The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences (original) (raw)
Abstract
Background
A series of Rps (resistance to Pytophthora sojae) genes have been protecting soybean from the root and stem rot disease caused by the Oomycete pathogen, Phytophthora sojae. Five Rps genes were mapped to the Rps1 locus located near the 28 cM map position on molecular linkage group N of the composite genetic soybean map. Among these five genes, _Rps1_-k was introgressed from the cultivar, Kingwa. _Rps1_-k has been providing stable and broad-spectrum Phytophthora resistance in the major soybean-producing regions of the United States. _Rps1_-k has been mapped and isolated. More than one functional _Rps1_-k gene was identified from the _Rps1_-k locus. The clustering feature at the _Rps1_-k locus might have facilitated the expansion of _Rps1_-k gene numbers and the generation of new recognition specificities. The _Rps1_-k region was sequenced to understand the possible evolutionary steps that shaped the generation of Phytophthora resistance genes in soybean.
Results
Here the analyses of sequences of three overlapping BAC clones containing the 184,111 bp _Rps1_-k region are reported. A shotgun sequencing strategy was applied in sequencing the BAC contig. Sequence analysis predicted a few full-length genes including two _Rps1_-k genes, _Rps1_-k-1 and _Rps1_-k-2. Previously reported _Rps1_-k-3 from this genomic region [1] was evolved through intramolecular recombination between _Rps1_-k-1 and _Rps1_-k-2 in Escherichia coli. The majority of the predicted genes are truncated and therefore most likely they are nonfunctional. A member of a highly abundant retroelement, SIRE1, was identified from the _Rps1_-k region. The _Rps1_-k region is primarily composed of repetitive sequences. Sixteen simple repeat and 63 tandem repeat sequences were identified from the locus.
Conclusion
These data indicate that the Rps1 locus is located in a gene-poor region. The abundance of repetitive sequences in the _Rps1_-k region suggested that the location of this locus is in or near a heterochromatic region. Poor recombination frequencies combined with presence of two functional Rps genes at this locus has been providing stable Phytophthora resistance in soybean.
Background
Many plant disease resistance (R) genes from different plant species have been isolated and characterized; but are grouped into a limited number of classes [2,3]. R loci are usually organized in clusters, and genes within one cluster are mostly derived from a common ancestor [4]. The clustering feature can facilitate the expansion of R gene number and the generation of new R gene specificities through recombination and positive selection [5]. Long contiguous sequences containing several R genes or resistance gene analogues (RGA) have been determined [6-8]. These sequences provided insights into the mechanisms of R gene evolution and generation of novel recognition specificity. Insertions of retroelements in genomic regions containing R genes or RGAs have been documented in these studies. Retroelements are suggested to create variability among paralogous R gene members [9].
Soybean (Glycine max L. Merr.) is a legume crop of great economic and agricultural importance across the world. Its estimated genome size is 1,115 Mb, of which approximately 40–60% is composed of repetitive sequence [10-12]. Repetitive DNA sequences have been shown to be the major determinant of plant genome sizes [13]. There are two main types of repetitive sequences, tandem repeat DNA sequences and dispersed DNA sequences such as retroelements [13]. Several tandem repeats, SB92, STR120 and STRR102 have been reported in soybean [14-16]. It has been suggested that soybean has experienced at least two rounds of genome-wide duplications [17-19]. Despite the availability of genomics resources such as densely saturated genetic maps, BAC and YAC libraries, large EST collections, BAC end sequences, a soybean genome database (SoyGD) browser, and the legume information system (LIS) [20], our knowledge of soybean genome structure is still largely limited [21-27].
Root and stem rot disease caused by Pytophthora sojae is one of the most destructive soybean diseases in the United States [28]. Use of Phytophthora resistance conferred by single dominant Rps genes has been providing reasonable protection of soybean against this pathogen. Five Rps genes including _Rps1_-k were mapped to the Rps1 locus located near the 28 cM map position on molecular linkage group N of the composite genetic soybean map [29,30]. Among these five genes, _Rps1_-k was introgressed from the cultivar, Kingwa. _Rps1_-k confers resistance to most races of P. sojae, and has been widely used for the past two decades [31]. By applying a positional cloning approach two classes of functional coiled coil-nucleotide binding-leucine rich repeat (CC-NB-LRR)-type resistance genes were isolated from the soybean _Rps1-_k locus [1]. A large cluster of highly polymorphic paralogous _Rps1_-k sequences is located at the adjacent _Rps1_-k region [32]. The _Rps1_-k locus was mapped to two overlapping BAC clones encompassing 184 kb, located at one end of an approximately 600 kb contiguous DNA spanned by several overlapping BAC clones [32]. CC-NB-LRR-type genes of the 184 kb _Rps1_-k region were evaluated and two classes of highly similar genes were shown to confer race-specific Phytophthora resistance [1]. To gain insights into the soybean genome organization and evolution of _Rsp1_-k genes, BAC clones encompassing the Rps1 locus were sequenced and analyzed.
Results
Sequence of three BAC clones spanning the _Rps1_-k locus
_Rps1_-k was previously mapped to a region flanked by two markers CG1 and 18R [30,32]. To understand the composition of the _Rps1_-k region, three overlapping BAC clones, GS_18J19, GS_43D16 and GS_99I16 that may encompass the Rps1 locus were chosen for sequencing [32]. Phytophthora resistance genes were previously identified from these three BAC clones through positional cloning [1].
A total of 4,093 reads (829, 1,189 and 2,065 reads for GS_18J19, GS_43D16 and GS_99I16, respectively) were generated from these BAC clones. GS_18J19, GS_43D16 and GS_99I16 were sequenced to a 14-, 12- and 9-fold redundancies, respectively. A single contig of 38,498 bp was obtained for GS_18J19 after the initial assembly (GenBank accession EU450800). Three and five contigs were obtained from assembling of sequences derived from GS_43D16 and GS_99I16, respectively. The resulting contigs of GS_43D16 and GS_99I16 were ordered into individual scaffolds manually, in which the order and orientation of the contigs were inferred by mate pairs (sequences obtained from both ends of a ~20 kb shotgun clone) [33]. The clones that span the gaps between two adjacent contigs were identified based on mate pairs and were used to obtain sequences of the gap regions. Gaps were filled out by applying the primer walking approach. Primers were designed based on the sequences of contig ends from which walking were initiated. To guarantee the high sequence quality, less sequenced regions were further sequenced by using suitable primers. After initial assembly and gap filling, 70,829 and 164,411 bp sequences were obtained from GS_43D16 and GS_99I16, respectively (GenBank accession EU450800). The assembled GS_18J19 sequences represent one end of the GS_43D16.
Directional sequencing of GS_43D16
Earlier, partial sequencing of the three BAC clones had allowed us to identify candidate genes underlying _Rps1_-k. The functional identities of _Rps1_-k genes were confirmed through stable transformation in soybean [1]. Two classes of _Rps1_-k genes were identified. The three Class I _Rps1_-k genes were identical in their ORF sequences. The Class I gene, _Rps1_-k-3, showed a recombination breakpoint at the 3' untranslated region originating from sequence exchange between members of both classes of genes [1].
