Mouse consomic strains: Exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies (original) (raw)
Abstract
Consomic (chromosome substitution) strains (CSs) represent the most recent addition to the mouse genetic resources aimed to geneticaly analyze complex trait loci (QTLs). In this study, we report the development of a set of 28 mouse intersubspecific CSs. In each CS, we replaced a single chromosome of the C57BL/6J (B6) inbred strain (mostly Mus m. domesticus) with its homolog from the PWD/Ph inbred strain of the Mus m. musculus subspecies. These two progenitor subspecies diverged less than 1 million years ago and accumulated a large number of genetic differences that constitute a rich resource of genetic variation for QTL analyses. Altogether, the 18 consomic, nine subconsomic, and one conplastic strain covered all 19 autosomes, X and Y sex chromosomes, and mitochondrial DNA. Most CSs had significantly lower reproductive fitness compared with the progenitor strains. CSs homosomic for chromosomes 10 and 11, and the C57BL/6J-Chr X males, failed to reproduce and were substituted by less affected subconsomics carrying either a proximal, central, or distal part of the respective chromosome. A genome-wide scan of 965 DNA markers revealed 99.87% purity of the B6 genetic background. Thirty-three nonsynonymous substitutions were uncovered in the protein-coding regions of the mitochondrial DNA of the B6.PWD-mt conplastic strain. A pilot-phenotyping experiment project revealed a high number of variations among B6.PWD consomics.
Consomic strains (CSs), also known as chromosome (chr) substitution strains, represent the newest addition to the mouse genetic resources aimed at a genetic analysis of complex traits. To construct a CS, the genomes of two inbred strains are combined so that one chromosome pair of the host strain is replaced by the corresponding chromosome pair of the donor strain (Nadeau et al. 2000). Since the mouse genome is composed of 19 pairs of autosomes, and X and Y gonosomes, 21 CSs constitute the complete set.
Nadeau, Lander, and coworkers developed the first set of mouse CSs with the A/J strain as a chromosome donor and C57BL/6J (hereafter B6) as the recipient strain (Singer et al. 2004). Another series of 65 genome-wide congenic (subconsomic) strains was created by transferring segments of individual autosomes of the DBA/2J strain on the background of B6 (Davis et al. 2005) that covered 95% of the autosomal genome. The transfer of sex chromosomes was not reported. Most recently, the same laboratory reported the construction of ∼40 congenic (subconsomic) strains from a CAST/Ei inbred strain of Mus m. castaneus origin (Davis et al. 2007). These strains cover ∼80% of CAST/Ei autosomes. The transfer of sex chromosomes and mitochondrial DNA was not reported. In the accompanying paper in this issue of Genome Research, Takada et al. (2008) describe a full set of consomic strains, using B6 as a background strain and the MSM/Ms of Mus m. molossinus origin as the chromosome donor.
Here, we describe the development and characterization of the first set of consomic strains with Mus m. musculus chromosomes introgressed into a B6 genetic background. The PWD/Ph inbred strain (hereafter PWD) had been established from a pair of trapped wild mice of Mus m. musculus origin (Gregorova and Forejt 2000) and served as the chromosome donor. PWD is one of the 15 strains recently resequenced by the NIEHS (Frazer et al. 2007; Yang et al. 2007). The B6 inbred strain (mostly Mus m. domesticus) provided the genetic background. These parental strains represent two mouse subspecies that can still mix their genomes without fundamentally compromising the viability or reproductive functions of the hybrids.
Results
Construction of a complete panel of intersubspecific consomic strains
In this project, we transferred individual autosomes, the X and Y sex chromosomes, and the mitochondrial genome of the PWD donor strain on the genetic background of B6 strain. The undertaking took over 7 yr and generated over 17,000 mice, 95,000 simple sequence length polymorphism (SSLP) genotypes, and 4300 matings. All of the strains, prepared and maintained at the Institute of Molecular Genetics, Prague, Czech Republic, were provided to The Jackson Laboratory as a research resource to make them available to the scientific community. The updated information about characterization and utilization of this set of consomics can be found at http://www.img.cas.cz/mmg/.
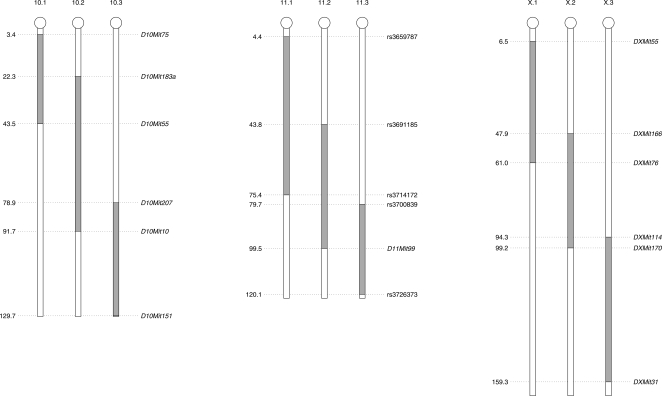
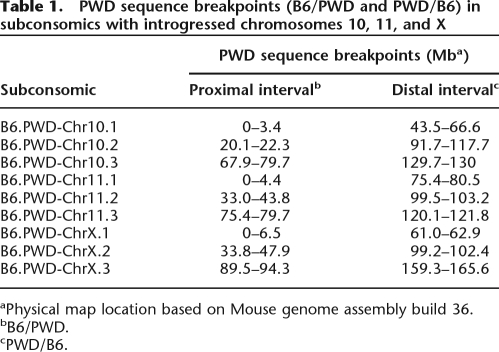
The mice of three consomic strains with PWD chromosomes reproduced very poorly (chr 10, chr 11) or were male sterile (chr X). Therefore, at N11 we developed three subconsomic strains carrying an overlapping proximal, middle, or distal PWD segment of a given chromosome, respectively (Fig. 1; Table 1). All but B6.PWD-Chr10.1 (for nomenclature, see Methods) subconsomics were viable and fertile. The proximal part of chr 10 was associated with the overgrowth of fetuses, the probable cause of the frequent death of females during delivery.
