A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans (original) (raw)
Abstract
Drosophila shibire and its mammalian homologue dynamin regulate an early step in endocytosis. We identified a Caenorhabditis elegans dynamin gene, dyn-1, based upon hybridization to the Drosophila gene. The dyn-1 RNA transcripts are _trans_-spliced to the spliced leader 1 and undergo alternative splicing to code for either an 830- or 838-amino acid protein. These dyn-1 proteins are highly similar in amino acid sequence, structure, and size to the Drosophila and mammalian dynamins: they contain an N-terminal GTPase, a pleckstrin homology domain, and a C-terminal proline-rich domain. We isolated a recessive temperature-sensitive dyn-1 mutant containing an alteration within the GTPase domain that becomes uncoordinated when shifted to high temperature and that recovers when returned to lower temperatures, similar to D. shibire mutants. When maintained at higher temperatures, dyn-1 mutants become constipated, egg-laying defective, and produce progeny that die during embryogenesis. Using a dyn-1::lacZ gene fusion, a high level of dynamin expression was observed in motor neurons, intestine, and pharyngeal muscle. Our results suggest that dyn-1 function is required during development and for normal locomotion.
Dynamin is a 100-kDa GTPase that regulates an early stage of endocytosis (1). Although it was first isolated from bovine brain (2), much of our understanding of dynamin has come from the analysis of the Drosophila dynamin mutant, shibire (3, 4). shibire mutations confer rapid and reversible temperature-sensitive paralysis (5) that results from a block in endocytosis and the subsequent depletion of synaptic vesicles (6). The overexpression of a similar dynamin mutation in mammalian cells inhibits clathrin-mediated endocytosis at a point after coat proteins assemble at the plasma membrane but before the coated pits become deeply invaginated (7, 8). Dynamin can self-assemble into rings in vitro, and dynamin rings are found at the neck of budding vesicles in the presence of GTPγS (9, 10). These observations suggest that dynamin might act to pinch off clathrin-coated vesicles from the plasma membrane.
Dynamin belongs to a growing family of GTP-binding proteins with diverse functions, such as the mammalian interferon-induced antiviral MX proteins (11), the yeast mitochondrial DNA replication protein MGM1 (12), the yeast membrane transport proteins VPS1 and DNM1 (13, 14), and the plant cell plate formation protein phragmoplastin (15). These proteins possess a conserved amino-terminal GTPase domain (43–66% identity) but are typically much less related in sequence outside of this region (<30% identity). Dynamin contains a pleckstrin homology domain, which is thought to mediate protein–protein or protein–lipid interactions (16), and a C-terminal proline-rich domain (PRD). Although in vitro association of the dynamin PRD with a variety of ligands (microtubules, acidic phospholipids, SH3 domains, and even mAbs) increases the rate of GTP hydrolysis (17–20), it is unclear whether such interactions regulate dynamin activity in vivo.
We have begun an analysis of dynamin function in the nematode Caenorhabditis elegans. We report here the cloning of a C. elegans dynamin gene, dyn-1, and the discovery of a temperature-sensitive dyn-1 mutant. Our results suggest that dyn-1 is essential for development and normal locomotion.
MATERIALS AND METHODS
Molecular Cloning and Sequence Analysis.
A library of wild-type C. elegans (var. Bristol strain N2) genomic DNA in the λ 2001 vector (21) was probed at low stringency with radiolabeled Drosophila shibire cDNA (4) by hybridizing for 16 h at 55°C in 5X standard saline phosphate/EDTA (SSPE) (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 5X Denhardt, and 50 μg/ml single-stranded DNA and by washing for 2 h at 55°C in 5X SSPE and 1% SDS and then for 1 h at 60°C in 5X SSPE and 0.1% SDS. Genomic DNA fragments from hybridizing phages were subcloned into pBluescript (Stratagene). These clones were used to screen two mixed stage C. elegans cDNA libraries (22) (Stratagene). We determined the sequences of genomic DNAs and cDNAs using custom primers and Sequenase (United States Biochemical). A cDNA for the 5′-end of the dyn-1 mRNA was obtained by reverse transcriptase PCR with a primer corresponding to spliced leader 1 (23) and primers complementary to dyn-1 sequences. These PCR products were cloned into the TA vector (Invitrogen) and sequenced. The DNA sequences of the dyn-1(ky51) mutant were determined by amplifying the dynamin coding regions from genomic DNA prepared as described (24). The PCR products were cloned into pBluescript, and several independent isolates were sequenced. The physical map position of dyn-1 was ascertained by hybridizing the dyn-1 genomic DNA to a filter containing an array of yeast artificial chromosomes that cover the C. elegans genome (kindly provided by R. Waterston, Washington University, St. Louis).