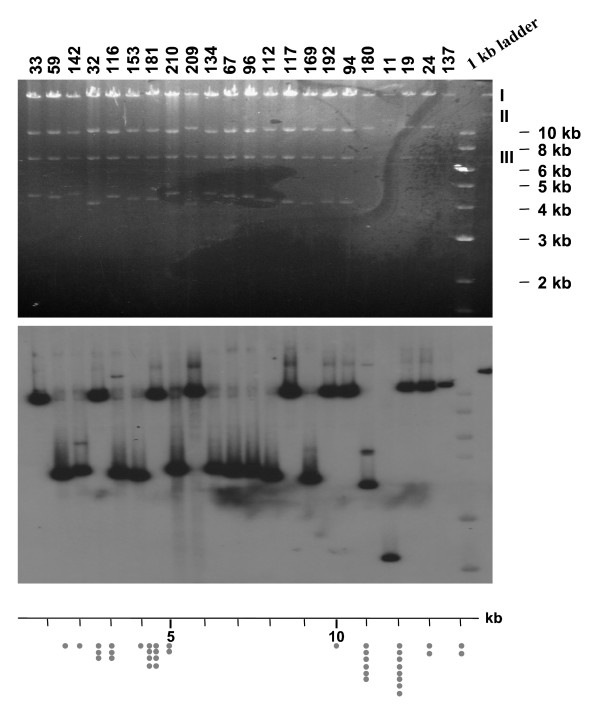
_Rps1_-k-1, _Rps1_-k-2 and _Rps1_-k-3 were isolated from GS_43D16 [1]. The existence of abundant repetitive sequences made it difficult to assemble the sequences of the BAC clones. To avoid any misassembly, GS_43D16 containing _Rps1_-k-1, _Rps1_-k-2 and _Rps1_-k-3 was subjected to directional sequencing using the EZ::TN <_Not_I/KAN-3> transposon of the EZ::TN in-Frame Linker Insertion Kit (Epicentre, Madison, WI). Two hundred and twenty-four EZ::TN <_Not_I/KAN-3> transposon insertion GS_43D16 clones were randomly selected for further analysis. This approach allowed us to map physically the individual sequence reads onto the GS_43D16 sequence as follows. Transposon insertion sites of individual transposition events were utilized to select EZ::TN <_Not_I/KAN-3> transposon containing GS_43D16 clones. Each clone was digested with _Not_I and hybridized to GS_43D16 end-specific probes in Southern analyses (Figure 1). There are three _Not_I sites in GS_43D16; one in the insert soybean genomic DNA and two in the pBeloBAC11 vector flanking the cloning _Hin_dIII site. Therefore, _Not_I digestion of GS_43D16 resulted in three _Not_I fragments (Figure 1); (I) a large DNA fragment of ~55 kb, (II) a small DNA fragment of ~15 kb, and (III) the pBeloBAC11 vector sequence. There are two _Not_I sites flanking the kanamycin resistance gene in the EZ::TN <_Not_I/KAN-3> transposon. Therefore, if there is a single transposon insertion in the GS_43D16 clone, then five fragments including the ~1.2 kb transposon, should be generated following _Not_I digestion (Figure 1).
Figure 1.
Physical mapping of the locations of individual EZ::TN <_Not_I/KAN-3> transposon insertions in a soybean bacterial artificial chromosome. Individual GS_43D16 clones containing the EZ::TN <_Not_I/KAN-3> transposon were digested with _Not_I. Three _Not_I fragments, I, II and III released from _Not_I digestion of GS_43D16 are shown in the last lane. Note that fragment III is comprised of the pBeloBAC11 vector sequence. The top panel shows the gel of _Not_I digested DNA of GS_43D16 clones carrying the transposon in the _Not_I Fragment II. The middle panel shows the Southern hybridization data of the gel shown in the top panel. The 245 bp probe for Southern analysis was obtained by PCR of the GS_43D16 end that overlaps with GS_18J19, but not GS_99I16. The lower panel shows the distribution of clones carrying the transposon at various regions of the _Not_I Fragment II. One dot represented one clone containing the transposon at that particular location of the _Not_I Fragment II.
Of the analyzed 224 random transposon-inserted GS_43D16 clones, 162 were shown to contain the transposon in the large fragment; 40 of them in the small fragment; and 22 in the pBeloBAC11 vector. Clones containing transposon insertions in the vector pBeloBAC11 were not considered for further study. Approximate physical locations of transposon insertions in individual _Not_I genomic DNA fragments were determined by Southern analyses as shown in Figure 1. Based on the physical location of transposon insertions, 114 GS_43D16 clones containing transposon insertions in either the 15 or 55 kb _Not_I fragment were selected for sequencing by using transposon end-specific primers. Only about 50% percent of the clones produced sequences that were readable. Pairwise sequence comparison between the assembled GS_43D16 sequence and sequences obtained from individual transposon inserted GS_43D16 clones revealed the transposon insertion sites in GS_43D16.
Among the randomly picked 224 transposon-inserted clones, the number of transposon insertions was proportional to the size of _Not_I fragments. However, there were no insertions in two regions, one of about 5 kb in the ~15 kb fragment and the other one is about 10 kb in the ~55 kb fragment. Whether this was due to bias in transposon insertion or due to sampling variance is yet to be determined.
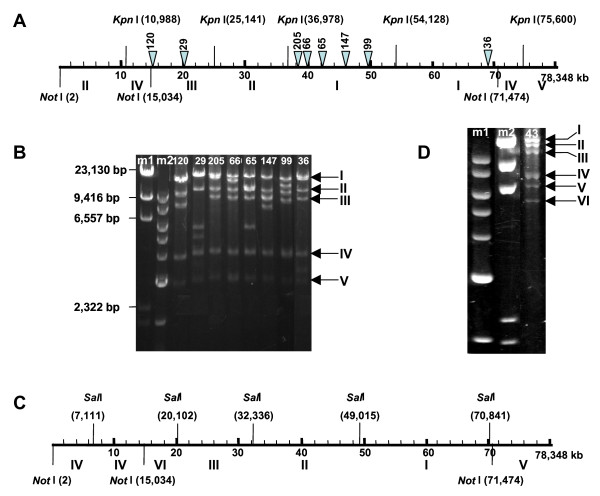
The quality of 78,313 bp assembled GS_43D16 sequence was verified through restriction mapping as follows. Clones carrying transposon insertions at various regions were selected and double digested with _Kpn_I and _Not_I. The predicted _Kpn_I – _Not_I restriction maps based on the assembled GS_43D16 sequence is shown in Figure 2A. Eight fragments are expected from _Kpn_1 and _Not_I double digestion of GS_43D16. Only five fragments were resolved in the gel analyses, because some of the fragments are of similar sizes. For example, there are two 17 kb fragments termed Fragment I. Following digestion of clones carrying single transposons with both enzymes released two additional fragments and the 1.2 kb transposon. Depending upon the position of the transposon in a given _Kpn_I or _KpnI_-_Not_I fragment two fragments of variable sizes were produced (Table 1). Comparison of observed fragment sizes with that of expected fragment sizes showed that there is general agreement between the observed and expected fragment sizes. _Sal_I-_Not_I map (Figure 2C) based on the assembled sequence was also verified by digesting GS_43D16 with _Sal_I and _Not_I. Eight fragments were expected from the double digestion. Two fragments, 7.9 kb and 7.11 kb were not resolved and termed Fragment IV (Figure 2C and 2D). Smallest fragment (0.6 kb) is not shown in Fig 2D. Taking these data together, it was concluded that the generated GS_43D16 sequence represents the physical distance of the soybean DNA present in that clone and no large fragments were remained to be sequenced.
Figure 2.
