Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates (original) (raw)
. Author manuscript; available in PMC: 2009 Feb 28.
Abstract
ClpX, an archetypal proteolytic AAA+ unfoldase, must engage the ssrA tags of appropriate substrates prior to ATP-dependent unfolding and translocation of the denatured polypeptide into ClpP for degradation. Here, specificity-transplant and disulfide-crosslinking experiments reveal that the ssrA tag interacts with different loops that form the top, middle, and lower portions of the central channel of the ClpX hexamer. Our results support a two-step binding mechanism, in which the top loop serves as a specificity filter and the remaining loops form a binding site for the peptide tag relatively deep within the pore. Crosslinking experiments suggest a staggered arrangement of pore loops in the hexamer and nucleotide-dependent changes in pore-loop conformations. This mechanism of initial tag binding would allow ATP-dependent conformational changes in one or more pore loops to drive peptide translocation, force unfolding, and mediate threading of the denatured protein through the ClpX pore.
Introduction
AAA+ unfoldases pull substrate proteins apart by a mechanism that requires engaging a peptide tag and then using ATP-fueled conformational changes to translocate this tag and the attached protein through a narrow central pore (for review, see Prakash and Matouschek, 2004; Sauer et al., 2004). These enzymes, which typically consist of ring hexamers of AAA+ ATPases, play important roles in protein degradation, in disaggregation, and in the disassembly of macromolecular complexes in all organisms (Neuwald et al., 1999; Hanson and Whitehart, 2005). In energy-dependent proteases—such as ClpXP, ClpAP, Lon, FtsH, HslUV, and the 26S proteasome—AAA+ machines unfold substrates and then translocate them into an internal proteolytic chamber of a compartmental peptidase for degradation.
In the translocation model of unfolding, recognition of the peptide tag is the critical step in establishing the geometry necessary to allow substrate engagement by the AAA+ translocation machinery and discrimination among potential substrates. One of the clearest examples is recognition of the ssrA tag by the AAA+ ClpX unfoldase of the ClpXP protease. In Escherichia coli, the ssrA tag (AANDENYALAA) is added to the C terminus of nascent proteins on stalled ribosomes as part of a quality-control system (Keiler et al. 1996). Importantly, addition of this tag, whether naturally or by cloning, appears to make any protein a substrate for ClpXP (Gottesman et al., 1998; Kim et al., 2000; Singh et al., 2000; Burton et al., 2001; Lee et al., 2001; Kenniston et al., 2003; 2004). ClpX specifically recognizes the C-terminal LAA and α–carboxylate of the ssrA tag (Flynn et al., 2001). The N-terminal portion of the ssrA peptide can be bound by SspB, an adaptor protein that tethers substrates to ClpXP and enhances binding and degradation (Levchenko et al., 2000; 2003; Flynn et al., 2001; Wah et al. 2002; 2003; Dougan et al., 2003; Song and Eck, 2003).
Current evidence suggests that the ssrA tag binds in the central pore of the ClpX hexamer. For example, mutations in each of the three loops that form this channel disrupt recognition of ssrA substrates (Fig. 1A; Siddiqui et al., 2004; Farrell et al., 2007; Martin et al., 2007). The basic RKH loop, located at the entry to the pore, appears to interact with the negatively charged α-carboxylate at the C-terminus of the tag. Precise roles in ssrA-tag recognition for the pore-1 loop in the center of the translocation channel and the pore-2 loop at the bottom of the channel have yet to be established, but mutations in both loops cause large decreases in apparent affinity for ssrA-tagged substrates (Siddiqui et al., 2004; Farrell et al., 2007; Martin et al., 2007). Crosslinking and stoichiometry experiments also indicate that the ssrA tag binds in the central pore of hexamers of ClpA, a relative of ClpX (Hinnerwisch et al., 2005; Piszczek et al., 2005).
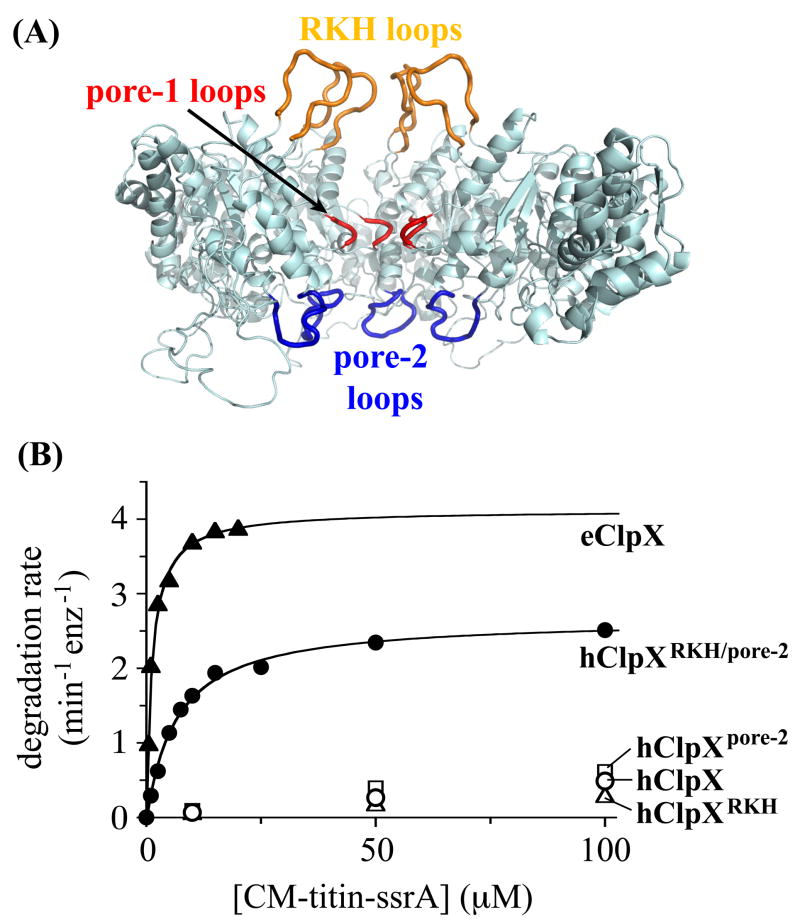
Figure 1.