Isolation of a dyn-1 Mutant.
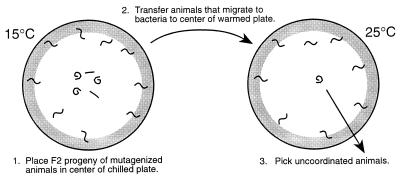
We performed a screen for mutants with an uncoordinated phenotype induced rapidly by an increase in temperature (see Fig. 3). Specifically, the F2 progeny of wild-type animals treated with ethylmethanesulfonate (25) were raised at 15°C. Animals were collected, washed with S Basal, and placed in the center of a precooled (15°C) 100-mm NGM plate, which had concentrated Escherichia coli spread around the edge. After 60–90 min of incubation at 15°C, animals that had migrated to the food were recovered and placed in the center of a prewarmed (25°C) NGM plate with E. coli around the edge. After 60–90 min of incubation at 25°C, uncoordinated animals remaining in the center of the plate were picked. Mutants were mapped using standard procedures (25).
Figure 3.
Screen for rapidly reversible, temperature-sensitive, uncoordinated mutants. The F2 progeny of animals treated with the mutagen EMS were placed in the center of a plate cooled to 15°C with E. coli around the edge (see Materials and Methods). Uncoordinated or paralyzed worms tended to remain in the center of the plate whereas worms that moved well at the nonrestrictive temperature migrated toward the bacteria. The mobile animals were recovered and placed in the center of a plate warmed to 25°C with E. coli spread around the edge. Animals that became uncoordinated at the restrictive temperature tended to remain in the center of the plate and were picked for further study.
To document motility, the tracks made by an individual worm in a lawn of E. coli were photographed at 1-min intervals for a total of 5 min after the worm was transferred from a 15°C to a prewarmed plate. The tracks were digitized by tracing them onto a graphics tablet and then measured using canvas software (Deneba, Miami). Pharyngeal pumping rates were determined by counting the number of contractions per minute in 20 young adult animals that had been incubated at 25°C for 2 h. The lengths of the defecation cycles were determined by measuring 10 cycles in each of 10 young adult animals that had been incubated at 25°C for 2 h.
Germ Line Transformation.
Germ line transformation of ky51 was done by coinjecting the 8-kb _Xba_I–_Hin_dIII dyn-1 genomic fragment (plasmid pCDG1) at 20 μg/ml with the dominant rol-6(su1006) roller marker (plasmid pRF4) at 50 μg/ml as described (26). A dyn-1::lacZ gene fusion was constructed by cloning a 3.2-kb _Xba_I–XhoI genomic fragment in-frame to the LacZ coding sequences of the pPD16.51 expression vector (Fig. 1c)(27). This genomic fragment contained the coding sequences for the first 214 amino acids of dyn-1 and 2375 nt upstream of the ATG. The expression construct contained the simian virus 40 nuclear localization signal to facilitate the identification of the β-galactosidase-expressing cells. Germ line transformation was accomplished by injecting the dyn-1::lacZ construct at 50 μg/ml with the roller marker (plasmid pRF4) at 50 μg/ml. Transgenic animals were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (27).