Verification of the restriction maps of GS_43D16. A, _Kpn_I and _Not_I map of the assembled GS_43D16 sequence. B, _Kpn_I and _Not_I double digestion of selected GS_43D16 clones carrying the EZ::TN <_Not_I/KAN-3> transposon insertions. Eight fragments were expected from _Kpn_I and _Not_I digestion of GS_43D16 carrying no transposons (A). Only five fragments were observed, because some of the fragments had similar mobilities in the gel. Some of these fragments were resolved after transposon insertion. A close relationship was observed between the restriction fragment sizes determined by gel electrophoresis and that by sequence data and location of transposon insertions (Table 1). m1, λ_Hin_dIII ladders, m2, 1 kb DNA ladder (New England Biolabs Inc., Beverly, MA). C, _Sal_I-_Not_I map of the assembled GS_43D16 sequence. D, _Sal_I and _Not_I digestion of GS_43D16. Eight fragments were expected from the double digestion of GS_43D16 (Figure 2C). Six fragments were resolved from the digestion of the clone (43 in 2D). 7.9 kb and 7.11 kb fragments were not resolved (Fragment IV, twice the intensity of either Fragment III or Fragment V) and 0.6 kb _Sal_I-_No_tI fragment is not included in 2D.
Table 1.
Restriction fragments produced from the _Kpn_I-_Not_I double digestion of GS_43D16 clones carrying the EZ::TN <_Not_I/KAN-3> transposon.
| Fragment size from Kpn I/_Not_I double digestion2 | |||
|---|---|---|---|
| Clone ID | Location of the transposon1 | Observed | Expected3 |
| 120 | 15,171 | ~10 kb, -4 | 0.1 kb, 10.1 kb |
| 29 | 20,255 | ~5.5 kb, ~4.5 kb | 5.2 kb, 4.9 kb |
| 205 | 38,493 | ~15 kb, - | 15.6 kb, 1.5 kb |
| 66 | 39,577 | ~14.6 kb, 2.6 kb with Fragment 3. | 14.6 kb, 2.6 kb |
| 65 | 42,203 | ~11.9 kb, 6 kb | 11.9 kb, 5.2 kb |
| 147 | 45,914 | ~10 kb with Fragment 3, ~8 kb | 8.9 kb, 8.2 kb |
| 99 | 49,522 | ~12.5 kb, 4.6 kb with Fragment 7. | 12.5 kb, 4.6 kb |
| 36 | 68,935 | ~14.5 kb, ~3 kb | 14.8 kb, 2.5 kb |
Genes underlying the Rps1-k
GS_18J19 overlaps with one end of GS_43D16. GS_99I16 comprised 51,109 bp sequences of GS_43D16 (Figure 3). There were 99.99%, 99.85% and 99.96% identities between the overlapping sequences of GS_18J19 and GS_43D16, GS_43D16 and GS_99I16, and GS_18J19 and GS_99I16, respectively. These results indicate high quality of the assembled sequences. High identity of GS_43D16 sequence with the overlapping regions of GS_18J19 and GS_99I16 suggested that there was no rearrangement in GS_43D16, from which _Rps1_-k-1, _Rps1_-k-2 and _Rps1_-k-3 were previously isolated [1].
Figure 3.
Molecular characterization of the _Rps1_-k region. A) The overlapping three BAC clones containing two CC-NB-LRR genes (green box) of the _Rps1_-k region are depicted. Sequences of individual BACs were utilized to show their overlapping regions. The composite _Rps1_-k region, shown at the top of the figure, is based on the sequences of these three BACs. GS_99I16 does not carry the 5'-end of _Rps1_-k-1, which is shown with a truncated green box. B) Arrangement of predicted genes and retrotransposons in the Rps1-k region. The green colored boxes represent full-length genes (_Rps1_-k-1 and _Rps1-_k-2); the red colored boxes represent partial genes; the blue colored boxes represent retroelements; white boxes represent introns in the predicted genes. Boxes above the ruler represent genes that have coding sequence on the forward strand, whereas the boxes under the ruler indicate the genes that are on the reverse strand. Detailed annotation data are presented in Table 2.
The gene content of an 184,111 bp contig sequence (GenBank Accession Number EU450800) carrying the _Rps1_-k locus derived from the GS_43D16 and GS_99I16 sequences was determined. Genes were predicted with GeneScan and GeneMark.hmm ES-3.0 programs [34]. To get a better gene prediction, genes predicted by GeneScan and GeneMark.hmm, and/or sequences having similarities to soybean ESTs were further analyzed by different NCBI Blast programs and sequence alignment programs. Putative annotations of the predicted genes were accomplished by BlastP searches. The gene content in the _Rsp1_-k region appears to be poor. Only a few full-length genes were predicted. These include two coiled coil-nucleotide binding-leucine rich repeat (CC-NB-LRR)-type _Rps_1-k genes and retrotransposons (Figure 3, Table 2).
Table 2.
Gene annotations of the _Rps1_-k region1
| Gene ID | Position2 | Predicted gene annotation | Closest protein homolog | BLASTP E value | Soybean ESTs3 (E ≤ e-50) |
|---|---|---|---|---|---|
| 1 | 18019–21708 (+) | _Rps1_-k-1 | Glycine max AY963292 | 0 | 14 |
| 2 | 42452–46201 (+) | _Rps1_-k-2 | Glycine max AY963293 | 0 | 14 |
| 3 | 63302–62705 (-) | CBL-Interacting protein kinase 15 | Arabidopsis thaliana NP_195801 | 6e-69 | 4 |
| Serine/threonine Kinase (partial seudogene) | Persea Americana AAL23677 | 3e-68 | |||
| 4 | 69126–68921 (-) | Ribosomal protein S6 | Glycine max AAS47511 | 4e-7 | 17 |
| 5 | 76950–78815 (+) | Conserved hypothetical protein | Medicago truncatula ABD32262 | 3e-40 | 4 |
| 6 | 79282–86280 (+) | Gag/pol polyprotein | Pisum sativum AAQ82033 | 0 | 19 |
| 7 | 90317–92266 (+) | Hypothetical 65 kDa avirulence protein in avrBs3 region | Xanthomonas campestris pv. vesicatoria P14729 | 5e-5 | 7 |
| 8 | 92658–96559 (+) | Gag-pol polyprotein | Zea Mays AAM94350 | 2e-147 | 21 |
| 9 | 113419–113916 (+) | NADH dehydrogenase subunit 1 (only the N-terminal 70 aa) | Trichosurus vulpecula NP_149931 | 3.9 | 77 |
| 10 | 114088–115118 (+) | MAD2 (only the N-terminal 65 aa) | Triticum aestivum BAD90977 | 3e-17 | 5 |
| 11 | 117048–116804 (-) | Cytochrome c oxidase subunit II (the N-terminal 40 aa) | Cynomys ludovicianus AAK52712 | 5.1 | 2 |
| 12 | 117709–119789 (+) | Cysteine proteinase Vacuolar processing enzyme precursor (the N-terminal 118 aa) | Glycine max BAA06030 P49045 | 3e-49 | 13 |
| 13 | 123937–123409 (-) | Unknown protein (partial pseudogene) | Arabidopsis thaliana NP_190603 | 2e-26 | 3 |
| 14 | 127141–126821 (-) | L-lactate dehydrogenase (partial pseudogene) | Lycopersicon esculentum CAA71611 | 9e-27 | 7 |
| 15 | 131753–138850 (-) | Glycoside hydrolase Integrase, catalytic region (partial pseudogene) | Medicago truncatula ABD33337 Medicago truncatula ABD32527 | 0 | 6 |
| 16 | 139054–139575 (+) | Unnamed protein product (C-terminal 173 aa) | Oryza sativa NP_912905 | 5e-60 | 3 |
| 17 | 140361–140014 (-) | Gag-pol polyprotein | Glycine max AAQ73529 | 1e-34 | 3 |
| 18 | 145152–148184 (+) | Dynein | Oncorhynchus mykiss CAA33503 | 1e-10 | 4 |
| 19 | 145722–145277 (-) | Prion-like Q/N-rich domain protein PQN-33 | Gallus gallus XP_428546 | 6e-48 | 3 |
| 20 | 154355–155745 (+) | Oxidoreductase (pseudogene) | Arabidopsis thaliana NP_201530 | 2e-21 | 7 |
| 21 | 159487–160392 (+) | Gag/pol polyprotein | Pisum sativum AAQ82037 | 5e-28 | 9 |
| 22 | 165713–166447 (+) | Glycoside hydrolase, family 1, Zinc finger, CCHC-type; Ribonuclease H fold | Medicago truncatula ABD333337 | 6e-29 | 1 |
| 23 | 167488–176781 (-) | SIRE1–8 retroelement | Glycine max AY205610 | 0 | 5 |
Most of the identified genes are truncated. Genes were considered truncated when their predicted reading frames are partial. For example, the predicted cysteine proteinase shares an 88% identity with the first 126 amino acids of a soybean cysteine proteinase protein (BAA06030) followed by a premature stop codon. BlastN search against the soybean EST database was performed to support our gene prediction. ESTs showing high similarities but no complete identities to all predicted genes were identified (Table 2).
