PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells (original) (raw)
Abstract
Earlier work has shown that the transcription factor C/EBPα induced a transdifferentiation of committed lymphoid precursors into macrophages in a process requiring endogenous PU.1. Here we have examined the effects of PU.1 and C/EBPα on fibroblasts, a cell type distantly related to blood cells and akin to myoblasts, adipocytes, osteoblasts, and chondroblasts. The combination of the two factors, as well as PU.1 and C/EBPβ, induced the up-regulation of macrophage/hematopoietic cell surface markers in a large proportion of NIH 3T3 cells. They also up-regulated these markers in mouse embryo- and adult skin-derived fibroblasts. Based on cell morphology, activation of macrophage-associated genes, and extinction of fibroblast-associated genes, cell lines containing an attenuated form of PU.1 and C/EBPα acquired a macrophage-like phenotype. The lines also display macrophage functions: They phagocytose small particles and bacteria, mount a partial inflammatory response, and exhibit strict CSF-1 dependence for growth. The myeloid conversion is primarily induced by PU.1, with C/EBPα acting as a modulator of macrophage-specific gene expression. Our data suggest that it might become possible to induce the transdifferentiation of skin-derived fibroblasts into cell types desirable for tissue regeneration.
Keywords: cell reprogramming, differentiation plasticity, hematopoiesis
It has long been assumed that differentiation is an irreversible process. This view has been called into question by the fact that mammals can be cloned by the transfer into oocytes of nuclei from differentiated cells, including those of B and T cells (1). Moreover, it was recently shown that mouse and human fibroblasts can be reprogrammed into cells with an embryonic stem cell phenotype by the ectopic expression of four transcription factors. These cells are capable of generating cells of all three germ layers (2, 3). Earlier work showed that within the hematopoietic system ectopic expression of a single transcription factor can induce transdifferentiation of committed cells from one lineage into another at very high frequencies. For example, GATA-1 can convert myeloid cells into megakaryocyte/erythroid cells by antagonizing the Ets family factor PU.1 whereas expression of PU.1 in megakaryocyte/erythroid cells can induce their conversion into myeloid cells (4). Likewise, enforced expression of either C/EBPα or C/EBPβ in pre-B or pre-T cells induces their transdifferentiation into functional macrophages at high frequencies. This process requires endogenous PU.1 and involves inactivation of key lymphoid regulators (5, 6). It has also been shown that C/EBPα or PU.1 are necessary for the formation of myeloid cells because mice lacking either gene lack macrophages and granulocytes (7). Together these results indicate that the combination of C/EBPα/β and PU.1 specifies a macrophage phenotype in a hematopoietic context, consistent with their ability to bind to and activate regulatory regions of myelomonocytic genes (8) and to form a protein complex (9).
An explanation for the ease with which a single transcription factor can convert committed hematopoietic cells into one another is that blood cells are closely developmentally related, probably sharing transcriptional networks and chromatin configurations. Implied in this assumption is the prediction that hematopoietic instructive transcription factors are unable to reprogram more distantly related cells. Based on the unexpected finding that PU.1 and C/EBPα up-regulate Mac-1 expression in fibroblasts (5), here we have asked whether their expression is sufficient to activate a myeloid program in fibroblasts. The latter cells are derived from mesenchymal stem cells and are thus more closely related to adipocytes, myocytes, osteoblasts, and chondrocytes than to blood cells (10). Surprisingly, we found that the two transcription factors induce a macrophage-like phenotype in a fibroblast cell line as well as in primary embryonic and adult fibroblasts.
Results
PU.1 and C/EBPα Up-Regulate Hematopoietic Cell Surface Antigens in NIH 3T3 Cells and in Primary Fibroblasts.
To test whether myeloid instructive transcription factors can activate hematopoietic genes in fibroblasts, NIH 3T3 cells were coinfected with PU.1 and C/EBPα viruses tagged with GFP and hCD4, respectively. FACS analysis of the infected cells distinguished four populations: uninfected, single infected, and double infected cells [supporting information (SI) Fig. S1]. PU.1 induced the up-regulation of the myelomonocytic marker Mac-1 and the panhematopoietic marker CD45 in 35–40% of the infected cells after 5 days. Coexpression of C/EBPα, while not altering the level of CD45, increased the number of Mac-1+ cells to 90%. No activation of Mac-1 and CD45 was seen with C/EBPα alone or with control GFP and hCD4 viruses (Fig. 1 A and B). Infection of cells with PU.1 and C/EBPβ yielded results similar to those with C/EBPα (Fig. S2). The presence of two distinct CD45 populations raised the possibility that the parental 3T3 cell line is heterogenous. However, this is not the case because both Mac-1 and CD45 expression could be induced in all of 12 3T3 subclones tested (Fig. S3).
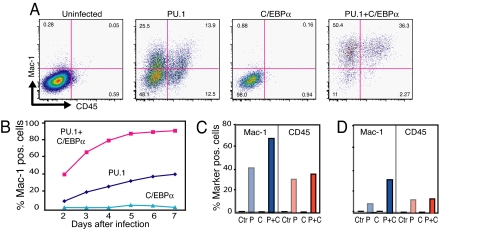
Fig. 1.