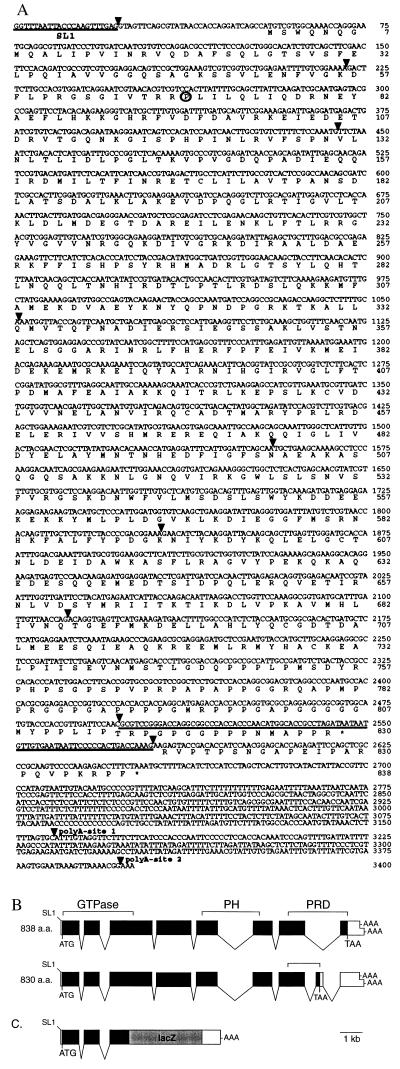
Figure 1.
(A) Nucleotide and predicted amino acid sequences of the dyn-1 3.4-kb cDNA. Nucleotides are numbered beginning at the first nucleotide of the spliced leader 1 _trans_-spliced leader. Amino acids are numbered from the first predicted methionine. The proline altered in the ky51 mutant is circled. Spliced leader 1 and exon 8, which is present in some dyn-1 transcripts, are underlined. Splice sites are marked by triangles. Polyadenylation sites are as marked. The termination codons are marked by asterisks. (B) dyn-1 genomic structure derived from cDNA and genomic DNA sequences. Coding regions are denoted by solid boxes; the 5′ and 3′ untranslated regions are shown as open boxes. The boundaries of the GTPase, pleckstrin homology and PRDs are marked. SL1, the _trans_-spliced leader; AAA, sites of polyadenylation; ATG and TAA, the predicted start and stop sites of translation, respectively. (C) dyn-1::lacZ gene fusion. LacZ coding sequences were fused in-frame to an _Xho_I site in the third exon of dyn-1 (see Materials and Methods).
RESULTS
Molecular Analysis of a C. elegans Dynamin Homologue.
We identified a C. elegans dynamin gene by screening a C. elegans genomic DNA library under low stringency hybridization conditions using a Drosophila dynamin cDNA as a probe. An 8-kb genomic fragment was recovered from one of seven positive hybridizing clones and was found by DNA sequence analysis to contain regions coding for a protein related in sequence to both Drosophila and mammalian dynamins. We named this dynamin gene “dyn-1.”
We used the 8-kb dyn-1 genomic fragment to isolate 18 cDNA clones from two different mixed stage cDNA libraries. In addition, we performed reverse transcriptase PCR to obtain full length cDNAs. We identified cDNAs representing two dyn-1 transcripts that differed by either the absence (17 clones) or presence (1 clone) of an 82-bp exon (exon 8). Transcripts lacking exon 8 encode an 838-amino acid (94.3 kDa) protein whereas transcripts containing exon 8 encode an 830-amino acid (93.3- kDa) protein (Figs. 1 and 2). The two dyn-1 proteins are identical in sequence for their first 815 amino acids but differ in their C-terminal sequences because exon 8 contains an in-frame termination codon. The last 15 amino acids of the smaller protein are encoded by exon 8, and the last 23 amino acids of the larger protein are encoded by exon 9. The two dyn-1 products are consistent in size, with a 100-kDa polypeptide band detected in crude C. elegans extracts by probing a Western blot with antisera raised against D. shibire protein (7) (data not shown). The dyn-1 proteins are highly similar in amino acid sequence to human dynamin-1 and D. shibire proteins (61% and 62% identical to the 838-amino acid dyn-1 protein, respectively; Fig. 2). The high degree of sequence conservation among these proteins extends throughout their lengths and includes the N-terminal GTPase, the pleckstrin homology, and the C-terminal PRDs.
Figure 2.