The _Rps1_-k region is composed of repetitive sequences
The major portion of the contiguous 184,111 bp sequence of the _Rps1_-k region is comprised of repetitive sequences including simple repeat sequences, tandem repeats and retroelements. The simple repeat and tandem repeat sequences were identified using Sputnik and tandem repeats finder. Sixteen simple repeat sequences were identified (Table 3). Sixty-three tandem repeats were revealed with copy numbers ranging from 1.8 to 72 and unit length varying from 7 to 310 bp (Table 4). The consensus motif length of the tandem repeat containing 72 copies is 24 bp. Sequence data from individual reads confirmed that they are tandem repeats in head-to-tail orientation. This sequence was used to query the soybean GSS (genomic survey sequence) database and a number of sequences with high identities were revealed. The one (CL868124) showing highest identity to the consensus 24 bp tandem sequence came from the project on characterization of the heterochromatic, gene-poor centric regions of soybean chromosomes.
Table 3.
Simple repeat sequences in the _Rps1_-k region
| Position | Repeat Unit | Copy Number |
|---|---|---|
| 7619–7663 | AT | 22 |
| 9814–9851 | AT | 19 |
| 24196–24231 | AT | 18 |
| 34682–34732 | AT | 25 |
| 38898–38960 | AAT | 21 |
| 41328–41354 | AAT | 9 |
| 51716–51901 | AT | 93 |
| 53915–53944 | AT | 15 |
| 59145–59168 | TC | 12 |
| 64934–64989 | AT | 28 |
| 110292–110313 | AT | 22 |
| 112406–112477 | AT | 36 |
| 116097–116116 | AT | 10 |
| 116665–116714 | AT | 25 |
| 127258–127281 | AG | 12 |
| 181688–181759 | AT | 36 |
Table 4.
Tandem repeat sequences in the _Rps1_-k region
| Position | Consensus sequence of tandem repeat unit | Copy number |
|---|---|---|
| 4872–4907 | TTAATAAATTTATT | 2.6 |
| 5279–5311 | TTTATT | 2.5 |
| 7219–7253 | TTTTATTATTTAAATAT | 2 |
| 7328–7366 | TTTTAAGTTAACATAAATT | 2 |
| 13986–14041 | CTTATATTTTTTTTAT | 3.5 |
| 14069–14121 | TTTAAATCTTTTATTTTTACC | 2.5 |
| 28228–28272 | TTTATTTATAAGATTATTTAAT | 2 |
| 34767–34826 | ATGCAAACATATATACATGC | 2.9 |
| 65181–65235 | TCATTACTAAAAAAAAATAG | 2.8 |
| 65966–66017 | GCCAGCATGCATGTATATC | 2.7 |
| 70677–70718 | TAAAAAGTTGAATAGATAC | 2.2 |
| 72634–72694 | CATTAAGTTCTTTTAATTCCTAGGTTAGTGG | 2 |
| 75090–75128 | CGTTCTTCAT | 3.8 |
| 87791–87926 | TGAATATATATAGCATGAAAATGCCTTGCAAAATA | 3.9 |
| 89787–89849 | AAATAGAAAAGGAAAGAAAATG | 2.9 |
| 90350–90511 | AAAAAGAAAAGAAAGGAAATTCCCAATCAAAGAGAAAGC | 3.8 |
| 90381–90538 | GAGAAAGCAAAAAGAAAAGAAAGGAAATTCCCAATCAAAGAGTGG | 3.5 |
| 91333–92076 | TACGCGGAGATACCTTACGGTTATCCGCACCCCCTTTGCCATTCAGACACAGTCGTGTCCGTTGGCAAGCAGAGACCAAGTTTGGTCATTCTGCACACATGA | 7.3 |
| 92743–92779 | GCTCGCCTGGGCGAGCTGA | 1.9 |
| 98273–98333 | CATTAAGTTCTTTCAATTCATAGGTTAGTGG | 2 |
| 113516–113540 | AAAAACCGTCTTA | 1.9 |
| 120828–120857 | TTTTTTTTTCC | 2.7 |
| 122442–122512 | ATCAAATAAAATGCTTGCAGATCA | 3 |
| 124367–124513 | AAAAAAAATTGAAGATTCTAAGACAGTTTTTAGGGAAAACCGTCTTAGAATGTCTTATTTTAAATAAAAAAAAATT | 2 |
| 133966–134004 | AATCAAAGAACAACTCAAGTG | 1.9 |
| 134057–134089 | TCAAGAA | 4.9 |
| 135918–136076 | GATCCACAAGGGATGTACCCTCCCTTATTCTCATTACAACAACCCAAGTAGATGTACCCTCCACT | 2.3 |
| 136235–136365 | AAGGGAGAAGAGAGACACAAAAAGAATTCAGGCGGTTAGTCCTTGTCGATTCTTTTTGGAA | 2.2 |
| 137034–137101 | TCTTCTCTTGAATCTTGAATTCAA | 2.9 |
| 144892–144919 | AGAAAAGGAAAAA | 2 |
| 145379–147112 | GGACTACACGTCCTCGCCTTCAGA | 72 |
| 147479–147972 | GGGATCGCGCCCACAAGACACCCAGTGGACCCGAAGGAGTCCAACAGGGCCCTGGGGTTTCCAGCTCTGGTTACGGGCCTCTGTCAGTCCTACAGGGTGCCCGTCCCCCCCAGCAAGGTCACCCCATCGTAACATAGGTAACTATGCACATCTCTCAACTGATTTCTGATGCCATCCAATATTTGCA | 2.6 |
| 148467–148657 | AAAAATACCTCACAAAATATATATATATTATGTTTAGGTAGCAAGATACCTTGGATACACATGTATATAGC | 2.7 |
| 149273–149361 | AAAGAAAGTTCCCGATCAAAGATCGAAAGAAAACAAAGAGAAAA | 2 |
| 150401–150651 | GTATGGTTATCAGCACCTGTCGTCAACCAGGGGCAAACGAGCCCGTTGACGCGCAGAGACTAACGTCATCTTCTGCACCTTTTGTCAACCAGAGACAGCGAGTCCAATGACATGTGGAGATACCCAAGCGATTATCC | 1.8 |
| 150612–151127 | GCACCTTTTGTCATCCAGAGACAGCGAGTCCGATGACATGCGAGGGTACCGTATGGTTATCC | 8.3 |
