Crystal structure of Yersinia enterocolitica type III secretion chaperone SycT (original) (raw)
Abstract
Pathogenic Yersinia species use a type III secretion (TTS) system to deliver a number of cytotoxic effector proteins directly into the mammalian host cell. To ensure effective translocation, several such effector proteins transiently bind to specific chaperones in the bacterial cytoplasm. Correspondingly, SycT is the chaperone of YopT, a cysteine protease that cleaves the membrane-anchor of Rho-GTPases in the host. We have analyzed the complex between YopT and SycT and determined the structure of SycT in three crystal forms. Biochemical studies indicate a stoichometric effector/chaperone ratio of 1:2 and the chaperone-binding site contains at least residues 52–103 of YopT. The crystal structures reveal a SycT homodimer with an overall fold similar to that of other TTS effector chaperones. In contrast to the canonical five-stranded anti-parallel β-sheet flanked by three α-helices, SycT lacks the dimerization α-helix and has an additional β-strand capable of undergoing a conformational change. The dimer interface consists of two β-strands and the connecting loops. Two hydrophobic patches involved in effector binding in other TTS effector chaperones are also found in SycT. The structural similarity of SycT to other chaperones and the spatial conservation of effector-binding sites support the idea that TTS effector chaperones form a single functional and structural group.
Keywords: chaperone, effector, SycT, type III secretion, Yersinia, YopT
Many Gram-negative pathogens of animals, plants, or insects, including the human-pathogenic Yersinia spp. (Y. pestis, Y. pseudotuberculosis, Y. enterocolitica), use the type III secretion pathway to deliver effectors into the eukaryotic host cell (Cornelis and Van Gijsegem 2000; Ghosh 2004). A 70-kb virulence plasmid (pYV) codes for the Ysc TTS system in Y. enterocolitica (Cornelis et al. 1998; Cornelis 2002), a food-borne pathogen causing gastroenteritis and mesenteric lymphadenitis (Bottone 1999). The syringe-shaped Ysc injectisome spans the inner membrane, the peptidoglycan layer, and the outer membrane of the bacterium (Marlovits et al. 2004). The pathogen uses this injectisome to translocate effector Yops (Y ersinia outer proteins) directly into the host cell, where they interfere with host proteins and impair cell functions including transcription, inflammatory signaling, and actin cytoskeleton dynamics. Six translocated Yop effectors have been described (for review, see Navarro et al. 2005). YopH dephosphorylates phospho-tyrosine residues in focal adhesion complexes and in a signaling complex in macrophages. YopE and YopT target small Rho-GTPases controlling actin polymerization. YopE acts as a GTPase activating protein (GAP) and YopT as a cysteine protease (see below). YopP (YopJ in Y. pseudotuberculosis and Y. pestis) impairs the MAP kinase and NF-κB signalling pathways, thereby impeding the release of proinflammatory cytokines and inducing apoptosis in macrophages. The substrate of the serine/threonine kinase YopO (YpkA in Y. pseudotuberculosis and Y. pestis) is still unknown, as well as the targets of YopM, which locates to the nucleus of the host cell. Effective translocation of some of the TTS effectors and components is regulated by a family of specialized chaperones, which form specific complexes with their cognate substrate in the bacterial cytoplasm. These TTS chaperones, in contrast to other chaperones such as GroEL or heat shock proteins, do not bind or hydrolyze nucleotides (Wattiau et al. 1996). Despite several common properties, such as a low molecular mass (Mr~15,000) and an acidic isoelectric point, they have low amino acid sequence similarity (Wattiau et al. 1994). Functions proposed for TTS chaperones include preventing effector agglomeration by binding the aggregation-prone N-terminal region of the effector; keeping the effector in a partially unfolded, secretion-competent state; and helping to regulate the TTS system (for reviews, see Francis et al. 2002; Feldman and Cornelis 2003; and Parsot et al. 2003). Based on their substrate, TTS chaperones can be grouped into three categories. The first class guides the transfer of one or more effector proteins, and several members have been characterized structurally. A second class promotes the transport of TTS components that form a pore in the host cell membrane (translocators). The third category comprises chaperones binding to other proteins of the injection apparatus. The first structure of a member of the third category of chaperones has recently been solved (Yip et al. 2005).
Yop effectors are modular proteins. They bear an N-terminal secretion/translocation signal of ~20 amino acids (Sory et al. 1995) not cleaved after translocation. The chaperone-binding domain in chaperone-complexed effectors generally lies within residues 50–150 (Woestyn et al. 1996; Tampakaki et al. 2004). In Yersiniae three of the translocated effectors (YopE, YopH, and YopT) possess a specific chaperone (Wattiau and Cornelis 1993; Wattiau et al. 1994; Iriarte and Cornelis 1998), whereas no chaperones have as yet been identified for YopM, YopP, or YopO (Trülzsch et al. 2003).
The effector YopT (322 amino acids, 36 kDa) (Iriarte and Cornelis 1998) is a member of a new family of papain-like cysteine proteases with the conserved catalytic triad comprising Cys139, His258, and Asp274 (Shao et al. 2002; Zhu et al. 2004). Residues 75–318 are required for proteolytic activity (Sorg et al. 2003). After translocation into the host cell, YopT cleaves the C-terminal, prenylated cysteine of membrane-anchored Rho-GTPases, thereby releasing the GTPase into the host cytosol (Zumbihl et al. 1999; Shao et al. 2002; Äpfelbacher et al. 2003; Sorg et al. 2003). The lack of Rho-GTPase–induced actin polymerization signal leads to the disintegration of the host actin stress fibres (Iriarte and Cornelis 1998) and contributes to the yersinial resistance to phagocytosis by inhibiting the formation of the actin-rich phagocytotic cup (Äpfelbacher et al. 2003). The specific effector chaperone for YopT is SycT (Mr~15,000; pI 4.6) (Iriarte and Cornelis 1998).
In this work, we present a structural and functional characterization of the chaperone SycT from Y. enterocolitica, including complex formation studies with its effector YopT.