Alignment of the C. elegans dyn-1 amino acid sequence (C. elegans) with D. shibire (Drosophila) (4) and human dynamin-1 sequences (Homo sapiens) (7). Conserved elements (I, II, and II) within GTPase domain are boxed. The pleckstrin homology and PRDs are indicated by dotted lines. The alternative C-terminal amino acid sequences for each dynamin isoform are boxed.
The dyn-1 gene structure was determined by comparing the cDNA and genomic DNA sequences (Fig. 1B). There are nine exons, including the alternatively spliced eighth exon (see above). Like many C. elegans transcripts (28), dyn-1 transcripts are modified by the addition of spliced leader 1 sequences by _trans_-splicing. Polyadenylation can occur at either nucleotide 3158 or 3397 of the cDNA. Alternatively spliced transcripts encoding dynamin proteins with different C-terminal proline-rich sequences have been documented for both Drosophila and mammalian dynamin genes. D. shibire mRNAs can contain one of two possible final exons coding for different C-terminal sequences whereas human dynamin-1 mRNAs can contain additional short exons that alter the reading frame (3, 4, 7). Further study is needed to determine whether the dyn-1 splice variants are developmentally regulated or cell-type-specific.
Identification of a Temperature-Sensitive Dynamin Mutant.
Mutations conferring a rapidly reversible, temperature-sensitive paralysis have been found for many genes in Drosophila (e.g., shibire and para), yet similar mutations have been recovered for only two C. elegans genes: cha-1 (29) and mah-2 (30). To identify new rapidly reversible, temperature-sensitive, uncoordinated C. elegans mutants, we devised a screen to enrich for mutants that become uncoordinated shortly after being shifted to a higher temperature (Fig. 3; see Materials and Methods). From screening the progeny of 35,000 F1 animals, we recovered five mutants (ky48, ky49, ky50, ky51, and ky52) that were wild-type or nearly wild-type when maintained at 15°C and that became severely uncoordinated when shifted to 25°C. When returned to 15°C, the animals recovered their ability to move normally. The ky48, ky49, ky50, and ky52 mutations were found to be alleles of cha-1, which encodes a choline acetyltransferase (31). The rapid onset of uncoordinated behavior caused by these and other cha-1 alleles (29, 30) suggests that acetylcholine is turned over at a high rate.
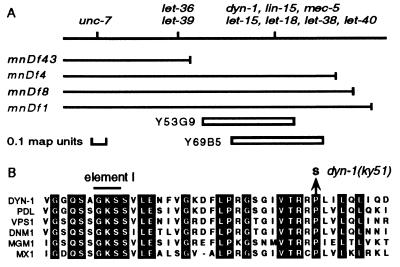
The remaining mutant, ky51, was recessive and defined a new Unc gene mapping near lin-15 on the X chromosome (Fig. 4A). ky51 was uncovered by the deficiencies mnDf1, mnDf4, and mnDf8 but not by the deficiency mnDf43. This genetic map position coincides with the location of the dyn-1 gene on the physical map, as evidenced by hybridization to the yeast artificial chromosome clones Y69B5 and Y53G9 (Fig. 4A). To investigate whether ky51 is an allele of dyn-1, we tested the 8-kb dyn-1 genomic fragment for the rescue of the uncoordinated and low fertility phenotypes of ky51 mutants at 25°C. One hundred percent of ky51 mutants were uncoordinated at 25°C whereas fewer than half of the ky51 animals that carried a dyn-1 extrachromosomal array were uncoordinated at 25°C. The dyn-1 array also rescued the low fertility phenotype of ky51 at 25°C (see below).
Figure 4.
(A) Genetic and physical maps of the dyn-1 region of the X chromosome. The positions of genes are marked and the extents of deficiencies are denoted by solid lines. The approximate positions of YAC Y53G9 and Y69B5 are shown below. Other than for dyn-1, we have not determined if other genes within this interval are contained within these yeast artificial chromosomes. (B) Alignment of amino acid sequences near element I from dynamin-related GTPases showing the substitution of the invariant proline at position 70 by a serine in the dyn-1(ky51) mutant. C. elegans dyn-1 sequences are from positions 39–78.