Induction of hematopoietic cell surface antigen expression in NIH 3T3 cells coinfected with PU.1-GFP and C/EBPα-hCD4 retroviruses. (A) FACS profiles of day-7-infected cells showing Mac-1 and CD45 expression in uninfected, single infected, and double infected cells, gated for GFP and hCD4 expression (Fig. S1). Numbers in quadrants represent the percentages of cells. (B) Kinetics of Mac-1 expression in cells expressing PU.1 and C/EBPα (red squares), PU.1 only (dark blue diamonds), and C/EBPα only (light blue triangles). (C) Mac-1 and CD45 expression in primary mouse embryo fibroblasts infected for 11 days with PU.1 (P) and C/EBPα (C) or both (P+C) viruses. Ctr, control. (D) As in C but using skin biopsy-derived fibroblasts.
To test whether expression of hematopoietic/macrophage antigens can also be induced in primary fibroblasts, cultures were prepared from mouse embryos as well as from a skin biopsy of an adult mouse. As with the cell line, GFP+ cells with an altered morphology could be observed. Approximately 70% of the embryo-derived fibroblasts coinfected with PU.1 and C/EBPα expressed Mac-1+ (Fig. 1C), with adult-derived cells showing percentages approximately two times lower (Fig. 1D). A proportion of the infected primary fibroblasts also expressed CD45 (Fig. 1 C and D). Between 30% and 40% of the Mac-1-positive cells also expressed CD45 (data not shown).
NIH 3T3-Derived Cell Lines Express Hematopoietic Cell Surface Antigens and Resemble Macrophages.
To determine which domains of PU.1 are required for its Mac-1-inducing activity, we tested three deletion mutants. Mutants lacking the transactivation and DNA binding domain did not activate Mac-1 in 3T3 cells. In contrast, PU.1ΔPEST did, although at a reduced efficiency compared with wild-type PU.1 (PU.1wt). However, PU.1ΔPEST and PU.1wt coexpressed with C/EBPα yielded comparable efficiencies (Fig. S4_A_). In an attempt to obtain cell lines stably expressing the factors, 3T3 infected with PU.1wt or PU.1ΔPEST with or without C/EBPα were sorted singly into 96 wells in growth medium containing 20 ng/ml CSF-1 (M-CSF), and colonies were scored after 3 weeks. PU.1wt-expressing colonies were small and did not develop into stable lines. In contrast, PU.1ΔPEST cell colonies were larger (Fig. S4_B_), and several of these could be grown into lines. Colonies H2 and G5, containing both PU.1ΔPEST and C/EBPα, were subcloned and characterized further. These lines were designated PC2.3, PC2.6, and PC5.3.
All three lines expressed Mac-1 and CD45 antigens, with Mac-1 levels varying over a wider range than in control macrophages (Figs. S4_C_ and S5_A_). The cells were also more adherent than 3T3 cells and more refractory to brightfield illumination and often formed long cytoplasmic extensions. They occasionally contained large vacuoles and became multinucleated (Fig. 2A). Staining for F-actin and an activated form of paxillin revealed that PC2.3 cells had fewer actin stress fibers and smaller but more numerous focal adhesion sites than 3T3 cells (Fig. 2B), resembling those seen in BAC macrophages. In conclusion, the converted cells more closely resembled the BAC1.2F5 macrophage cell line used as a control than the original 3T3 cells.
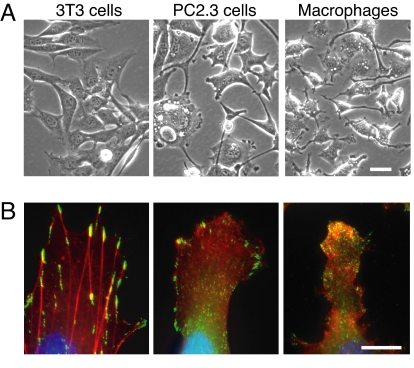
Fig. 2.
Morphological characterization of a cell line (PC2.3) coexpressing PU.1ΔPEST and C/EBPα. (A) Phase contrast images of 3T3 and PC2.3 cells and BAC macrophages. (B) Cells stained with phalloidin to reveal F actin (red), antibodies against paxillin Y118 to reveal focal contacts (green), and DAPI to reveal nuclei (blue). (Scale bars: 10 μm.)
The Converted Cell Lines Up-Regulate Macrophage-Restricted Genes and Down-Regulate Fibroblast-Associated Genes.
We next performed a gene expression array analysis of the PC2.3 cell line using 70-mer mouse probes. For this, cDNAs from PC2.3 cells were labeled with a yellow-green fluorophore, mixed with red fluorophore-labeled cDNAs from either 3T3 fibroblasts or BAC macrophages and hybridized to the arrays. This resulted in 8,042 informative genes. As shown in Tables S1 and S2, when comparing BAC macrophages with 3T3 cells, 960 genes were expressed at least two times more highly. Of these, 620 genes (65%) were also up-regulated in PC2.3 compared with 3T3 cells. A total of 1,052 genes were expressed at least two times lower in BAC macrophages than in 3T3 cells, of which 212 (20%) were also down-regulated in PC2.3 cells. When comparing PC2.3 cells with 3T3 cells, 953 genes were up-regulated, of which 598 (63%) were also expressed more highly in BAC macrophages than in 3T3 cells. Conversely, of 400 genes down-regulated in PC2.3 cells relative to 3T3 cells, 231 (58%) were also expressed at lower levels in BAC macrophages compared with 3T3 cells. The top 250 up- or down-regulated genes in each of the two comparisons are illustrated in Fig. S6.