Three pore loops of ClpX are involved in ssrA-tag binding. (A) The RKH loops are colored gold, the pore-1 loops red, and the pore-2 loops blue in a ClpX hexamer model based on the subunit structure of H. pylori ClpX and the structure of the HslU hexamer (Kim and Kim, 2003; Bochtler et al., 2000). Three subunits of the hexamer were removed to allow visualization of the pore loops. These loops are not resolved in the crystal structure and the conformations shown are hypothetical. (B) A variant of human ClpX (hClpXRKH/pore-2; 0.3 μM) containing the RKH and pore-2 loops of E. coli ClpX supported efficient degradation of CM-titin-ssrA by human ClpP (0.9 μM). Values of KM and Vmax derived from Michaelis-Menten analysis were 5μM and 2.7 min−1 enz−1 for hClpXRKH/pore-2, and 1μM and 4.4 min−1 enz−1 for E. coli ClpXP. At equivalent enzyme concentrations, hClpX or variants with just the RKH loop (hClpXRKH) or the pore-2 loop (hClpXpore-2) from E. coli ClpX supported only slow hClpP degradation of the ssrA-tagged substrate. Vmax may be lower for hClpXeRKH/pore-2/hClpP than for E. coli ClpXP because the E. coli pore-2 loops do not interact properly with human ClpP. For example, E. coli ClpP binding represses the ATPase activity of E. coli ClpX in a pore-2 dependent manner (Martin et al., 2007), whereas the ATPase activity of hClpXeRKH/pore-2 was not affected by hClpP binding (data not shown).
In related AAA+ enzymes, loops similar to the RKH and pore-2 loops of ClpX are frequently absent or poorly conserved. By contrast, the pore-1 loop of ClpX contains a GYVG sequence that is absolutely invariant in orthologs and highly conserved in other AAA+ unfolding machines. Indeed, pore-1 mutations inactivate or severely damage the HslU, FtsH, Yme1, ClpA, and ClpB unfoldases (Song et al., 2000; Yamada-Inagawa et al., 2003; Schlieker et al., 2004; Weibezahn et al., 2004; Park et al., 2005; Hinnerwisch et al., 2005; Okuno et al., 2006; Graef and Langer, 2006). At present, however, there are no structures that show how any of these enzymes bind cognate substrates. Moreover, the mutational data available for ClpX are puzzling, as the RKH and pore-2 loops are roughly 30 Å apart in models of the hexamer and yet both loops play key roles in recognition of a short portion of the ssrA tag.
Here, we present specificity-transplant studies and disulfide-crosslinking experiments that probe the importance of ClpX pore loops in ssrA-tag recognition and binding. Our results support a two-step mechanism of ssrA-tag binding with the RKH, pore-1, and pore-2 loops playing specific roles, reveal nucleotide-dependent changes in contacts between individual pore loops and the ssrA tag, and suggest a staggered arrangement of equivalent pore loops in different subunits of the ClpX hexamer. The ssrA tag of a substrate, either alone or in complex with the SspB adaptor, binds deeply enough in the ClpX pore to allow ATPase-driven loop movements to initiate translocation of the tag and to force subsequent substrate unfolding and threading of the denatured protein through the ClpX pore.
Results
A specificity transplant
Unlike E. coli ClpX, human mitochondrial ClpX does not recognize ssrA-tagged substrates (Kang et al., 2002). These ClpX orthologs share a common pore-1 loop but have different RKH and pore-2 loops. It is known that mutations in the latter loops of E. coli ClpX disrupt binding to the ssrA tag of substrates (Farrell et al., 2007; Martin et al., 2007), but loss-of-function mutations can act by a variety of mechanisms. Hence, to establish a positive role for the E. coli ClpX RKH and pore-2 loops in ssrA-tag recognition, we sought to transplant this binding specificity into the human enzyme.
We constructed variants of human ClpX in which just the RKH loop or just the pore-2 loop were replaced with the corresponding loop from E. coli ClpX. Both hybrid enzymes were similar to human ClpXP in supporting very low rates of degradation of the unfolded substrate CM-titin-ssrA (KM>300 μM; Fig. 1 B). Importantly, however, a variant of the human enzyme containing both the E. coli RKH and pore-2 loops degraded this ssrA-tagged substrate with a KM (5 μM) only slightly higher than the value for E. coli ClpXP (Fig. 1B). Likewise, only the human variant with both the E. coli RKH and pore-2 loops mediated efficient degradation of the natively folded substrate GFP-ssrA (KM=6.5 μM; data not shown). This gain of function confirms that the RKH and pore-2 loops of E. coli ClpX are critical for binding of ssrA-tagged substrates.
Crosslinking strategy
The proximity of specific residues in two molecules can be accurately assessed by the ability of cysteines at these positions to form a disulfide bond, which requires both atomic contact and the correct orientation. Hence, to obtain detailed information about the geometry and mechanism of binding, we introduced cysteines into specific pore loops of ClpX and into ssrA-tag peptides. We worried that introducing cysteines into every subunit of a ClpX hexamer would prevent ssrA-tag binding or result in disulfide bonds across the pore. To avoid these problems, we used covalently linked single-chain ClpX hexamers, which lack the N-domain but recognize ssrA-tagged substrates normally (Martin et al., 2005). Cysteines were then placed in just one subunit of a hexamer, either in the RKH loop (R228C), the pore-1 loop (Y153C), or the pore-2 loop (R200C), at positions that have been shown previously to be important for ssrA binding (Siddiqui et al., 2004; Farrell et al., 2007; Martin et al., 2007). All ClpX subunits in these experiments also contained either an E185Q mutation (E subunit), which eliminates ATP hydrolysis but allows subunits to adopt an ATP-state conformation, or an R370K mutation (R subunit), which prevents ATP hydrolysis and traps the subunit in an empty-state conformation (Joshi et al., 2004, Hersch et al., 2005; Martin et al., 2005). A mixture of E and R subunits was used to prevent translocation of the ssrA peptide during crosslinking and to mimic the normal asymmetric state of the ClpX hexamer, in which only a subset of subunits binds ATP. To test for crosslinking differences that depend upon nucleotide state, we performed separate experiments with the cysteine mutation placed in either an E or an R subunit.