To confirm that ky51 is an allele of dyn-1, we sequenced the dyn-1 coding regions of the ky51 allele. ky51 contains a G:C to A:T transition that results in the substitution of a serine for a proline at residue 70 within the GTPase domain (Fig. 4B). This proline is invariant in all members of the dynamin-related GTPase family. The two temperature-sensitive D. shibire mutations are also in the GTPase domain, corresponding to positions 148 and 275 in the dyn-1 sequence (4). However, dyn-1(ky51) is recessive, and the Drosophila mutations are semi-dominant. ky51 complemented the four known lethal mutations [let-15(mn127), let-18(mn122), let-38(mn141), and let-40(mn150)] located within the interval defined by the right ends of the deficiencies mnDf43 and mnDf4 (Fig. 4A) (32). We were unable to rescue these lethal mutations either by germ line transformation with the 8-kb dyn-1 genomic fragment or by crossing in an extrachromosomal dyn-1 array. We concluded that dyn-1 represents a new gene.
Characterization of dyn-1(ky51).
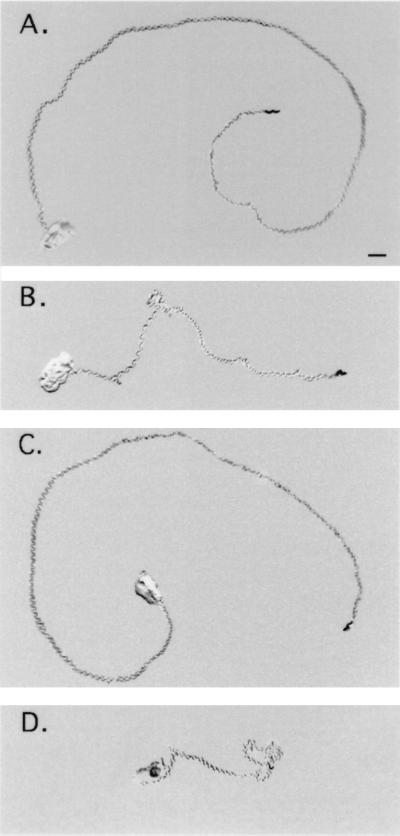
When dyn-1(ky51) animals were maintained at 15°C, their movement, defecation rate, egg-laying, and fertility were similar to those of wild-type animals. Less than a minute after being shifted to 25°C, the mutants exhibited a striking uncoordinated phenotype: They became sluggish and assumed a kinked posture. Adult animals initially displayed a loopy movement defect whereas larvae were more rapidly and severely affected by the shift in temperature. Typically, mutant animals stopped moving and assumed a kinked posture within 20 s of transfer. Over the next few minutes, the animals began to move at a rate of 6.8 mm/min for freely moving dyn-1(ky51) (n = 10), and wild-type animals moved at a rate of 8.6 mm/min (n = 10)(Fig. 5). The difference in rates of movement was more extreme at 30°C: 2.8 mm/min for dyn-1(ky51) (n = 10) and 8.2 mm/min for wild-type animals (n = 10). When the mutants were returned to 15°C, their rate of movement returned to normal with recovery times ranging from a few minutes to hours, depending on their age and on the length and severity of the heat treatment.
Figure 5.
Mobility of wild-type and dyn-1(ky51) animals. The rate of movement of a single animal was analyzed by recording the tracks left in a bacterial lawn over time. Animals raised at 15°C were transferred to the center of a bacterial lawn on a warmed plate. Photographs were taken at 1-min intervals, and the distance each animal traveled was calculated. Tracks shown are 5-min time points for wild-type animals at (A) 25°C and (C) 30°C and for dyn-1(ky51) animals at (B) 25°C and (D) 30°C. (Bar = 1 mm.)
dyn-1(ky51) animals exhibited a reduced pharyngeal pumping rate (160 pumps per minute, n = 20, SD = 52, compared with 259 for wild-type animals, n = 20, SD = 13), a prolonged defecation cycle (62 s per cycle, n = 10, SD=14, compared with 44 for wild-type animals, n = 10, SD=4), and an egg-laying defect when maintained at 25°C for many hours. The brood size for dyn-1(ky51) animals at 25°C was greatly diminished, with an average of 25 progeny for dyn-1(ky51) animals (n = 20, SD = 8), compared with 161 for wild-type animals (strain N2; n = 20, SD = 24). The reduction in the dyn-1(ky51) brood size was caused by a decrease in egg production and by embryonic and early larval lethality.