Figure 1.
Scheme of introgressed regions in subconsomic strains. The borders of PWD intervals (gray) were mapped to the closest neighboring single nucleotide polymorphism (SNP) or SSLP. Megabase scale.
Table 1.
PWD sequence breakpoints (B6/PWD and PWD/B6) in subconsomics with introgressed chromosomes 10, 11, and X
Search for transmission-ratio and sex-ratio distortion in nascent and established consomic strains
In Drosophila and other species, introgression of a chromosome or chromosomal segment from a closely related species can result in meiotic drive (de Villena and Sapienza 2001; Hurst and Werren 2001; Orr 2005). We compared the expected and observed frequencies of transgressed PWD chromosomes in N4–N11 backcross generations. Sex ratio was examined in 24 consomic strains (homosomic for the substituted PWD chromosome, Supplemental Tables 1–3). A highly significant deficiency of nonrecombinant PWD chr 7 and chr 18 (P < 0.0002) was found in the observed frequencies of nonrecombinant PWD chromosomes. This could be explained by a lower fitness of mice with a PWD-substituted chromosome, rather than by meiotic drive. All other observed deviations in transmission ratio of substituted PWD chromosomes or in sex-ratio did not reach the magnitude known from the studies on the _t_-complex in mice (Bauer et al. 2007) or on the sex-ratio segregation distortion in Drosophila (Tao et al. 2007) (for more details, see Supplemental data and Supplemental Tables 1–3).
Reproductive fitness of intersubspecific CSs
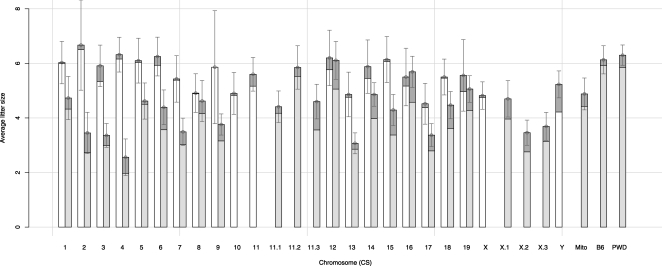
The PWD autosomes and the Y chromosome did not compromise the reproductive performance of heterosomic carriers even after 10 generations of backcrosses. The average litter size of heterosomic mice was 5.8 (range 4.9–6.7) and their postnatal losses were <4% (range 0%–9.7%; see Fig. 2). The introgression of the PWD X chromosome resulted in the complete sterility of B6.PWD-X males (Storchova et al. 2004), but the heterosomic XPWD/XB6 females were viable and fertile.
Figure 2.
Reproductive performance of mice heterosomic (white columns) or homosomic (gray columns) for PWD introgressed chromosome. The offspring lost before weaning are shown in dark gray on the top of each column.
In the progeny of N11 intercross, highly significant deficiency of homosomic offsprings was observed (expected 4.27%, observed 3.05%, P = 0.0069; see Supplemental Table 4). The reproductive performance deteriorated in homosomic mice, depending on a particular introgressed PWD chromosome. The B6.PWD-Chr10 and B6.PWD-Chr11 consomic strains could not be maintained because of their low fecundity. In the remaining CSs, the average litter size was reduced to 4.5 (range 2.7–6.1). The poor reproductive performance was enhanced by fivefold higher postnatal losses compared with heterosomics (17.9%, range 2.6%–32.5%; Fig. 2). Most losses occurred within the first 24 h of postnatal life.
Quality control of substituted chromosomes and genetic background of CSs
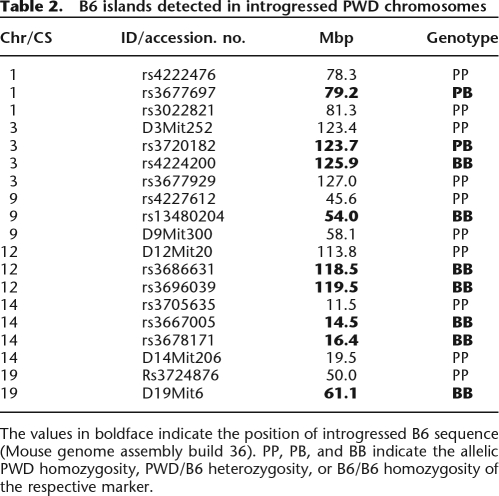
An ideal CS carries an intact chromosomal pair of donor origin on a pure host strain genetic background. In practice, the donor chromosome can carry hidden islands of the host sequence due to unrecognized double crossing-overs or gene conversion events, while the host genetic background can be contaminated by residual donor sequences. With an increasing number of backcross generations, the risk of hidden double crossovers increases, while the background contamination decreases. We evaluated the quality of newly established CSs with a set of 279 SSLP (MIT) markers and 686 SNP markers, all polymorphic between B6 and PWD (Supplemental Tables 5, 6). Altogether, six B6 islands were found on introgressed PWD chromosome numbers 1, 3, 9, 12, 14, and 19 (Supplemental Fig. 1; Table 2). In B6.PWD-Chr1, one SNP marker at 79.2 Mb was in a heterozygous state, the remaining five CSs displayed one or two SNPs already fixed to B6 homozygosity. The terminal markers on chr 12 and chr 19 were of B6 origin.
