Nucleosome formation with the testis-specific histone H3 variant, H3t, by human nucleosome assembly proteins in vitro (original) (raw)
Abstract
Five non-allelic histone H3 variants, H3.1, H3.2, H3.3, H3t and CENP-A, have been identified in mammals. H3t is robustly expressed in the testis, and thus was assigned as the testis-specific H3 variant. However, recent proteomics and tissue-specific RT-PCR experiments revealed a small amount of H3t expression in somatic cells. In the present study, we purified human H3t as a recombinant protein, and showed that H3t/H4 forms nucleosomes with H2A/H2B by the salt-dialysis method, like the conventional H3.1/H4. We found that H3t/H4 is not efficiently incorporated into the nucleosome by human Nap1 (hNap1), due to its defective H3t/H4 deposition on DNA. In contrast, human Nap2 (hNap2), a paralog of hNap1, promotes nucleosome assembly with H3t/H4. Mutational analyses revealed that the Ala111 residue, which is conserved among H3.1, H3.2 and H3.3, but not in H3t, is the essential residue for the hNap1-mediated nucleosome assembly. These results suggest that H3t may be incorporated into chromatin by a specific chaperone-mediated pathway.
INTRODUCTION
In eukaryotes, the huge amount of genomic DNA is compacted into the nucleus by the chromatin structure (1,2). The nucleosome is the fundamental repeating unit of chromatin, and is composed of a histone octamer and a 146 base-pair DNA, which is wrapped 1.7 times around the histone octamer (3,4). The histone octamer contains two each of the core histones, H2A, H2B, H3 and H4. In the nucleosome, the core histones are composed of flexible N- and/or C-terminal tails and the globular domain, which contains the histone fold (3,4). The histone-fold domain is involved in the formation of dimers between histones H2A and H2B (H2A/H2B), and between histones H3 and H4 (H3/H4) (5).
Three non-allelic isoforms of histone H3, H3.1, H3.2 and H3.3, have been identified in higher eukaryotes (6,7), in addition to the centromere-specific H3 homolog, CENP-A (8,9). These four H3 variants increase the diversity of nucleosomes, and are considered to have distinct functions in the organization of chromatin structure (6,7,10–12). H3.1 and H3.2 are expressed at S phase, and are incorporated into chromatin in a replication-dependent fashion (7,10). On the other hand, H3.3 is constitutively expressed, and is incorporated into transcriptionally active chromatin regions in a replication-independent fashion (7,10). Histone H3t was found as the testis-specific H3 variant in mammals, and it may play an important role in the chromatin reorganization required for spermatogenesis (13–15).
Comprehensive proteome analyses revealed the presence of H3t in the nucleoli of HeLa cells (16,17). This finding suggested that H3t may also function in somatic cells, or that it may be expressed due to an extensive modification of the chromatin structure leading to malignant transformation. In addition, an RT-PCR experiment revealed H3t expression in the brain and in embryos (18). However, the nucleosomes containing H3t probably comprise only a small proportion of the bulk chromatin in somatic cells, if H3t is present. Therefore, the mechanisms controlling the assembly and function of H3t in chromatin have not been elucidated.
The incorporation of histone variants into chromatin is mediated by histone chaperones. In human cells, the H3.1 and H3.3 variants reportedly form large pre-assembled complexes including the histone chaperones CAF1 and HIRA, respectively (19). CAF1 and HIRA are known to be required for the DNA-synthesis-dependent and -independent nucleosome assembly pathways, respectively (20,21), suggesting their specific functions in H3.1 and H3.3 incorporation into nucleosomes. Consistently, in Drosophila, H3.3 is deposited throughout the chromosomes except at the centromeres, in a DNA-synthesis-independent manner, whereas the conventional H3 was excluded from the DNA-synthesis-independent nucleosome-assembly pathway (10). Therefore, the specific histone-chaperone activity may be required for nucleosome assembly with the specific H3 variant.
Nucleosome assembly protein 1, Nap1, is a prominent histone chaperone, which facilitates nucleosome assembly in vitro (22,23). Nap1 reportedly interacts with H2A/H2B in vitro and in vivo (23–25), suggesting that it functions as the histone chaperone for H2A/H2B. However, biochemical studies indicated that Nap1 prefers to bind H3/H4 in the presence of all four core histones (26–28). Consistently, Nap1 has the potential to alter the H3/H4 dimer–tetramer equilibrium within the tetramer nucleosomes (tetrasome) without H2A/H2B (29). These facts suggest that Nap1 also functions for H3/H4. On the other hand, Nap2 (hNap2), which shares about 60% amino acid sequence identity with hNap1 (30), has been identified in humans, and may also function as a histone chaperone (31).
In the present study, we purified human H3t as a recombinant protein, and established a method for the preparation of the H3t/H4 complex. We also purified hNap1 and hNap2, and tested their nucleosome-formation activities with H3t/H4 in vitro.
MATERIALS AND METHODS
Overexpression of recombinant human histones
Human H2A, H2B, H3.1 and H4 were overexpressed in Escherichia coli cells as N-terminal His6-tagged proteins, as described previously (32). DNA fragments containing the coding sequences of the human H3t and H3 mutants were constructed from the H3.1 expression vector by mutagenesis using a Quik-Change kit (Stratagene). The His6 tag sequence is as follows: Met-Gly-Ser-Ser-His-His-His-His-His-His-Ser-Ser-Gly-Leu-Val-Pro-Arg-Gly-Ser-His, and it also contains the recognition sequence (Leu-Val-Pro-Arg-Gly-Ser) for thrombin protease. Escherichia coli cells freshly transformed with the expression vectors [strain BL21(DE3) for H2A, H2B, H3.1, H3t and H3 mutants, or JM109(DE3) for H4] were grown on LB plates containing ampicillin (100 μg/ml) at 37°C. After a 16 h incubation, 5–20 colonies grown on the LB plates were collected and inoculated into LB medium (2.5 l) containing ampicillin (50 μg/ml). Cells were grown at 37°C for 16–20 h in the absence of isopropyl-β-d-thiogalactopyranoside.