Results and Discussion
SycT forms a dimer in solution
Full-length SycT (SycT1–130) was produced heterologously in Escherichia coli as a GST fusion protein. After cleavage with the site-specific tobacco etch virus (TEV) protease, the molecular mass of SycT was found to be ~30 kDa (gel permeation chromatography) and ~36 kDa (dynamic light scattering). SycT thus forms a homodimer in solution (theoretical mass, 30.6 kDa). Dimerization is a common property of TTS effector chaperones (Wattiau and Cornelis 1993; Parsot et al. 2003; Singer et al. 2004; Van Eerde et al. 2004). MALDI-TOF analysis of native SycT stored at 4°C for 4 wk indicated a molecular mass of 14,478 Da rather than 15,342 Da, the theoretical mass of TEV-cleaved SycT. N-terminal sequencing revealed an intact N terminus, implying that SycT had been cleaved C-terminally, yielding a chaperone truncated to residues 1–122 (theoretical mass 14,483 Da). For structural analysis and to facilitate crystallization, the _syc_T gene was accordingly truncated to produce SycT1-122.
Structure determination
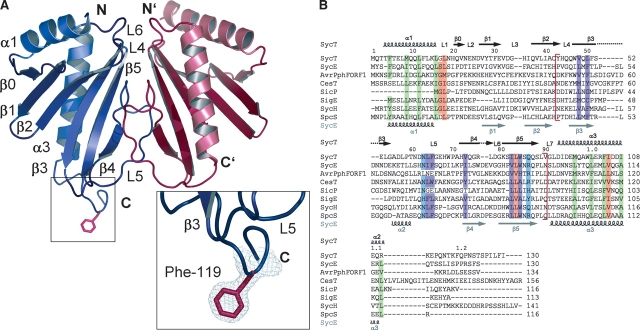
Native full-length SycT crystallized in 5% isopropanol, 0.1 M sodium citrate (pH 5.6), and 21% polyethylene glycol 4000 at 4°C as rhombic plates (100 μm) in the monoclinic space group P21. Attempts to solve the structure of SycT by molecular replacement using the structures of other TTS chaperones failed. Soaking crystals with mercury, platinum, osmium, or bromide caused crystal cracking and loss of diffraction or crystals did not incorporate the heavy atom. Seleno-methionine (SeMet)–substituted SycT122 was crystallized in 1.6 M ammonium sulfate, 0.1 M CAPS (pH 10.5), and 0.15 M lithium sulfate (plate-like crystal form #1, space group P212121). An unrelated crystal form #2 (P62 ) was obtained from 1.8 M ammonium sulfate, 0.1 M sodium bicarbonate (pH 10.5), and 0.05 M magnesium chloride with hexagonal rod-like crystals growing to a size of 200 × 150 μm. The structure of SycT122 was determined in crystal form #2 by selenium single-wavelength anomalous dispersion (SAD) and refined to a resolution of 1.9 Å. SycT122 crystal form #1 and the structure of native SycT were then solved by molecular replacement to a resolution of 2.0 Å and 1.8 Å, respectively. In all three crystal forms, the asymmetric unit bears one homodimer (Table 1; Fig. 1A ▶). The three crystal structures are very similar; e.g., the Cα-atoms of SycT122 monomers have a root mean square deviation (RMSD) of 0.6 Å in #1 and 0.7 Å in #2. Differences are confined to the flexible N- and C-terminal parts with the exception of residues 20–28 of one of the native SycT monomers. Well-defined electron density is observed for residues 3–130 (A), 2–117, and 125–130 (B) of native SycT; residues 1–114 of both monomers in SycT122 crystal form #1; and residues 3–122 (A) and 1–113 (B) in #2.
Table 1.
Data collection and refinement statistics
| SycT122 | |||
|---|---|---|---|
| native | crystal form #1 | crystal form #2 | |
| Data collection statistics | |||
| Space group | P21 | P212121 | P62 |
| Unit cell dimensions (Å), (°) | a=34.1, b=79.6, c=52.1 | a=51.5, b=63.1, c=74.4 | a=b=92.0, c=55.4 |
| α=γ=90, β=101.21 | α=β=γ=90 | α=β=90, γ=120 | |
| Data set | Peak | ||
| Wavelength (Å) | 1.05 | 0.9795 | 0.97957 |
| Resolution (Å) | 79.5–1.83 (1.86–1.83) | 48.1–2.0 (2.1–2.0) | 79.0–1.9 (2.0–1.9) |
| Mosaicity (°) | 0.5 | 0.25 | 0.14 |
| Completeness (%) | 97.6 (83.5) | 98.4 (96.4) | 99.5 (96.4) |
| (Anomalous) redundancy | 2.8 | 3.9 | 5.8 |
| Observations | 65,888 | 118,050 | 237,251 |
| Unique reflections | 23,529 | 16,331 | 21,134 |
| I/σ (I) | 14.8 (2.1) | 12.3 (4.6) | 16.6 (5.4) |
| Rsym, Rmeas | 5.6 (31.6)a | 8.0 (37.3)b | 7.2 (39.8)b |
| Phasing | Molecular replacement | Molecular replacement | SAD (selenium) |
| Number of sites | 6 sites in a dimer | ||
| Molecules per asymmetric unit | 2 | 2 | 2 |
| Solvent content (%) | 46 | 41 | 45 |
| Refinement statistics | |||
| R / Rfreec (%) | 18.4 / 23.3 | 21.9 / 28.4 | 19.6 / 23.6 |
| Atoms protein/solvent | 2131 / 504 | 1950 / 358 | 1908 / 477 |
| RMSD | |||
| Bonds (Å) / Angles (°) | 0.015 / 1.5 | 0.018 / 1.85 | 0.019 / 1.98 |
| Ramachandran plot | |||
| Residues in allowed / additionally allowed / generously allowed regions (%) | 94.1 / 5.9 / 0.0 | 86.1 / 13.5 / 0.5 | 88.2 / 10.4 / 1.4 |
| Residues in disallowed regions (%) | 0.0 | 0.0 | 0.0 |
| B factor (Å2) average / Wilson B | 28.2 / 28.5 | 20.6 / 31.6 | 32.2 / 32.1 |
| Protein / solvent | 27.8 / 33.8 | 21.0 / 26.2 | 31.0 / 40.5 |
Figure 1.