To allow disulfide crosslinking to the ssrA tag, a set of peptides was synthesized with cysteine replacing each individual tag residue. These peptides also carried an N-terminal extension that contained a fluorescein for quantification of crosslinking after non-reducing SDS-PAGE and a biotinylated residue (Fig. 2A). Avidin was present during crosslinking experiments, as the avidin•biotin-ssrA complex mimics presentation of the ssrA tag from a native protein substrate (Fig. 2A). Indeed, this complex is a substrate for ClpX or ClpXP, which can pull the biotin out of its binding site in avidin in an ATP-dependent reaction (not shown). Crosslinking experiments were performed in the presence of ClpP and with ATPγS, which supports binding of ClpX to the ssrA tag, or without nucleotide as a control for non-specific interactions (Bolon et al., 2004a). Before crosslinking, pore-loop cysteines in ClpX were reacted with Ellman’s reagent to form an activated mixed disulfide and facilitate disulfide-bond formation with ssrA-peptide variants.
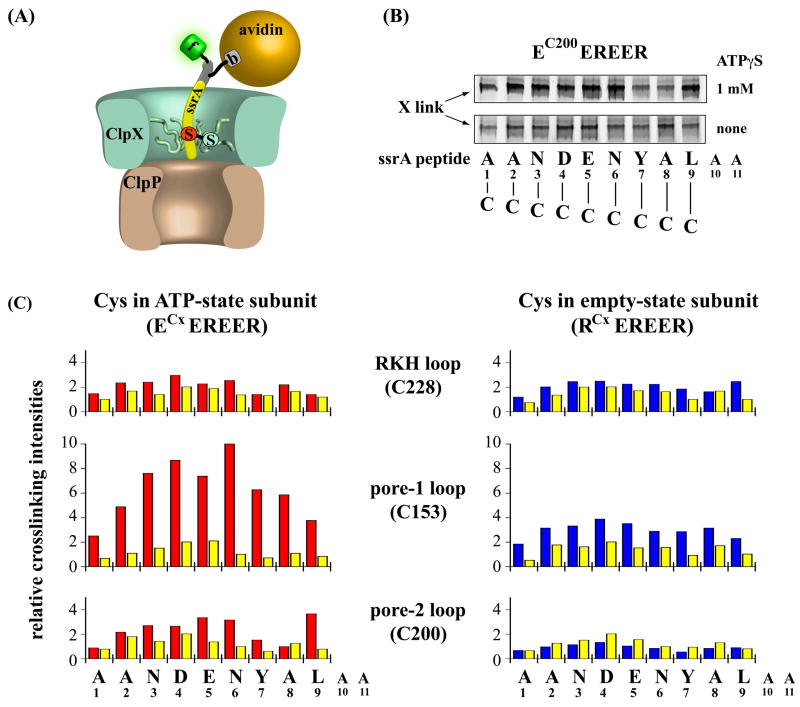
Figure 2.
Crosslinking of ssrA peptides to ClpX pore loops. (A) Cartoon depicting disulfide crosslinking between a cysteine-containing ssrA peptide and a cysteine introduced into a pore loop of ClpX. Peptides contained a single cysteine in the ssrA-tag portion of the molecule (red), a biotinylated residue (b) in an N-terminal extension (yellow), and a N-terminal fluorescein (f). The avidin-biotin complex mimics a native protein substrate and orients the ssrA peptide for binding to ClpX. (B) Non-reducing SDS-PAGE of fluorescent cysteine-substituted ssrA peptides crosslinked the single-chain ClpX hexamer EC200EREER with a cysteine in the pore-2 loop. Peptides with residues 1–9 of the ssrA tag individually replaced by cysteine were crosslinked to ClpX in the presence of ATPγS (upper panel) or in the absence of nucleotide to test for non-specific crosslinking (lower panel). Peptides A10C and A11C did not bind ClpX. (C) Bar graphs show the relative amounts of crosslinked product formed between cysteine-substituted ssrA peptides and a cysteine in the RKH loop (top panel), pore-1 loop (middle panel), or pore-2 loop (bottom panel) of a ClpX subunit trapped in the ATP state (ECEREER, red bars) or empty state (RCEREER, blue bars). Yellow bars show the intensities of non-specific crosslinking for each mutant in the absence of ATPγS. All intensities are averages from three independent crosslinking reactions; the highest mean intensity was assigned a value of 10. Standard deviations are not shown but were less than 10% of the mean.
Crosslinking of the ssrA tag to pore loops
Most cysteine substitutions in the ssrA tag were compatible with binding to ClpX. For example, cysteines at ssrA-tag positions 1 to 9 showed distinct patterns and strengths of crosslinking with cysteines in the three ClpX pore loops (Fig. 2B, 2C). However, neither binding nor specific crosslinking to ClpX was detected for the A10C or A11C peptides (not shown). Indeed, other substitutions for these residues also decrease ClpX binding dramatically (Flynn et al., 2001; Farrell et al., 2007), indicating that alanines at the two C-terminal tag positions are critical determinants of binding.
In the pore-1 loop, C153 crosslinked to many cysteines in the ssrA-peptide variants (Fig. 2C). Importantly, crosslinking to all positions was stronger in the presence of ATPγS (red or blue bars), than in the absence of nucleotide (yellow bars). Because ATP or ATPγS is required for ClpX to bind the ssrA tag, this result establishes that most of the observed crosslinking is specific. C153 crosslinked with high efficiency to the middle part of the ssrA tag when in an ATP-state subunit, but showed overall lower crosslinking intensities and shifted maxima when in an empty-state subunit. Hence, the “nucleotide” state of a ClpX subunit influences the geometry of its pore-1 loop relative to different parts of a bound ssrA tag.