Preparation of the H2A/H2B and H3/H4 complexes
The E. coli cells producing recombinant histones were collected and disrupted by two rounds of sonication for 200 s each in 50 ml of Tris(pH8.0) buffer [50 mM Tris–HCl (pH 8.0), 500 mM NaCl, 1 mM PMSF and 5% glycerol]. After centrifugation (27 216 g; 20 min; 4°C), the pellet containing His6-tagged histones as insoluble forms was re-suspended in Tris(pH8.0)–urea buffer [Tris(pH8.0) buffer containing 6 M urea] and was cleared by centrifugation (27 216 g; 20 min; 4°C). The supernatants containing the His6-tagged histones were combined with 4 ml (50% slurry) of nickel–nitrilotriacetic acid (Ni–NTA) agarose resin (Qiagen), and were mixed by rotation for 1 h at 4°C. The agarose beads were then washed with 20 ml Tris(pH8.0)–urea buffer, packed into an Econo-column (Bio-Rad), and further washed with 100 ml Tris(pH8.0)–urea buffer containing 5 mM imidazole, at a flow rate of 0.8 ml/min. The His6-tagged histones were eluted by a 100 ml linear gradient of imidazole from 5 to 300 mM in Tris(pH8.0)–urea buffer. His6-tagged H3.1, H3t and the H3 mutants were each mixed with His6-tagged H4 in a 1:1 molar ratio in Tris(pH8.0)–urea buffer, and the mixtures were dialyzed for 16 h against 1 l of 50 mM Tris–HCl buffer (pH 8.0) containing 10 mM DTT, 2 mM EDTA and 7 M guanidine-HCl. His6-tagged H2A and H2B were mixed in a 1:1 molar ratio in Tris(pH8.0)–urea buffer. These H2A/H2B and H3/H4 samples were dialyzed overnight against 1 l of reconstitution buffer [20 mM Tris–HCl (pH 8.0), 5 mM DTT, 1 mM EDTA, 1 mM PMSF and 5% glycerol] containing 2 M NaCl. The NaCl concentration was then reduced by stepwise dialysis against the reconstitution buffer at 4°C; 1 M NaCl for 4 h, 0.5 M NaCl for 4 h and 0.1 M NaCl overnight. The N-terminal His6 tags were removed from the H2A/H2B and H3/H4 complexes by thrombin protease treatment (1 unit/mg of histones, GE Healthcare Biosciences) at room temperature for 3 h. The removal of the His6 tags was confirmed by SDS-16% polyacrylamide gel electrophoresis (PAGE); the recombinant histones without the His6 tag migrated faster than the His6-tagged histones. The reconstituted H3.1/H4, H3t/H4 and H2A/H2B complexes were fractionated using Superdex 200 resin (GE Healthcare Biosciences) packed in an Econo-column (1.6 × 13.5 cm; 0.5 ml/min flow rate; 0.5 ml fractions) with reconstitution buffer containing 0.1 M NaCl. H3.1/H4 and H3t/H4 were eluted from the column in two peaks. The first peak contained aggregates of histones, and the H3.1/H4 and H3t/H4 in the second peak were used in this study.
Preparation of a DNA fragment for nucleosome reconstitution
A 195 base-pair (bp) DNA fragment containing the Lytechinus variegates 5S ribosomal RNA gene was amplified by polymerase chain reaction (PCR) using the following primers:
5S rDNA-FW 5′-CAACGAATAACTTCCAGGGATTTATAAGCCG-3′ and 5S rDNA-REV 5′-AATTCGGTATTCCCAGGCGGTCTCC-3′.
After the PCR reaction, the DNA fragment was extracted by phenol/chloroform, precipitated by ethanol and further purified by Superdex 75 gel filtration chromatography to remove the primers and dNTPs.
Nucleosome reconstitution by the salt-dialysis method
Purified H2A/H2B (7.5 μg) was mixed with H3t/H4 (7.5 μg) at 4°C in the presence of the 195 bp 5S rDNA fragment (10 μg) in a solution containing 2 M NaCl (100 μl). The mixture was first dialyzed against 300 ml dialysis buffer (20 mM Tris–HCl buffer [pH 8.0], 2 mM EDTA and 20 mM 2-mercaptoethanol) containing 2 M NaCl for 4 h at 4°C. The concentration of NaCl was gradually reduced from 2 M to 0.6 M by adding dialysis buffer containing 0.1 M NaCl, using a peristaltic pump (0.8 ml/min flow rate). The sample was further dialyzed against dialysis buffer containing 0.1 M NaCl for 16 h at 4°C. The nucleosomes reconstituted with the recombinant histones were analyzed by 6% PAGE in 0.2 × TBE buffer (18 mM Tris base, 18 mM boric acid and 0.4 mM EDTA) at 6.25 V/cm for 3 h, followed by ethidium bromide staining.
Purification of recombinant hNap1 and hNap2
The DNA fragments encoding hNap1 and hNap2 were ligated into the _Nde_I and _Bam_HI sites of the pET15b vector (Novagen), which harbors the His6 tag and the PreScission protease recognition sequence (GE Healthcare Biosciences) at the N-terminus. Freshly transformed E. coli cells, Rosetta-gami B(DE3) (Novagen), were grown on LB plates containing ampicillin (100 μg/ml) at 37°C. After a 16 h incubation, 5–20 colonies on the LB plates were collected and inoculated into LB medium (5 l) containing ampicillin (100 μg/ml), and the cultures were incubated at 37°C. When the cell density reached an OD600 = 0.4, 1 mM isopropyl-β-d-thiogalactopyranoside was added to induce the expression of hNap1 or hNap2, and the cultures were further incubated at 18°C for 12 h. The cells producing hNap1 or hNap2 were harvested, re-suspended in 50 ml Tris (pH7.5) buffer [50 mM Tris–HCl (pH 7.5) and 1 mM PMSF] containing 10% glycerol and 0.5 M NaCl, and disrupted by sonication. The cell debris was removed by centrifugation (27 216 g; 20 min), and the lysate was mixed gently with 4 ml (50% slurry) of Ni-NTA beads at 4°C for 1 h. The hNap1 or hNap2-bound Ni-NTA beads were then packed into an Econo-column and washed with 100 ml Tris(pH7.5) buffer containing 150 mM NaCl and 20 mM imidazole, at a flow rate of about 0.8 ml/min. The His6-tagged hNap1 or hNap2 was eluted by a 100 ml linear gradient of imidazole from 20 to 300 mM in Tris(pH7.5) buffer containing 150 mM NaCl. The His6 tag was removed from the hNap1 or hNap2 portion by PreScission protease (3 units/mg of protein).
