ChIPping away at gene regulation (original) (raw)
Abstract
The coordinated regulation of gene expression in higher eukaryotes is complex and poorly understood. Recent technological advances have allowed the first insights into these networks on a genome-wide scale. These investigations have identified transcription factor target sites in the genome and successfully predicted cooperative interactions with other factors. However, a detailed understanding of the processes that coordinate gene expression remains elusive. Here, we highlight the advances that have been made using current methods, and the need for new technologies to address the gaps in our knowledge and to map these complex pathways further.
Keywords: chromatin immunoprecipitation, gene expression, transcription factor
Introduction
All biological processes rely on the coordinated expression of genes, the products of which act together to mediate cellular function. Understanding the processes that control gene expression is essential to our comprehension of development and disease. However, transcriptional regulation in higher eukaryotes is complex. This is exemplified by the large number of proteins that control gene expression; there are more than 3,000 transcription factors (TFs) representing approximately 10% of all human genes (Babu et al, 2004), as well as the vast areas of the genome that participate in transcription by acting as scaffolds on which regulatory complexes assemble (Carroll et al, 2006). Therefore, considerable effort has been made to identify transcriptional networks and to map the regions of the genome that participate in the control of gene expression (Collas & Dahl, 2008; Kim & Ren, 2006; Wu et al, 2006).
Initial efforts to unravel these complex problems used nuclease-protection assays, and in vitro DNA-binding and reporter assays. These tools have allowed the identification of regulatory elements proximal to candidate genes and have identified direct targets of candidate TFs. However, these approaches are limited by the requirement for candidate target genes and by the relatively small area of the genome that can be analysed. It is also clear that not all direct targets of a given TF are regulated in the same way: the TF might activate some direct target genes but repress others under the same cellular conditions. For example, 44 oestrogen receptor-α (ERα) target genes are upregulated after 3 h of stimulation with oestrogen, whereas 24 ERα targets are downregulated under the same conditions (Carroll et al, 2006). Therefore, examination of individual candidate genes in the study of TF biology might not adequately represent the full range of direct target genes.
The use of recombinant TF DNA-binding domains to enrich sequences from libraries of random DNA sequences—for example, through systematic evolution of ligands by exponential enrichment (SELEX) or cyclic amplification and selection of targets (CASTing; Roche et al, 1992; Wright et al, 1991)—has defined the preferred in vitro binding motifs for several TFs (834 position-weight matrices in the TRANSFAC database). In combination with complete genome sequences, these preferred DNA-binding motifs allow the in silico mapping of candidate regions of the genome that might be involved in TF recruitment and, therefore, transcriptional regulation. Although this approach has been used successfully to identify TF-binding sites in the genome, it is clear that these in vitro sites might differ from those preferred in vivo (Barbulescu et al, 2001; Verrijdt et al, 2003). There are also examples of perfect matches to _in vitro_-derived TF-binding sequences that are not bound by a TF in vivo (Horie-Inoue et al, 2006). Therefore, in vitro approaches to identify TF-binding sites in the genome are hampered by both false negatives and false positives. These differences are probably partly a result of protein–protein interactions recruiting TFs to sequences in the genome that do not correspond to the optimal in vitro DNA-binding element, as well as conformational differences between the recombinant DNA-binding domain and the native conformations of these domains when they are expressed endogenously. The in vivo appearance of TF-binding sites is also controlled by the packaging of genomic DNA in chromatin, the compaction and relaxation of which might mask some consensus TF-binding sites and reveal others. Therefore, alternative approaches are required to map TF-binding sites in the genome accurately.