Table 2.
B6 islands detected in introgressed PWD chromosomes
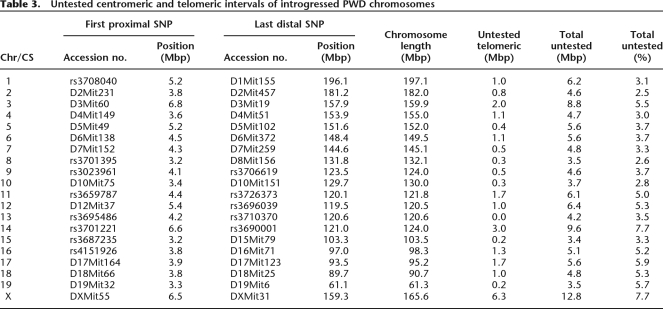
The maximum sum of B6 islands in introgressed PWD chromosomes (intervals up to the first adjacent PWD/PWD markers) was 33.5 Mb, representing 1.3% of the mouse genome sequence. The length of untested intervals on the introgressed chromosomes at centromeric and telomeric ends ranged from 0.0 to 6.7 Mb (total 113.7 Mb; see Table 3).
Table 3.
Untested centromeric and telomeric intervals of introgressed PWD chromosomes
We tested the residual contamination of the B6 background by PWD sequences by genome-wide screen with 965 markers, with the average distance between any two adjacent markers 2.82 Mb or 1.76 cM. Nine out of 20 tested CSs did not reveal any PWD contamination, the remaining 11 CSs were heterozygous in 19 markers, and PWD/PWD homozygous in five markers. Hence, the average purity of the genetic background was >99.87% (Supplemental Table 7).
Intersubspecific exchange of mitochondrial DNA in B6.PWD-mt
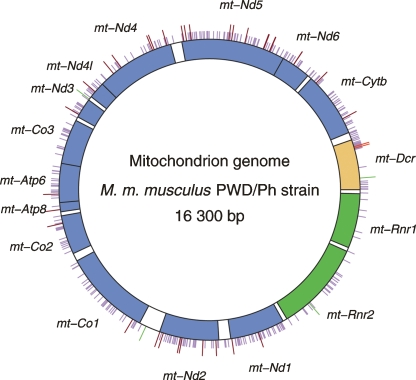
Swapping of B6 for the PWD mitochondrial genome resulted in creation of the B6.PWD-Mit conplastic strain. To facilitate its characterization, we completely sequenced the PWD mitochondrial DNA (mtDNA, GenBank accession no. DQ874614). The sequence is 100% identical to the PWD/PhJ sequence reported recently (Goios et al. 2007) and 99.67% identical to Mus m. molossinus. The total identity of B6 and PWD mtDNAs is 97.6% (Fig. 3; Supplemental Table 8). The comparison of the protein-coding regions of PWD and B6 mtDNAs revealed 33 nonsynonymous substitutions. The nonsimilar amino acid substitutions are not in the positions containing known mitochondrial mutations (Supplemental Table 9).
Figure 3.
Map of the PWD mitochondrial DNA sequence (mitochondrial chromosome). (Blue) Protein-coding genes; (green) 12S RNA genes; (beige) the D-loop region; (short bars) the SNP positions between PWD and B6; (long brown bars) nonsynonymous substitutions; (long green bars) insertions in PWD; (long red bar) the deletion in PWD mtDNA, compared with B6.
The PWD origin of mtDNA in the conplastic strain was confirmed by the presence of the Mus m. musculus diagnostic BamHI restriction site in the mt-Nd1 gene (Bozikova et al. 2005).
Phenome preview
Just as B6 and PWD mouse inbred strains represent great genetic diversity, they also display considerable phenotypic variation. In the first series of phenotypic measurements, eight males and females from 11 CSs and B6 and PWD parental strains were examined for 24 phenotypes that included complete blood counts and other measures of blood formation and blood chemistries. The initial characterization of each strain identified 97 QTCs (quantitative trait chromosome—carrying one or more QTLs) in female consomics and 119 QTC in males. Only 44 QTC were shared by both sexes. The data have been deposited in the Mouse Phenome Database (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home).
Discussion
Only 1% out of 2000 QTLs have been identified with a particular gene in mice and rats, mainly because the QTL regions were too broad (Flint et al. 2005) or because the independently segregating modifiers blurred the phenotype (Nadeau 2001). Systems genetics integrating transcriptional networks with complex phenotypes would profit from more focused positioning of QTLs. A more efficient genetic mapping could be achieved by reducing the genetic complexity in a defined way. Derivatives of inbred strains such as congenic, or recombinant congenic strains, and most recently consomics strains, have been developed for this purpose (Peters et al. 2007).