(A) Ribbon diagram of the structure of Yersinia enterocolitica chaperone SycT (SycT122 crystal form #2, monomer A in blue, monomer B in red). The α-helix and β-strand numbering corresponds to that of earlier TTS chaperone structures. (Insets) The well-defined electron density of the protruding Phe119 side chain (red sticks) of monomer A. (B) Multiple sequence alignment of SycT with related TTS effector chaperones. Conserved hydrophobic residues (green) in SycT helices α1 (Phe5, Met9, Leu12, and Leu16) and α3 (Met97, Leu101, Phe104, and Ile108); SycT dimerization interface (red brackets) involves residues Tyr43, His44, Trp47, Gln49, Phe51, Asp62, Asn63, Leu64, Phe65, Trp69, Pro70, Ala71, Val73, Gly75, Arg76, Leu77, Trp84, Gln87, and Val90. The sequence section containing the dimerization-mediating residues includes also conserved polar residues (Asn63 and Gln86, in blue) and hydrophobic residues (Val48, Leu50, Leu64, Phe65, Val73, Ile82, and Trp84, in purple); other conserved residues (Gly17, Leu18, Leu83, and Ile105) are highlighted in orange. Sequences were aligned with ClustalW and adjusted by analyzing the SycT structure. Secondary structure elements for SycT are indicated above and those for SycE below the sequences. Y. enterocolitica SycT (GenBank AAD16809), Yersinia pseudotuberculosis SycE (NC_006153), Pseudomonas syringae AvrPphF ORF1 (AAF67148), Escherichia coli CesT (P58233), Salmonella typhimurium SicP (AAC38655), Salmonella enterica SigE (NP455587), Yersinia pestis SycH (NP052425), and putative chaperone Orf1 from Pseudomonas aeruginosa, abbreviated to SpcS (AAA66490).
SycT broadly shares the fold of other effector-binding TTS chaperones
Structurally, a SycT monomer comprises two α-helices packed against one side of a twisted six-stranded, anti-parallel β-sheet (SycT122 #2) (Fig. 1A ▶). It thereby shares most characteristic features of the globular fold common to effector-binding TTS chaperones. Despite low sequence identity of 9%–21%, the RMSD for common Cα-positions (79–109 aligned residues) is <2.7 Å compared with other TTS chaperones such as SicP and SigE from Salmonella sp. (Luo et al. 2001; Stebbins and Galan 2001), SycE and SycH from Yersinia sp. (Birtalan and Ghosh 2001; Evdokimov et al. 2002; Trame and McKay 2003; Phan et al. 2004), CesT from enterohemorrhagic E. coli (Luo et al. 2001), and AvrPphF Orf1 from Pseudomonas syringae (Singer et al. 2004). SycT shares the highest sequence identity with the structurally uncharacterized Orf1 from Pseudomonas aeruginosa (abbreviated as SpcS) believed to be the chaperone of ExoS (Frithz-Lindsten et al. 1997). Apart from the overall similarity, SycT displays some unique and distinct features. The depression between the N-terminal α-helices α1 and the adjoining loops L4 and L6, typical for other effector chaperones, is less pronounced in the SycT dimer (Fig. 1A ▶). Instead, SycT exhibits a pronounced groove between loops L5 on the opposite dimer face. Overall, SycT is thus quite compact compared with the heart-shaped outline of other chaperones (Fig. 2 ▶; Fig. 4A ▶, below).
Figure 2.
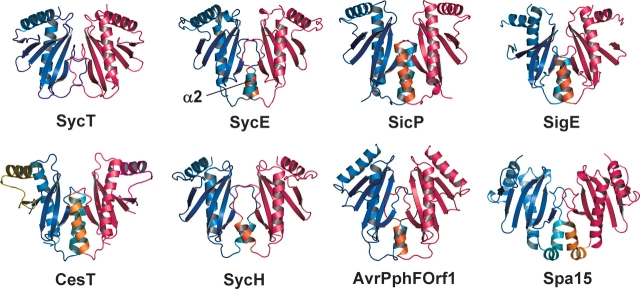
SycT lacks the dimerization helix α2. Ribbon models of Yersinia enterocolitica SycT, Yersinia pseudotuberculosis SycE (PDB identifier 1L2W; Birtalan et al. 2002), Salmonella typhimurium SicP (1JYO; Stebbins and Galan 2001), Salmonella enterica SigE (1K3S; Luo et al. 2001), Yersinia pestis SycH (1TTW; Phan et al. 2004), Pseudomonas syringae pv. phaseolicola AvrPphF ORF1 (1S28; Singer et al. 2004), and Shigella flexneri Spa15 (1R9Y; Van Eerde et al. 2004). The monomers in a dimer are colored blue and red. The helices α2 are highlighted in orange and turquoise.
Figure 4.
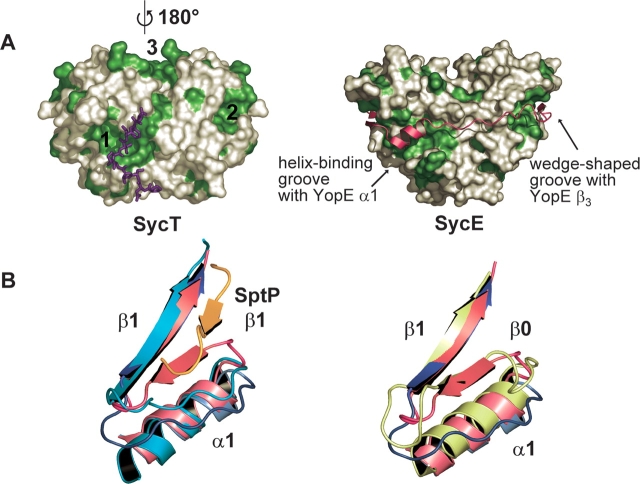
Effector-binding sites in SycT. (A) Two extended hydrophobic patches (1, 2) located on the surface of SycT (left, SycT122 crystal form #1) are duplicated by symmetry to yield four patches per dimer. In the native SycT crystal, patch 1 forms a hydrophobic interaction with residues 125–130 of the C-terminal peptide (violet sticks). Patches 1 and 2 correspond to those of a SycE dimer involved in effector binding (right; SycE in complex with the CBD of YopE [YopE23–78, red]). SycT has an additional hydrophobic patch (3) not observed in SycE. Surfaces corresponding to hydrophobic side chains are depicted in green. (B) A shallow groove alongside hydrophobic patch 2 is involved in effector binding in SicP. In SycT and Spa15, this groove is filled by the additional strand β0 formed by residues connecting α1 and β1. Both figures show the two superimposed monomers of native SycT: Monomer one (red) displays the additional strand β0, while in the other monomer (blue) β0 has changed into a loop and moved toward α1, opening a shallow groove. (Left) The SicP/SptP complex is superimposed on SycT (SicP in green, SptP in orange). In SicP, β1 and the loop connecting α1 with β1 form a groove occupied by strand β2 of the effector SptP, extending the chaperone β-sheet. Should YopT follow the same path on SycT as SptP does on SicP, it would partially overlap with strand β0 of the red SycT monomer but not with the conformationally rearranged blue monomer. (Right) Overlay of Spa15 (light green) and SycT. Although the connecting loop between α1 and β1 is four residues longer in Spa15 than in SycT, the structural organization is similar to the red SycT monomer.