Chromatin immunoprecipitation (ChIP) allows the identification of in vivo direct TF-binding sites in the context of chromatin and therefore avoids many of the problems mentioned above (Orlando & Paro, 1993; Solomon et al, 1988). ChIP involves chemical crosslinking of DNA–protein interactions in living cells to ‘fix' TFs to their cognate binding sites in the genome (Fig 1). Crosslinked chromatin is then fragmented and specific antibodies are used to immunoprecipitate TFs together with their bound DNA fragments. DNA–protein crosslinks are reversed, and enriched DNA fragments are then purified for downstream analysis. In theory, ChIP could be used to investigate any target on chromatin against which an antibody can be raised and, consequently, it has successfully been used to identify regions of the genome associated with specific TFs, cofactors, histone modifications and DNA methylation (Orlando & Paro, 1993; Solomon et al, 1988; Weber et al, 2005). Standard ChIP assays use Southern blotting, polymerase chain reaction (PCR) or quantitative real-time PCR (qPCR) as a read-out; however, these approaches are also limited by the requirement for candidate regions. Combining ChIP with cloning and sequencing—for example, in sequence tag analysis of genomic enrichment (STAGE), serial analysis of chromatin occupancy (SACO) and ChIP-paired end tag (ChIP-PET), genomic microarrays (ChIP-chip), or direct sequencing using 454 or Solexa G1 sequencing platforms (ChIPseq)—allows large-scale identification of TF-binding sites in the genome (Carroll et al, 2006; Impey et al, 2004; Robertson et al, 2007).
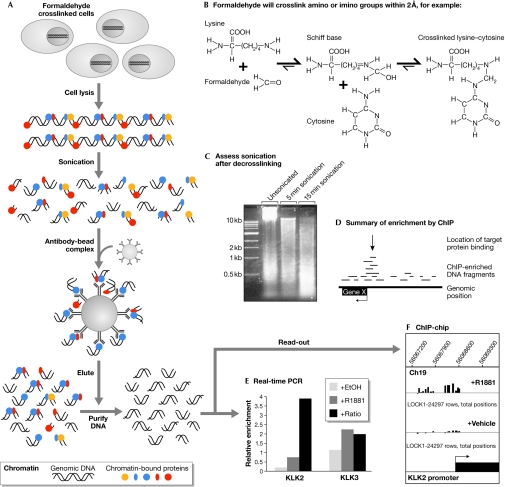
Figure 1.
Overview of chromatin immunoprecipitation strategies. (A) Summary of chromatin immunoprecipitation (ChIP) methodology. (B) Formaldehyde crosslinking chemistry. (C) Example of gel-electrophoresis analysis of chromatin before and after sonication. (D) Summary of DNA-fragment enrichment by ChIP, showing the large number of low-abundance non-specific DNA fragments and high-abundance specific ChIP-enriched DNA fragments. (E) Analysis of androgen receptor (AR) ChIP enrichment by real-time quantitative PCR for the Kallikrein (KLK)2 and KLK3 promoters. Values shown are relative to input material and β-actin control PCR. (F) Example of AR ChIP-chip data showing enrichment of the KLK2 promoter region in the androgen (+R1881) versus control (+Vehicle) treated conditions. Genomic positions are shown above ChIP-chip data ‘tracks' from the androgen (+R1881) and control (+Vehicle) conditions. The level of enrichment is shown by the height of the vertical bars. The location of the gene encoding KLK2 relative to the array probes is shown on the bottom line by a rectangle and the arrow indicates the direction of transcription.
Comparison of ChIP methodologies
There are three main categories of ChIP methodologies used in TF-binding site location analysis: ChIP-tag library and sequencing, ChIP-chip, and ChIPseq. The library and sequencing-based methods include SACO and STAGE, both of which involve linker-mediated amplification, digestion, concatamerization of sequence ‘tags' and sequencing (Bhinge et al, 2007; Impey et al, 2004). SACO and STAGE are both variations of the serial analysis of gene expression (SAGE) method (Bhinge et al, 2007; Impey et al, 2004). ChIP-PET differs in that ChIP fragments are cloned before concatenation of fragment ends and sequencing, thereby allowing both ends of a fragment to be mapped (Wei et al, 2006). These methods are not limited by probe performance or array coverage issues and therefore offer true genome-wide analysis; however, they do require the generation of sequence ‘tag' libraries, a large number of sequence reads (∼40,000 reads producing ∼512,000 tags), deconvolution of concatamerized tag sequences and mapping of tags to the genome (Wei et al, 2006). Binding sites are defined by the enrichment of a genomic region as assessed by the number of sequence tags corresponding to that locus in the ChIP samples. Depth of coverage—that is, the number of binding sites and the number of tags per binding site—is determined by the number of sequence reads performed; therefore, the specificity of the ChIP enrichment is directly linked to the number of binding sites identified—that is, a higher background level will decrease the proportion of enriched tags sequenced.