To purify hNap1 using Heparin Sepharose (GE Healthcare Biosciences), the NaCl concentration of the protein preparation was decreased by dialysis against Tris(pH7.5) buffer containing 2 mM 2-mercaptoethanol at 4°C, before mixing with 2.5 ml bead suspension (containing 0.7 g of resin). After mixing by rotation at 4°C for 1 h, the beads were packed into an Econo-column and washed with 100 ml Tris(pH7.5) buffer containing 2 mM 2-mercaptoethanol. hNap1 was eluted by a 100 ml linear gradient of KCl from 0 to 200 mM in the same buffer. hNap1 was then purified using a Mono Q (GE Healthcare Biosciences) column, by elution with a 30 ml linear gradient of KCl from 150 to 600 mM in the same buffer, and finally by using a Superdex 200 column eluted with 1.2 column volumes of the same buffer containing 150 mM KCl. hNap2 was purified essentially using the same procedure as for hNap1, but without the Heparin Sepharose step. The purified hNap1 and hNap2 were concentrated using a Mono Q column, and the proteins were dialyzed against 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 0.1 mM PMSF and 10% glycerol. The concentrations of purified hNap1 and hNap2 were determined by the Bradford method (33) with BSA as the standard protein.
Nucleosome reconstitution by hNap1 or hNap2, and micrococcal nuclease (MNase) assay
H2A/H2B (8 ng/μl) and either H3.1/H4 or H3t/H4 (8 ng/μl) were pre-incubated with hNap1 (45 or 182 ng/μl) or hNap2 (43 or 171 ng/μl) at 23°C for 10 min. The nucleosome assembly reaction was initiated by the addition of 5S rDNA (8 ng/μl) into the 10 μl mixture in 20 mM Tris–HCl (pH 8.0), 80 mM NaCl and 1 mM DTT, and was continued at 23°C for 60 min, After the reaction, the samples were incubated at 42°C for 60 min, to eliminate non-specific DNA binding by the histones, and then were analyzed by 6% PAGE. In some cases, the samples were treated with 1.2 units of MNase (Takara) in 15 μl of 20 mM Tris–HCl (pH 8.0), 70 mM NaCl, 2.5 mM CaCl2, 1 mM DTT and 50 μg/ml BSA. After a 5 min incubation at 23°C, the reaction was stopped by the addition of 60 μl of proteinase K solution [20 mM Tris–HCl (pH 8.0), 20 mM EDTA, 0.5% SDS and 0.5 mg/ml proteinase K (Roche)]. After a 15 min incubation at 23°C, the DNA was extracted with phenol/chloroform, precipitated with ethanol, and then analyzed by 10% PAGE in 0.2 × TBE buffer (8.33 V/cm for 3 h).
Ni-NTA bead pull-down assay
Purified His6-tagged hNap1 or His6-tagged hNap2 (9 μg) was mixed with H3.1/H4 or H3t/H4 (2. 5 μg) in 500 μl of 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 30 mM imidazole and 10 μg/ml BSA, and Ni-NTA-agarose beads (3 μl, 50% slurry) were added. After an incubation for 1 h at 4°C, the beads were pelleted and washed three times with 500 μl of 20 mM Tris–HCl (pH 8.0), 200 mM NaCl, 50 mM imidazole and 0.2% Tween 20. The proteins pelleted with the beads were analyzed by SDS-15% PAGE and CBB staining. The reciprocal experiments were performed using His6-tagged H3.1/H4 or His6-tagged H3t/H4 (6 μg) and hNap1 or hNap2 (4.6 μg).
Gel electrophoretic mobility shift assay
hNap1 (2.3 μg) was mixed with H3.1/H4 or H3t/H4 (0.15–1.5 μg) in 9 μl of 20 mM Tris–HCl (pH 8.0), 100 mM NaCl and 1 mM DTT, incubated for 1 h at 23°C, and analyzed by 5% PAGE in 0.5 × TBE buffer (45 mM Tris base, 45 mM boric acid and 1 mM EDTA; 10.4 V/cm for 1.5 h) followed by CBB staining.
DNA competition assay
hNap1 (2.3 μg) was mixed with H3.1/H4 (0.75 μg) or H3t/H4 (1.2 μg), incubated for 10 min at 23°C, and combined with supercoiled plasmid DNA (pGSAT4; 40, 80, 120 or 160 ng). The samples were further incubated for 1 h and analyzed by 5% PAGE in 0.5 × TBE buffer (10.4 V/cm for 1.5 h) followed by CBB staining.
RESULTS
Nucleosome assembly with H3.1/H4 and H3t/H4 by hNap1 and hNap2
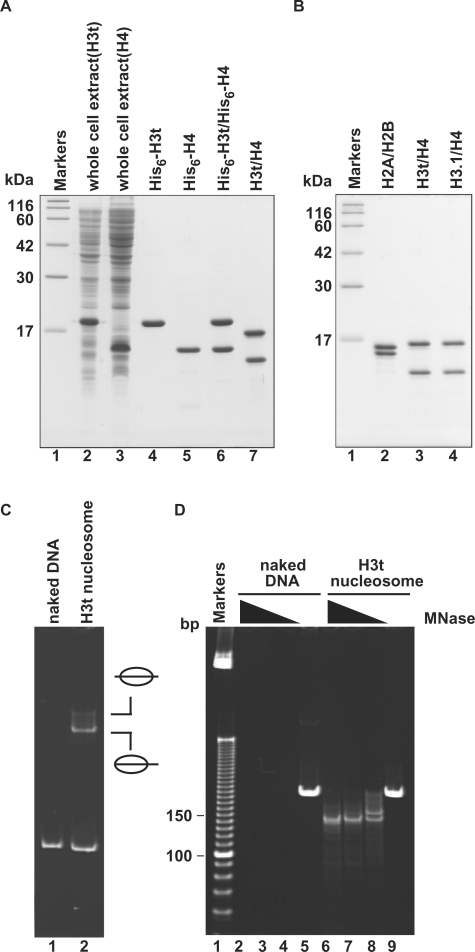
We prepared recombinant human H3t and H4 from E. coli cells under denaturing conditions, and the H3t/H4 complex was reconstituted by stepwise salt dialysis against the buffer without urea (Figure 1A). After this reconstitution step, the His6 tags were removed from H3t/H4 using thrombin protease, and the H3t/H4 sample was subjected to Superdex 200 gel filtration chromatography, to remove the aggregates (Figure 1B, lane 3). The H2A/H2B and H3.1/H4 samples were also prepared by the same or similar procedures (Figure 1B, lanes 2 and 4). As shown in Figure 1C, H3t/H4 formed nucleosomes with H2A/H2B by the salt-dialysis method, as revealed by a gel electrophoretic mobility shift assay (EMSA); the DNA fragment exhibited two retarded bands, corresponding to the middle and end translational positions of nucleosomes (34,35). To confirm the integrity of the nucleosomes formed by these recombinant histones, we performed an MNase-treatment assay. In this assay, a 146 bp DNA fragment, which is tightly wrapped around the histone octamer, was protected from the MNase digestion when the nucleosomes were properly formed. As shown in Figure 1D, the 146 bp DNA fragments protected from the MNase digestion were clearly observed with the H3t-containing nucleosomes, indicating proper nucleosome formation by H3t/H4 with H2A/H2B.