SycT lacks the dimerization helix
All previously determined structures of effector-binding TTS chaperones display three α-helices packing against the β-sheet. Of these, α2 is extensively involved in dimerization (Fig. 2 ▶) (Parsot et al. 2003). Strikingly, α-helix α2 is consistently absent in all three crystal forms of SycT. Instead, dimerization involves hydrophobic and polar residues exclusively located in loops and β-strands (Fig. 1B ▶). The SycT dimerization interface involves some residues of the region between residues 43 and 90. In particular, residues from strands β4 and β5 are involved as are loops L4, L6, and the extended loop L5 (Fig. 1 ▶). The accessible surface area buried per monomer is 1100 Å2 or ~15% of the total surface area. This is similar to the buried surface of other TTS effector chaperones ranging from 900 to 1200 Å2.
A closed cavity at the dimer interface
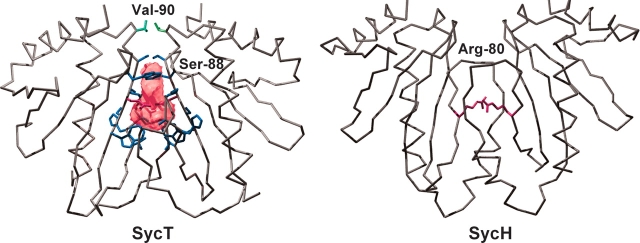
The dimerization interface of SycT harbors a cavity of 150 Å3 (Fig. 3 ▶). Five (native SycT, SycT122 #2) or six (#1) water molecules trapped within the cavity form H-bonds to polar residues. Symmetrically positioned Ser88 residues from both monomers restrict entry to the cavity and are mutually H-bonded in the native structure and #1. Both Val90 residues in loop L6 shield the cavity from the outside (Fig. 3 ▶). A polar cavity located at the dimerization interface of TTS effector chaperones was first described for SycE (Evdokimov et al. 2002). Fully enclosed cavities, as in SycT, are also present in Spa15 and AvrPphF Orf1 dimers (Singer et al. 2004; Van Eerde et al. 2004). The dimerization interfaces of other TTS effector chaperones, e.g., SycE, YscB/SycN (Schubot et al. 2005), or SycH (Phan et al. 2004), reveal similar yet open, solvent-accessible cavities (Fig. 3 ▶, right). Compared with other solvated intersubunit cavities (Hubbard and Argos 1994) that of SycT is quite large, with ~100 Å2 or 9% of the dimerization interface per monomer not being involved in direct protein –protein interactions. Conservation of the cavity, open or closed, among TTS effector-binding chaperones is strongly suggestive of a biological role. In SycE, SycH, and SycN/YscB, the three chaperones from Y. pestis for which structural information is available, two arginines create a positively charged patch inside the cavity (Evdokimov et al. 2002; Phan et al. 2004; Schubot et al. 2005). These arginines are conserved among many of the effector chaperones and were therefore proposed to be functionally relevant, possibly for interaction with the type III secretion machinery (Phan et al. 2004; Schubot et al. 2005). The lack of charged residues in the cavity of SycT means that they are unlikely to be a general point of interaction between TTS effector chaperones and the secretion machinery.
Figure 3.
Cavity in the SycT dimerization interface. A polar cavity is enclosed in the dimerization interface of SycT (crystal form #1, cavity as pink surface model and blue side chains of residues Trp47, Pro70, Ala71, His72, Val73, Leu83, Trp84, Ser85, and Ser88, and red side chain for Gln86). Both Val90 residues (green) shield the cavity from the outside. In contrast, the cavity in SycH is accessible and possesses a positive charge (Arg80, red side chain).
Effector-binding sites in SycT are spatially conserved
The surface of the SycT homodimer displays three types of hydrophobic patches. Patches 1 and 2, distal to the twofold symmetry axis, are symmetrically duplicated on the dimer surface. Patch 1, near the dimer interface, comprises residues Leu64, Phe65, Gly66, and Trp69 of one monomer, and Val27, Leu39, Ile40, Ala41, Tyr43, Trp47, Gln49, Phe51, Gly75, and Ile82 of the other. Patch 2, composed of residues Leu16, Gly17, Leu18, Phe30, Val32, and Ile105, is formed by the two amphipathic α-helices. A third hydrophobic patch (patch 3) stretches between both N-terminal helices α1 (residues Met0, Leu8, Leu89, Val90, Gly91, Leu92, and Ile94) (Fig. 4A ▶, left). Patches 1 and 2 are present at roughly the same position in other effector chaperones and were initially predicted to be involved in effector binding (Birtalan and Ghosh 2001). This hypothesis was later confirmed by the crystal structures of several effector/chaperone complexes such as YopE23-78/SycE, SptP36-139/SicP, and YopN23-273/SycN-YscB complexes (Stebbins and Galan 2001; Birtalan et al. 2002; Schubot et al. 2005). In these structures, the N-terminal region of the effector wraps around the chaperones, displaying only secondary but no tertiary structure. In the YopE/SycE complex, both patches 1 bind α-helices, while both patches 2 bind β-strands of the YopE chaperone-binding domain (Fig. 4A ▶, right). The conserved position of patches 1 and 2 in SycT suggests a similar mode of effector binding. The details of YopT/SycT interaction will presumably differ from those of other effector–chaperone pairs. The third hydrophobic patch of SycT may indicate an enlarged contact area and the effector YopT may follow a different path wrapping around SycT.