Here, we report the development of a set of 28 chromosome substitution strains composed of 18 consomic, nine subconsomic strains, and one conplastic strain; the PWD inbred strain served as the donor of chromosomes, and B6 as the background strain. The B6 strain was established by C.C. Little in 1921 (Beck et al. 2000) from fancy mice predominantly of Mus m. domesticus ori gin (Frazer et al. 2007; Yang et al. 2007), while the PWD strain originated in 1974 from a pair of wild mice of a closely related subspecies Mus m. musculus (Pavljukova and Forejt 1981; Gregorova and Forejt 2000). The reported panel of interspecific CSs represents a unique snapshot of genetic variation between the two subspecies (∼1% of SNPs versus ∼0.1% of SNPs in human), and reflects 350,000–1,000,000 yr of their independent evolution (Guenet and Bonhomme 2003).
We backcrossed all nascent consomic strains for 10 generations to clean the B6 genetic background. Subsequent testing showed the average background purity better than 99.87%. The PWD-substituted chromosomes have two kinds of imperfections: First, the PWD/B6 origin of centromeric and telomeric ends could not be determined up to the first polymorphic SSLP marker used in the construction of the strains. A posteriori genome-wide genotyping revealed B6/B6 homozygosity for terminal markers in introgressed chr 12 and chr 19, causing an irreversible loss of 3 Mb and 11 Mb of PWD variation, respectively. Moreover, the PWD polymorphism was lost at the pseudoautosomal region of the X chromosome (Perry et al. 2001). Second, the islands of B6 sequence inside substituted PWD chromosomes occurred by double crossing-over between two adjacent markers, or by gene conversion events. B6 islands were found in six introgressed chromosomes after a whole-genome screening of 965 DNA markers, but their total maximum size did not exceed 1.3% of the mouse genome.
The introgression of a chromosome to another subspecies can provoke the preferential recovery of one allelic form from gametes of a heterozygous parent, the phenomenon known as meiotic drive (Hurst and Werren 2001; Dyer et al. 2007). We detected a significant transmission ratio distortion during backcrosses of PWD chromosomes 7 and 18. In both cases, the transmission of a PWD chromosome was lower than the expected 50%. This finding and the reduced fecundity of many consomic strains are in agreement with the concept of a strong selection for coadapted allelic forms of gene clusters among classical mouse inbred strains (Petkov et al. 2005). The transmission ratio distortion of whole chromosomes or their parts was reported in interspecific backcrosses of B6 × Mus spretus (Siracusa et al. 1991; Montagutelli et al. 1996) or in interspecies hybrids of wild Mus musculus musculus (Agulnik et al. 1993). Sex ratio distortion was described as a high transmission ratio of a proximal half of M. spretus X chromosome in an interspecific backcross (Montagutelli et al. 1996). In the present set of CSs, the observed sex-ratio distortion of the proximal region of X (B6.PWD-ChrX.1) did not reach statistical significance.
The majority of established consomic strains displayed lower reproductive performance than their parental strains. In particular, strains carrying PWD chromosomes 2, 3, 4, 7, 8, 9, 13, 17, and X.3 had average litter size less than four. Moreover, cannibalism of newborn mice by their consomic mothers was more frequent than in parental strains. The lower fitness of consomics could be explained by abnormal gene interactions caused by the independent evolution of the two subspecies for ∼500,00 yr (Abe et al. 2004). A similar deficit in breeding performance was reported for most B6.D2 subconsomics (Churchill et al. 2004), a finding that supports the idea (Petkov et al. 2005) of intense selection pressures for coadapted sets of alleles during the formation of inbred strains.
The main justification for the intersubspecies consomic project was to generate a new resource for systems genetics. Our pilot analyses of 24 phenotypes of blood counts and blood chemistry reveal considerable variation among strains; these data were integrated in the Mouse Phenome Database (Paigen and Eppig 2000; Bogue et al. 2007).
Furthermore, the present set of CSs can serve to seek out the speciation-related genes. Hybrid male sterility of some (PWD × laboratory strain) F1 hybrids (Forejt 1996; Gregorova et al. 1996) models the Dobzhansky-Muller genomic incompatibility (Wu and Hollocher 1998) governed by the epistatic interactions of several independently coadapted genes (Forejt 1996). Two hybrid sterility genes, Hst1 and Hstx1 have already been mapped (Forejt and Ivanyi 1974; Trachtulec et al. 1997, 2005; Storchova et al. 2004). The reconstitution of the hybrid sterility phenotype can be attempted by combining the consomic strains with relevant PWD chromosomes.
Takada et al. (2008, this issue) have constructed another series of intersubspecific CSs by using the MSM/Ms strain derived from Japanese wild mice (Mus m. molossinus) as the chromosome donor and B6 as the host. Some of these strains were then used to partially rescue the hybrid sterility caused by the X-linked Sha1, Sha2, and Sha3 loci (Oka et al. 2007). Because the Japanese Mus m. molossinus is genetically very similar to the East European Mus m. musculus (Abe et al. 2004), it will be important to compare the genetic architectures of their reproductive isolation from the Mus. m. domesticus subspecies.