The ChIP-chip method involves the amplification or pooling of ChIP samples, fluorescent labelling of ChIP-enriched DNA and hybridization to genomic microarrays, either alone or in competition with a control DNA sample, such as total genomic DNA. Genomic binding sites for a TF are defined as regions that have a significantly higher fluorescent signal intensity than the control DNA sample—that is, they are enriched in the ChIP sample. In contrast to ChIP-library and ChIPseq methods, the resolution and coverage of ChIP-chip assays are defined by the choice of array platform. Whole-genome tiling arrays are available for all non-repetitive regions of the genome, although they are costly and, for the human genome, consist of a set of roughly 38 arrays (Carroll et al, 2006). However, many ChIP-chip studies have used promoter arrays, encyclopaedia of DNA elements (ENCODE) arrays, candidate region arrays or chromosome 21–22 tiling arrays, to successfully identify TF-binding sites and to gain insights into TF cooperation, target site selection and downstream-regulated pathways (Bolton et al, 2007; Massie et al, 2007; Takayama et al, 2007; Wang et al, 2007). Indeed ChIP-chip platforms with lower coverage, such as promoter arrays, have in many cases allowed the identification of similar numbers of ‘functional' TF target genes (supplementary Table 1 online), possibly because of the difficulties involved in mapping the functions of distal TF-binding sites.
ChIPseq involves the direct sequencing of ChIP-enriched DNA, after limited amplification and size selection of DNA fragments to set the binding-site resolution (Robertson et al, 2007). Binding sites are defined by significant enrichment of sequence reads above the background levels at genomic loci. As is the case for ChIP-library methods, there are no array-based limitations for ChIPseq, which therefore offers unbiased whole-genome coverage for sequence reads that can be mapped to single-copy regions of the genome. ChIPseq has several advantages over other ChIP-based approaches including the following: simplicity (no requirement for library generation); smaller amounts of starting material (and therefore less amplification); high-throughput G1 or 454 sequencing platforms that generate deeper coverage than is practical with ChIP-library methods (∼24 × 106 reads of 27 base pairs (bp) and two to three orders of magnitude more reads than ChIP-PET; Euskirchen et al, 2007; Robertson et al, 2007; Wei et al, 2006); identification of single-nucleotide polymorphisms or mutations in TF-binding sites; and increased resolution of binding sites (± 50 bp owing to size selection of ChIP fragments). These features promise to improve the identification of TF-binding sites in the genome and to improve TF-binding motif analysis as a result of the higher resolution. However, access to Solexa G1 or 454 sequencing technologies and the depth of coverage for each sequencing run might limit the use of ChIPseq in defining TF-binding sites.
In vivo TF-binding site selection
Libraries of TF-binding sites generated by ChIP are useful to identify not only candidate target genes of a given TF, but also in vivo DNA sequences that recruit TFs. These sequences can be mined for known TF-binding motifs or used to generate new consensus binding motifs. Most groups have shown that _in vitro_-derived TF consensus binding motifs for their targets are relatively enriched in ChIP data; for example, NOTCH (P = 1 × 10−13), androgen receptor (AR; P < 2 × 10−6) and glucocorticoid receptor (GR; P < 0.05; supplementary Table 1 online; Massie et al, 2007; Palomero et al, 2006; Phuc Le et al, 2005). However, these _in vitro_-derived motifs rarely account for more than a small subset of the total TF-binding sites identified by ChIP, although the figure is variable. For example, in two independent AR ChIP-chip studies, 73–90% of AR-binding sites did not contain the consensus ARE motif (Massie et al, 2007; Wang et al, 2007), and in a recent E2F ChIP-chip study of three E2F factors, 90–96% of binding sites did not contain the _in vitro_-derived E2F consensus (Xu et al, 2007). By contrast, only about 20% of hepatocyte nuclear factor 4a (HNF4a)-binding sites, 10% of repressor element 1-silencing transcription factor (REST)-binding sites and approximately 30% of avian erythroblastosis virus E26 homologue 1 (ETS1)/ETS-like factor 1(ELF1) binding sites lacked sequences resembling their respective _in vitro_-derived consensus motifs (Hollenhorst et al, 2007; Johnson et al, 2007; Rada-Iglesias et al, 2005). Consequently, for some TFs, in vivo binding sites might resemble the in vitro situation more closely (for example, HNF4a, REST and ETS1/ELF1), whereas for others, most genomic binding sites seem to be influenced by the presence of additional factors in vivo (for example, AR and E2F).