Significant differences between SycT and the other effector- binding chaperones (except possibly Spa15) are observed in the vicinity of patch 2. In most chaperones studied to date, patch 2 consists of a pocket, formed by conserved hydrophobic residues from the α-helices α1 and α3 and a shallow groove between β-strand β1 and the loop connecting the N-terminal α-helix α1 with β1. The effectors YopE, SptP, and YopN bind to patch 2 of their cognate chaperone, filling this shallow groove and extending the chaperone β-sheet by an additional strand along side strand β1 (Fig. 4B ▶, left). In five of the six crystallographically independent monomers of SycT, the residues connecting α1 and β1 form an additional β-strand β0 anti-parallel to β1 that occupies the shallow groove (Fig. 4B ▶, left). This strongly resembles the situation in Spa15 (Van Eerde et al. 2004) (Fig. 4B ▶, right). In one monomer of the native SycT dimer, these connecting residues do not form β0. Instead they form a loop flipped toward α1, thereby opening the abovementioned groove. This conformational rearrangement of β0 may be required for binding of YopT, although no conformational changes are observed upon effector binding in SycE (Birtalan et al. 2002).
Interaction of the flexible C terminus with a hydrophobic patch
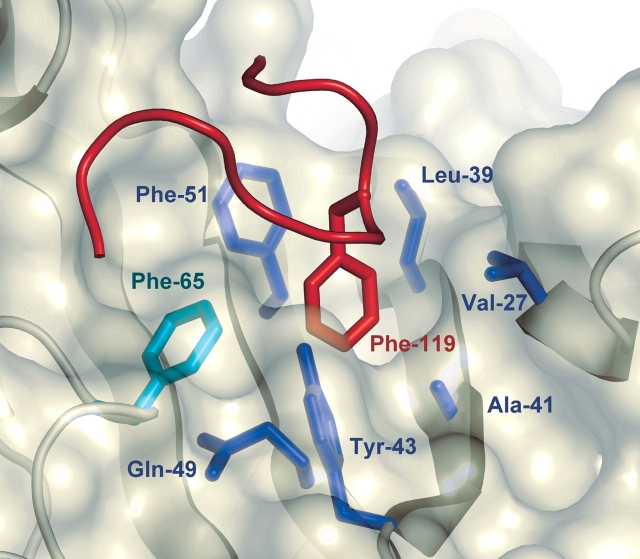
In full-length SycT, the hydrophobic peptide SPILFI (residues 125–130) of both monomers interacts with one hydrophobic patch 1 of the primary dimer and of a symmetry-related dimer at the same time, thereby mediating extensive crystal contacts (Fig. 4A ▶, left). Hydrophobic patch 1 harbors a groove (Phe65 in loop L5 from one monomer; Val27 from β2; Leu39, Ala41, and Tyr43 from β3; Gln49 and Phe51 from β4 in the other monomer). In crystal form #2 of truncated SycT122, this hydrophobic groove of one monomer is occupied by Phe119 from the C-terminal peptide of a neighboring monomer in the crystal (Figs. 1 ▶, 5 ▶), sustaining an important crystal contact. These C-terminal residues (115–122) are disordered in monomer B (#2) and in both monomers of #1. Residues 115–122 thus seem to be intrinsically flexible. Depending on the crystal packing environment, these C-terminal stretches in SycT are ordered by fortuitously binding to patch 1 of a neighboring molecule. This resembles the situation in one of the SycE structures where the flexible C terminus similarly binds to the hydrophobic patch 1 of an adjacent dimer (Trame and McKay 2003).
Figure 5.
Hydrophobic interactions of the C terminus in SycT. Hydrophobic pocket in patch 1. Phe119 from the C-terminal peptide of a symmetry-related monomer of SycT122 (#2; backbone of loop in red) fills a hydrophobic pocket in patch 1 (translucent surface) created by residues Phe65 (green, monomer one) and Val27, Leu39, Ala41, Tyr43, Gln49, and Phe51 (blue, monomer two).
Mapping of the YopT chaperone-binding domain
Complex formation between YopT and full-length SycT was experimentally verified by gel permeation chromatography: Two YopT species were copurified with SycT, full-length YopT, and a stable fragment truncated N-terminally by 13 amino acids. Full-length and truncated YopT coeluted with SycT as one complex with an apparent molecular mass of ~65 kDa (data not shown). This indicates a YopT:SycT stoichiometry of 1:2, similar to that of other effector/chaperone complexes such as YopE/SycE (Birtalan et al. 2002). The molecular mass of ~84 kDa determined by dynamic light scattering suggests an elongated shape of the YopT/SycT complex (data not shown).
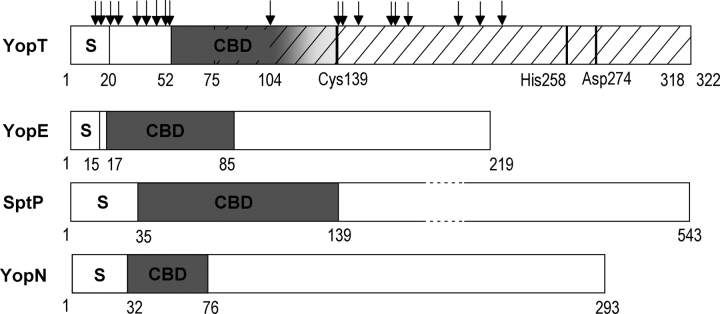
To determine the chaperone-binding site in YopT, the YopT/SycT complex was subjected to a protease-protection assay and digested with different proteases (trypsin, thermolysin, subtilisin, papain, chymotrypsin, and endoproteinase lys-C). Under the conditions used, SycT was stable except for the eight C-terminal residues. YopT was cleaved, and 28 degradation fragments detectable on SDS gels were N-terminally sequenced. Mapping of the protease cleavage sites revealed numerous sites in the region N-terminal of residue 52 and C-terminal of residue 140 with a single site at residue 104 (Fig. 6 ▶). Protection of residues 52–103 of YopT from proteolytic digestion in complex with SycT suggests this region to be protected by binding to the chaperone. This is supported by similar experiments defining the chaperone-binding domains of YopE and SptP (Stebbins and Galan 2001; Birtalan et al. 2002). The chaperone-binding domain of YopT most likely does not include residues N-terminal of amino acid 52, while the C-terminal boundary may be located after residue 104. Based on our findings, we propose a more detailed domain organization for YopT: The N-terminal ~20 amino acids of the effector comprise the secretion/translocation signal. The chaperone- binding domain of YopT covers at least residues 52– 103. The minimal YopT fragment capable of substrate binding and proteolytic cleavage includes residues 75–318 (Fig. 6 ▶) (Sorg et al. 2003).