In the pore-2 loop, C200 also formed specific ATP dependent and nucleotide-state dependent disulfide crosslinks with cysteines in the ssrA tag, although crosslinking intensities were generally weaker than observed for the pore-1 loop (Fig. 2C). When present in the empty-state subunit, little specific crosslinking of C200 to any of the peptide variants was observed, suggesting that the pore-2 loop is not close to the ssrA tag in this subunit conformation. In the ATP-state subunit, however, the most intense crosslinking of C200 was detected to cysteines near the middle of the tag and to cysteine at position 9 near the C-terminus of the ssrA tag (Fig. 2B, 2C). Position 9 is the first residue of the LAA motif that is specifically recognized by ClpX in the wild-type ssrA tag. Thus, it appears that the critical C-terminal portion of the ssrA tag binds deeply enough in the ClpX pore to contact the lowest pore loop. Importantly, conformational changes in ClpX driven by ATP hydrolysis were not required for the ssrA tag to reach this binding site. Maximum crosslinking was observed in about 1 min, whereas the rates of ATPγS hydrolysis by the ATPase deficient ClpX variants used for crosslinking were significantly less than 1 min−1 (not shown).
In the RKH loop, C228 crosslinked relatively poorly to all of the ssrA-peptide variants with efficiencies being largely independent of nucleotide state and only slightly higher in the presence of ATPγS than in the absence of nucleotide (Fig. 2C). In combination with previous experiments that implicate the RKH loop in ssrA-tag recognition, these crosslinking results suggest that the interaction between this loop and the ssrA tag probably occurs transiently during complex formation (see Discussion).
Interactions between SspB•ssrA complexes and ClpX pore loops
During SspB-mediated substrate delivery, the ssrA tag is bound both to the adaptor protein and to ClpX (Bolon et al., 2004a), potentially restricting or altering contacts between ClpX and the ssrA tag. It seemed possible, for example, that steric occlusion might prevent the C-terminal portion of the SspB-bound ssrA tag from reaching deeply enough into the central channel of ClpX to contact the pore-2 loop. To address this question, we asked if a pore-2 loop mutation altered binding of a covalently linked SspB-ssrA-tag complex. This covalent complex binds ClpX tightly (KD ≈ 15 nM) and has a structure nearly identical to the wild-type SspB•ssrA complex (Bolon et al., 2004a). Notably, we found that deletion of the ClpX pore-2 loop decreased affinity for the covalent SspB-ssrA complex more than 60-fold as measured by fluorescence anisotropy (Fig. 3a). This result cannot be explained by dramatic changes in ClpX structure, as the same pore-2 deletion mutant supports normal rates of ClpP degradation of other substrates (Martin et al., 2007). Thus, the pore-2 loop of ClpX plays important roles in binding the ssrA tag by itself or when bound to SspB.
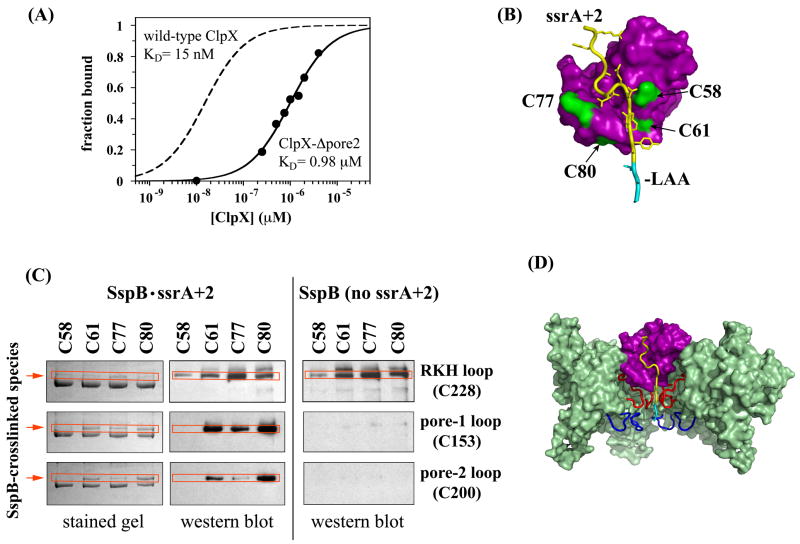
Figure 3.
Interaction of SspB•ssrA complexes with ClpX pore loops. (A) Binding of ClpX or ClpX-Δpore2 to a fluorescein-labeled disulfide-bonded SspB-ssrA complex (25 nM) was assayed by fluorescence anisotropy. The dashed binding curve for wild-type ClpX is from Bolon et al. (2004a). The solid binding curve for ClpX-Δpore2 reveals an approximate 65-fold decrease in affinity, implicating the pore-2 loop in SspB-mediated substrate delivery. (B) Surface-exposed positions around the ssrA-binding site in SspB were mutated individually to cysteine to probe potential disulfide crosslinking with ClpX. Most of the structure shown is based on the 1OU9 cocrystal structure (Levchenko et al., 2003), but the C-terminal six residues of the extended ssrA + 2 peptide were modeled. (C) Disulfide crosslinking between cysteine-substituted variants of SspB and single-chain variants of ClpX (ECEREER) with cysteines at position 228 (RKH loop), 153 (pore-1 loop), or 200 (pore-2 loop) was performed in the presence or absence of GFP-ssrA+2. Crosslinked products were detected by anti-SspB western blotting (middle and right panels) or by coomassie staining (left panel) after non-reducing SDS-PAGE. Crosslinking reactions contained 1μM cysteine-substituted single-chain ClpX, 10μM cysteine-substituted SspB, and 4 μM GFP-ssrA + 2. (D) In this model of ClpX (green; two subunits removed to allow visualization of the pore), equivalent pore-loops (pore-1 red; pore-2 blue) are at the same axial level (Bochtler et al., 2000; Kim and Kim, 2003) and SspB (purple) with a bound ssrA tag (yellow) enters the upper part of the ClpX channel. This model accounts well for direct SspB handoff of ssrA-tagged substrate to ClpX and for the role of the pore-2 loop in recognition of SspB•ssrA complexes, but it is difficult to rationalize strong crosslinking between the pore-2 loops and SspB.