To investigate further the stage at which dyn-1 function was required for viability, we collected eggs laid over a 4-h period at 15°C from five young adult wild-type animals and from five young adult dyn-1(ky51) animals. Half of the eggs from each strain were transferred to 25°C, and the remainder was kept at 15°C. The numbers of dead progeny (unhatched eggs and young larvae) and surviving progeny were counted. We found that 96% of dyn-1(ky51) eggs laid and maintained at 15°C grew to adulthood (132/138). Similarly, 97% of the dyn-1(ky51) eggs laid at 15°C and transferred to 25°C grew to adulthood (105/108) although these animals took at least twice as long to become adults and were very sick. Under either condition, all wild-type eggs hatched and grew to adulthood (237/237 at 15°C and 231/231 at 25°C). By contrast, dyn-1(ky51) eggs laid by hermaphrodites maintained at 25°C did not recover when they were transferred to 15°C. Because dyn-1(ky51) animals retained their eggs at 25°C, these eggs were typically older than those recovered at 15°C. Eggs are typically laid between the gastrulation stage and morphogenesis (33), suggesting that maternal or early zygotic dynamin function is essential for viability.
dyn-1 Expression in Neurons, Intestine, and Pharyngeal Muscle.
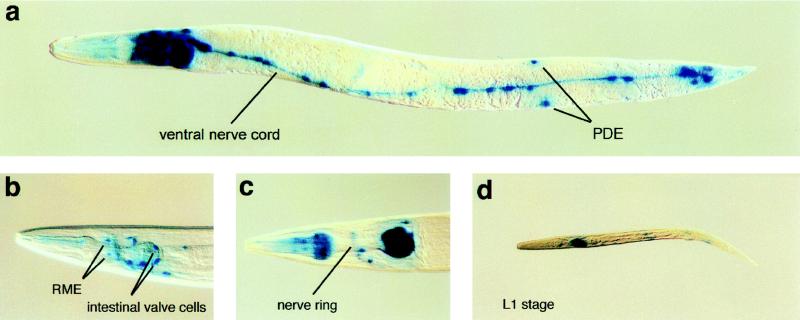
To investigate the expression pattern of dynamin, we constructed a dyn-1::lacZ gene fusion (Fig. 1C). In hermaphrodites containing the dyn-1::lacZ extrachromosomal array, we detected β-galactosidase staining in early embryos, all larval stages, and adults (Fig. 6). Strong staining was observed in motor neurons located in the head and ventral cord (Fig. 6 B and C). Weak staining was found in sensory neurons and interneurons located in the head around the nerve ring and in the tail. These observations suggest that motor neurons, which innervate body wall muscles required for locomotion and head musculature required for head movements, contain a higher level of dynamin than sensory neurons and interneurons, perhaps reflecting a higher level of synaptic activity.
Figure 6.
Expression pattern of a dyn-1::lacZ gene fusion. (A) Adult animal showing neuronal staining in the head region, along the ventral nerve cord and in preanal and lumbar ganglia. (B and C) Close-ups of heads showing staining in motor neurons around the pharynx. (D) A larva showing staining along the ventral nerve chord.
The pharyngeal–intestinal valve, intestinal–rectal valve, and intestinal cells also exhibited strong staining (Fig. 6B). The pharyngeal–intestinal valve connects the pharynx to the intestine and the intestinal–rectal valve connects the intestine to the rectum (34). The intestinal cells and specific cells within the valves contain microvilli on their apical surfaces, consistent with a need for high levels of dynamin for nutrient uptake via endocytosis. Strong staining was observed in pharyngeal muscle cells, especially m3 and m4, but not in body wall and vulval muscles. We also failed to detect staining in the hypodermis.
The dyn-1 expression pattern was similar to that of D. shibire, which is expressed at high levels in the nervous system and at lower levels in most other tissues (35). By contrast, the three mammalian dynamin genes have distinct expression patterns: Dynamin-1 is expressed in brain (7, 36), dynamin-2 is expressed in all tissues (36, 37), and a third dynamin is expressed in testis and at low levels in other tissues (38, 39).