Experimental evidence has been accumulated on the high efficiency of CSs to detect QTL for various complex traits, such as anxiety, prepulse inhibition, pubertal timing, airway hyper-responsiveness, diet-induced obesity, susceptibility to germ-cell tumors, transcriptional variation in liver, or male infertility (Matin et al. 1999; Krewson et al. 2004; Oka et al. 2004; Singer et al. 2004, 2005; Storchova et al. 2004; Ackerman et al. 2005; Petryshen et al. 2005; Shockley and Churchill 2006; Zhu et al. 2007). The identification of the genes responsible for QTLs will be facilitated by the availability of the PWD genomic BAC library (Jansa et al. 2005) and by the partial genomic resequencing of the PWD genome in Perlegen's Mouse SNP Discovery Project developed in conjunction with the NIEHS (Frazer et al. 2007; Yang et al. 2007; http://mouse.perlegen.com/mouse/index.html).
Methods
The origin of mouse strains
The PWD/Ph inbred strain was created from a single pair of wild mice of the Mus musculus musculus subspecies. The wild mice were trapped in 1972 in Kunratice, in the central part of the Czech Republic (Gregorova and Forejt 2000). On August 28, 2007, they were at the F87 generation of brother × sister mating. The C57BL/6J inbred strain (B6) was imported from The Jackson Laboratory in 1998. The CSs have been produced and maintained in the Specific Pathogen Free barrier facility on a 12 h light/12 h dark cycle. The mice had ad libitum access to a standard rodent diet (VELAS, ST-1, 3.4% fat) and acidified water. Principles of laboratory animal care followed the Czech Republic Act Animal Protection no. 246/92 Sb. fully compatible with the corresponding Directive 806/609/EEC of the Council of Europe Convention ETS123. The consomic strains were prepared by selection for nonrecombinant PWD chromosome in 10 or more consecutive backcrosses to the B6 host strain.
Construction of intersubspecific consomic and subconsomic strains
The backcrosses started with (B6 × PWD) F1 females to avoid the partial meiotic arrest and resulting subfertility of F1 males (Forejt 1996). The exception was the line dedicated to transfer the chromosome YPWD. The conplastic B6.PWD-mt strain, carrying mitochondrial PWD DNA and the B6 nuclear genome was generated by 10 backcrosses, starting with (PWD × B6) × B6, always using backcross females and B6 males. After each backcross generation (_N_2–_N_11), the mice with a particular nonrecombinant PWD chromosome were selected for further backcross to B6. To reduce the risk of hidden double crossovers, a genotyping panel was used of 279 evenly spaced (average 6.0 cM) SSLPs (Dietrich et al. 1994) preselected for distinct B6/PWD polymorphism (Supplemental Table 5). To ensure the B6 origin of the mitochondrial DNA, B6 was used as a female parent in one backcross generation. The conplastic B6.PWD-mt strain was produced by 10 generations of backcrosses of Nn females to B6 males without genotyping.
The nomenclature for consomic and subconsomic strains
Following the recommendation of the International Committee on Standardized Genetic Nomenclature for Mice, the designation is C57BL/6J-Chr #PWD for a consomic strain carrying chromosome # on genetic background of C57BL/6J. Subconsomics with proximal, middle, and distal segments are termed C57BL/6J-Chr #.1PWD, C57BL/6J-Chr #.2PWD, and C57BL/6J-Chr #.3PWD. The abbreviated form, used throughout this study, is B6.PWD-Chr.#, e.g., B6.PWD-Chr10.1 for C57BL/6J-Chr 10.1PWD. The term chromosome substitution strain is considered as a common designation for consomic, subconsomic, and conplastic strains.
Genome-wide genotyping and sequence analysis
Genomic DNA was obtained from mouse-tail biopsies. Two millimeters of a tail were added to 600 μL of 50 mM NaOH, heated to 95°C for 90 min, vortexed, and neutralized with 50 μL of 1 M Tris (pH 8). One-half microliter of DNA solution was used for SSLP amplification as described (Storchova et al. 2004). PCR products were separated on a 4% agarose gel and visualized with ethidium bromide. SSLP markers were selected from a database available at http://www.genome.wi.mit.edu/cgi-bin/mouse/index (see also Dietrich et al. 1994). A total of 269 SSLP markers were validated as polymorphic between PWD and B6 (Supplemental Table 1). Sixty were selected, three for each chromosome, as markers for the proximal, middle, and distal regions. These SSLPs were used in the first step of selecting nonrecombinant donor chromosomes. Only animals with PWD allele at all three markers were tested for additional markers to exclude double recombinants.
To ascertain the residual contamination of the host background by donor sequences and to uncover possible islands of B6 sequences in transferred PWD chromosomes not revealed by SSLP genotyping, 686 SNPs, chosen from a validated panel (Petkov et al. 2004) and tested for PWD/B6 polymorphism were used. The SNP genotyping was done by the Allele Typing Service at The Jackson Laboratory in conjunction with Kbiosciences.
Phenotyping
The Heart, Lung, and Blood Mutagenesis Program at The Jackson Laboratory measured PWD and B6 strains for multiple traits (for Methods, see http://pga.jax.org/protocols.html).
CONSOMIC database and statistical analyses
The data about mice, crosses, litters, and SSLP markers and genotypes were maintained in a web database CONSOMIC developed on purpose for this project. The Database CONSOMIC was powered by mySQL relational database system running on Sun Solaris. The web interface was created using CGI scripts written in Perl. Statistical analysis was performed in Microsoft Excel. The observed and expected numbers of mice were compared using χ2-test for independence with one degree of freedom (built-in functions CHITEST and CHIINV). All _P_-values were reported unadjusted in order to be clear about the scope of multiple testing.