Macrophages perform important functions in host defense, innate immunity, and tissue remodeling. They clear pathogens, cellular debris, and apoptotic cells and can be activated by inflammatory stimuli to secrete cytokines and chemokines (11). To obtain an indication of the functionality of the converted cells we therefore plotted the relative expression levels of selected genes relevant for macrophage functions. As shown in Fig. 3A, macrophage-associated genes involved in adhesion, motility, and invasiveness were up-regulated, such as those encoding the αM and β2 integrin chains of Mac-1, the actin cytoskeleton modulator WAS, and the tyrosine phosphatases PTPRC (CD45) and PTP-Phi (data not shown). Likewise, genes involved in phagocytosis, such as Fcγ receptor IIB, CD36 (LDL receptor), CDC42, and Rac2, as well as the tyrosine kinases Syk and Lyn and the chemokine/receptor genes CCl2, CCl3, CCl7, CCl9, and CXCL4 were up-regulated. Four genes encoding respiratory burst proteins were up-regulated, namely p22-phox, p47-phox, p67-phox, and p40-phox. Of genes involved in innate immunity, those encoding the Toll-like receptors Tlr-2 and Tlr-4, the LPS coreceptor CD14, and the adaptor MyD88 were also up-regulated. Finally, of genes involved in growth control, those encoding CSF-1R and GM-CSFRβ were up-regulated, whereas CSF-1 and PDGFRα were down-regulated. The data also show that the differences in expression levels of up- or down-regulated genes in the converted cells were typically less pronounced than in BAC macrophages. This is also seen when plotting the fold expression changes of BAC macrophages versus 3T3 cells and PC2.3 cells versus 3T3 cells (Fig. 3B).
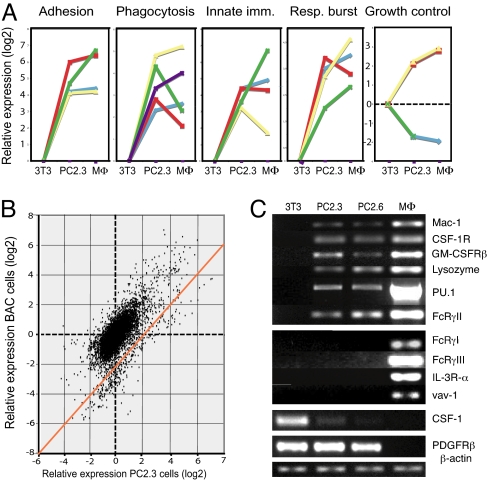
Fig. 3.
Gene expression analysis of PC2.3 cells. (A) Graphs showing expression of selected functional genes in PC2.3 cells and in BAC macrophages (MΦ) relative to 3T3 cells (delta log2 values). Adhesion and motility genes: β2 integrin (itgb2, blue) and αM integrin chains (itgam, red) of Mac-1, Was (was, yellow), and PTPRC (ptprc, green). Respiratory burst genes: p22-phox (cyba, green), p47-phox (ncf1, blue), p67-phox (ncf2, red), and p40-phox (ncf4, yellow). Innate immunity genes: tlr2 (blue), tlr4 (red), CD14 (yellow), CD80 (green), and CD37 (purple). Growth control genes: CSF-1R (csf1r, yellow line), GM-CSFRβ (csf2rb, red), CSF-1 (csf1, blue), and PDGFRα (pdgfra, purple). (B) Relative expression of BAC macrophage genes and PC.3 genes versus 3T3 fibroblasts plotted against each other. The broken lines correspond to positions of genes where no change in expression was observed. The orange line shows the positions of genes that show a perfect correlation between the expression of BAC macrophage and PC2.3 cell genes. (C) Semiquantitative RT-PCR of 3T3 cells, PC2.3/2.6 cells, and bone marrow-derived macrophages. cDNA in the samples was adjusted to comparable levels with β-actin cDNA. The PU.1 probe used detects endogenous PU.1.
Expression of selected genes was also tested by semiquantitative RT-PCR (Fig. 3C). Both PC2.3 and PC2.6 lines expressed mRNA for the αM chain of Mac-1, CSF-1 receptor, the GM-CSF-1 receptor β-chain, and lysozyme M, as well as that of endogenous PU.1, confirming the array data. We also found expression of mRNAs for CD32 (Fcγ receptor IIb), but not for CD64 and CD16 (Fcγ receptors I and III) or for the IL-3 receptor α-chain and the panhematopoietic marker vav-1. As expected from the arrays, CSF-1 was down-regulated in PC2.3/6 cells and undetectable in macrophages. In contrast, PDGFRβ was expressed in both 3T3 and PC2.3/6 cells but not in macrophages. Similar results were also obtained with the PC5.3 line (Fig. S5_B_). FACS analyses confirmed up-regulation of CSF-1R and down-regulation of CSF-1 at the protein level (Fig. S7).