‘Non-conforming' TF-binding sites might be indirect binding sites—through chromatin looping or recruitment by another factor—or direct binding sites that diverge from the in vitro consensus sequences. ChIP-chip has revealed that the AR can bind not only to the _in vitro_-derived 15 bp bipartite ARE sequence (AGAACAnnnTGTTCT), but also to 6 bp half-sites (AGAACA) and bipartite motifs with spacer sequences of 0–8 bp (Massie et al, 2007; Wang et al, 2007). Even among ‘conformist' TFs, such as REST and ETS1, a subset of divergent binding sites has been identified—for example, although the majority of REST-binding sites contain canonical non-identical half-sites spaced by 11 bp, many contain such half-sites separated by 4–25 bp (Hollenhorst et al, 2007; Johnson et al, 2007).
_In vitro_-derived TF-binding sites have proved informative in predicting modular or co-dependent recruitment of TF clusters based on over-representation of motif classes. This method is prone to both false-positive and false-negative results, although it provides testable models and insights into the cooperative function of certain TFs (supplementary Table 1 online). For example, sequence analysis of ERα ChIP-chip binding sites revealed enrichment of forkhead (FKHD) TF motifs, leading to the identification of Forkhead box A1 (FOXA1) as a ‘pioneer factor', which is required to mark regions of the genome for ERα recruitment (Carroll et al, 2006; Laganiere et al, 2005). Another intriguing example of this approach was recently reported from de novo sequence analysis of 87 genomic fragments that were bound by ETS1 and Runt-related transcription factor 1 (RUNX1) in ChIP-chip experiments (Hollenhorst et al, 2007). The resulting sequence motif was divergent from both the ETS1 and RUNX1 _in vitro_-derived motifs, and ETS1 binding to this motif was shown to be strongly dependent on RUNX1, suggesting that these factors bind cooperatively (supplementary Table 1 online; Hollenhorst et al, 2007). Co-dependent recruitment of TFs at certain genomic binding sites creates the possibility of therapeutically targeting one class of TFs to affect the activation or recruitment of another. For example, FOXA1 could be targeted to regulate ER activity in breast cancer and ETS TFs could be targeted to regulate AR activity in prostate cancer.
Location of regulatory elements
Traditionally, gene promoter regions, which are considered to be upstream and proximal to transcription start sites (TSSs), were thought to be the most important regulatory elements in the genome. However, until recently this had not been assessed in an unbiased manner. ChIPseq, ChIP-cloning and ChIP-chip using arrays that are not limited to proximal promoter regions—for example, whole-genome tiling, chromosome-wide, ENCODE or candidate arrays—allows the identification of regions of the genome that are enriched for TF binding and might therefore have an important role in transcriptional regulation. The relative importance of proximal and distal regions to TF binding varies (supplementary Table 1 online). ChIP-chip analysis of three E2F factors in three different cell lines using ENCODE arrays (representing 1% of the human genome) revealed that 50–85% of E2F-binding sites were within 2 kb of a TSS (see ENCODE Consortium at http://www.genome.gov/12513391; Xu et al, 2007). ChIP-PET analysis of P493 cells revealed that 63% (372 out of 593) of the cellular counterpart of the transforming gene of the avian myelocytomatosis virus MC29 (cMYC)-binding sites were within 10 kb of a known TSS (Zeller et al, 2006). Analysis of Krüppel-associated box-associated protein 1 (KAP1) binding sites in Ntera2 cells by ChIP-chip using whole-genome tiling arrays showed that approximately 40% of KAP1-binding sites were more than 5 kb away from a known TSS (O'Geen et al, 2007). It is also clear from these and other studies that binding sites for TFs are evenly distributed upstream and downstream of the TSS, with binding sites additionally occurring in transcribed regions (O'Geen et al, 2007; Xu et al, 2007; Zeller et al, 2006). These data indicate that, for at least some TFs, there is a strong bias towards proximal promoter binding, both upstream and downstream of the TSS.