The previous results and the fact that the ClpX- and SspB-binding determinants in the ssrA tag are closely spaced (Flynn et al., 2001; Levchenko et al., 2003; Song and Eck, 2003) suggest that SspB itself must enter at least the top part of the ClpX channel during substrate delivery. To probe crosslinking of SspB to ClpX, we substituted cysteines for individual surface residues of SspB (positions 58, 61, 77, or 80), which surround the region where the ssrA tag exits the SspB peptide-binding groove (Fig. 3B). These mutations did not interfere with the folding or function of SspB, and all Cys variants had normal activity in delivering ssrA-tagged substrate to wild-type ClpX (not shown). In the wild-type case, interactions between the C-terminal tail of SspB and the N-domain of ClpX tether the adaptor to the enzyme and help overcome clashes during substrate delivery (Wojtyra et al., 2003; Bolon et al., 2004b; Hersch et al., 2004; Martin et al., 2005; McGinness et al., 2007). Our single-chain ClpX variants for crosslinking lack the N-domain, and bringing SspB into their proximity only relies on the simultaneous binding of the ssrA tag to the adaptor and the enzyme. Considering the clash between ClpX and SspB, we were unsure if SspB-bound ssrA would interact with ClpX strongly enough to deliver SspB for crosslinking. However, an extended ssrA-peptide variant that relieves the adaptor-enzyme clash (AANDENYNYALAA; called ssrA+2; Hersch et al. (2004)) facilitated formation of ternary complexes with SspB and ClpX. Single-chain ClpX constructs containing a cysteine in one pore loop of an ATP-state subunit were activated for crosslinking by formation of a mixed disulfide and incubated, in the presence or absence of ssrA+2 peptide, with cysteine-substituted SspB and ClpP. Crosslinking was analyzed by non-reducing SDS-PAGE and anti-SspB western blotting (Fig. 3C).
C228 in the RKH loop of ClpX crosslinked to each of the four cysteine variants of SspB in the presence but also in the absence of ssrA+2 peptide (Fig. 3C). Thus, formation of the SspB•ssrA+2•ClpXP complex does not significantly enhance crosslinking between the RKH loop and SspB, suggesting that this loop is rather exposed and accessible to free SspB in solution. By contrast, crosslinking of the SspB variants to cysteines in the pore-1 and pore-2 loops was only observed in the presence of the ssrA+2 peptide (Fig. 3C). C153 in pore-1 loop of ClpX crosslinked to three of the four cysteine variants of SspB, whereas C200 in pore-2 loop of ClpX crosslinked efficiently to two of the SspB mutants and weakly to a third variant (Fig. 3C). Hence, formation of an SspB•ssrA+2•ClpXP complex brings SspB into the central channel of ClpX and close enough to the pore-1 and pore-2 loops to allow formation of disulfide bonds.
Control experiments established that crosslinking of cysteine-substituted SspB to the pore-1 and pore-2 loops of ClpX required formation of an SspB•ssrA+2•ClpXP complex and was not a consequence of conformational changes in SspB or ClpX upon binding to the ssrA peptide (not shown). For example, no disulfide-bond formation was observed when we used an extended ssrA tag with a C-terminal DAS sequence (AANDENYNYADAS), which has 100-fold lower affinity to ClpX but binds normally to SspB (McGinness et al., 2006; 2007). Similarly, no crosslinking was detected when we used an ssrA peptide (ACEDENYNYALAA) that binds normally to ClpX but fails to bind SspB (Levchenko et al., 2000; Bolon et al., 2004a).
Specific crosslinking was observed when we used the non-extended wild-type ssrA peptide to deliver SspB. However, as expected, the yields of crosslinked product were significantly lower than for ssrA+2, consistent with a potential clash between SspB and ClpX and therefore lower stability of the SspB•ssrA•ClpXP complex (Hersch et al., 2004).
Taken together, these results suggest that SspB enters at least the top portion of the ClpX pore during handoff of ssrA-tagged substrates (Fig. 3D). Moreover, if the pore-1 and pore-2 loops of ClpX can contact cysteines near the SspB binding groove, then they should be able to contact the LAA portion of the ssrA tag, which protrudes from the SspB groove. Interestingly, contacts between the ClpX pore-2 loop and SspB were largely restricted to the two cysteines (SspB positions 61 and 80) that would be closest to the LAA of the SspB-bound ssrA tag.
Discussion
Protein substrates must be engaged by proteolytic ATPases, like ClpX, before they can be unfolded and the denatured polypeptide can be translocated into the degradation chamber. Our results show that the ssrA tag of a substrate is initially bound in the central channel of the ClpX hexamer, where it interacts with loops that form the top, middle, and lower portions of this pore. Subsequent conformational changes in one or more of these loops, driven by cycles of ATP hydrolysis, could then translocate the tag, force unfolding of the attached native protein, and mediate translocation of the rest of the denatured substrate into ClpP for degradation.
A two-step model for ssrA-tag binding in the ClpX pore
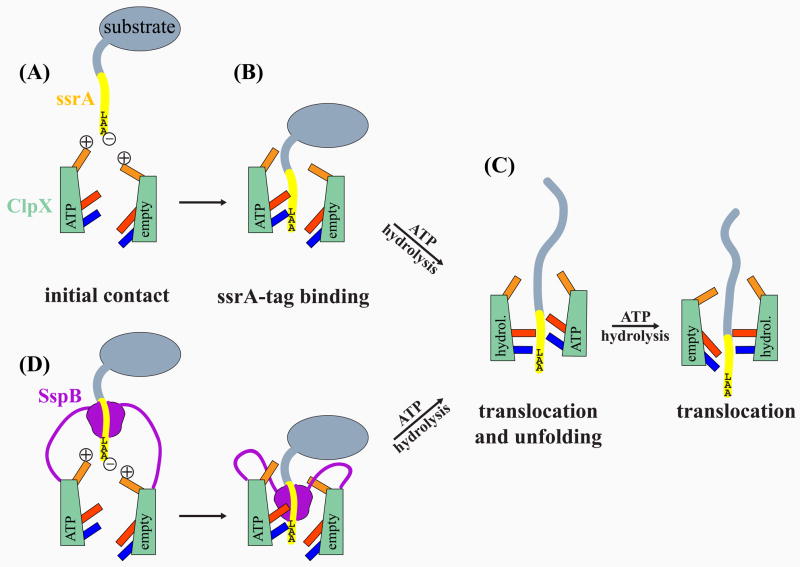
Although ClpX recognizes just the LAA-COO− portion of the ssrA tag (Flynn et al., 2001), the RKH, pore-1, and pore-2 loops all play roles in binding. Farrell et al. (2007) initially showed that mutations in the RKH loops, which surround the entrance to the ClpX channel, dramatically weaken binding to the ssrA tag, apparently by removing favorable electrostatic interactions with the negatively charged α-carboxylate. Consistent with a critical role of the RKH loop in recognition, we find that human ClpX variants bind the ssrA tag only when they contain the E. coli RKH loop.