| 150799–150931 | CACCTTTCGTCAACCAGGGGCAAACGAGCCCATTGACGCGCAGAGACTAACGTCGTCTTCTG | 2.1 |
| 150550–151365 | GCACCTTTCGTCAACCAGGGGCAAGCGAGCCCGTTGACGCGCAGAGACTAACGTCGTCTTCTGCACCTTTTGTCAACCAGAGATAGCGAGTCCGATGACATGCGAGGGTAACGTATGGTTATCCGCACCTTTTTTCATCCAGAGACAGCGAGTCCGATGACATGCGGGGGTACCGTATGGTTATCCGCACCTTTTGTCATCCACAGACGGCAAGTCCGATGACACGCGGAGGTACCGTATGGTTATCCACACCTTTCGTCAACCAGGGGCAAACGAGCCCATTGACGCACAGAGACTAACGTCGTCTTCC | 2.6 |
| 150536–151060 | CCGTATGGTTATCACACCTTTCGTCAACCAGGGGCAAACGAGCCCATTGACGCGCAGAGACTAACGTCGTCTTCTGCACCTTTCGTCAACCAGAGAGAGCGAGCCCAATGAATGCGAGGCTAACGATCGTTATCCGCACCTTTTATCATCCAGAGACGGCTAGTCCGATGACATGCGGGGGTACCGTATGGTTATCCGCACCTTTTGTCATCCACAGACAGCAAGTCCGATAACACGCAGGGGTA | 2.1 |
| 150901–151386 | CGCAGAGACTAACGTCGTCTTCCGCACCTTTTGTCATCCAGAGATAGCGAGTCCGATGACATGCGGAGGTACCGTATGGTTATCCGCACCTTTTGTCAACCAGAGGCAAGCGAGTCCGTTGACA | 3.9 |
| 151985–152116 | AATCCGTAAAGTTTCGCAACATTCTGGAAGTCAAAACAAGTATTGCTGCAC | 2.6 |
| 152550–152596 | TTCTTCATCG | 4.6 |
| 152558–152597 | CGTTCTTCATCGTTCTTCGTT | 1.9 |
| 153216–153398 | CCAAGAGATCGTTAATGGTCCAACGCCTTAACGTTTCTCTCCTTTCAAAA | 3.6 |
| 153555–153596 | AAAAAAGACAAAAAACAT | 2.3 |
| 156483–156603 | ATCAAACATCACTTGAGATCGTTTCAAGGTCCAACGCCTTAACCATTCTCTCCGCTTTTC | 2 |
| 161707–161856 | ACATCTGAGAAGAAAACTCATTCGACCAGGAGCTCATGGAAAATTCCCAAAGACAATTGTGATAGTAGGGT | 2.1 |
| 162626–162804 | TTTTAGAGGACTCAAAGTCCTCACCTTTATC | 5.8 |
| 163722–163780 | ATCAAAGAACAACTCAAGTGA | 2.9 |
| 163762–163815 | GAATCAAGAACAAGTCAAGACTCAA | 2.1 |
| 163765–163818 | TCAAGAATCAAGAAGAAT | 2.9 |
| 163826–163911 | AATCAAG | 12.3 |
| 164281–164330 | TTCAAAAAGGTTTTAACTTT | 2.5 |
| 164461–164485 | TTGAATCTCT | 2.5 |
| 166799–166839 | AGTATTTTCAAAAAT | 2.9 |
| 168846–168891 | TCATAAATCATGCATAATATCCT | 2 |
| 172682–172713 | TTTTCTGCA | 3.4 |
| 178161–178219 | ATCAAAGAACAACTCAAGTGA | 2.9 |
| 178201–178254 | GAATCAAGAACAAGTCAAGACTCAA | 2.1 |
| 178204–178257 | TCAAGAATCAAGAAGAAT | 2.9 |
| 178265–178343 | AATCAAG | 11.3 |
| 178713–178762 | TTCAAAAAGGTTTTAACTTT | 2.5 |
| 180693–180809 | AAAGGCACGCTAAGCCCAATTCCAACCGAGAGGAAGTGCACTGAGCGGCCC | 2.3 |
| 183350–183378 | AATTTATGGAGCCA | 2.1 |
Another abundant tandem repeat contains the consensus AATCAAG sequence. There are 12.3 copies of this repeat sequence between positions 163,795 and 163,880 and 11.3 copies between 178,234 and 178,312. Several soybean tandem repeat sequences, SB92, STR120 and STR102, have been identified [14-16]. Seven copies of a tandem repeat sequence with 102 bp unit length were also found in the _Rps1_-k locus, but it shares no similarity with STR120 or STR102.
The ~20 kb intergenic sequence between _Rps1_-k-1 and _Rps1_-k-2 is primarily made up of repetitive sequences. Four simple repeat sequences were localized in this interval. Notably, a 220 bp sequence was found at two locations, one between positions 24,318 and 24,537 and the other one between positions 29,963 and 30,182. This sequence encodes part of a protein sharing high similarity to the receptor-like protein kinase, Xa21 (BAD27933).
A copia/_Ty1_-like retroelement, SIRE1–8, was identified from the assembled 184,111 bp sequence of the _Rps1_-k region [35]. The 9.5 kb sequence encoding the SIRE1–8 element was used to query the soybean EST database. Two ESTs (CB063565 and CO983516) showed 99% identity to part of the gag-pol encoding sequence, one EST showed 92% identity to the LTRs and one EST exhibited similarities to the envelope-like sequence.
The CC-NB-LRR-type gene, _Rps1_-k-3 evolved through intramolecular recombination in Escherchia coli
Previously it was reported that a CC-NB-LRR gene, _Rps1_-k-3 was evolved from recombination between _Rps1_-k-1 and _Rps1_-k-2 [1]. The gene was comprised of 5' end from _Rps1_-k-1 and 3'-untranslated region from _Rps1_-k-2. This gene was isolated form GS_43D16 but not from either GS_18J19 or GS_99I16. This observation suggested that these two BAC clones did not overlap [1]. Therefore, it was concluded that the two Class I CC-NB-LRR genes isolated from GS_18J19 and GS_99I16 were unique.