PC2.3 Cells Are Phagocytic and Mount a Partial Inflammatory Response.
Three assays were used to test the phagocytic capacity of the converted cells. The first consisted of incubating the cells with inactivated dsRed-labeled Escherichia coli and measuring uptake by flow cytometry. As can be seen in Fig. 4A, 96% of PC2.3 cells were positive for the uptake of bacteria, compared with 3% for 3T3 cells. Similar results were obtained with the PC5.3 line (Fig. S5_C_). In another assay, PC2.3 and control cells were incubated with red fluorescent carboxylated beads (1 μm) and imaged by confocal microscopy to ensure that the particles were not merely attached to the cells. As illustrated in Fig. 4B, there was a significant increase in the number of intracellular beads in PC2.3 cells versus 3T3 cells. Scoring cells with sections containing more than five beads, 83% of PC2.3 cells were particle-positive versus 3% of 3T3 cells and 100% of BAC macrophages. Primary mouse embryo fibroblasts coinfected with PU.1ΔPEST and C/EBPα viruses were also tested in this assay. The majority of the GFP-positive cells showed a dramatic accumulation of the fluorescent beads (Fig. S8 A–C). To determine whether PC2.3 cells are also capable of exhibiting Fc receptor-dependent phagocytosis they were incubated with sheep RBCs (SRBCs) coated with rabbit anti-SRBC antibodies. Most of the cells incubated with opsonized SRBCs formed Fcγ receptor-mediated rosettes (Fig. S8_D_). Rosetting could be prevented by omitting the antibody coating or by preincubation of PC2.3 cells with antibodies to FcγRII and III (data not shown). After a second incubation to permit internalization of opsonized SRBCs, cells were treated with a buffer that lyses noningested SRBCs. No SRBCs could be detected inside the PC2.3 cells after lysis (Fig. S8_E_), whereas BAC macrophages were highly positive (Fig. S8 F–H). The capacity of PC2.3 cells to form Fc rosettes and their inability to phagocytose opsonized cells can be explained by the observed expression of FcγRIIb and the absence of FcγRI and FcγRIII, because it is known that only the latter receptors mediate cell internalization (12).
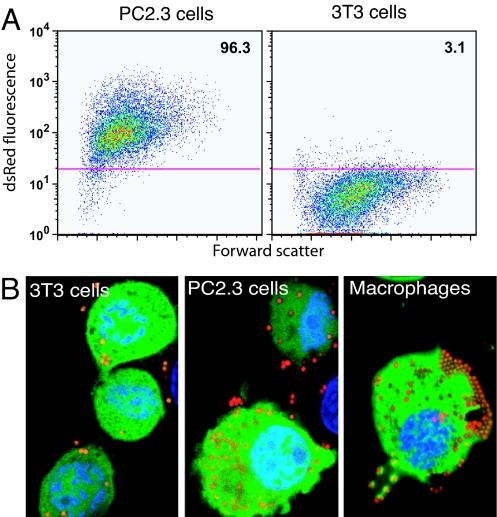
Fig. 4.
Phagocytic capacity of PC2.3 cells. (A) Infection with dsRed E. coli and measure of the bacteria uptake by FACS analysis, comparing PC2.3 cells to 3T3 cells. (B) Capacity to ingest beads. 3T3 cells, PC2.3 cells, and BAC1.2F5 macrophages were incubated with 1-μm carboxylated red fluorescent beads and washed. The confocal images (planes of 0.01 μm) show GFP in green, beads in red-orange, and nuclei in blue.
To test whether PC2.3 cells can mount an inflammatory response, they were incubated with 10 μg/ml LPS for 6 h and analyzed for the expression of TNFα, IL-6, IL-1β, and MIP1α by quantitative RT-PCR. Whereas BAC macrophages mounted the expected response, PC2.3 cells showed only a modest up-regulation of IL-6, IL-1β, and MIP1α and no change for TNFα. 3T3 cells showed no response (Fig. S9).
PC2.3 Cells Have Acquired CSF-1 Growth Dependence.
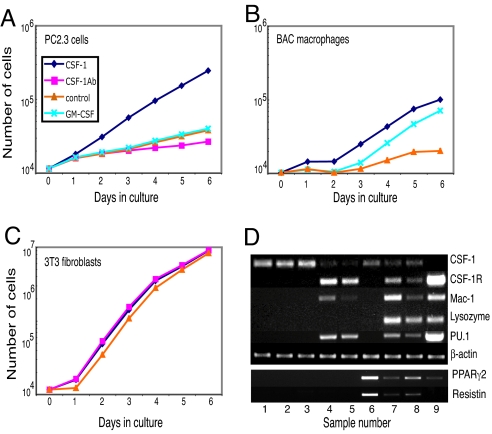
The expression array, RT-PCR, and FACS results showed that converted cells down-regulate the expression of CSF-1 and up-regulate the CSF-1 receptor. To measure CSF-1 production in the converted cells, PC2.3 and PC2.6 cells were washed and incubated with cytokine-free medium for 10 h. The CSF-1 activity in the supernatants was then tested by using RIA. Whereas 3T3 cells produced 9 ng/ml CSF-1, the converted lines produced 0.4–0.6 ng/ml CSF-1 and the BAC macrophages were negative (Fig. S10). We next tested whether the converted cells acquired CSF-1 dependence for growth by seeding them with and without the cytokine. PC2.3 cultures in the presence of CSF-1 had a population doubling time of ≈30 h (Fig. 5A), similar to that of BAC macrophages (Fig. 5B). In the absence of CSF-1 the doubling time was significantly extended to ≈100 h, similar to what was observed with BAC macrophages (Fig. 5A). CSF-1-neutralizing antibody further reduced the residual growth of PC2.3 cells in the absence of CSF-1, and CSF-1 addition could not be replaced by GM-CSF (Fig. 5A). The population doubling time of 3T3 cells was ≈11 h under all conditions (Fig. 5C). Finally, similar to BAC macrophages, PC2.3 cells formed clusters of disperse cells when grown in semisolid medium containing CSF-1 but formed no colonies without the cytokine (data not shown).