Strong disulfide crosslinking between the RKH loop and the ssrA tag was not detected, however. We propose, therefore, that the interaction between these elements, although important, may be transient. For example, the RKH loops could function as a specificity filter, attracting the negatively charged C-terminus of ssrA-tagged substrates to the pore entrance, where it would then be positioned to move more deeply into the ClpX pore and to interact with the pore-1 and pore-2 loops (Fig. 4). This two-step binding model would explain why mutations in the pore-1 and pore-2 loops weaken or abolish ClpX binding to the ssrA tag (Siddiqui et al., 2004; Martin et al., 2007) and why human ClpX variants need both the E. coli RKH and pore-2 loops to recognize the ssrA tag efficiently. Moreover, this model is consistent with the efficient disulfide crosslinking we observed between the ssrA tag and the pore-1 and pore-2 loops, and it explains how the RKH and pore-2 loops, which are distant in the ClpX structure (Kim and Kim, 2003), can both be required for tight binding to a small region of the ssrA tag.
Figure 4.
Two-step model for ssrA-tag binding to the ClpX pore. The cartoons show a schematic ClpX complex with two subunits in ATP-bound or empty nucleotide states and their associated RKH loops (gold), pore-1 loops (red), and pore-2 loops (blue). (A) The -carboxylate of ssrA is attracted by transient electrostatic interactions with the positively charged RKH loop at the entrance of the ClpX channel. (B) Binding of the LAA motif to the pore-1 and pore-2 loops moves the ssrA tag deep into the pore. (C) ATP-hydrolysis driven conformational changes in the loops and/or entire subunits (“up” conformation in ATP state; “down” conformation in empty state) translocate the ssrA tag, unfold the substrate, and thread the unfolded chain through the ClpX pore. (D) SspB delivery of ssrA-tagged substrate. A staggered orientation of equivalent loops in different subunits may allow SspB to access the pore-2 loop for direct substrate handoff.
Cysteine substitutions for the C-terminal alanines of the ssrA tag prevented ClpX binding and therefore disulfide crosslinking. We found, however, that an antepenultimate cysteine in the tag (CAA-COO−) could be crosslinked efficiently to cysteines introduced into either the pore-1 or pore-2 loops of ClpX. In fact, the pore-2 loop crosslinked most intensely and specifically to this antepenultimate cysteine. Hence, the pore-2 loop may interact not only with the corresponding position in the wild-type ssrA tag but also with the entire LAA motif. This model is supported by our loop-transplant results and the previous finding that deletion or mutation of the pore-2 loop eliminates ClpX binding of the ssrA-tag (Martin et al., 2005), which is recognized primarily through its C-terminal alanines (Flynn et al., 2001; Farrell et al., 2007). Thus, we propose that the pore-1 and pore-2 loops of ClpX form much, if not all, of the binding site for the LAA-COO− of a fully engaged ssrA tag.
Implications for ClpX hexamer structures
The crystal structure of a ClpX subunit is known (Kim and Kim, 2003), but the arrangement of subunits in ClpX hexamers is uncertain. Hexameric structures are known for the unfoldase HslU (Bochtler et al., 2000; Sousa et al., 2000; Wang et al., 2001), which shares ≈50% sequence homology with the large and small AAA+ domains of ClpX. In a model of the ClpX hexamer based on HslU (Kim and Kim, 2003), equivalent pore loops from different subunits occupy similar positions along the pore axis. In other words, the RKH loops are all at the top of the pore, the pore-1 loops are all in the middle where the channel is most constricted, and the pore-2 loops are all near the bottom (Fig. 1A). We refer to this structure as a “level” hexamer. If the ssrA tag of a substrate bound to this ClpX conformation, then the C-terminal LAA of the tag would have to extend past the pore-1 loops almost to the bottom of the central channel. This seems possible as the 11-residue ssrA tag in a fully extended conformation could span a distance of roughly 22 Å and an attached native protein could plausibly enter the top portion of the pore, which is relatively wide. It is more difficult, however, to rationalize crosslinking between the pore-2 loops of ClpX and SspB in terms of a level hexamer model. Although a complex of SspB with an ssrA-tagged substrate could enter the upper part of the pore, the pore-1 loops and/or ssrA tag would be expected to block access of the pore-2 loops to SspB, and the pore-2 loops would be too far from SspB to allow disulfide crosslinking (Fig. 3D). Because both unassisted and SspB-mediated delivery of ssrA-tagged substrates to ClpX share common requirements for the LAA of the tag and for the pore-2 loop of ClpX, our results suggest that it is unlikely that ClpX initially recognizes substrates as a level hexamer.
We favor an initial binding model in which equivalent pore loops in different subunits in the ClpX hexamer are axially staggered with respect to each other (Fig. 4), as observed in some hexamers of the AAA+ protease FtsH and in several hexameric ATP-dependent helicases (Singleton et al., 2000; Bieniossek et al., 2006; Enemark and Joshua-Tor, 2006). In a staggered conformation, some pore-2 loops of ClpX would be higher up in the central channel, allowing contacts with SspB as well as the ssrA tag during substrate delivery (Fig. 4). This arrangement would enable direct handoff, in which the N-terminal portion of an ssrA tag remained bound to SspB, while the C-terminal part of the tag contacted the pore-1 and pore-2 loops of ClpX during substrate delivery. A staggered hexamer conformation could also explain our finding that cysteines in both the pore-1 and pore-2 loops of ClpX can crosslink to positions in the ssrA tag that span roughly 8–10 residues, showing several local crosslinking maxima.