DISCUSSION
C. elegans dyn-1 encodes two proteins that each contain an amino-terminal GTPase, a pleckstrin homology domain, and a carboxy-terminal PRD. The striking similarities in sequence, structure and size of the dyn-1 proteins with D. shibire (3, 4) and mammalian dynamins indicate that dyn-1 is a dynamin gene. Like other dynamin genes, dyn-1 transcripts undergo alternative splicing to code for two proteins that differ in their proline-rich sequences. The proline-rich regions have been shown to mediate interactions with SH3 domain-containing proteins, so these isoforms might have distinct functions either within the same cell or in different cells.
The genomic sequence analysis by the C. elegans Sequencing Consortium and the identification of only one dynamin gene by DNA hybridization suggests that only one dynamin gene is present in C. elegans. If true, dyn-1 would be expected to be expressed in all cells. A high level of dyn-1 expression was detected in neurons, intestine, and pharyngeal muscle. Surprisingly, we did not detect dyn-1 expression in the hypodermis, which secretes the cuticle that covers the animal, nor in coelomocytes, which have many coated pits and vesicles in their cytoplasm. Our dyn-1::lacZ gene fusion possessed only part of the dyn-1 gene, so it might lack regulatory sequences essential for expression at detectable levels in all tissues.
We recovered dyn-1(ky51) in a screen for mutants with an uncoordinated phenotype induced rapidly by an increase in temperature. Although ky51 animals appear wild-type when maintained at lower temperatures, ky51 animals have a kinked posture, a sluggish movement, a reduced pharyngeal pumping rate, a prolonged defecation cycle, an egg-laying defect, and embryonic lethality at higher temperatures. dyn-1(ky51) animals become severely uncoordinated in less than 1 min when shifted to the restrictive temperature and recover within minutes, depending on the severity and duration of heat treatment, when returned to lower temperatures. The rapid onset and recovery of locomotion defects seen in dyn-1(ky51) animals is comparable to the kinetics of paralysis and recovery seen in D. shibire mutants (5, 40). The paralysis observed in shibire mutants is caused by a depletion of synaptic vesicles resulting from a block in endocytosis (6). The locomotion and other defects seen in dyn-1 mutants likely also result from a defect in endocytosis.
The dyn-1(ky51) mutation confers a recessive phenotype and alters a conserved proline within the GTPase domain. By contrast, the two shibire mutants, ts1 and ts2, cause a dominant phenotype and alter two other residues within the GTPase domain. The overexpression of dynamin containing the ts1 mutation in mammalian cells blocks clathrin-mediated endocytosis at a point after coat proteins assemble at the plasma membrane but before the coated pits become deeply invaginated. It will be interesting to determine how the dyn-1(ky51) mutation affects the GTPase activity of dynamin and how endocytosis is blocked by this defect in dynamin function.
All characterized D. shibire mutants, even the weak mutants, ultimately become paralyzed at higher temperatures (5, 40). Although dyn-1(ky51) animals become rapidly uncoordinated when first shifted to higher temperatures, they never become fully paralyzed and eventually begin to move slowly. This distinction between the C. elegans and Drosophila dynamin mutants might simply reflect a difference in either the anatomy and physiology or the types of dynamin mutations. Alternatively, the ability of dyn-1(ky51) animals to move slowly in the absence of dynamin activity might indicate up-regulation of nonclathrin-mediated endocytosis. Previous experiments have shown that nonclathrin-mediated endocytosis can be induced in transfected mammalian cells when clathrin-mediated endocytosis is blocked by mutant dynamin (41).
Acknowledgments
A. M.v.d.B. thanks Paul Sternberg and the members of his lab for their advice and for sharing reagents. We are grateful to Erik Jorgensen for his valuable comments on this manuscript. Some C. elegans strains were obtained from the Caenorhabditis Genetics Center stock center. S.G.C. was supported by the Helen Hay Whitney Foundation and by the American Cancer Society (California Division). This work was supported by National Institutes of Health Program Project Grant GM40499 to E.M.M., by Public Health Services Grant DC01393 to C.I.B., and by Muscular Dystrophy Association Grant WB931203 and National Institutes of Health Grant GM51866 to A.M.v.d.B. C.I.B. is an Assistant Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
PRD
proline-rich domain
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. L29031).