By contrast, genome-wide ERα ChIP-chip revealed that only 4% of ERα-binding sites were located within 1 kb of a TSS; consequently, most ERα-binding sites are within regions of the genome that had not previously been associated with transcriptional regulation (Carroll et al, 2006). In a previous study, one distal ERα-binding site (144 kb from the TSS) was shown to interact with the closest promoter region through chromosome looping (3C) and a transcriptional enhancer function was confirmed for about 75% of distal elements by using reporter assays (Carroll et al, 2005). These data indicate that at least some distal ERα-binding sites have a role in long-range control of gene transcription, although there are clear technical challenges in validating their contribution based on an inability to robustly mimic DNA looping and distal protein recruitment using reporter assays. Chromosome-wide ChIP-chip analysis of tumour protein 53 (TP53), c-MYC and SV40 early promoter transcription factor 1 (SP1) indicated that only 22% of binding sites were within 1 kb of a TSS (Cawley et al, 2004). An alternative hypothesis was proposed for the distal binding sites in this study, according to which the non-promoter TF-binding sites were proximal to new transcripts rather than being enhancers for distal genes (Cawley et al, 2004). Complementary to this hypothesis, data from the ENCODE Consortium identified 1,393 ‘regulatory clusters' in the human genome by integrating 129 ChIP-chip data sets, only 25% of which were within 2.5 kb of a known TSS, whereas 65% were within 2.5 kb of a known or new TSS (http://www.genome.gov/12513391). Together, these data indicate that, although there are genuine distal TF-binding sites that function as enhancers, the current status of genome annotation, and the limited data on new and non-coding transcripts, mean that many TF-binding sites are currently misclassified as distal binding sites. These issues could be resolved by using high-throughput chromosome-conformation assays (for example, 4C or 5C; Dostie et al, 2006; Zhao et al, 2006), direct sequencing of transcriptomes using next-generation sequencing platforms (Solexa G1 or 454) to identify all expressed transcripts in a given cell type, and better annotation of TSSs for all commonly used cell types (for example, cap analysis of gene expression (CAGE); Kodzius et al, 2006).
Functional binding sites
One of the main aims of ChIP-based studies is to identify functionally direct target genes of a TF, to gain insights into its biology and the downstream pathways that it activates. These approaches have been applied successfully to several important areas of biology, including: the contribution of octamer-binding protein 4 (OCT4), SRY box-containing gene 2 (SOX2) and NANOG to pluripotency in embryonic stem cells; myoblast differentiation focusing on myoblast determination protein 1 (MyoD), myogenin and myocyte-enhancer factor 2 (MEF2); and epigenetic marks (Blais et al, 2005; Boyer et al, 2005; Fischer et al, 2008). Epigenetic data are useful as an additional tier of analysis to correlate TF binding with transcriptional activation, as there is often no robust way of predicting this process based on TF-binding profiles alone. Nonetheless, the complexity of these data makes network analysis a huge challenge. Many TFs drive the expression of others, which complicates the integration of expression-array and ChIP data to decipher direct and indirect targets for a given TF. The conservation of the coding sequences for certain TFs and their binding motifs across species has led groups to compare TF-binding profiles in tissues across species. For example, the cross-species binding and expression correlation for HNFs in a comparison of mouse and human liver samples was significantly lower than predicted (Odom et al, 2007). This has clear implications for groups using mouse models to predict human TF biology and strongly suggests a significant epigenetic component in determining TF function.
However, most ChIP studies compare the TF-binding profiles under various stimuli—for those that can be activated or inhibited by stimuli such as hormones, mitogens or DNA damage—or with RNA-interference knockdown, and relate these changes to the expression profiles of genes adjacent to TF-binding sites (Carroll et al, 2006; Johnson et al, 2007; Zeller et al, 2006). For example, nuclear hormone receptors, such as the AR, ER and GR, can be inhibited by the depletion of circulating hormones and stimulated by the addition of the specific ligand for the receptor of interest. This methodology has successfully identified AR-regulated target genes in several ChIP-chip studies, including 206 using a candidate array, 92 using a promoter array, 34 using chromosome-wide arrays and 8 functional AR targets using ENCODE arrays (Bolton et al, 2007; Massie et al, 2007; Takayama et al, 2007; Wang et al, 2007).