Fig. 5.
Growth factor dependence of PC2.3 cells and gene expression changes induced early by PU.1 and C/EBPα in 3T3 cells. (A–C) Ten thousand washed PC2.3 cells, BAC macrophages, and 3T3 cells were seeded in duplicate with or without CSF-1, GM-CSF, or anti-CSF-1 antibodies, and cell numbers were determined at the times indicated. (D) 3T3 cells were infected with PU.1 and C/EBPα viruses and uninfected as well as singly and doubly infected cells (Fig. S11) sorted 4 days later. cDNAs calibrated for β-actin expression were tested by RT-PCR. (Upper) Lanes 1, 2, and 3, control fibroblasts expressing GFP, GFP/hCD4, and hCD4; lane 4, PU.1wt; lane 5, PU.1ΔPEST; lane 6, C/EBPα; lane 7, PU.1wt plus C/EBPα; lane 8, PU.1ΔPEST plus C/EBPα; lane 9, bone marrow-derived macrophages. The genes tested are indicated on the right. (Lower) Expression of PPARγ2 and resistin within the above samples. Quantification of the data are shown in Fig. S12.
Complex Regulation of Macrophage and Fibroblast Genes by PU.1 and C/EBPα.
To determine the relative contributions of PU.1 and C/EBPα for the regulation of gene expression in fibroblasts, 3T3 cells were infected with PU.1 and C/EBPα viruses alone or in combination, and 4-day-infected cells (Fig. S11) were sorted and tested by semiquantitative RT-PCR. As expected, control virus-infected cells were CSF-1-positive but negative for macrophage genes. In contrast, both PU.1wt and PU.1ΔPEST viruses induced a 5- to 10-fold down-regulation of CSF-1 and an up-regulation of CSF-1R, Mac-1, and endogenous PU.1. C/EBPα on its own induced a weak down-regulation of CSF-1 but did not induce a detectable expression of any of the macrophage genes tested. However, C/EBPα both positively and negatively modulated the PU.1-induced changes in CSF-1, CSF-1R, and Mac-1 and endogenous PU.1 expression. In addition, neither factor alone activated lysozyme expression, but the combination of the two factors did.
PU.1 Inhibits the Capacity of C/EBPα to Up-Regulate Adipocyte Genes.
C/EBPα and C/EBPβ are essential mediators of hormone-induced adipocyte differentiation (13). To determine whether C/EBPα activates adipocyte genes in 3T3 cells and, if so, whether expression of these genes is affected by PU.1, the above cDNA samples were also tested for the expression of PPARγ2 and resistin. PPARγ2 is highly expressed in adipocytes (13) and at lower levels in macrophages, where it plays a role in the inflammatory response mediated by the uptake of lipids through the CD36 receptor (14). C/EBPα induced in 3T3 cells PPARγ2 expression to a level above that seen in macrophages. In addition, C/EBPα, but not PU.1wt or PU.1ΔPEST, induced the expression of the preadipocyte gene resistin. Finally, both PU.1wt and PU.1ΔPEST inhibited the expression of C/EBPα-induced PPARγ2 and resistin expression (Fig. 5D). A quantification of the data is shown in Fig. S12.
Discussion
Here we have shown that NIH 3T3 cells as well as primary mouse embryo- and skin-derived fibroblasts can be converted into macrophage-like cells by retroviral expression of PU.1 and C/EBPα. Cell lines stably expressing an attenuated form of PU.1 together with C/EBPα exhibited several functions of macrophages, including phagocytic capacity, partial inflammatory response, and specific CSF-1 dependence for growth, while losing the capacity to grow in medium containing fetal serum alone. Together with the observation that MyoD induces muscle cell differentiation in fibroblasts and other cell types (15), our data suggest that it might become possible to transdifferentiate skin-derived fibroblasts into desired cell types of potential therapeutic value.