Figure 1.
Preparation and nucleosome formation ability of H3t/H4. (A) SDS–PAGE analysis of proteins from each preparation step for H3t/H4. Lanes 2 and 3: the whole cell lysates of the E. coli cells expressing His6-tagged H3t and His6-tagged H4, respectively. Lanes 4 and 5: the peak Ni-NTA agarose fractions of His6-tagged H3t and His6-tagged H4, respectively. Lanes 6 and 7: His6-tagged H3t/H4 before and after the removal of the hexahistidine tag, respectively. The positions of the size markers (lane1) are indicated on the left. (B) SDS–PAGE analysis of purified histones used in this study. Lane 1: size markers. Lanes 2–4: histones; H2A/H2B (lane 2), H3t/H4 (lane 3) and H3.1/H4 (lane 4). (C) Nucleosome reconstitution analyzed by non-denaturing 6% PAGE. Lane 1: naked DNA. Lane 2: nucleosomes reconstituted with H2A/H2B and H3t/H4 by the salt-dialysis method. Nucleosomes with different translational positions are indicated. (D) MNase assay analyzed by non-denaturing 10% PAGE. Naked DNA (lanes 2–5) and H3t-containing nucleosomes reconstituted by the salt-dialysis method (lanes 6–9) were treated with MNase, and the resulting DNA fragments were analyzed. Lane 1: the molecular mass markers (10 bp DNA ladder). Amounts of MNase (unit/μl) were 0.18 (lanes 2 and 6), 0.13 (lanes 3 and 7), 0.08 (lanes 4 and 8) and 0 (lanes 5 and 9). In H3t-containing nucleosomes, 146 bp DNA, which is tightly wrapped around the nucleosomes and resistant to MNase, is detected (lanes 6–8).
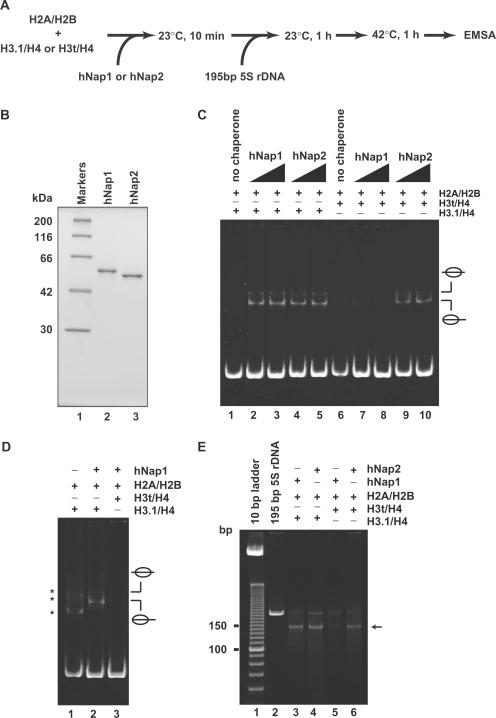
We then tested the formation of H3t-containing nucleosomes (Figure 2A) using the histone chaperones hNap1 and hNap2, which were expressed in and purified from E. coli (Figure 2B). In this assay, hNap1 or hNap2 was pre-incubated with the H2A/H2B and H3.1/H4 or H3t/H4 complexes, and then 195 bp 5S rDNA was added into the reaction mixture. As shown in Figure 2C (lanes 1–5), both hNap1 and hNap2 promoted the formation of nucleosomes containing the conventional H2A, H2B, H3.1 and H4, indicating that the hNap1 and hNap2 prepared in this study had nucleosome assembly activity. When the histones were incubated with the DNA at 23°C for 1 h in the absence of hNap1 or hNap2, non-specific DNA binding by histones was observed (Figure 2D, lane 1). Therefore, the 42°C incubation step, which stripped the non-specifically bound histones from the naked DNA, was included in this assay (Figure 2A). In striking contrast, we found that hNap1 exhibited quite low nucleosome formation activity when H3t was used instead of H3.1 (Figure 2C, lanes 7 and 8, and Figure 2D, lane 3), whereas hNap2 efficiently promoted the formation of the H3t-containing nucleosomes (Figure 2C, lanes 9 and 10).
Figure 2.
Nucleosome assembly with H2A/H2B and H3t/H4 or H3.1/H4 by hNap1 and hNap2. (A) Schematic representation of the nucleosome-reconstitution assay by hNap1 or hNap2. (B) SDS–PAGE analysis of purified hNap1 (lane 2) and hNap2 (lane 3) lacking the His6 tag. (C) Nucleosome-reconstitution experiments with the 42°C incubation, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with histones without (lanes 1 and 6) or in combination with hNap1 (lanes 2–3 and 7–8), or hNap2 (lanes 4–5 and 9–10). To assemble nucleosomes containing the histone octamer, H2A/H2B was used with H3.1/H4 (lanes 1–5) or H3t/H4 (lanes 6–10). (D) Nucleosome-reconstitution experiments without the 42°C incubation, analyzed by non-denaturing 6% PAGE. Reactions were performed as in (C). Lane 1 indicates the experiment with H3.1/H4/H2A/H2B in the absence of hNap1. Lanes 2 and 3 indicate the experiments with H3.1/H4/H2A/H2B and H3t/H4/H2A/H2B, respectively, in the presence of hNap1. Bands corresponding to non-specific DNA binding of histones are indicated by asterisks. (E) MNase assay analyzed by non-denaturing 10% PAGE. The samples incubated with the indicated combinations were treated with MNase, and the resulting DNA fragments were analyzed. A total of 146 bp DNA fragments in nucleosomes (arrow) are found in the combinations of H3.1/H4 with hNap1 (lane 3) or hNap2 (lane 4), and H3t/H4 with hNap2 (lane 6), but not H3t/H4 with hNap1 (lane 5).