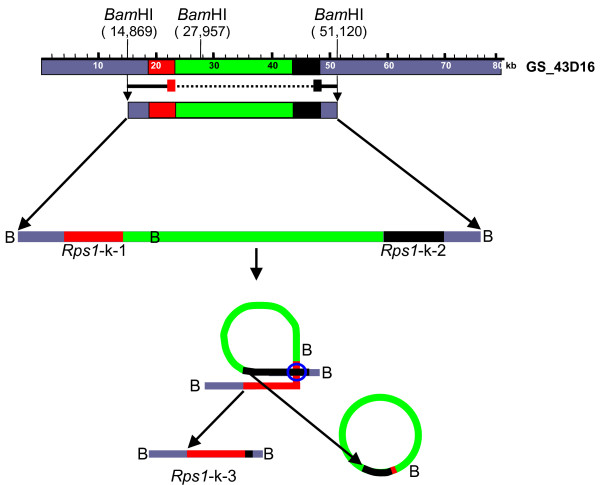
Following sequencing, _Rps1_-k-3 was not identified from GS_43D16. Physical mapping of GS_43D16 (Figure 2) and high identities between overlapping sequences of three independent BAC clones (Figure 3) suggested strongly that the complete sequence of GS_43D16 was obtained and this clone did not go through any rearrangement. _Rps1_-k-3 was identified from a binary clone, p43-10 cloned in the pTF101.1 vector [1]. The clone was isolated from a library of binary clones prepared from GS_43D16 DNA partially digested with _Bam_HI. Since this gene was not found in GS_43D16, the gene must have evolved through intramolecular recombination in Escherichia coli. Comparison of the insert sequence of p43-10 with GS_43D16 sequence revealed that the insert of the binary clone carries sequences identical to a _Bam_HI fragment that contains an internal _Bam_HI site (Figure 4). However, p43-10 insert DNA does not contain a segment of the GS_43D16 including the internal _Bam_HI site. This gene presumably originated from recombination in E. coli. By looking at the recombinant breakpoint in _Rps1_-k-3, it was hypothesized that two identical 174 bp sequences (21,980 through 22,154 and 46,473 through 46,647 of the _Rps1_-k region shown in Figure 3B) of the 3'-end of both _Rps1_-k-1 and _Rps1_-k-2 were involved in the RecA-independent recombination process as shown in Figure 4.
Figure 4.
Generation of the _Rps1_-k-3 through intramolecular recombination. Locations of _Rps1_-k-1 and _Rps1_-k-2 on the GS_43D16 sequence are shown. Partial _Bam_HI digested GS_43D16 DNA was cloned into the binary vector pTF101.1 and a library of binary clones was obtained. The library was screened for LRR sequences. Binary clone p43-10 contained the _Rps1_-k-3 gene, which is not present in GS_43D16 (Figure 3). This gene was presumably originated from intramolecular recombination in E. coli. Three _Bam_HI (B) sites involved in generation of the _Rps1_-k-3 [1] are shown on the map. Solid line shows the region cloned in p43-10 and broken line indicates the region not found in p43-10. Presumably this region was lost during the recombination process in E.coli. The possible recombination process involved in the evolution of _Rps1_-k-3 is shown at lower part of the figure. The two identical 174 bp sequences of _Rps1_-k-1 (red line) and _Rps1_-k-2 (black line) involved in the recombination process are shown within the blue open circle (21,980 through 22,154 of _Rps1-_k-1 and 46,473 through 46,647 of _Rps1_-k-2 in the _Rps1_-k region shown in Figure 3B). The proposed model for the recombination event in E. coli is based on the article by Weisberg and Adhya [65].
The major recombination pathway in E. coli requires RecA [36]. Therefore, recA- strains, such as DH10B used in our experiment, are considered for recombinant gene cloning experiments to avoid any recombination events. Unfortunately, RecA-independent intraplasmid recombination does occur in these recA- strains. The frequency of recombination is however much lower as compared to that observed in _recA_+ strains [37,38]. A recent study suggested that RecA-independent recombination is suppressed by single-strand DNA exonuclease (ssExos) activity. In absence ssExos, the extent of RecA-independent recombination in recA- strains is comparable to that in recA+ strains [39].
Discussion
Genomes of higher plants vary significantly in their size and complexity. Repetitive DNA sequences have been shown to be the major determinant of genome sizes in higher plants [13]. The prevalence of transposable elements and retroelements can promote unequal crossing-over leading to transposon-mediated rearrangements and gene duplications [40]. It has been hypothesized that transposable elements play a major role in the expansion and diversification of transmembrane receptor kinase-type disease resistance Xa21 gene family [9]. The abundance of retroelements has been observed in several genomic regions containing R genes or RGA loci, such as barley powdery mildew resistance gene, Mla, and Citrus virus resistance gene, Ctv [7,8]. The variability among 14 rice Xa21 gene members has been considered to be generated mainly from the rearrangements mediated by transposon-like elements [9]. _Rps1_-k genes are arranged closely. About 38 copies of _Rps1_-k-like sequences were predicted to exist in the soybean genome. Most of the copies are clustered in the _Rps1_-k region [32]. A copia_-like retroelement, Tgm_r, has previously been reported from the _Rps1_-k region [41]. It is possible that retrotransposons facilitated the amplification of the _Rps1_-k gene family.
In many plant species such as Arabidopsis thaliana and Medicago tuncatula, chromosome arms are differentiated into euchromatic and heterochromatic regions [42-44]. Recently, Lin et al. [14] showed that in soybean heterochromatic regions are also delimited from euchromatin. Studies in Arabidopsis, Medicago and Lycopersicon esculentum have shown that the euchromatin has a high gene density, whereas pericentromeric heterochromatin is largely comprised of repetitive sequences [44-46]. The _Rps1_-k region is composed of mostly tandem repeat sequences and retroelements (Figure 3; Table 2). The gene content is very similar to that of a soybean BAC clone identified from the pericentromeric heterochromatin [14]. FISH mapping showed that SIRE1 and other retroelements are sequestered to the heterochromatic and/or pericentromeric regions [14]. The tandem repeat sequences and retroelements including SIRE1 are commonly abundant in heterochromatic and/or pericentromeric regions of the soybean genome. Therefore, the _Rps1_-k region could be located in heterochromatic region which may be pericentromeric.
The possible microcollinearities of the _Rps1_-k locus with genomic sequences of plant species such as Arabidopsis, Medicago and Lotus japonicus were investigated. An R protein-like sequence of Medicago genomic clone, MTH2-138E10, showed 65% identities to Rps1-k-2. However, no synteny was observed between the _Rps1_-k region and MTH2-138E10 sequence. A limited synteny of the _Rps1_-k locus was observed with the Lotus genome. Two copies of a _Lotus Rps1_-k homolog located five kb apart showed 54%–58% identity with the Rps1-k-2 protein. These two genes are located in two overlapping Lotus BAC clones, LjT02F05 and LjT20J15. Apart from the _Lotus Rps1_-k homolog, no nucleic acid sequences of these two Lotus BAC clones showed similarity to sequences of the _Rps1_-k region. In order to identify susceptible haplotype (rps1), the _Rps1_-k contig sequence was compared with the available BAC end sequences from the SoyGD database that contains sequences of the cultivar Forrest carrying the susceptible rps1 gene [27]. No sequence from LG N of the Forrest haplotype was identified that showed similarity to the _Rps1_-k contig reported here (Figure 3B).
It has been reported that plant disease resistance gene loci exhibit extensive loss of synteny. R gene-like sequences frequently lack syntenic map locations between the cereal species rice, barley, and foxtail millet [47]. An effort to clone the rice homolog of the barley Rpg1 gene was unsuccessful; because, although the DNA markers flanking Rpg1 were syntenic between rice and barley, the region containing the gene is absent in the syntenic rice genome [48]. These observations imply that R gene loci evolve faster than the rest of the genomes. This is further supported by comparative sequence analysis conducted in crucifers and grasses [49]. R genes may be located in less stable regions of the genome such as telomeric or pericentromeric regions where synteny is poorly conserved [50]. The tomato Tm-2 gene resides in a heterochromatic region near the centromere of chromosome 9 [51]. The Rpg1 gene is located near the telomere of the short arm of barley chromosome 1 [48]. The tomato Mi-1 gene is located at the border region between euchromatin and heterochromatin [52]. The lack of microsynteny of the _Rps1_-k region with the currently available genome sequences and abundance of repeat sequences including retroelements suggested that _Rps1_-k is located in a heterochromatic region which could be pericentromeric.