However, when reviewing the published data, it is notable that most TF-binding sites identified using ChIP do not seem to have any effects on the transcription of adjacent genes in the cell type tested (supplementary Table 1 online). This suggests that most TF-binding sites are not functional in a given cell type, but that there are several factors that might confound the identification of directly regulated genes. TFs with a large number of binding sites and that are far from gene-coding regions might have inherent false negatives, as it is often difficult to attribute transcriptional function over long genomic distances (Carroll et al, 2006). It is also likely that feedback or feed-forward signalling and the interdependency of transcriptional networks contribute to the difficulties in identifying functional TF targets. For example, a strong enrichment of ERα ChIP-chip binding sites was observed around genes with delayed transcriptional regulation in response to oestrogen stimulation (for >12 h), indicating that the expression of these genes requires ERα to regulate the expression of another cooperating TF or cofactor (Carroll et al, 2006). The suggestion is that ERα modulates its own transcriptional activity at certain targets by regulating the expression of cooperating TFs and/or cofactors. This feed-forward loop might confound the identification of direct functional targets using this approach, as the initial depletion of hormones not only directly inhibits ERα transcriptional activity, but also indirectly inhibits the capacity of ERα to transactivate a subset of its direct target genes by downregulating essential cofactors. Hormone depletion also results in the accumulation of cells in the G0/G1 phase of the cell cycle, which might impinge further on TF function. It is therefore possible that direct functional targets of ERα have been missed because they are not regulated under the conditions tested.
Functional redundancy might also contribute to the difficulty involved in identifying directly regulated TF target genes. For example, ChIP-chip analysis of E2F1, E2F4 and E2F6 showed that 55–75% of binding sites were shared between any two of these three E2Fs (Xu et al, 2007). A similar picture was also revealed in ChIP-chip analysis of ETS1, ELF1 and GA-binding protein-α (GABPα), in which 50–60% of binding sites were common between any two ETS factors (Hollenhorst et al, 2007). These insights into the redundant occupation of TF-binding sites raise many questions as to whether the TFs bind together on the same genomic fragment and to individual alleles or occupy the same loci in different cells as a result of cell-cycle regulation, recruitment by cooperating factors or competition for binding sites. Irrespective of the dynamics, these data highlight the problems inherent in dissecting the functional targets of TFs in the context of mammalian cells. Such cells express hundreds of TFs, many of which recognize similar genomic binding sites, and assemble on regulatory regions of the genome that bind to many cooperating and competing TFs and cofactors.
There is also the possibility that these sites might not be functionally significant in a traditional sense. The initial site of occupancy for a TF on recruitment to DNA might not be its actual site of function, as there remains the largely untested possibility that a TF might move along the DNA or be repositioned once other factors bind. False positives in the ChIP-based assays could also account for a poor correlation between TF binding and effects on gene expression, and antibody crossreactivity—resulting in the antibody precipitating unrelated proteins—can contribute greatly to this. We address strategies to overcome this issue in the final section of the review. In analysing data, a less direct but perhaps more meaningful alternative to these attempts to analyse TF function in cell lines is therefore to make the jump directly to gene-expression profiles from relevant tissue samples and disease states. This method has been successfully used in TP53 ChIP-PET and AR ChIP-chip studies to show that direct target genes of these TFs are sufficient to cluster clinical expression-array data from breast and prostate cancers, respectively (Massie et al, 2007; Wei et al, 2006). The gene-expression profiles for the direct TP53 target genes allowed 251 breast cancer samples to be clustered into two groups based on disease-free survival (Wei et al, 2006).
The need for new technologies and approaches
The ChIP-based approaches outlined above have provided many important insights into transcriptional regulation and TF biology. Extending these studies and carefully integrating the available data will doubtless lead to further insights into transcriptional biology. However, these approaches also have their limitations, not least of which is the requirement for high-quality antibodies against candidate TFs that have been implicated in the disease or biological pathway under investigation. Young and colleagues made pioneering progress towards understanding transcriptional networks by using epitope tagging of TFs in more than 200 strains of Saccharomyces cerevisiae (Harbison et al, 2004; Lee et al, 2002). Epitope tagging of genomic loci in eukaryotic cell lines has been hampered by the delivery systems available and the low rates of recombination in many lines. Therefore, there have been many difficulties in determining similar gene-regulatory networks in mammalian cells. The challenges posed include low homologous-recombination efficiencies and diploid genomes in eukaryotic cells. Recently, the approach used to make somatic gene knockouts in cell lines has been successfully adapted to insert epitope tags in a targeted manner into cell-line genomes (Kohli et al, 2004; Zhang et al, 2008). This provides hope that the challenge of antibody specificity can be overcome and variations on this theme are being developed by two international consortia: the European Transcriptome, Regulome and Cellular Commitment Consortium (www.eutracc.eu) and the International Regulome Consortium (www.internationalregulomeconsortium.ca).