To characterize the nucleosomes formed by hNap1 and hNap2, we performed the MNase-treatment assay. Consistent with the EMSA analyses, the 146 bp DNA fragments were protected from the MNase digestion when H3t was used with hNap2, indicating the formation of H3t-containing nucleosomes (Figure 2E, lane 6). In contrast, the 146 bp DNA fragments were scarcely detected, when the reaction was performed using H3t and hNap1 (Figure 2E, lane 5). These results indicate that H3t/H4 is not a proper substrate for hNap1-mediated nucleosome formation. Therefore, hNap1 may discriminate H3t from the conventional H3.1 during the nucleosome assembly process, whereas hNap2 possesses the ability to form the H3t-containing nucleosome.
H3.1/H4 and H3t/H4 binding by hNap1 and hNap2
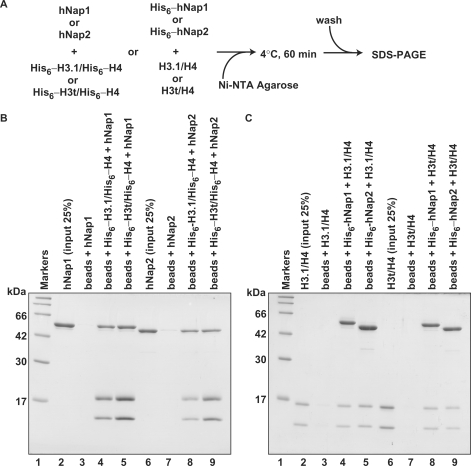
We next performed the pull-down assay with the Ni-NTA beads, to test whether hNap1 and hNap2 physically interact with H3.1/H4 and H3t/H4 (Figure 3A). For this assay, we prepared His6-tagged versions of the histones and chaperones to mix with the untagged proteins. As shown in Figure 3B, hNap1 and hNap2 without the His6 tag were co-pelleted by either His6-tagged H3.1/H4 or His6-tagged H3t/H4, indicating that hNap1 and hNap2 both physically interacted with H3.1/H4 and H3t/H4. Consistently, the reciprocal pull-down assay with His6-tagged hNap1 and His6-tagged hNap2 also showed that both proteins bind to H3.1/H4 and H3t/H4 (Figure 3C). Therefore, hNap1 binds to H3t/H4 as well as H3.1/H4, although it is inefficient for the H3t-containing nucleosome formation.
Figure 3.
The Ni-NTA-agarose pull-down assay. (A) A schematic diagram of the assay. (B) Pull-down assay using His6-tagged histones. hNap1 (lanes 3–5) or hNap2 (lanes 7–9) was mixed with His6-tagged H3.1/H4 (lanes 4 and 8) or His6-tagged H3t/H4 (lanes 5 and 9), and the proteins bound to the Ni-NTA agarose beads were analyzed by 15% SDS–PAGE with CBB staining. Lanes 2 and 6: the inputs (25%) of hNap1 and hNap2, respectively. (C) Pull-down assay using His6-tagged hNap1 and hNap2. H3.1/H4 (lanes 3–5) or H3t/H4 (lanes 7–9) was mixed with His6-tagged hNap1 (lanes 4 and 8) or His6-tagged hNap2 (lanes 5 and 9), and the proteins bound to the Ni-NTA agarose beads were analyzed by 15% SDS–PAGE with CBB staining. Lanes 2 and 6: the inputs (25%) of H3.1/H4 and H3t/H4, respectively.
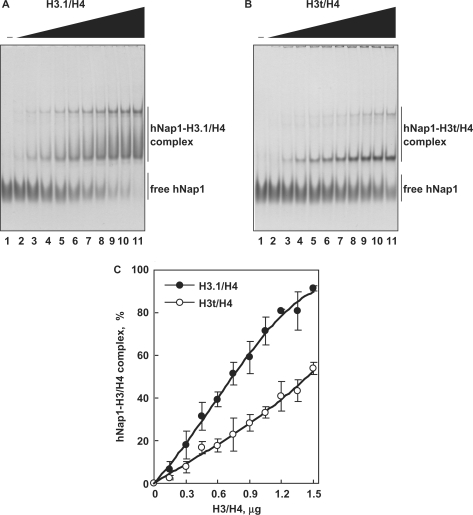
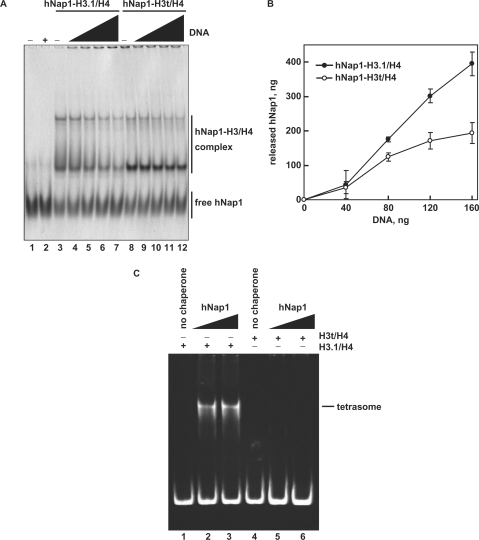
To compare the apparent affinities of hNap1 to H3.1/H4 and H3t/H4, we performed the EMSA analyses. In this assay, H3.1/H4 and H3t/H4 did not enter the gels, since they are highly positively charged (26); therefore, free hNap1 and the hNap1-H3.1/H4 and hNap1-H3t/H4 complexes were detected. As shown in Figure 4A and B, hNap1 bound to both H3.1/H4 and H3t/H4, and formed two complexes, like the yeast Nap1-H3/H4 complexes, as previously reported (26). These complexes may have different hNap1:histone stoichiometries or different multimerization states. A graphic representation indicated that hNap1 bound to H3.1/H4 with higher affinity than to H3t/H4 (Figure 4C).