Most of the up- or down-regulated genes in PC2.3 cells are expressed proportionally to the corresponding genes in the BAC macrophage cell line used as reference, and some of these genes, such as ptprc (CD45), are known to be direct PU.1 targets (16). However, the majority of up-regulated genes are expressed at lower levels than in macrophages relative to 3T3 cells (the genes above the 45° line in Fig. 3). Our data suggest that the reprogrammed fibroblasts represent intermediates that are stabilized by the continuous expression of PU.1 and C/EBPα. Thus, after prolonged culture of the PC2.3 and PC2.5 lines GFP-negative subpopulations emerged. These cells down-regulated the expression of CSF-1R and CD45 and, after some delay, also of Mac-1 antigen (unpublished data), indicating that the continuous expression of exogenous PU.1 is required to maintain a macrophage phenotype. In addition, the ectopic expression of PU.1 and C/EBPα did not establish a stable autoregulatory loop of PU.1 because the level of endogenous PU.1 in the partially reprogrammed cell lines was well below that seen in control macrophages. A number of alternatives could explain the observed incomplete reprogramming of fibroblasts induced by PU.1 and C/EBPα. First, chromatin domains might have become irreversibly altered during development of fibroblasts from a mesenchymal precursor, limiting transcription factor accessibility. If true, this might impose a serious restriction in further attempts to achieve full transcription factor-induced transdifferentiation. Second, fibroblasts might lack a transcription factor that acts early and perhaps transiently during blood cell specification. Such a factor might be Runx1, which has recently been shown to regulate PU.1 (17). In support of this possibility are the observations that PC2.3 cells show reduced levels of both Runx1 and endogenous PU.1 (Fig. 3C and data not shown). A third possibility is that fibroblasts lack a late-acting transcription factor required for the establishment of a full macrophage phenotype, such as MafB, IRF8, or Egr1/2 (18–20), which either are absent or are expressed at reduced levels in PC2.3 cells compared with macrophages (data not shown). The postulated missing factors would have to be present or fully activatable in B cells, because these cells show an apparently complete reprogramming into macrophages in response to C/EBPα (5).
Our results suggest that PU.1 is a primary regulator of macrophage genes in mesenchymal cells and that it can coopt C/EBPα as a hematopoietic cofactor. This might be achieved by sequestering C/EBPα away from putative complexes of proteins involved in adipogenesis. However, PU.1 is also known to be required for the formation of B cells and dendritic cells (21). The observation that neither the B lineage-specific PU.1 target B220 (22) nor the dendritic marker CD11c was found to be expressed in the reprogrammed cells suggests that PU.1 interacts with different lineage-associated transcription factors in different cellular contexts. Such a mechanism (23) would be reminiscent of the transcription factor mixture party model proposed earlier (24).
Recent studies indicate that, during hematopoiesis, PU.1 up-regulates the CSF-1R gene in a two-step mechanism. According to this model it first binds to the gene's promoter in hematopoietic stem cells inducing low-level receptor expression and then to the gene's enhancer during myeloid differentiation. The latter step would be mediated by synergizing with other factors, such as C/EBPα, causing high-level expression in macrophages (25). The observed modest PU.1-induced activation of Mac-1 in fibroblasts might recapitulate the first step, and the increased up-regulation induced by C/EBPα might recapitulate the second. However, the situation is more complex because different hematopoietic genes differed in their response: expression of CD45 is C/EBPα-independent; CSF-1R and endogenous PU.1 are negatively affected by C/EBPα; and lysozyme expression requires both PU.1 and C/EBPα.
In conclusion, our results indicate that fibroblasts exhibit a surprisingly high degree of differentiation plasticity in response to a transcription factor combination that specifies macrophages within the hematopoietic system. The ability to readily induce differentiated functions of distantly related cells is an encouraging step toward custom-designing cells for tissue regeneration directly from patients' cultured biopsies. Such a procedure would obviate the necessity to generate embryonic stem cells before inducing their differentiation along desired pathways.
Experimental Procedures
Cell Lines and Primary Mouse Fibroblast Cultures.
The 3T3 cell line was obtained from Robert Weinberg (Whitehead Institute, Boston). The isolation and characterization of the BAC1.2F5 macrophage cell line was described previously (26). Primary mouse embryo fibroblasts were prepared from day-15 C57BL/6J mouse embryos by collagen treatment. Contaminating Mac-1+ myeloid cells were removed with an AutoMACS device (Miltenyi Biotec) by using the POSSELD program after incubation with magnetic beads coated with Mac-1 antibodies. The depleted cells were reanalyzed by FACS and found to contain <0.02% Mac-1+ cells. The growth medium consisted of DMEM with 10% FCS plus antibiotics. Infected cells and reprogrammed cell lines were cultured in growth medium in the presence of 10 ng/ml human recombinant CSF-1, a kind gift from Chiron, and BAC macrophages in 30 ng/ml. Dermal fibroblasts were obtained by preparing a skin biopsy from the back of a 3-month-old C57BL/6J mouse. The cells were cultured for 2 weeks before they were used for infection, and they contained no detectable Mac-1-expressing cells.
Retrovirus Vectors.
The PU.1-GFP, C/EBPα-hCD4, and C/EBPβ-hCD4 retroviruses encoding murine transcription factors and reporter genes placed downstream of an IRES element have been described, as have the empty virus controls GFP and hCD4 (5). The plasmids of PU.1 lacking the transactivation, the PEST, and the DNA binding domains [obtained from Art Skoultchi (Albert Einstein College of Medicine)] (27) were inserted into the murine stem cell virus IRES GFP vector as described earlier (5). Viruses were produced by transfecting the Phoenix packaging cell line, and infections were done as reported earlier (5).