The _Rps1_-k-1 and _Rps1_-k-2 genes are about 20 kb apart (Figure 3). Most frequently R genes are arranged in clusters, and genes within one cluster are mostly derived from a common ancestral gene [4]. This clustering feature is considered to facilitate the expansion of R gene numbers and race-specificities through recombination and positive selection [5]. Study of multiple, genetically linked R gene families has provided insight into the molecular mechanisms of R gene evolution and the generation of novel recognition specificity. Seven family members of Xa21 were identified within a 230-kb region [9]. Similarly, seven members of the tomato I2 gene family reside in a 90 kb region [53]. The tomato Cf-2 locus contains two nearly identical resistance genes in a 17 kb fragment [54]. Both tomato Cf4 and Cf9 loci comprised of four additional tandemly duplicated paralogous copies within a 36-kb region [55]. Paralogous R gene sequences have also been reported to map more distantly. For example, members of the lettuce Dm3 family span at least 3.5 Mb with at least 120 kb distance between two gene members [56]. Although in these examples members are evolved from one progenitor gene [4], three distinct CC-NB-LRR gene families were identified in the Mla locus within a 240-kb region [7,57]. The potato R1 locus also contains three fast evolving CC-NB-LRR genes that undergo frequent sequence exchanges among members of individual groups [58].
Plants have to generate novel resistance specificities to combat the quickly evolved pathogens. This clustering feature can facilitate the expansion of R gene numbers and the generation of new R gene specificities through recombination and positive selection [5]. Paralogous R gene sequences were most likely evolved through unequal recombination. The maize Rp1 locus, carrying nine paralogues, is probably the best example of unequal recombination for evolution of tandem paralogous R gene sequences [59]. An unequal crossing over between Arabidopsis RPP8 and its paralog, RPHA8 most likely resulted in rpp8 [60]. An unequal crossing over event was detected at the _Rps1_-k region leading to tandem duplication [32].
Conclusion
Genomes of higher plants vary significantly in their size and complexity because of the existence of a large amount of repetitive sequences. It was observed that the _Rps1_-k region is composed of mostly tandem repeat sequences and retroelements. Several disease resistance genes have been found in the less stable regions of the genome such as telomeric or pericentromeric regions where synteny is poorly conserved. The lack of microsynteny of the _Rps1_-k region with the currently available genome sequences and abundance of repeat sequences in the locus suggest that _Rps1_-k is located in a heterochromatic region that could be pericentromeric.
Methods
BAC DNA sequencing
The details of sequencing strategies of the three BACs, GS_18J19, GS_43D16 and GS_99I16 were described previously [1]. The sequence reads generated were assembled using the Phred/Phrap/Consed package [61,62].
Directional sequencing of GS_43D16
The EZ::TN <_Not_I/KAN-3> transposon insertion BAC clones were generated using the EZ::TN in-Frame Linker insertion kit (Epicentre, Madison, WI). The transposon insertion sites were mapped by _Not_I digestion. Southern hybridization was carried out to physically map the position of transposon insertion in each clone. Both ends of GS_43D16 were used as probes. The 5'-end sequence of GS_43D16 was amplified with primers: (i) GS_43D16 end1F: CTGTAAATTATAAACACATGCCAT and (ii) GS_43D16-end1R: GCTGAATTTCAGTGTAGTGGCGTTTAC. The 3'-end sequence of GS_43D16 was amplified with primers: (i) GS_43D16 end2F: CCCATCCTCATTAATACTTCACACCAC and (ii) GS_43D16 end2R: GTAGTGGAAGTCTATAGTTGTATACCTCTC. BAC DNA was prepared using the alkaline lysis minipreparation procedure. The clones were sequenced in a 96-well plate using either _Not_I/KAN-3 FP-2 or _Not_I/KAN-3 RP-2 primer provided in the EZ::TN in-Frame Linker insertion kit (Epicentre, Madison, WI). Sequencing was conducted at the Iowa State University DNA Facility.
Gene prediction and sequence analysis
Two gene prediction software packages were used in analyzing the BAC sequences: GENSCAN and GeneMark.hmm ES-3.0 (E – eukaryotic; S – self-training; 3.0 – the version) [34]. The Arabidopsis-based scoring matrix was applied when using GENSCAN. Arabidopsis, maize, rice and Medicago were used as model species when GeneMark.hmm was applied. To more accurately predict gene content in the _Rps1_-k region, the predicted genes were further analyzed using different BLAST programs of the NCBI Basic Local Alignment Search Tool (Blast) server [63]: (i) discontiguous Mega Blast program with entrez query limited to Arabidopsis, lotus, Medicago and soybean; (ii) Blastn against the soybean EST database; (iii) BlastX and (iv) BlastP. Soybean EST distribution on the BAC sequence was evaluated using the BlastN program. The simple repeat sequences and tandem repeat sequences were identified using Sputnik and tandem repeats finder program [64], respectively.
Authors' contributions
HG carried out all the studies presented in the paper. MKB conceived the study and participated in the experiment design and coordination, and helped with drafting and finalizing the manuscript.
Acknowledgments
Acknowledgements
We are grateful to Drs. Phil Becraft, Adam Bogdanove, Randy C. Shoemaker and Steven Whitham for invaluable discussion and James Baskett for critically reading this manuscript. This research has been supported by USDA-NRI Grant No. 2001-35301-10577 and a grant from Iowa Soybean Association and ISU Agronomy Department Endowment Fund.
Contributor Information
Hongyu Gao, Email: gaohy@iastate.edu.
Madan K Bhattacharyya, Email: mbhattac@iastate.edu.