Nucleotide-dependent loop movements
We found that the pore-2 loop in an ATP-state subunit of ClpX could be crosslinked to the ssrA tag, but the same loop in an empty-state subunit showed no substantial crosslinking. This behavior suggests that this loop assumes different conformations depending upon the presence or absence of bound nucleotide in the host subunit. For example, the pore-2 loop in an ATP-state subunit could move up towards a bound ssrA tag, whereas this loop in a empty subunit might move down, away from the tag and towards the ClpP-binding interface of ClpX. Previous crosslinking studies between the pore-2 loop of ClpX and ClpP also support the existence of “up” and “down” conformations of the pore-2 loop (Martin et al., 2007). The distinct patterns of ssrA-tag crosslinking to the ClpX pore-1 loop in ATP-state and empty-state subunits also suggest that this loop adopts different nucleotide-dependent conformations. During the normal ATPase cycle of ClpX, such changes in the conformations of the pore-1 loops may produce the power stroke and drive substrate unfolding and translocation.
General principles for substrate recognition
SsrA-tagged substrates are also degraded by the ClpAP protease (Gottesman et al., 1998), albeit ClpA does not contain a RKH loop and strongly differs in its pore-2 loop sequence from ClpX. It is known, however, that ClpA recognizes the ssrA tag differently from ClpX and that the bound determinants do not include the C-terminal residue or the α-carboxylate of ssrA (Flynn et al., 2001). Yet, mutational and substrate crosslinking studies indicated that the pore-1 and pore-2 loops of ClpA are involved in ssrA binding (Hinnerwisch et al., 2005). Thus, although the details of ssrA-tag binding to ClpX and ClpA differ, both enzymes use similar general strategies to engage the ssrA tag of a bound substrate in their central pores. It seems likely that substrate engagement by AAA+ unfoldases will generally require an initial binding step, in which part of the polypeptide is brought deeply enough into the pore to allow interaction with the translocation machinery. For ssrA-tag binding to ClpX, interactions of the LAA motif with the pore-2 loop would position a segment of the tag past the pore-1 loop so that ATP-dependent conformational changes in the pore-1 loop could initiate translocation and subsequent unfolding of an attached substrate. The pore-1 loop of ClpX has highly conserved counterparts in all AAA+ unfoldases (Neuwald et al., 1999). Hence, the key event in substrate recognition appears to be the binding of the degradation tag in a manner that allows appropriate interactions with this conserved pore loop.
Experimental Procedures
Protein expression and purification
Genes for human mitochondrial ClpX and ClpP (hClpX; hClpP) were kindly provided by Dr. Michael Maurizi (NIH). An hClpX variant (residues 154-633; hClpX-ΔN) lacking its N-terminal domain and hClpX-ΔN mutants with RKH and/or pore-2 loops transplanted from E. coli ClpX (eClpX) were generated by PCR and cloned into pACYCDuet-1 (Novagen). In the pore-2 mutant, hClpX residues 364-GSVPGIHQL-372 were replaced with eClpX residues 190-SRKSDNPSIT-199; in the RKH mutant, hClpX residues 397-EKNSRKLRGET-407 were replaced with eClpX residues 224-PQGGRKHPQQEF-235. All hClpX-ΔN variants contained an N-terminal His6 tag. Linked single-chain hexamers of E. coli ClpX-ΔN (residues 62–424) had a C-terminal His6 tag and were constructed by PCR and cloned into pACYCDuet-1 (Martin et al., 2005).
Variants of hClpX, variants of single-chain eClpX, and hClpP were expressed for 3 h at 22°C in the _recA_− E. coli strain BLR (DE3) and purified as described (Martin et al., 2007). Unlinked wild-type eClpX, E. coli ClpP-His6, SspB, and SspB variants were expressed and purified as described (Kim et al., 2000; Burton et al., 2003; Bolon et al., 2004a). GFP-His6 variants were constructed with AANDENYALAA (wild-type ssrA), AANDENYNYALAA (ssrA+2), ACEDENYALAA (ssrA-CE), or AANDENYADAS (ssrA-DAS) C-terminal tags, expressed from pACYC in E. coli BLR (DE3), and purified by Ni++-NTA affinity and ion-exchange chromatography (Kim et al., 2000; Bolon et al., 2004a). Unlabeled or 35S-labeled titin-I27-ssrA was expressed and purified as described (Kenniston et al., 2003, 2005). To obtain unfolded carboxymethylated (CM) titin-I27-ssrA, the protein was alkylated for 3 h at 22°C with a 100-fold excess of iodoacetic acid at pH 8.8 in 6 M GuHCl. The SspB Y44C/D147C variant was labeled at C147 with 5-iodoacetamidofluorescein and disulfide-bonded via C44 to an ssrA A2C peptide as described (Bolon et al., 2004a).
Variant ssrA peptides used for disulfide-crosslinking experiments were derivatives of the sequence fl-βKKGRE*HGAANDENYALAA (fl, fluorescein; β, β–alanine; E*, glutamate with a biotin linked to the side chain via a PEG linker) in which the underlined residues in the ssrA tag were replaced individually by cysteine. Peptides were synthesized by the MIT Biopolymers Laboratory, reduced by treatment with 50 mM DTT for 1 h at 22°C (pH 8.0), purified by HPLC on a C18 reverse-phase column, and dissolved in 60 μM HCl, 300 mM KCl, 10% glycerol for storage. Peptide concentrations were determined based on the absorbance of fluorescein in basic ethanol (extinction coefficient 91989 M−1cm−1 at 500 nm) and/or the amount of biotin using the HABA assay (Pierce).
Biochemical assays
Degradation of 35S-labeled CM-titin-ssrA by eClpX or hClpX-ΔN variants (0.3 μM pseudo-hexamer equivalents) and eClpP14 or hClpP14 (0.9 μM) was performed at 30°C in PD buffer (25 mM HEPES (pH 7.6), 100 mM KCl, 20 mM MgCl2, 1 mM EDTA, 10% glycerol) with an ATP regeneration system (5 mM ATP, 16 mM creatine phosphate, 6 μg/ml creatine phosphokinase) and was assayed by the release of acid-soluble peptides (Kenniston et al., 2003). The degradation of GFP-ssrA was monitored by the loss of GFP fluorescence (Kim et al., 2000) using a QM-2000-4SE spectrofluorimeter (Photon Technology International).