Antibodies, FACS Analysis, and Cell Sorting.
The following antibodies were used: Mac-1 PE and CD45-PECy7 from BD Pharmingen and F4/80-PE from Caltag; biotinylated antibodies to IAb were kindly obtained from Laura Santambroglio (Albert Einstein College of Medicine). Biotinylated monoclonal antibodies against CSF-1 and CSF-1R were as described in refs. 28 and 29. Streptavidin-PECy7 was from BD Pharmingen and Abcam. Neutralizing rabbit polyclonal antibody against CSF-1 was described earlier (29). Antibodies against Gr1 and Msr1 were from BD Pharmingen. For cell surface staining, cells were removed from the plate with nonenzymatic cell dissociation solution (Sigma), and 105 to 106 cells were suspended in 100 μl of PBS with 4% FCS and preincubated with 0.1 μg of Fc-block for 10 min on ice. Samples were then treated as described (5) and analyzed on an LSR-E flow cytometer (Becton Dickinson) using FlowJo software (Tree Star). Sorting of virus-infected fibroblasts was done with a MoFlow machine (Cytomation) using a single-cell deposition device.
Analysis of Actin Skeleton and Focal Adhesion Contacts.
Cells were grown on fibronectin-coated coverslips (BD Biosciences), fixed, and stained with rhodamine-phalloidin (Invitrogen) to visualize F-actin and with antibodies to paxillinY118-FITC (BioSource), a constitutively active form of the molecule that reveals adhesion structures (23). Cells were also stained with DAPI to reveal the nuclei. Images shown were taken with an Olympus 1 × 70 inverted microscope, and images were recorded by a Photometrics CH1 CCD camera and processed with Photoshop Element 2.0 software.
Gene Expression Analysis by Microarray.
RNA was extracted from 3T3, PC2.3, and BAC1.2F5 cell samples, and the quality of the RNA was verified with a bioanalyzer. A total of 2 μg of cDNA of each sample was amplified by using a LabelStar Kit (Qiagen), with cDNA from the PC2.3 cells labeled with a yellow fluorophore and that of 3T3 and BAC cells labeled with a red fluorophore. These probes were hybridized in pairs (3T3 and PC2.3; PC2.3 and BAC) to three arrays each (Qiagen V3.0 chips, containing 31,769 mouse probes plus controls, consisting of 70-mer oligonucleotides). The average values for the differential gene expression were used to calculate the fold differences in expression between 3T3 cells and macrophages. Only probes in which all three samples gave an interpretable signal were taken into consideration. The standard error of the mean of up-regulated genes was ≈10%, and that of down-regulated genes was ≈11%.
Phagocytosis Assays.
Bacterial phagocytosis assay.
Cells were seeded 24 h before infection in 24-well plates. One hundred dsRed E. coli per cell were added, and plates were centrifuged at 2,000 rpm for 5 min. The cells were then incubated for 1 h at 37°C, followed by a 90-min incubation at 37°C with 400 μg/ml gentamycin to eliminate extracellular bacteria. Cells were then washed three times, trypsinized, and analyzed by FACS. The dsRed E. coli was obtained from R. Copin and J.J. Letesson (University of Namur, Namur, Belgium).
Bead-uptake assay.
Cells were incubated with 1-μm red fluorescent carboxylated microspheres (Molecular Probes) at 37°C in DMEM plus 10% FCS and 10 ng/ml CSF-1. The cells were then thoroughly washed, stained with Hoechst 33342, and photographed with a Nikon TE200 using bright field and fluorescence illumination with Endow GFP, TRITC, and cyan GFP filters (Chroma Scientific). Images were acquired with a confocal microscope (Leica). Cells were scored as particle-positive if more than five intracellular beads were seen per section, with 100 cells evaluated for each sample.
Fc-dependent phagocytosis assay.
Cells were incubated with either untreated or antibody-coated SRBCs at 37°C for 60 min and washed, and brightfield images were taken. The cells were then incubated for another 60 min and treated with lysis buffer (0.2% NaCl) for 1 min at room temperature. Then, an equal volume of 1.6% NaCl was added and the cells were immediately washed. Cultures were scored microscopically and photographed under brightfield as well under fluorescence mode using the Endow filter, imaging the same areas photographed after the first incubation.
Inflammatory Response.
To test the cell's ability to mount an inflammatory response, cells were deprived of CSF-1 for 18 h and then treated with LPS (10 ng/ml; Sigma) for 6 h. After two washes with cold PBS, total RNA was extracted and reverse transcription was performed after DNase treatment. Quantitative PCR was performed using a final volume of 12.5 μl of SYBR Green Reaction Mix (Applied Biosystems). Annealing was performed at 60°C for 30 s. Real-time monitoring of PCR amplification was performed by using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Data were expressed relative mRNA to β-actin expression in each sample. A control sample without RNA was included in each reaction.
Semiquantitative RT-PCR and Primers.