Figure 4.
The gel mobility shift assay. (A) hNap1-H3.1/H4 complex formation. hNap1 (2.3 μg) was incubated with different amounts of H3.1/H4, and the hNAP1-H3.1/H4 complex was detected by non-denaturing 5% PAGE with CBB staining. The amounts of H3.1/H4 (μg) used in this study were: 0 (lane 1), 0.15 (lane 2), 0.3 (lane 3), 0.45 (lane 4), 0.6 (lane 5), 0.75 (lane 6), 0.9 (lane 7), 1.05 (lane 8), 1.2 (lane 9), 1.35 (lane 10) and 1.5 (lane 11). (B) hNap1-H3t/H4 complex formation. hNap1 (2.3 μg) was incubated with different amounts of H3t/H4, and the hNap1-H3t/H4 complex formation was analyzed as in (A). (C) Graphic representation of the hNap1-H3/H4complex formation. The relative amounts of hNap in the complex with H3.1/H4 (closed circles) and H3t/H4 (open circles) were plotted as the averages of three independent experiments, as in (A) and (B), with the SD values.
The hNap1-H3t/H4 complex is not efficiently disrupted by DNA
When Nap1 deposits histones on DNA during nucleosome assembly, Nap1 is released from the histone complex (36). To study the histone-deposition activity of hNap1 on DNA, we next tested whether hNap1 was released from the hNap1-H3.1/H4 or hNap1-H3t/H4 complex in the presence of DNA. As shown in Figure 5A (lanes 3–7) and B, hNap1 was efficiently released from the hNap1-H3.1/H4 complex in the presence of DNA. However, the disruption of the hNap1-H3t/H4 complex by DNA was minimal (Figure 5A, lanes 8–12, and B), as compared to that of the hNap1-H3.1/H4 complex. These results suggested that hNap1 deposits H3.1/H4 much more efficiently than H3t/H4 on DNA. To test this possibility more directly, we performed the tetrasome-formation assay with H3.1/H4 or H3t/H4 and hNap1 in the absence of H2A/H2B. Consistently, hNap1 efficiently formed tetrasomes containing H3.1/H4, but exhibited quite low activity for tetrasome formation with H3t/H4 (Figure 5C).
Figure 5.
The H3/H4 deposition assay. (A) The deposition of H3.1/H4 or H3t/H4 from the hNap1-H3.1/H4 or hNap1-H3t/H4 complex onto DNA, analyzed by non-denaturing 5% PAGE with CBB staining. hNap1 (2.3 μg) was incubated without (lanes 1–2) or with H3.1/H4 (0.75 μg; lanes 3–7) or H3t/H4 (1.2 μg; lanes 8–12) to form the complex; about half of the hNap1 remained free under these conditions (lanes 3 and 8). After the incubation with supercoiled DNA, the samples were analyzed. The amounts of competitor DNA (ng) were 0 (lanes 3 and 8), 40 (lanes 4 and 9), 80 (lanes 5 and 10), 120 (lanes 6 and 11) and 160 (lanes 2, 7 and 12). (B) Graphic representation of hNap1 release. The amounts of hNap1 released from hNap1-H3.1/H4 (open circles) or hNap1-H3t/H4 complex (closed circles) are plotted as the averages of three independent experiments, performed as in (A), with the SD values. (C) Tetrasome-reconstitution with H3.1/H4 or H3t/H4 by hNap1. A total of 195 bp 5S DNA was incubated with hNap1 in combination with H3.1/H4 (8 ng/μl; lanes 1–3) or H3t/H4 (8 ng/μl; 4–6) (in the absence of H2A/H2B). The tetrasome formation was analyzed by non-denaturing 6% PAGE. The amounts of hNap1 (ng/μl) were 0 (lane 1), 45 (lane 2) and 182 (lane 3). The amounts of hNap2 (ng/μl) were 0 (lane 4), 43 (lane 5) and 171 (lane 6).
Mutational analyses of histones H3.1 and H3t
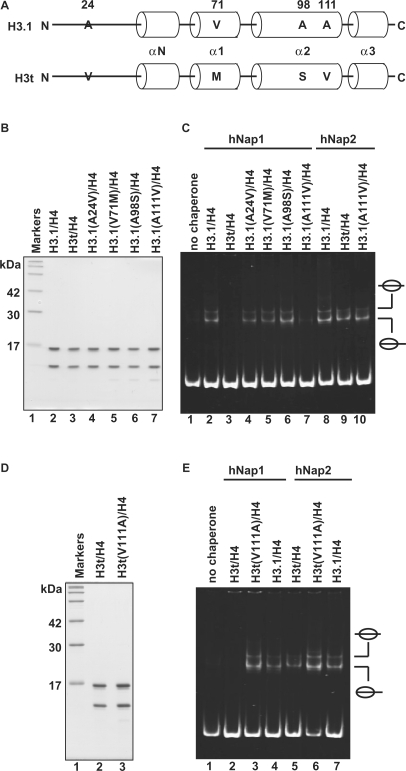
There are four amino acid differences between H3.1 and H3t (Figure 6A). The Ala24, Val71, Ala98 and Ala111 residues of H3.1 are naturally replaced by Val24, Met71, Ser98 and Val111 in H3t, respectively. To identify the amino acid residue(s) responsible for the H3t discrimination by hNap1, we constructed H3.1 mutants, H3.1(A24V), H3.1(V71M), H3.1(A98S) and H3.1(A111V), in which each H3.1 residue, Ala24, Val71, Ala98 or Ala111, was replaced by the corresponding amino acid residue of H3t. These H3.1 mutants were prepared as the complex forms with H4 (Figure 6B).
Figure 6.