References
- Gao H, Narayanan NN, Ellison L, Bhattacharyya MK. Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol Plant Microbe Interact. 2005;18:1035–1045. doi: 10.1094/MPMI-18-1035. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003;14:177–193. doi: 10.1016/S0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol. 2002;19:76–84. doi: 10.1093/oxfordjournals.molbev.a003984. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Graham MA, Marek LF, Shoemaker RC. Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics. 2002;162:1961–1977. doi: 10.1093/genetics/162.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing RA, Wise RP. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell. 2002;14:1903–1917. doi: 10.1105/tpc.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZN, Ye XR, Molina J, Roose ML, Mirkov TE. Sequence analysis of a 282-kilobase region surrounding the citrus Tristeza virus resistance gene (Ctv) locus in Poncirus trifoliata L. Raf. Plant Physiol. 2003;131:482–492. doi: 10.1104/pp.011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Pi LY, Wang GL, Gardner J, Holsten T, Ronald PC. Evolution of the rice Xa21 disease resistance gene family. Plant Cell. 1997;9:1279–1287. doi: 10.1105/tpc.9.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- Goldberg RB. DNA sequence organization in the soybean plant. Biochem Genet. 1978;16:45–68. doi: 10.1007/BF00484384. [DOI] [PubMed] [Google Scholar]
- Gurley WB, Hepburn AG, Key JL. Sequence organization of the soybean genome. Biochim Biophys Acta. 1979;561:167–183. doi: 10.1016/0005-2787(79)90500-8. [DOI] [PubMed] [Google Scholar]
- Kubis S, Schmidt T, Heslop-Harrison JS. Repetitive DNA Elements as a Major Component of Plant Genomes. Annals of Botany. 1998;82:45–55. doi: 10.1006/anbo.1998.0779. [DOI] [Google Scholar]
- Lin JY, Jacobus BH, SanMiguel P, Walling JG, Yuan Y, Shoemaker RC, Young ND, Jackson SA. Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics. 2005;170:1221–1230. doi: 10.1534/genetics.105.041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Jurman I, Shi L, Zhu T, Keim P, Rafalski JA. The STR120 satellite DNA of soybean: organization, evolution and chromosomal specificity. Chromosome Res. 1997;5:363–373. doi: 10.1023/A:1018492208247. [DOI] [PubMed] [Google Scholar]
- Vahedian MA, Shi L, Zhu T, Okimoto R, Danna K, Keim P. Genomic organization and evolution of the soybean SB92 satellite sequence. Plant Mol Biol. 1995;29:857 –8862. doi: 10.1007/BF00041174. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, Kochert G, Boerma HR. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (LIS) LIS. http://www.comparative-legumes.org/
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, Specht JE. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- Danesh D, Penuela S, Mudge J, Denny RL, Nordstrom H, Martinez JP, Young ND. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor Appl Genet. 1998;96:196–202. doi: 10.1007/s001220050727. [DOI] [Google Scholar]
- Marek LF, Shoemaker RC. BAC contig development by fingerprint analysis in soybean. Genome. 1997;40:420–427. doi: 10.1139/g97-056. [DOI] [PubMed] [Google Scholar]
- Meksem K, Zobrist K, Ruben E, Hyten D, Quanzhou T, Zhang HB, Lightfoot D. Two large-insert soybean genomic libraries constructed in a binary vector: applications in chromosome walking and genome wide physical mapping. Theor Appl Genet. 2001;101:747–755. doi: 10.1007/s001220051540. [DOI] [Google Scholar]
- Salimath SS, Bhattacharyya MK. Generation of a soybean BAC library, and identification of DNA sequences tightly linked to the Rps1-k disease resistance gene. Theor Appl Genet. 1999;98:712–720. doi: 10.1007/s001220051125. [DOI] [Google Scholar]
- Santra DK, Sandhu D, Tai T, Bhattacharyya MK. Construction and characterization of a soybean yeast artificial chromosome library and identification of clones for the Rps6 region. Funct Integr Genomics. 2003;3:153–159. doi: 10.1007/s10142-003-0092-8. [DOI] [PubMed] [Google Scholar]
- Shultz JL, Kurunam D, Shopinski K, Iqbal MJ, Kazi S, Zobrist K, Bashir R, Yaegashi S, Lavu N, Afzal AJ, Yesudas CR, Kassem MA, Wu C, Zhang HB, Town CD, Meksem K, Lightfoot DA. The Soybean Genome Database (SoyGD): a browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res. 2006;34:D758–65. doi: 10.1093/nar/gkj050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrather JA, Stienstra WC, Koenning SR. Soybean disease loss estimates for the United States from 1996 to 1998. Can J Plant Pathol. 2001;23:122–131. [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Kasuga T, Salimath SS, Shi J, Gijzen M, Buzzell RI, Bhattacharyya MK. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol Plant-Microbe Interact. 1997;10:1035–1044. doi: 10.1094/MPMI.1997.10.9.1035. [DOI] [Google Scholar]
- Schmitthenner AF, Hobe M, Bhat RG. Phytophthora sojae races in Ohio over a 10-year interval. Plant Dis. 1994;78:269–276. [Google Scholar]
- Bhattacharyya MK, Narayanan NN, Gao H, Santra DK, Salimath SS, Kasuga T, Liu Y, Espinosa B, Ellison L, Marek L, Shoemaker R, Gijzen M, Buzzell RI. Identification of a large cluster of coiled coil-nucleotide binding site-leucine rich repeat-type genes from the Rps1 region containing Phytophthora resistance genes in soybean. Theor Appl Genet. 2005;111:75–86. doi: 10.1007/s00122-005-1993-9. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten HM, Havecker ER, Farmer LM, Voytas DF. SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol Biol Evol. 2003;20:1222–1230. doi: 10.1093/molbev/msg142. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Margulies AD. Isolation and Characterization of Recombination-Deficient Mutants of Escherichia Coli K12. Proc Natl Acad Sci U S A. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Liu LF. recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J Mol Biol. 1994;235:414–423. doi: 10.1006/jmbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- Sosio M, Bossi E, Donadio S. Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:E37. doi: 10.1093/nar/29.7.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra BE, Sutera VA, Jr., Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci U S A. 2007;104:216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. Transposons and genome evolution in plants. Proc Natl Acad Sci USA. 2000;97:7002–7007. doi: 10.1073/pnas.97.13.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya MK, Gonzales RA, Kraft M, Buzzell RI. A copia-like retrotransposon Tgmr closely linked to the Rps1-k allele that confers race-specific resistance of soybean to Phytophthora sojae. Plant Mol Biol. 1997;34:255–264. doi: 10.1023/A:1005851623493. [DOI] [PubMed] [Google Scholar]
- Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998;13:867–876. doi: 10.1046/j.1365-313X.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- Fransz PF, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell. 2000;100:367–376. doi: 10.1016/S0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook D, Huguet T, de Jong JH, Fransz PF, Bisseling T. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 2001;27:49–58. doi: 10.1046/j.1365-313x.2001.01057.x. [DOI] [PubMed] [Google Scholar]
- Initative AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang X, Cheng Z, Mueller L, Giovannoni J, Tanksley SD. Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics. 2006;172:2529–2540. doi: 10.1534/genetics.106.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P. Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acady Sci U S A. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Kilian A, Chen JP, Kudrna D, Steffenson B, Yamamoto K, Matsumoto T, Sasaki T, Kleinhofs A. Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome. 1999;42:1071–1076. doi: 10.1139/gen-42-6-1071. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- Michelmore R. Genomic approaches to plant disease resistance. Curr Opin Plant Biol. 2000;3:125–131. doi: 10.1016/S1369-5266(99)00050-3. [DOI] [PubMed] [Google Scholar]
- Motoyoshi F, Ohmori T, Murata M. Molecular characterization of heterochromatic regions around the Tm-2 locus in chromosome 9 of tomato. Symp Soc Exp Biol. 1996;50:65–70. [PubMed] [Google Scholar]
- Zhong XB, Bodeau J, Fransz PF, Williamson VM, van Kammen A, de Jong JH, Zabel P. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor ApplGenet. 1999;98:365–370. doi: 10.1007/s001220051081. [DOI] [Google Scholar]
- Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, Van der Lee T, Bleeker M, Onstenk J, de Both M, Haring M, Mes J, Cornelissen B, Zabeau M, Vos P. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell. 1996;84:451–459. doi: 10.1016/S0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/S0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell. 1998;10:1817–1832. doi: 10.1105/tpc.10.11.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang H, Wei F, Marano MR, Wirtz U, Wang X, Liu J, Shum WP, Zaborsky J, Tallon LJ, Rensink W, Lobst S, Zhang P, Tornqvist CE, Tek A, Bamberg J, Helgeson J, Fry W, You F, Luo MC, Jiang J, Robin Buell C, Baker B. The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 2005;44:37–51. doi: 10.1111/j.1365-313X.2005.02506.x. [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- NCBI Basic Local Alignment Search Tool (Blast) http://www.ncbi.nlm.nih.gov/BLAST/ [DOI] [PubMed]
- Sputnik http://tandem.bu.edu/trf/trf.submit.options.html
- Weisberg RA, Adhya S. Illegitimate recombination in bacteria and bacteriophage. Annu Rev Genet. 1977;11:451–473. doi: 10.1146/annurev.ge.11.120177.002315. [DOI] [PubMed] [Google Scholar]