Acknowledgments
We thank Marta Landikova, Marie Dzur-Gajdosova, David Janak, Phyllis Magnani, and Susan Sheehan for technical assistance. This work was supported in part by NIH grant 1R01 HG00318, by FP6 project AnEUploidy 037627, by the Ministry of Education, Youth, and Sports of the Czech Republic grant no. 1M6837805002—Center for Applied Genomics, and by the CSF grant no. 301/07/1264. Jiri Forejt was supported as International Scholar of the Howard Hughes Medical Institute grant 55000306.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession no. DQ874614.]
References
- Abe K., Noguchi H., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Noguchi H., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Ezawa K., Saitou N., Hattori M., Sakaki Y., Saitou N., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y., et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman K.G., Huang H., Grasemann H., Puma C., Singer J.B., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Huang H., Grasemann H., Puma C., Singer J.B., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Grasemann H., Puma C., Singer J.B., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Puma C., Singer J.B., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Singer J.B., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Hill A.E., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Lander E., Nadeau J.H., Churchill G.A., Drazen J.M., Nadeau J.H., Churchill G.A., Drazen J.M., Churchill G.A., Drazen J.M., Drazen J.M., et al. Interacting genetic loci cause airway hyperresponsiveness. Physiol. Genomics. 2005;21:105–111. doi: 10.1152/physiolgenomics.00267.2004. [DOI] [PubMed] [Google Scholar]
- Agulnik S.I., Sabantsev I.D., Orlova G.V., Ruvinsky A.O., Sabantsev I.D., Orlova G.V., Ruvinsky A.O., Orlova G.V., Ruvinsky A.O., Ruvinsky A.O. Meiotic drive on aberrant chromosome 1 in the mouse is determined by a linked distorter. Genet. Res. 1993;61:91–96. doi: 10.1017/s0016672300031189. [DOI] [PubMed] [Google Scholar]
- Bauer H., Veron N., Willert J., Herrmann B.G., Veron N., Willert J., Herrmann B.G., Willert J., Herrmann B.G., Herrmann B.G. The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes & Dev. 2007;21:143–147. doi: 10.1101/gad.414807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J., Festing M., Fisher E., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J., Festing M., Fisher E., Hafezparast M., Lennon-Pierce M., Eppig J., Festing M., Fisher E., Lennon-Pierce M., Eppig J., Festing M., Fisher E., Eppig J., Festing M., Fisher E., Festing M., Fisher E., Fisher E. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bogue M.A., Grubb S.C., Maddatu T.P., Bult C.J., Grubb S.C., Maddatu T.P., Bult C.J., Maddatu T.P., Bult C.J., Bult C.J. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozikova E., Munclinger P., Teeter K.C., Tucker P.K., Macholan M., Pialek J., Munclinger P., Teeter K.C., Tucker P.K., Macholan M., Pialek J., Teeter K.C., Tucker P.K., Macholan M., Pialek J., Tucker P.K., Macholan M., Pialek J., Macholan M., Pialek J., Pialek J. Mitochondrial DNA in the hybrid zone between Mus musculus musculus and Mus musculus domesticus: A comparison of two transects. Biol. J. Linnean Soc. 2005;84:363–378. [Google Scholar]
- Churchill G.A., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Beavis W.D., Belknap J.K., Bennett B., Berrettini W.,, Belknap J.K., Bennett B., Berrettini W.,, Bennett B., Berrettini W.,, Berrettini W., The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Davis R.C., Schadt E.E., Smith D.J., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Schadt E.E., Smith D.J., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Smith D.J., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Rosales M., Doss S., Meng H., Allayee H., Doss S., Meng H., Allayee H., Meng H., Allayee H., Allayee H., et al. A genome-wide set of congenic mouse strains derived from DBA/2J on a C57BL/6J background. Genomics. 2005;86:259–270. doi: 10.1016/j.ygeno.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Davis R.C., Jin A., Rosales M., Yu S., Xia X., Ranola K., Schadt E.E., Lusis A.J., Jin A., Rosales M., Yu S., Xia X., Ranola K., Schadt E.E., Lusis A.J., Rosales M., Yu S., Xia X., Ranola K., Schadt E.E., Lusis A.J., Yu S., Xia X., Ranola K., Schadt E.E., Lusis A.J., Xia X., Ranola K., Schadt E.E., Lusis A.J., Ranola K., Schadt E.E., Lusis A.J., Schadt E.E., Lusis A.J., Lusis A.J. A genome-wide set of congenic mouse strains derived from CAST/Ei on a C57BL/6 background. Genomics. 2007;90:306–313. doi: 10.1016/j.ygeno.2007.05.009. [DOI] [PubMed] [Google Scholar]
- de Villena F.P.M., Sapienza C., Sapienza C. Nonrandom segregation during meiosis: The unfairness of females. Mamm. Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- Dietrich W., Miller J., Steen R., Merchant M., Damron D., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Miller J., Steen R., Merchant M., Damron D., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Steen R., Merchant M., Damron D., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Merchant M., Damron D., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Damron D., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Nahf R., Gross A., Joyce D., Wessel M., Dredge R., Gross A., Joyce D., Wessel M., Dredge R., Joyce D., Wessel M., Dredge R., Wessel M., Dredge R., Dredge R., et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat. Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Dyer K.A., Charlesworth B., Jaenike J., Charlesworth B., Jaenike J., Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc. Natl. Acad. Sci. 2007;104:1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Valdar W., Shifman S., Mott R., Valdar W., Shifman S., Mott R., Shifman S., Mott R., Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- Forejt J., Ivanyi P., Ivanyi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet. Res. 1974;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., Montgomery J., Morenzoni M.M., Nilsen G.B., Morenzoni M.M., Nilsen G.B., Nilsen G.B., et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Goios A., Pereira L., Bogue M., Macaulay V., Amorim A., Pereira L., Bogue M., Macaulay V., Amorim A., Bogue M., Macaulay V., Amorim A., Macaulay V., Amorim A., Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova S., Forejt J., Forejt J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies—A valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. (Praha) 2000;46:31–41. [PubMed] [Google Scholar]
- Gregorova S., Mnukova-Fajdelova M., Trachtulec Z., Capkova J., Loudova M., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Mnukova-Fajdelova M., Trachtulec Z., Capkova J., Loudova M., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Trachtulec Z., Capkova J., Loudova M., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Capkova J., Loudova M., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Loudova M., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Hoglund M., Hamvas R., Lehrach H., Vincek V., Klein J., Hamvas R., Lehrach H., Vincek V., Klein J., Lehrach H., Vincek V., Klein J., Vincek V., Klein J., Klein J., et al. Sub-milliMorgan map of the proximal part of mouse chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome. 1996;7:107–113. doi: 10.1007/s003359900029. [DOI] [PubMed] [Google Scholar]
- Guenet J.L., Bonhomme F., Bonhomme F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Hurst G.D.D., Werren J.H., Werren J.H. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- Jansa P., Divina P., Forejt J., Divina P., Forejt J., Forejt J. Construction and characterization of a genomic BAC library for the Mus m. musculus mouse subspecies (PWD/Ph inbred strain) BMC Genomics. 2005;6:161. doi: 10.1186/1471-2164-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewson T.D., Supelak P.J., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Supelak P.J., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Hill A.E., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Singer J.B., Lander E.S., Nadeau J.H., Palmert M.R., Lander E.S., Nadeau J.H., Palmert M.R., Nadeau J.H., Palmert M.R., Palmert M.R. Chromosomes 6 and 13 harbor genes that regulate pubertal timing in mouse chromosome substitution strains. Endocrinology. 2004;145:4447–4451. doi: 10.1210/en.2004-0543. [DOI] [PubMed] [Google Scholar]
- Matin A., Collin G., Asada Y., Varnum D., Nadeau J., Collin G., Asada Y., Varnum D., Nadeau J., Asada Y., Varnum D., Nadeau J., Varnum D., Nadeau J., Nadeau J. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nat. Genet. 1999;23:237–240. doi: 10.1038/13874. [DOI] [PubMed] [Google Scholar]
- Montagutelli X., Turner R., Nadeau J.H., Turner R., Nadeau J.H., Nadeau J.H. Epistatic control of non-Mendelian inheritance in mouse interspecific crosses. Genetics. 1996;143:1739–1752. doi: 10.1093/genetics/143.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.H. Modifier genes in mice and humans. Nat. Rev. Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- Nadeau J., Singer J., Matin A., Lander E., Singer J., Matin A., Lander E., Matin A., Lander E., Lander E. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Oka A., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Toshimori K., Moriwaki K., Shiroishi T., Moriwaki K., Shiroishi T., Shiroishi T. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics. 2004;166:913–924. doi: 10.1534/genetics.166.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Yamamoto H., Kuriki S., Takagi N., Kuriki S., Takagi N., Takagi N., et al. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics. 2007;175:185–197. doi: 10.1534/genetics.106.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A. The genetic basis of reproductive isolation: Insights from Drosophila. Proc. Natl. Acad. Sci. 2005;102 (Suppl 1):6522–6526. doi: 10.1073/pnas.0501893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., Eppig J.T., Eppig J.T. A mouse phenome project. Mamm. Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- Pavljukova H., Forejt J., Forejt J. Private communication. Mouse News Lett. 1981;65:44. [Google Scholar]