Figure 6.
Domain organization of YopT. Domain borders are designated by residue numbers. S indicates secretion signal; CBD, chaperone- binding domain. YopT: YopT region (residues 75–318, hatched) capable of substrate binding and proteolytic cleavage (catalytic triad Cys139, His258, and Asp274). Sites susceptible to protease cleavage (arrows) were identified by N-terminal sequencing of fragments produced by limited proteolysis of the complex YopT/SycT. The accumulation of sites N-terminally of amino acid 52 and C-terminally of 140 indicates the CBD to comprise at least residues 52–103. The C-terminal border of the CBD may lie after the single cleavage site at residue 104 (indicated as a gradient of gray). For comparison, the CBD of other TTS effectors are plotted below. YopE from Yersinia pseudotuberculosis, SptP from Salmonella typhimurium, and YopN from Y. pestis (Stebbins and Galan 2001; Birtalan et al. 2002; Schubot et al. 2005).
Conclusions
The crystal structure of SycT, the chaperone of the TTS effector YopT, reveals that it shares the global fold of other effector-binding chaperones. Nevertheless, some distinct differences such as lacking the dimerization α-helix set SycT apart from other chaperones. Although the location of two hydrophobic patches involved in effector binding in other chaperones is conserved in SycT, the precise mode of interaction may be different, as SycT possesses an additional strand β0 that extends the β-sheet alongside β1. Other chaperones bind their effectors (YopE, SptP, and YopN) by allowing these to extend the chaperone β-sheet by a β-strand adjacent to strand β1. In one crystal packing of SycT β0 undergoes a conformational change and opens a groove. This may indicate a mechanism for effector binding.
Despite low sequence conservation, TTS effector-binding chaperones share a common fold and effector binding mode. In addition to binding multiple substrates (Parsot et al. 2003), Spa15 is structurally distinct from other effector-binding chaperones because of the relative orientation of the monomers in the dimer and the presence of an additional β-strand (Van Eerde et al. 2004). SycT shares the additional strand β0 with Spa15 yet binds a single effector. This indicates that chaperones such as Spa15 that bind multiple effectors are structurally not set apart from other TTS chaperones. Instead effector-binding chaperones form a single functional and structural group.
Materials and methods
Expression vector construction
Gene sequences of _syc_T and _yop_T were amplified from the Yersinia enterocolitica virulence plasmid pYVe227 by PCR. The _syc_T sequence was cloned into pET-M-30 (from G. Stier, EMBL Heidelberg, Germany) using restriction endonucleases NcoI and NotI, introducing an additional glycine codon (residue Gly1) after the _syc_T start codon ATG (residue Met0). N-terminal sequencing and MALDI-TOF analysis of SycT revealed that the eight C-terminal residues are susceptible to proteolysis, resulting in the stable fragment SycT1-122. Hence, for a second construct used for crystallization, only codons 1–122 of sycT were cloned into the expression vector. To coexpress _yop_T and _syc_T, a bicistronic construct was generated. The DNA sequence encoding full-length SycT was cloned into pET-M-30 as a glutathion-S-transferase (GST) fusion followed by a consensus ribosome binding site and the untagged _yop_T sequence. Both expression vectors were checked by sequencing.
Protein expression and purification
YopT and full-length GST-tagged SycT were produced by coexpression in E. coli BL21(DE3) codon plus RIL (Stratagene). Native, full-length SycT was produced as a GST-fusion protein in the same strain. SeMet-substituted GST-tagged SycT122 protein was produced as described using the strain from above (Guerrero et al. 2001). Cells were grown at 37°C to an OD600 of 0.7. Recombinant gene expression was induced with 1 mM isopropyl-β-D-thiogalactoside overnight at 20°C. Cells were resuspended in ice-cold lysis buffer (50 mM phosphate [pH 8.0], 150 mM NaCl, 10 mM β-mercaptoethanol) and lysed using a French press (SLM Aminco). Clarified lysates were applied to glutathione-loaded Sepharose resin (Amersham Biosciences) and washed with 20 mM Tris-HCl (pH 8.0), 150 NaCl, and 5 mM dithiothreitol (DTT). The GST-tag was cleaved on column at 16°C overnight using 1:60 TEV protease. His-tagged TEV protease was removed by Ni2+ loaded agarose (Qiagen). The TEV protease cleavage site (Glu-Asn-Leu-Tyr-Phe-Gln | Gly-Ala) introduces two additional amino acids (Gly-Ala) at the N terminus of the cleaved protein. The YopT/SycT complex was further purified by gel permeation chromatography (Superdex-75, Amersham Biosciences) with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM DTT as running buffer. Native SycT and SycT122 were purified by anion exchange chromatography (MonoQ, Amersham Biosciences) using a linear NaCl gradient in 20 mM Tris-HCl (pH 8.0) and gel permeation chromatography as above. Native SycT and SycT122 were dialyzed against 20 mM Tris-HCl (pH 8.0), 10 mM NaCl, and 1 mM DTT; subjected to MALDI-TOF analysis to check the protein size and complete incorporation of SeMets (three per molecule), respectively; and concentrated to ~12 mg/mL for crystallization trials.
Molecular mass determination
Gel permeation chromatography as described above, calibrated with standard proteins from low- and high-molecular-mass gel filtration calibration kits (Amersham Biosciences), and dynamic light scattering (DynaPro 801TC system, ProteinSolutions) were used to determine the molecular mass of SycT and the complex YopT/SycT.