References
- 1.Baba T, Damke H, Hinshaw J, Ikeda K, Schmid S L, Warnock D E. Cold Spring Harbor Symp Quant Biol. 1995;60:235–242. doi: 10.1101/sqb.1995.060.01.027. [DOI] [PubMed] [Google Scholar]
- 2.Shpetner H S, Vallee R B. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen M S, Obar R A, Schroeder C C, Austin T W, Poodry C A, Wadsworth S C, Vallee R B. Nature (London) 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 4.van der Bliek A M, Meyerowitz E M. Nature (London) 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 5.Grigliatti T A, Hall L, Rosenbluth R, Suzuki D T. Mol Gen Genet. 1973;120:107–114. doi: 10.1007/BF00267238. [DOI] [PubMed] [Google Scholar]
- 6.Kosaka T, Ikeda K. J Cell Biol. 1983;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herskovits J S, Burgess C C, Obar R A, Vallee R B. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinshaw J E, Schmid S L. Nature (London) 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 10.Takei K, McPherson P S, Schmid S L, DeCamilli P. Nature (London) 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 11.Arnheiter H, Meier E. New Biol. 1990;2:851–857. [PubMed] [Google Scholar]
- 12.Jones B A, Fangman W L. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- 13.Rothman J H, Raymond C K, Gilbert T, O’Hara P, Stevens T. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- 14.Gammie A E, Kurihara L J, Vallee R B, Rose M D. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu X, Verma D P S. EMBO J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw G. Bioessays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 17.Shpetner H S, Vallee R B. Nature (London) 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- 18.Tuma P L, Stachniak M C, Collins C A. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- 19.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, Campbell I D, Waterfield M D. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 20.Warnock D E, Terlecky L J, Schmid S L. EMBO J. 1995;14:1322–1328. doi: 10.1002/j.1460-2075.1995.tb07118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karn J, Matthes H W, Gait M J, Brenner S. Gene. 1984;32:217–224. doi: 10.1016/0378-1119(84)90049-0. [DOI] [PubMed] [Google Scholar]
- 22.Barstead R J, Waterston R H. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 23.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 25.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 28.Blumenthal T. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 29.Rand J B. Genetics. 1989;122:73–80. doi: 10.1093/genetics/122.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosono R, Kuno S, Midsukami M. J Exp Zool. 1985;235:409–421. doi: 10.1002/jez.1402350313. [DOI] [PubMed] [Google Scholar]
- 31.Alfonso A, Grundahl K, McManus J R, Rand J B. J Neurosci. 1994;14:2290–2300. doi: 10.1523/JNEUROSCI.14-04-02290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meneely P M, Herman R K. Genetics. 1981;97:65–84. doi: 10.1093/genetics/97.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood W B. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 215–241. [Google Scholar]
- 34.White J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 81–122. [Google Scholar]
- 35.Chen M S, Burgess C C, Vallee R B, Wadsworth S C. J Cell Sci. 1992;103:619–628. doi: 10.1242/jcs.103.3.619. [DOI] [PubMed] [Google Scholar]
- 36.Sontag J-M, Fykse E M, Ushkaryov Y, Liu J-P, Robinson P J, Sudhof T C. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- 37.Cook T A, Urrutia R, McNiven M A. Proc Natl Acad Sci USA. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook T, Mesa K, Urrutia R. J Neurochem. 1996;67:927–931. doi: 10.1046/j.1471-4159.1996.67030927.x. [DOI] [PubMed] [Google Scholar]
- 39.Nakata T, Takemura R, Hirokawa N. J Cell Sci. 1993;105:1–5. doi: 10.1242/jcs.105.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y-T, Wu C-F. J Neurogenetics. 1990;7:1–14. doi: 10.3109/01677069009084149. [DOI] [PubMed] [Google Scholar]
- 41.Damke H, Baba T, van der Bliek A M, Schmid S L. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]