To obtain total RNA, cell lines or different sorted populations of infected cells were extracted by using TRIzol (Life Technologies), digested with RNase free DNaseI (Roche) to remove contaminating genomic DNA, and then processed as described (5). The cDNA concentration in different samples was normalized to β-actin cDNA. The PCR was run at 94°C for 2 min (denaturation) followed by 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 25–33 cycles (amplification) depending on the relative mRNA abundance. PCR products were resolved on (1%) agarose gels and visualized by ethidium bromide staining. Images were taken by ChemiImager 4400 (Alpha Innotech) and quantitated with ImageQuant V1.2 software (Molecular Dynamics). Published primers were as follows: αM chain of Mac-1, CSF-1R, G-CSFR, GM-CSFRa, lysozyme M, PU.1, FcγRI, FcγRII, FcγRIII (5), and CSF-1 (28). The primers were 5′-gtataacgtggaggtcaagc and 3′-ggaggactggagaaaatcag for vav, 5′-ctcagagccacaggaatac and 3′-gtcacctcgcagtcttcaag for IL-3Ra, 5′-ATGCTACTGTTGCAAGCTCTC and 3′-tcagttggtatcatggtagag for PPARγ, and 5′-atgaagaacctttcatttcccc and 3′-tcaggaagcgacctgcagc for resistin.
CSF-1 Production and Cell Growth Assays.
To measure the amounts of CSF-1 produced, cells were thoroughly washed and 1 × 106 cells were seeded in duplicate in 12-well dishes in 1 ml of growth medium overnight. The conditioned media were tested in a CSF-1 RIA (29). To test their growth requirement, 104 cells were seeded in duplicate in 12-well plates with 1 ml of growth medium in the absence or presence of 20 μg/ml recombinant human CSF-1, 20 μg/ml recombinant murine GM-CSF (R & D Systems), or 1:100 diluted rabbit antibodies against CSF-1. Cells were trypsinized and counted with a Coulter Counter (Beckman Coulter).
Supplementary Material
Supporting Information
Acknowledgments.
We thank D. Cox for help with phagocytosis assays; S. Nandi and V. Chitu with CSF-1 assays; D. Reynolds, W. Tran, and J. Lozano for help with arrays and bioinformatic analyses; and M. Stadtfeld for discussions. This work was supported by the Albert Einstein College of Medicine, the Center for Genomic Regulation, the Institució Catalana de Recerca i Estudis Avançats (T.G.), and National Institutes of Health Grants K08 CA097348 (to F.P.), R01s CA 32551 (to E.R.S.), and CA 26504 (to E.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 2.Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol. 2007;18:467–473. doi: 10.1016/j.copbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Graf T. Differentiation plasticity of hematopoietic cells. Blood. 2002;99:3089–3101. doi: 10.1182/blood.v99.9.3089. [DOI] [PubMed] [Google Scholar]
- 5.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells in to macrophages. Cell. 2004;117:1–14. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 6.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Koschmieder S, Rosenbauer F, Steidl U, Owens BM, Tenen DG. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J Hematol. 2005;81:368–377. doi: 10.1532/ijh97.05051. [DOI] [PubMed] [Google Scholar]
- 8.Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005;33:131–143. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Reddy VA, et al. Granulocyte inducer C/EBPalpha inactivates the myeloid master regulator PU.1: Possible role in lineage commitment decisions. Blood. 2002;100:483–490. doi: 10.1182/blood.v100.2.483. [DOI] [PubMed] [Google Scholar]
- 10.Marion NW, Mao JJ. Mesenchymal stem cells and tissue engineering. Methods Enzymol. 2006;420:339–361. doi: 10.1016/S0076-6879(06)20016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 12.Nimmerjahn F, Ravetch JV. Fcgamma receptors: Old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Chawla A. Role of PPARgamma in macrophage biology and atherosclerosis. Trends Endocrinol Metab. 2004;15:500–505. doi: 10.1016/j.tem.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Weintraub H, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson KL, et al. PU.1 is a lineage-specific regulator of tyrosine phosphatase CD45. J Biol Chem. 2001;276:7637–7642. doi: 10.1074/jbc.M009133200. [DOI] [PubMed] [Google Scholar]
- 17.Huang G, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2007;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 18.Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 20.Laslo P, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 21.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 22.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med. 2005;202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pixley FJ, Lee PS, Condeelis JS, Stanley ER. Protein tyrosine phosphatase phi regulates paxillin tyrosine phosphorylation and mediates colony-stimulating factor 1-induced morphological changes in macrophages. Mol Cell Biol. 2001;21:1795–1809. doi: 10.1128/MCB.21.5.1795-1809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieweke MH, Graf T. A transcription factor party during blood cell differentiation. Curr Opin Genet Dev. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 25.Krysinska H, et al. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol. 2007;27:878–887. doi: 10.1128/MCB.01915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan C, Pollard JW, Stanley ER. Isolation and characterization of a cloned growth factor dependent macrophage cell line, BAC12F5. J Cell Physiol. 1987;130:420–427. doi: 10.1002/jcp.1041300316. [DOI] [PubMed] [Google Scholar]
- 27.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai XM, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 29.Stanley ER. The macrophage colony-stimulating factor, CSF-1. Methods Enzymol. 1985;116:564–587. doi: 10.1016/s0076-6879(85)16044-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information