Limited proteolysis and domain mapping
Purified YopT/SycT complex (~8 mg/mL) was digested on ice with 1:70 trypsin, chymotrypsin, papain, and endoproteinase Lys-C, as well as with 1:350 subtilisin and thermolysin. SycT mostly resisted proteolytic cleavage. Fragments were separated by SDS-PAGE, blotted onto PVDF membrane, and subjected to N-terminal sequencing.
Crystallization, data collection, and processing
Optimizing birefringent spherulites of native SycT using hanging- drop vapor diffusion at 4°C and a crystallization cocktail of 5% isopropanol, 0.1 M sodium citrate (pH 5.6), and 21% polyethylene glycol 4000 finally yielded orthorhombic crystals. For SeMet-substituted SycT122, initial screening in 96-well format using diverse commercially available screens and nano-drop pipetting resulted in a single promising condition. Optimized SycT122 crystals grew at 4°C using hanging-drop vapor diffusion by mixing equal volumes of SycT122 and 1.6 M ammonium sulfate, 0.1 M CAPS (pH 10.5), and 0.15 M lithium sulfate, revealing crystal form #1. Changing the crystallization condition to 1.8 M ammonium sulfate, 0.1 M sodium bicarbonate (pH 10.5), and 0.05 M magnesium chloride and using a protein/reservoir ratio of 2:1 yielded crystals with a different crystal form (#2). Data of native SycT and SycT122 were collected at 100 K using 19%–25% glycerol in the crystallization mix as cryoprotectant. SAD data of SycT122 at the peak region of the selenium edge were collected at beamline BL14.1 (BESSY). Data of SycT122 were processed with XDS and scaled with XSCALE (Kabsch 1993). Data of native SycT were collected at BW6 (DESY) and processed with DENZO/SCALEPACK (Otwinowski and Minor 1997). Data collection statistics are given in Table 1.
Structure determination
The heavy-atom search, phasing, and solvent flattening programs SHELXD (Uson and Sheldrick 1999) and SHELXE (Sheldrick 2002) run through the graphical user interface HKL2MAP (Pape and Schneider 2004) located all six selenium atoms in the hexagonal packing (#2) and provided interpretable electron density. Phases for #2 as output by SHELXE were used with the same anomalous data set to trace an initial model (80% complete) in ARP/wARP (Perrakis et al. 1999). The final model was obtained by several cycles of manual building with the program O (Jones et al. 1991) and TLS restrained refinement with REFMAC5 (Murshudov et al. 1997). Electron density was visible for SycT122 residues 1–113 in one monomer, and 3–122 in the second. The structure for the orthorhombic (#1) data set was solved by molecular replacement using the #2 structure as search model in the program PHASER (Storoni et al. 2004) and was completed as above. Electron density was not observed for SycT122 residues 115–122 in both monomers of model #1. The native structure was solved using both refined SycT122 structures as search ensemble for molecular replacement in PHASER, and was initially refined with CNS (Brunger et al. 1998) and then with REFMAC5. The refined native SycT model comprises residues 3–130 (A), 2–117, and 125–130 (B). Refinement statistics are given in Table 1. Ribbon diagrams and surfaces were produced with PyMOL (DeLano Scientific) and Swiss-PDBViewer (Guex and Peitsch 1997), secondary structure assignment was calculated using STRIDE (Heinig and Frishman 2004), buried surface areas were calculated with AREAIMOL (Lee and Richards 1971), cavity volumes were calculated with VOIDOO (Kleywegt 1994), pairwise and multiple sequence alignments were produced with EMBOSS (Pearson 1990) and ClustalW (Thompson et al. 1994), and structures were aligned with DALI (Holm and Sander 1993).
Coordinates
Coordinates and structure factors have been deposited in the Protein Data Bank. Accession codes are 2BSH (SycT122 crystal form #2), 2BSI (#1), and 2BSJ (native SycT).
Acknowledgments
We greatly appreciate the assistance of Dr. Uwe Müller (PSF, BESSY beamline BL14.1) during SAD data collection and acknowledge the use of beam time at DESY beamline BW6. We thank Dr. Hans-Jürgen Hecht for crystallographic advice, Dr. Wolf-Dieter Schubert for help in crystallography and critical reading of the manuscript, and Rita Getzlaff for N-terminal sequencing. This work was supported by the DFG priority program 1150—“Signalling pathways to the cytoskeleton and bacterial pathogenicity” (to H.N.).
Abbreviations
- #1 and #2, crystal form 1 (space group P212121) and crystal form 2 (P62) of SycT122, respectively
- CBD, chaperone-binding domain
- ORF, open reading frame
- TTS, Type III secretion
- Yop, Yersinia outer protein
- SAD, single-wavelength anomalous dispersion
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051474605.