Steady-state ATPase assays were used to monitor ClpX•substrate binding using 0.3 μM eClpX or hClpX (hexamer equivalents), 0.9 μM eClpP14 or hClpP14, and 20 μM ssrA-tagged substrate at 30°C in PD buffer with an NADH-coupled regeneration system as described (Burton et al., 2003). The binding of 50 μM cysteine-containing ssrA peptides in complex with avidin to 0.3 μM ClpX was assayed by repression of ClpX ATPase activity compared to control reactions without peptide.
Binding of the fluorescein-labeled disulfide-bonded SspB-ssrA complex (25 nM) to ClpX wild-type or to ClpX-Δpore2 was assayed by fluorescence anisotropy (excitation 477 nM; emission 515 nm) in PD buffer plus 5 mM ATPγS at 30°C using a QM-2000-4SE spectrofluorimeter (Photon Technology International).
Crosslinking
Cysteines introduced into the pore loops of single-chain ClpX hexamers were activated by formation of a mixed disulfide with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), following buffer exchange using Micro Bio-Spin columns (BIO-RAD) to remove DTT present during ClpX purification. ClpX variants (10 μM pseudo-hexamer) were incubated with 1.5 mM DTNB for 5 min at 22°C in XL-buffer (25 mM HEPES (pH 7.8), 300 mM KCl, 20 mM MgCl2, 1 mM EDTA, 10% glycerol). After separation of free DTNB by buffer exchange, the DTNB-activated ClpX (1 μM) was mixed with ClpP (2 μM) in the absence or presence of 1 mM ATPγS or 4 mM ATP; and cysteine-containing ssrA peptide (50 μM) in complex with avidin (50 μM) in XL-buffer was added. Crosslinking was allowed to proceed for 2 min at 22°C, before stopping the reaction by addition of ice-cold 8.5% TCA, 8 mM biotin. The protein precipitate was washed with ethanol and dissolved in buffer containing 600 mM Tris (pH 8.5), 9 M urea, 3 mM EDTA, and 200 mM iodoacetic acid. Alkylation of free cysteines was allowed to proceed for 30 min at 22°C, 15 mM biotin was added, SDS loading buffer was added, and samples were boiled for 2 min prior to separation by non-reducing SDS-PAGE. The amount of ssrA peptide crosslinked to the single-chain ClpX hexamer was determined by fluorescein-fluorescence using a Typhoon 9400 imager (Amersham Biosciences). Control experiments established that cysteines in different ssrA-peptide variants formed disulfide crosslinks with a test protein containing an activated cysteine with comparable reactivities.
Cysteine-substituted SspB mutants (10 μM) were premixed with GFP-ssrA or variants with altered ssrA tags (4 μM) and incubated with DTNB-activated single-chain ClpX hexamers (1 μM) in the presence of ATPγS (3 mM) for 15 min at 22°C to allow crosslinking. Reactions in the absence of ssrA-tagged substrate were performed to analyze the dependence of crosslinking on the formation of SspB•ssrA•ClpX complexes. Crosslinking reactions were stopped by alkylating free cysteines for 30 min with 150 mM iodoacetic acid in 400 mM Tris-HCl (pH 8.5), 6.0 M urea, 2 mM EDTA. Following non-reducing SDS-PAGE, disulfide-crosslinked SspB-ClpX complexes were detected by western blotting using an anti-SspB antibody.
Acknowledgments
This research was supported by NIH grant AI-16892. We thank M. Maurizi And D. Bolon for providing materials and XXX for helpful discussions. A.M. was supported by a Merck/MIT CSBi postdoctoral fellowship. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bieniossek C, Schalch T, Bumann M, Meister M, Meier R, Baumann U. The molecular architecture of the metalloprotease FtsH. Proc Natl Acad Sci. 2006;103:3066–71. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell. 2004a;16:343–50. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: a protein-design study. Mol Cell. 2004b;13:443–9. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: Linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 2003;12:893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell. 2003;12:373–80. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Mol Cell. 2007;25:161–166. doi: 10.1016/j.molcel.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;11:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Langer T. Substrate specific consequences of central pore mutations in the i-AAA protease Yme1 on substrate engagement. J Struct Biol. 2006;156:101–8. doi: 10.1016/j.jsb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nature Rev. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Baker TA, Sauer RT. SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc Natl Acad Sci. 2004;101:12136–41. doi: 10.1073/pnas.0404733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–41. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem. 2002;277:21095–102. doi: 10.1074/jbc.M201642200. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of a AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–6. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol Cell. 2003;12:365–72. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22:701–7. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- McGinness KE, Bolon DN, Kaganovich M, Baker TA, Sauer RT. Altered tethering of the SspB adaptor to the ClpXP protease causes changes in substrate delivery. J Biol Chem. 2007;282:11465–73. doi: 10.1074/jbc.M610671200. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Okuno T, Yamanaka K, Ogura T. Characterization of mutants of the Escherichia coli AAA protease, FtsH, carrying a mutation in the central pore region. J Struct Biol. 2006;156:109–14. doi: 10.1016/j.jsb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Park E, Rho YM, Koh OJ, Ahn SW, Seong IS, Song JJ, Bang O, Seol JH, Wang J, Eom SH, Chung CH. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–8. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- Prakash S, Matouschek A. Protein unfolding in the cell. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Piszczek G, Rozycki J, Singh SK, Ginsburg A, Maurizi MR. The molecular chaperone, ClpA, has a single high affinity peptide binding site per hexamer. J Biol Chem. 2005;280:12221–30. doi: 10.1074/jbc.M411733200. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–15. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, a AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HK, Eck MJ. Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine. Mol Cell. 2003;12:75–86. doi: 10.1016/s1097-2765(03)00271-5. [DOI] [PubMed] [Google Scholar]
- Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc Natl Acad Sci. 2003;97:14103–8. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell. 2003;12:355–63. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem Biol. 2002;9:1237–45. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–65. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wojtyra UA, Thibault G, Tuite A, Houry WA. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem. 2003;278:48981–90. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]
- Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem. 2003;278:50182–7. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]