- Perry J., Palmer S., Gabriel A., Ashworth A., Palmer S., Gabriel A., Ashworth A., Gabriel A., Ashworth A., Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L.L., Robledo R.F., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Robledo R.F., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Churchill G.A., Paigen B.J., Svenson K.L., Paigen B.J., Svenson K.L., Svenson K.L. The mouse as a model for human biology: A resource guide for complex trait analysis. Nat. Rev. Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Petkov P.M., Ding Y., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Ding Y., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., Asquith S., Crew V., Johnson K.A., Robinson P., Crew V., Johnson K.A., Robinson P., Johnson K.A., Robinson P., Robinson P., et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov P.M., Graber J.H., Churchill G.A., DiPetrillo K., King B.L., Paigen K., Graber J.H., Churchill G.A., DiPetrillo K., King B.L., Paigen K., Churchill G.A., DiPetrillo K., King B.L., Paigen K., DiPetrillo K., King B.L., Paigen K., King B.L., Paigen K., Paigen K. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 2005;1:e33. doi: 10.1371/journal.pbio.0050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen T.L., Kirby A., Hammer R.P., Purcell S., O'Leary S.B., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Kirby A., Hammer R.P., Purcell S., O'Leary S.B., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Hammer R.P., Purcell S., O'Leary S.B., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Purcell S., O'Leary S.B., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., O'Leary S.B., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Singer J.B., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Hill A.E., Nadeau J.H., Daly M.J., Sklar P., Nadeau J.H., Daly M.J., Sklar P., Daly M.J., Sklar P., Sklar P. Two quantitative trait loci for prepulse inhibition of startle identified on mouse chromosome 16 using chromosome substitution strains. Genetics. 2005;171:1895–1904. doi: 10.1534/genetics.105.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockley K.R., Churchill G.A., Churchill G.A. Gene expression analysis of mouse chromosome substitution strains. Mamm. Genome. 2006;17:598–614. doi: 10.1007/s00335-005-0176-y. [DOI] [PubMed] [Google Scholar]
- Singer J.B., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., Conti D.V., Witte J.S., Lander E.S., Witte J.S., Lander E.S., Lander E.S., et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Singer J.B., Hill A.E., Nadeau J.H., Lander E.S., Hill A.E., Nadeau J.H., Lander E.S., Nadeau J.H., Lander E.S., Lander E.S. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics. 2005;169:855–862. doi: 10.1534/genetics.104.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa L.D., Alvord W.G., Bickmore W.A., Jenkins N.A., Copeland N.G., Alvord W.G., Bickmore W.A., Jenkins N.A., Copeland N.G., Bickmore W.A., Jenkins N.A., Copeland N.G., Jenkins N.A., Copeland N.G., Copeland N.G. Interspecific backcross mice show sex-specific differences in allelic inheritance. Genetics. 1991;128:813–821. doi: 10.1093/genetics/128.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova R., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Buckiova D., Kyselova V., Divina P., Forejt J., Kyselova V., Divina P., Forejt J., Divina P., Forejt J., Forejt J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Takada T., Mita A., Maeno A., Sakai T., Shitara H., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Mita A., Maeno A., Sakai T., Shitara H., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Maeno A., Sakai T., Shitara H., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Sakai T., Shitara H., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Shitara H., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Kikkawa Y., Moriwaki K., Yonekawa H., Shiroishi T., Moriwaki K., Yonekawa H., Shiroishi T., Yonekawa H., Shiroishi T., Shiroishi T. Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res. 2008 doi: 10.1101/gr.7175308. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Masly J.P., Araripe L., Ke Y., Hartl D.L., Masly J.P., Araripe L., Ke Y., Hartl D.L., Araripe L., Ke Y., Hartl D.L., Ke Y., Hartl D.L., Hartl D.L. A sex-ratio meiotic drive system in Drosophilasimulans. I: An autosomal suppressor. PLoS Biol. 2007;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtulec Z., Mnukova-Fajdelova M., Hamvas R., Gregorova S., Mayer W., Lehrach H., Vincek V., Forejt J., Klein J., Mnukova-Fajdelova M., Hamvas R., Gregorova S., Mayer W., Lehrach H., Vincek V., Forejt J., Klein J., Hamvas R., Gregorova S., Mayer W., Lehrach H., Vincek V., Forejt J., Klein J., Gregorova S., Mayer W., Lehrach H., Vincek V., Forejt J., Klein J., Mayer W., Lehrach H., Vincek V., Forejt J., Klein J., Lehrach H., Vincek V., Forejt J., Klein J., Vincek V., Forejt J., Klein J., Forejt J., Klein J., Klein J. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm. Genome. 1997;8:312–316. doi: 10.1007/s003359900430. [DOI] [PubMed] [Google Scholar]
- Trachtulec Z., Mihola O., Vlcek C., Himmelbauer H., Paces V., Forejt J., Mihola O., Vlcek C., Himmelbauer H., Paces V., Forejt J., Vlcek C., Himmelbauer H., Paces V., Forejt J., Himmelbauer H., Paces V., Forejt J., Paces V., Forejt J., Forejt J. Positional cloning of the hybrid sterility 1 gene: Fine genetic mapping and evaluation of two candidate genes. Biol. J. Linnean Soc. 2005;84:637–641. [Google Scholar]
- Wu C.-I., Hollocher H., Hollocher H. Subtle is nature: The genetics of species differentiation and speciation. In: Howard D., Berlocher S., Berlocher S., editors. Species and speciation. Oxford University Press, Oxford; New York: 1998. pp. 339–351. [Google Scholar]
- Yang H., Bell T.A., Churchill G.A., de Pardo-Manuel Villena F., Bell T.A., Churchill G.A., de Pardo-Manuel Villena F., Churchill G.A., de Pardo-Manuel Villena F., de Pardo-Manuel Villena F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Zhu R., Ji Y., Xiao L., Matin A., Ji Y., Xiao L., Matin A., Xiao L., Matin A., Matin A. Testicular germ cell tumor susceptibility genes from the consomic 129.MOLF-Chr19 mouse strain. Mamm. Genome. 2007;18:584–595. doi: 10.1007/s00335-007-9036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]