References
- Äpfelbacher, M., Trasak, C., Wilharm, G., Wiedemann, A., Trulzsch, K., Krauss, K., Gierschik, P., and Heesemann, J. 2003. Characterization of YopT effects on Rho GTPases in _Yersinia enterocolitica_-infected cells. J. Biol. Chem. 278 33217–33223. [DOI] [PubMed] [Google Scholar]
- Birtalan, S. and Ghosh, P. 2001. Structure of the Yersinia type III secretory system chaperone SycE. Nat. Struct. Biol. 8 974–978. [DOI] [PubMed] [Google Scholar]
- Birtalan, S.C., Phillips, R.M., and Ghosh, P. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9 971–980. [DOI] [PubMed] [Google Scholar]
- Bottone, E.J. 1999. Yersinia enterocolitica: Overview and epidemiologic correlates. Microbes Infect. 1 323–333. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R. 2002. The Yersinia YSC-YOP “type III” weaponry. Nat. Rev. Mol. Cell Biol. 3 742–752. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R. and Van Gijsegem, F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54 735–774. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R., Boland, A., Boyd, A.P., Geuijen, C., Iriarte, M., Neyt, C., Sory, M.P., and Stainier, I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62 1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs, K. and Karplus, P.A. 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4 269–275. [DOI] [PubMed] [Google Scholar]
- Evdokimov, A.G., Tropea, J.E., Routzahn, K.M., and Waugh, D.S. 2002. Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis. Acta Crystallogr. D Biol. Crystallogr. 58 398–406. [DOI] [PubMed] [Google Scholar]
- Feldman, M.F. and Cornelis, G.R. 2003. The multitalented type III chaperones: All you can do with 15 kDa. FEMS Microbiol. Lett. 219 151–158. [DOI] [PubMed] [Google Scholar]
- Francis, M.S., Wolf-Watz, H., and Forsberg, A. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5 166–172. [DOI] [PubMed] [Google Scholar]
- Frithz-Lindsten, E., Du, Y., Rosqvist, R., and Forsberg, A. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III– dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol. Microbiol. 25 1125–1139. [DOI] [PubMed] [Google Scholar]
- Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero, S.A., Hecht, H.J., Hofmann, B., Biebl, H., and Singh, M. 2001. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl. Microbiol. Biotechnol. 56 718–723. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Heinig, M. and Frishman, D. 2004. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32 W500–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138. [DOI] [PubMed] [Google Scholar]
- Hubbard, S.J. and Argos, P. 1994. Cavities and packing at protein interfaces. Protein Sci. 3 2194–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriarte, M. and Cornelis, G.R. 1998. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol. Microbiol. 29 915–929. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26 795–800. [Google Scholar]
- Kleywegt, G.J. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50 178–185. [DOI] [PubMed] [Google Scholar]
- Lee, B. and Richards, F.M. 1971. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55 379–400. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Bertero, M.G., Frey, E.A., Pfuetzner, R.A., Wenk, M.R., Creagh, L., Kay, C., Haynes, C., Finley, B.B., and Strydnaka, N.C.J. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8 1031–1036. [DOI] [PubMed] [Google Scholar]
- Marlovits, T.C., Kubori, T., Sukhan, A., Thomas, D.R., Galan, J.E., and Unger, V.M. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306 1040–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Alto, N.M., and Dixon, J.E. 2005. Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr. Opin. Microbiol. 8 21–27. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pape, T. and Schneider, T.R. 2004. HKL2MAP: A graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37 843–844. [Google Scholar]
- Parsot, C., Hamiaux, C., and Page, A.-L. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6 7–14. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183 63–98. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6 458–463. [DOI] [PubMed] [Google Scholar]
- Phan, J., Tropea, J.E., and Waugh, D.S. 2004. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr. D Biol. Crystallogr. 60 1591–1599. [DOI] [PubMed] [Google Scholar]
- Schubot, F.D., Jackson, M.W., Penrose, K.J., Cherry, S., Tropea, J.E., Plano, G.V., and Waugh, D.S. 2005. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346 1147–1161. [DOI] [PubMed] [Google Scholar]
- Shao, F., Merritt, P.M., Bao, Z.Q., Innes, R.W., and Dixon, J.E. 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109 575–588. [DOI] [PubMed] [Google Scholar]
- Sheldrick, G.M. 2002. Macromolecular phasing with SHELXE. Zeitschrift für Kristallographie 217 644–650. [Google Scholar]
- Singer, A.U., Desveaux, D., Betts, L., Chang, J.H., Nimchuk, Z., Grant, S.R., Dangl, J.L., and Sondek, J. 2004. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12 1669–1681. [DOI] [PubMed] [Google Scholar]
- Sorg, I., Hoffmann, C., Dumbach, J., Aktories, K., and Schmidt, G. 2003. The C terminus of YopT is crucial for activity and the N terminus is crucial for substrate binding. Infect. Immun. 71 4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory, M., Boland, A., Lambermont, I., and Cornelis, G.R. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. 92 11998–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, C.E. and Galan, J.E. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414 77–81. [DOI] [PubMed] [Google Scholar]
- Storoni, L.C., McCoy, A.J., and Read, R.J. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60 432–438. [DOI] [PubMed] [Google Scholar]
- Tampakaki, A.P., Fadouloglou, V.E., Gazi, A.D., Panopoulos, N.J., and Kokkinidis, M. 2004. Conserved features of type III secretion. Cell. Microbiol. 6 805–816. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trame, C.B. and McKay, D.B. 2003. Structure of the Yersinia enterocolitica molecular-chaperone protein SycE. Acta Crystallogr. D Biol. Crystallogr. 59 389–392. [DOI] [PubMed] [Google Scholar]
- Trülzsch, K., Roggenkamp, A., Äpfelbacher, M., Wilharm, G., Ruckdeschel, K., and Heesemann, J.R. 2003. Analysis of chaperone-dependent Yop secretion/translocation and effector function using a mini-virulence plasmid of Yersinia enterocolitica. Int. J. Med. Microbiol. 293 167–177. [DOI] [PubMed] [Google Scholar]
- Uson, I. and Sheldrick, G.M. 1999. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9 643–648. [DOI] [PubMed] [Google Scholar]
- Van Eerde, A., Hamiaux, C., Perez, J., Parsot, C., and Dijkstra, B.W. 2004. Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity. EMBO Rep. 5 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau, P. and Cornelis, G.R. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol. Microbiol. 8 123–131. [DOI] [PubMed] [Google Scholar]
- Wattiau, P., Bernier, B., Deslee, P., Michiels, T., and Cornelis, G.R. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. 91 10493–10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau, P., Woestyn, S., and Cornelis, G.R. 1996. Customized secretion chaperones in pathogenic bacteria. Mol. Microbiol. 20 255–262. [DOI] [PubMed] [Google Scholar]
- Woestyn, S., Sory, M.P., Boland, A., Lequenne, O., and Cornelis, G.R. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20 1261–1271. [DOI] [PubMed] [Google Scholar]
- Yip, C.K., Finlay, B.B., and Strynadka, N.C.J. 2005. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12 75–81. [DOI] [PubMed] [Google Scholar]
- Zhu, M., Shao, F., Innes, R.W., Dixon, J.E., and Xu, Z. 2004. The crystal structure of Pseudomonas avirulence protein AvrPphB: A papain-like fold with a distinct substrate-binding site. Proc. Natl. Acad. Sci. 101 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbihl, R., Äpfelbacher, M., Andor, A., Jacobi, C.A., Ruckdeschel, K., Rouot, B., and Heesemann, J. 1999. The cytotoxin YopT of Yersinia enterocolitica induces modification and cellular redistribution of the small GTP-binding protein RhoA. J. Biol. Chem. 274 29289–29293. [DOI] [PubMed] [Google Scholar]