In addition to antibody specificity, chemical crosslinkers, such as formaldehyde, carry the risk of complexing DNA fragments or proteins that are in proximity to, but not necessary for, the assembly of specific transcription complexes based on the indeterminate kinetics of the crosslinking reaction. Typically, this step runs for several minutes creating the possibility of a crosslinked protein–protein and protein–DNA network of which direct DNA-binding proteins might be only a sub-fraction. Instant non-chemical crosslinking methods are now being developed based on laser illumination, and these approaches promise to increase speed and specificity (Zhang et al, 2004).
Other hurdles to be overcome include the requirement for large numbers of cells (typically ∼1 × 107), which has acted, in part, as a barrier to the application of this approach to primary cultures (for example, of neurons) or to clinical specimens. Consequently, it is unclear how best to extrapolate from ChIP data in cell lines to cancer specimens, given the impossibility of directly comparing DNA-binding profiles; therefore, the focus has remained downstream of TF recruitment—transcriptomics and immunohistochemistry. Another main challenge is computational. There is a lack of consensus as to the best way to define binding motifs from ChIP datasets and false-discovery rates (FDRs) in ChIP experiments (Johnson et al, 2006; Pyne et al, 2006). In part, this reflects the technical differences between platforms, as well as the lack of a coordinated multinational effort to define standards in the field. The integration of data from multiple ChIP experiments, reporter assays, small-interfering RNA screens, DNaseI hypersensitive sites, chromosome-capture assays and nucleosome mapping to define a transcriptional regulatory network is a huge challenge. The ENCODE Consortium provides a model for this that needs extending (http://www.genome.gov/12513391).
Increasingly, groups will seek to apply ChIP to study proteins that act as co-regulators but do not bind to DNA directly or for which antibodies have not been validated. In situ gene-tagging approaches, as discussed previously, and DNA adenine methyltransferase identification (DamID) might help. DamID detects potential DNA-binding sites for a protein by analysing DNA adenine methylation in cells that express the Escherichia coli DNA adenine methyltransferase fused to a protein of interest (van Steensel & Henikoff, 2000). The principle is that the enzymatic activity of the fusion protein causes local DNA methylation at the binding sites, and these regions can then be isolated based on their sensitivity to digestion by methylation-specific restriction endonucleases, and identified by hybridization to DNA microarrays. Although this approach could theoretically be used to study most proteins and to capture regions of protein binding within the cell without using highly specific antibodies, there are potential drawbacks. Not least of these is the fact that Dam-fusion proteins might have properties that are distinct from endogenous proteins, and so the physiological relevance of the results needs to be confirmed. In addition, fusion might have the effect of impairing the methyltransferase activity of the enzyme, although this is much easier to determine at an early stage.
The question of distal binding site function—that is, distal enhancers compared with new TSSs—can be addressed with current technologies, for example, 4C, 5C and Solexa G1 transcriptome sequencing; however, the requirement for candidates based on current knowledge and the availability of antisera are inherent limitations of the methodologies. There is clearly a need for new techniques that are not limited by our present understanding of the transcriptional machinery. For example, using tags or oligonucleotides to isolate specific regions of the genome coupled with mass spectrometry would allow the unbiased assessment of all chromatin-bound proteins at given loci. This could notably improve our understanding of transcriptional biology and, in light of the implication of several proteins with known alternative non-genomic functions—for example, Huntingtin interacting protein 1 (HIP1), clathrin heavy chain (CHC) and high osmolarity glycerol 1p (Hog1p)—in direct transcriptional regulation, it might also open the floodgates to a host of new transcriptional regulators (Enari et al, 2006; Mills et al, 2005; Pokholok et al, 2006).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary information Table 1 legend
Supplementary Table 1

Charles E. Massie

Ian G. Mills
Acknowledgments
C.E.M. is a postdoctoral researcher funded by a Cancer Research UK (CRUK) programme grant. I.G.M is a CRUK core-funded Associate Scientist. The authors would like to acknowledge the significant contributions made by many research groups to this field, and apologize for any omissions in the review caused by constraints of space and scope. We would also like to acknowledge the support of the University of Cambridge and Hutchison Whampoa Limited.
