PubMed] [Google Scholar]
- 27.Lund, O., O. S. Lund, G. Gram, S. D. Nielsen, K. Schonning, J. O. Nielsen, J. E. Hansen, and E. Mosekilde. 1997. Gene therapy of T helper cells in HIV infection: mathematical model of the criteria for clinical effect. Bull. Math. Biol. 59**:**725-745. [DOI] [PubMed] [Google Scholar]
- 28.Mariani, R., and J. Skowronski. 1993. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc. Natl. Acad. Sci. USA 90**:**5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzio, G., M. Vink, K. Verhoef, A. de Ronde, and B. Berkhout. 2002. Efficient human immunodeficiency virus replication requires a fine-tuned level of transcription. J. Virol. 76**:**3084-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mautino, M. R., N. Keiser, and R. A. Morgan. 2000. Improved titers of HIV-based lentiviral vectors using the SRV-1 constitutive transport element. Gene Ther. 7**:**1421-1424. [DOI] [PubMed] [Google Scholar]
- 31.Mautino, M. R., and R. A. Morgan. 2002. Gene therapy of HIV-1 infection using lentiviral vectors expressing anti-HIV-1 genes. AIDS Patient Care STDS 16**:**11-26. [DOI] [PubMed] [Google Scholar]
- 32.Mautino, M. R., and R. A. Morgan. 2002. Inhibition of HIV-1 replication by novel lentiviral vectors expressing transdominant Rev and HIV-1 env antisense. Gene Ther. 9**:**421-431. [DOI] [PubMed] [Google Scholar]
- 33.McKinney, D. M., D. A. Lewinsohn, S. R. Riddell, P. D. Greenberg, and D. E. Mosier. 1999. The antiviral activity of HIV-specific CD8+ CTL clones is limited by elimination due to encounter with HIV-infected targets. J. Immunol. 163**:**861-867. [PubMed] [Google Scholar]
- 34.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272**:**1167-1170. [DOI] [PubMed] [Google Scholar]
- 35.Mittler, J. E., M. Markowitz, D. D. Ho, and A. S. Perelson. 1999. Improved estimates for HIV-1 clearance rate and intracellular delay. AIDS 13**:**1415-1417. [DOI] [PubMed] [Google Scholar]
- 36.Mohri, H., A. S. Perelson, K. Tung, R. M. Ribeiro, B. Ramratnam, M. Markowitz, R. Kost, A. Hurley, L. Weinberger, D. Cesar, M. K. Hellerstein, and D. D. Ho. 2001. Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med. 194**:**1277-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, G. W., and A. S. Perelson. 1995. Modeling defective interfering virus therapy for AIDS: conditions for DIV survival. Math. Biosci. 125**:**127-153. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, P. W., J. E. Mittler, and A. S. Perelson. 2001. Effect of drug efficacy and the eclipse phase of the viral life cycle on estimates of HIV viral dynamic parameters. J. Acquir. Immune Defic. Syndr. 26**:**405-412. [DOI] [PubMed] [Google Scholar]
- 39.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282**:**103-107. [DOI] [PubMed] [Google Scholar]
- 40.Nowak, M. A., and R. M. May. 2000. Virus dynamics: mathematical principles of immunology and virology. Oxford University Press, Oxford, England.
- 41.Parkin, N. T., Y. S. Lie, N. Hellmann, M. Markowitz, S. Bonhoeffer, D. D. Ho, and C. J. Petropoulos. 1999. Phenotypic changes in drug susceptibility associated with failure of human immunodeficiency virus type 1 (HIV-1) triple combination therapy. J. Infect. Dis. 180**:**865-870. [DOI] [PubMed] [Google Scholar]
- 42.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387**:**188-191. [DOI] [PubMed] [Google Scholar]
- 43.Perelson, A. S., and P. W. Nelson. 1999. Mathematical analysis of HIV-1 dynamics in vivo. Siam. Rev. 41**:**3-44. [Google Scholar]
- 44.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271**:**1582-1586. [DOI] [PubMed] [Google Scholar]
- 45.Phillips, A. N. 1996. Reduction of HIV concentration during acute infection: independence from a specific immune response. Science 271**:**497-499. [DOI] [PubMed] [Google Scholar]
- 46.Podsakoff, G. M. 2001. Lentiviral vectors approach the clinic but fall back: National Institutes of Health Recombinant DNA Advisory Committee review of a first clinical protocol for use of a lentiviral vector. Mol. Ther. 4**:**282-283. [DOI] [PubMed] [Google Scholar]
- 47.Ramratnam, B., S. Bonhoeffer, J. Binley, A. Hurley, L. Zhang, J. E. Mittler, M. Markowitz, J. P. Moore, A. S. Perelson, and D. D. Ho. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354**:**1782-1785. [DOI] [PubMed] [Google Scholar]
- 48.Ribeiro, R. M., and S. Bonhoeffer. 2000. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl. Acad. Sci. USA 97**:**7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283**:**857-860. [DOI] [PubMed] [Google Scholar]
- 50.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9**:**727-728. [DOI] [PubMed] [Google Scholar]
- 51.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373**:**117-122. [DOI] [PubMed] [Google Scholar]
- 52.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73**:**2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]