Mutational analyses of H3.1 and H3t. (A) Sequence comparison between human H3.1 and H3t. The different amino acids between H3.1 and H3t are indicated by capital letters. Cylinders indicate α-helices found in the crystal structure of the human nucleosome core particle (39). (B) SDS–PAGE analysis of the H3.1 mutants complexed with H4. Lane 1: molecular mass markers. Lanes 2 and 3: purified H3.1/H4 and H3t/H4, respectively. Lanes 4–7: H3.1 mutants complexed with H4; lane 4: H3.1(A24V), lane 5: H3.1(V71M), lane 6: H3.1(A98S) and lane 7: H3.1(A111V). (C) Nucleosome-reconstitution with the H3.1 mutants by hNAP1 or hNAP2, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with hNap1 (lanes 2–7) or hNap2 (lanes 8–10) in combination with H3.1/H4 (lanes 2 and 8), H3t/H4 (lanes 3 and 9) and H3.1 mutants/H4 (lanes 4–7 and 10); all reactions were performed in the presence of H2A/H2B. (D) SDS–PAGE analysis of the H3t(V111A) mutant complexed with H4. (E) Nucleosome-reconstitution with H3t(V111A)/H4 by hNap1 or hNap2, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with hNap1 (lanes 2–4) or hNap2 (lanes 5–7) in combination with H3t/H4 (lanes 2 and 5), H3t(V111A)/H4 (lanes 3 and 6) and H3.1/H4 (lanes 4 and 7); all reactions were performed in the presence of H2A/H2B.
We then tested these H3.1 mutants for the hNap1 and hNap2-mediated nucleosome formation. Interestingly, when the H3.1(A111V) mutant was used for the experiment, the hNap1-mediated nucleosome formation was significantly decreased, whereas hNap1 efficiently promoted nucleosome formation with the H3.1(A24V), H3.1(V71M) and H3.1(A98S) mutants (Figure 6C). However, hNap2 still promoted nucleosome formation with the H3.1(A111V) mutant (Figure 6C, lane 10). Therefore, the Ala111 residue of H3.1 is important for the hNap1-mediated nucleosome formation.
We next prepared the complementary H3t(V111A) mutant, in which Val111 of H3t was replaced by Ala, and used it for nucleosome formation as the complex form with histone H4 (Figure 6D). As anticipated, in contrast to H3.1(A111V), H3t(V111A) was efficiently incorporated into nucleosomes by hNap1 (Figure 6E, lane 3). These results indicate that hNap1 does not efficiently promote nucleosome formation with the H3t variant containing the Val residue at position 111. Accordingly, the Val111 residue is responsible for the hNap1-mediated histone H3 discrimination.
DISCUSSION
The testis-specific H3 variant, H3t, has been identified in humans (15), but its nucleosome formation ability has not been reported. In the present study, we established a method for the reconstitution of H3t/H4, and showed that H3t/H4 is proficient in nucleosome formation with H2A/H2B by the salt-dialysis method. However, we found that H3t/H4 is not efficiently incorporated into nucleosomes by hNap1, which is a prominent nucleosome assembly protein in eukaryotes. H3t is the only H3 variant defective in the hNap1-mediated nucleosome assembly in vitro, because hNap1 promoted nucleosome assembly with other H3 variants, H3.2, H3.3 and CENP-A, in addition to H3.1, under our experimental conditions (data not shown). To form nucleosomes, Nap1 must deposit H3/H4 on DNA prior to the H2A/H2B deposition (36). Our results showed that hNap1 deposits H3t/H4 less efficiently than H3.1/H4 on DNA. Therefore, the defects in the H3t-containing nucleosome formation by hNap1 may be due to this inefficient H3t/H4 deposition. On the other hand, we found that hNap1 bound to H3.1/H4 with higher affinity than to H3t/H4, although the hNap1-H3.1/H4 complex is less stable than the hNap1-H3t/H4 complex in the presence of DNA. One possible explanation for this apparent discrepancy is that the hNap1-H3.1/H4 complex may have a more suitable conformation for the H3/H4 deposition than the hNap1-H3t/H4 complex. Further studies are required to clarify this issue.
Intriguingly, hNap2, which shares ∼60% amino acid sequence identity with hNap1, catalyzes the formation of H3t-containing nucleosomes in vitro. This finding suggests that the formation of the H3t-containing nucleosome may require a specific histone chaperone. hNap2 expression is reportedly three-fold higher in the testis, as compared to that in other somatic tissues (30), implying that it may have a specific function in chromatin reorganization during meiosis and/or the post-meiotic maturation of male germ cells. In germ cells, drastic chromatin reorganization, including the process of histone replacement by histone variants (37), occurs in association with meiotic events, such as homologous recombination, synaptonemal complex formation and chromosome segregation. After meiosis, the histones are finally replaced by protamines during sperminogenesis. To promote this post-meiotic maturation of male haploid germ cells, chromatin reorganization by the testis-specific histone variants may also be an essential process (38). It is intriguing to study how histone chaperones contribute to such global chromatin remodeling with histone variants during and after meiosis.
Our mutational analyses with recombinant H3 also revealed that the Ala111 residue of the conventional H3.1 plays an essential role in the hNap1-mediated nucleosome formation. H3t, which does not participate in hNap1-mediated nucleosome assembly, has a Val residue at position 111. Although the molecular mechanism by which the H3t-Val111 residue inhibits the hNap1-mediated nucleosome formation has not been elucidated, our results indicate the functional importance of residue 111 in the chaperone-mediated nucleosome assembly. In the crystal structure of the human nucleosome core particle (39), the β-CH3 group of the Ala111 residue of H3.1 is located close to the side chain of the Arg116 residue, which forms hydrogen bonds with the Asp123 residue within the H3.1 monomer. The Val residue includes two γ-CH3 groups, which do not exist in the Ala residue, and these additional masses at position 111 in H3t may cause a steric clash with Arg116. A mutation of the Arg116 residue, known as the Sin2 mutation, alleviates the transcriptional defects due to the inactivation of the SWI/SNF chromatin remodeling complex in yeast (40). An in vitro study revealed that the Arg116 mutation of H3 decreased the stability of the nucleosome core particle, and increased the nucleosome-sliding rate (41,42). These facts indicate that the Arg116 residue, which may sterically clash with the Val111 residue in the H3t-containing nucleosome, is important for the nucleosome structure. Therefore, the naturally occurring A111V substitution in H3t probably affects its tertiary structure, resulting in a hNap1-H3t/H4 complex that is less competent for nucleosome assembly. Structural studies of the H3t-containing nucleosome may be required to understand the molecular basis of H3t discrimination by hNap1.