References
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 [DOI] [PubMed] [Google Scholar]
- Barbulescu K, Geserick C, Schuttke I, Schleuning WD, Haendler B (2001) New androgen response elements in the murine pem promoter mediate selective transactivation. Mol Endocrinol 15: 1803–1816 [DOI] [PubMed] [Google Scholar]
- Bhinge AA, Kim J, Euskirchen GM, Snyder M, Iyer VR (2007) Mapping the chromosomal targets of STAT1 by sequence tag analysis of genomic enrichment (STAGE). Genome Res 17: 910–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR (2007) Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 21: 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA et al. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS et al. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Cawley S et al. (2004) Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Collas P, Dahl JA (2008) Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci 13: 929–943 [DOI] [PubMed] [Google Scholar]
- Dostie J et al. (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Ohmori K, Kitabayashi I, Taya Y (2006) Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev 20: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM et al. (2007) Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res 17: 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JJ, Toedling J, Krueger T, Schueler M, Huber W, Sperling S (2008) Combinatorial effects of four histone modifications in transcription and differentiation. Genomics 91: 41–51 [DOI] [PubMed] [Google Scholar]
- Harbison CT et al. (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ (2007) Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S (2006) Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun 339: 99–106 [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH (2004) Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B (2007) Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS (2006) Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Ren B (2006) Genome-wide analysis of protein–DNA interactions. Annu Rev Genomics Hum Genet 7: 81–102 [DOI] [PubMed] [Google Scholar]
- Kodzius R et al. (2006) CAGE: cap analysis of gene expression. Nat Methods 3: 211–222 [DOI] [PubMed] [Google Scholar]
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B (2004) Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res 32: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V (2005) Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA 102: 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, Mills IG (2007) New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep 8: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Gaughan L, Robson C, Ross T, McCracken S, Kelly J, Neal DE (2005) Huntingtin interacting protein 1 modulates the transcriptional activity of nuclear hormone receptors. J Cell Biol 170: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E (2007) Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ (2007) Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet 3: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R (1993) Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75: 1187–1198 [DOI] [PubMed] [Google Scholar]
- Palomero T et al. (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA 103: 18261–18266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH (2005) Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet 1: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313: 533–536 [DOI] [PubMed] [Google Scholar]
- Pyne S, Futcher B, Skiena S (2006) Meta-analysis based on control of false discovery rate: combining yeast ChIP-chip datasets. Bioinformatics 22: 2516–2522 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A et al. (2005) Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum Mol Genet 14: 3435–3447 [DOI] [PubMed] [Google Scholar]
- Robertson G et al. (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Roche PJ, Hoare SA, Parker MG (1992) A consensus DNA-binding site for the androgen receptor. Mol Endocrinol 6: 2229–2235 [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A (1988) Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53: 937–947 [DOI] [PubMed] [Google Scholar]
- Takayama K et al. (2007) Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 26: 4453–4463 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18: 424–428 [DOI] [PubMed] [Google Scholar]
- Verrijdt G, Haelens A, Claessens F (2003) Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol Genet Metab 78: 175–185 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Wei CL et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Wright WE, Binder M, Funk W (1991) Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol 11: 4104–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Smith LT, Plass C, Huang TH (2006) ChIP-chip comes of age for genome-wide functional analysis. Cancer Res 66: 6899–6902 [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ (2007) A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17: 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI et al. (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang K, Prandl R, Schoffl F (2004) Detecting DNA-binding of proteins in vivo by UV-crosslinking and immunoprecipitation. Biochem Biophys Res Commun 322: 705–711 [DOI] [PubMed] [Google Scholar]
- Zhang X et al. (2008) Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods 5: 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z et al. (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information Table 1 legend
Supplementary Table 1