H3t expression was also found in somatic cells. A proteome analysis revealed that H3t exists in the nucleoli of HeLa cells (16). The H3t mRNA was also detected in cells from brain and embryos (18). However, nucleosomes containing H3t may comprise only a small proportion of the bulk chromatin in somatic cells, because H3t expression in somatic cells was drastically lower than that in testis (18). This makes the study of endogenous H3t in somatic cells difficult. Hence, our in vitro analyses with the purified histones and histone chaperones provide useful new information related to the chaperone-mediated nucleosome assembly of H3 variants.
In the present study, we showed that hNap1 discriminates H3t in nucleosome formation. hNap1 reportedly mediated the assembly and disassembly of H2A.Bbd, a histone H2A variant, more efficiently with nucleosomes containing H3.3 than with those containing the conventional H3.1 in vitro (43). These facts suggest that hNap1 functions as a chromatin remodeling factor at distinct chromosome regions, where specific histone variants are located, and may regulate gene expression by its chromatin remodeling activity. Interestingly, the Nap1 deletion in yeast leads to up- and down-regulation of about 10% of all open reading frames (44). These transcriptional regulation changes in the Nap1-deletion strain may be associated with an altered chromatin structure, in which promoter activation and transcription elongation through the nucleosomal DNA may be perturbed in the absence of Nap1. Consistently, Nap1 stimulates transcription factor binding to DNA in vitro (45), and co-localizes in promoter regions with the CHD (chromo-helicase/ATPase DNA binding) remodeling factors (46), supporting the idea that Nap1 is involved in transcription processes. In addition, Nap1 reportedly associates with transcriptional regulators, such as p300/CBP and Alien (a potential co-repressor of the vitamin D receptor) (28,47,48). Therefore, Nap1 may function as a transcriptional regulator by remodeling chromatin in chromosome regions containing specific histone variants.
ACKNOWLEDGEMENTS
We thank K. Kawaguchi (Waseda University) for the preparation of recombinant histones. Funding for this work was provided by Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS), and the Ministry of Education, Culture, Sports, Science and Technology, Japan. H.T. was also supported by JSPS Research Fellowships for Young Scientists (JSPS Research Fellow). Funding to pay the Open Access publication charges for this article was provided by Waseda University.
Conflict of interest statement. None declared.
REFERENCES
- 1.van Holde KE. Chromatin. New York: Springer; 1989. [Google Scholar]
- 2.Wolffe AP. Chromatin: Structure & Function. London: Academic Press; 1998. [Google Scholar]
- 3.Richmond TJ, Finch JT, Rushron B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 Å resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl Acad. Sci. USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nature Struct. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 7.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the ‘H3 barcode hypothesis’. Proc. Natl Acad. Sci. USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc. Natl Acad. Sci. USA. 2002;99:16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. In and out: histone variant exchange in chromatin. Trends Biochem. Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 13.Franklin SG, Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 14.Albig W, Ebentheuer J, Klobeck G, Kunz J, Doenecke D. A solitary human H3 histone gene on chromosome 1. Hum. Genet. 1996;97:486–491. doi: 10.1007/BF02267072. [DOI] [PubMed] [Google Scholar]
- 15.Witt O, Albig W, Doenecke D. Testis-specific expression of a novel human H3 histone gene. Exp. Cell Res. 1996;229:301–306. doi: 10.1006/excr.1996.0375. [DOI] [PubMed] [Google Scholar]
- 16.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 17.Govin J, Caron C, Rousseaux S, Khochbin S. Testis-specific histone H3 expression in somatic cells. Trends Biochem. Sci. 2005;30:357–359. doi: 10.1016/j.tibs.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 2007;176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 20.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 21.Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 22.Ishimi Y, Hirosumi J, Sato W, Sugasawa K, Yokota S, Hanaoka F, Yamada M. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishimi Y, Kojima M, Yamada M, Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur. J. Biochem. 1987;162:19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- 26.McBryant SJ, Park TJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 27.Mazurkiewicz J, Kepert JF, Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J. Biol. Chem. 2006;281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 28.Eckey M, Hong W, Papaioannou M, Baniahmad A. The nucleosome assembly activity of NAP1 is enhanced by Alien. Mol. Cell Biol. 2007;27:3557–3568. doi: 10.1128/MCB.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson S, Danowit R, Wunsch A, Jackson V. NAP1 catalyzes the formation of either positive or negative supercoils on DNA on basis of the dimer-tetramer equilibrium of histones H3/H4. Biochemistry. 2007;46:8634–8646. doi: 10.1021/bi6025215. [DOI] [PubMed] [Google Scholar]
- 30.Hu RJ, Lee MP, Johnson LA, Feinberg AP. A novel human homologue of yeast nucleosome assembly protein, 65 kb centromeric to the p57KIP2 gene, is biallelically expressed in fetal and adult tissues. Hum. Mol. Genet. 1996;5:1743–1748. doi: 10.1093/hmg/5.11.1743. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, et al. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 1997;44:253–265. doi: 10.1006/geno.1997.4868. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, Yokoyama S. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5 S rDNA. J. Mol. Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 35.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa T, Bulger M, Muramatsu M, Ito T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J. Biol. Chem. 2001;276:27384–27391. doi: 10.1074/jbc.M101331200. [DOI] [PubMed] [Google Scholar]
- 37.Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr. Opin. Cell Biol. 2007;19:257–265. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 41.Kurumizaka H, Wolffe AP. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell Biol. 1997;17:6953–6969. doi: 10.1128/mcb.17.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuwaki M, Kato K, Shimahara H, Tate S, Nagata K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkuni K, Shirahige K, Kikuchi A. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2003;306:5–9. doi: 10.1016/s0006-291x(03)00907-0. [DOI] [PubMed] [Google Scholar]
- 45.Walter P, Owen-Hughes T, Cote J, Workman J. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asahara H, Tartare-Deckert S, Nakagawa T, Ikehara T, Hirose F, Hunter T, Ito T, Montminy M. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell Biol. 2002;22:2974–2983. doi: 10.1128/MCB.22.9.2974-2983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shikama N, Chan HM, Krstic-Demonacos M, Smith L, Lee CW, Cairns W, La Thangue NB. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell Biol. 2000;20:8933–8943. doi: 10.1128/mcb.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]