Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis (original) (raw)
Abstract
Small non-protein-coding RNAs (ncRNAs) have systematically been studied in various model organisms from Escherichia coli to Homo sapiens. Here, we analyse the small ncRNA transcriptome from the pathogenic filamentous fungus Aspergillus fumigatus. To that aim, we experimentally screened for ncRNAs, expressed under various growth conditions or during specific developmental stages, by generating a specialized cDNA library from size-selected small RNA species. Our screen revealed 30 novel ncRNA candidates from known ncRNA classes such as small nuclear RNAs (snRNAs) and C/D box-type small nucleolar RNAs (C/D box snoRNAs). Additionally, several candidates for H/ACA box snoRNAs could be predicted by a bioinformatical screen. We also identified 15 candidates for ncRNAs, which could not be assigned to any known ncRNA class. Some of these ncRNA species are developmentally regulated implying a possible novel function in A. fumigatus development. Surprisingly, in addition to full-length tRNAs, we also identified 5′- or 3′-halves of tRNAs, only, which are likely generated by tRNA cleavage within the anti-codon loop. We show that conidiation induces tRNA cleavage resulting in tRNA depletion within conidia. Since conidia represent the resting state of A. fumigatus we propose that conidial tRNA depletion might be a novel mechanism to down-regulate protein synthesis in a filamentous fungus.
INTRODUCTION
Cells from all organisms, studied to date, contain two different kinds of RNA species, the protein-encoding messenger RNAs (mRNAs) as well as non-protein-coding RNAs (ncRNAs). In contrast to mRNAs, ncRNAs are not translated into proteins, but have important cellular functions, either on their own or in complex with proteins (1–6). Functions of ncRNAs range from RNA processing, modification, transcriptional regulation, mRNA stability and translation up to protein secretion (2). Reported sizes of many known ncRNAs are generally well below sizes of mRNAs and range from 21–22-nt long microRNAs (7,8) to about 500 nt [e.g. telomerase RNA (9)]. In addition, also very large ncRNAs, including the 17-kb long human Xist RNA (10,11) or the 108-kb long mouse Air RNA (12) have been observed.
Recently, whole genome screens in eukaryal organisms have revealed a large number of ncRNAs which have been shown to regulate gene expression by novel mechanisms such as RNA interference, gene co-suppression, gene silencing, imprinting and DNA methylation (8,13–15). Evidence for the involvement of ncRNAs exerting critical functions during vegetative growth, development or cell differentiation as well as in diseases, such as carcinogenesis, is becoming increasingly clear (16,17).
Several single-cellular eukaryal organisms have been studied in the past, revealing a plethora of novel ncRNAs (18–20). A bioinformatical analysis of the fungal genomes from seven different yeast species provided a significant number of evolutionarily conserved, structured ncRNAs, suggesting their roles in post-transcriptional regulation (21). In contrast, identification and functions of ncRNAs in filamentous fungi, such as Aspergillus species, have not been studied.
Most filamentous fungi are saprophytes playing important roles in carbon and nitrogen recycling. Moreover, several members of this fungal group are well known for production of biotechnological important secondary metabolites, as producers of toxins, or as facultative pathogens for plants and animals. Infections with filamentous fungi have emerged as an increasing risk for immuno-suppressed patients. Aspergillus fumigatus accounts for most of these infections, termed invasive aspergillosis, and can be regarded as the most common airborne fungal pathogen. Specific diagnostics as well as therapeutic possibilities are limited (22–24). Hence, the mortality rate of invasive aspergillosis ranges between 30 and 90%, depending on the immune status of the host (22,23).
Its global ubiquity as well as the infectious cycle of this pathogen is perpetuated by prolific production of asexual spores (termed conidia) from specialized aeral hyphae (termed conidiophores). Conidial germination, e.g. in the human lung, following spore inhalation represents the initiating event of pulmonary disease. Three important steps can be distinguished during spore germination: activation of the resting spore to appropriate environmental conditions, isotropic growth that involves water uptake and wall growth (termed swelling) and polarized growth that results in the formation of a germ tube from which the new mycelium originates (25). Conidia are dormant, metabolically inactive cells, which can be stored for extended periods. The combined presence of air, water and a carbon source induces germination with the first measurable activities being trehalose breakdown and translation (26).
Aspergillus fumigatus cells contain a haploid nuclear genome of 28.9 Mb in size, distributed into eight chromosomes (27) and a circular mitochondrial genome exhibiting a size of 32 kb. Apart from ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), no other ncRNAs have yet been annotated and characterized within the A. fumigatus genome (27). However, knowledge on the number and functions of ncRNAs is vital for understanding cell functions in A. fumigatus and could potentially open up new avenues for the development of novel anti-fungal drugs. Thus, for the experimental identification of novel ncRNA species in A. fumigatus we generated a specialized cDNA library comprising small ncRNA species sized from 20 to –500 nt (28,29).
MATERIAL AND METHODS
Strain and growth conditions
Aspergillus fumigatus wild-type ATCC46645 (30) was maintained on solid Aspergillus minimal media (AMM) according to Pontecorvo et al. (31) containing 1% (wt/vol) glucose as carbon source and 20 mM glutamine as a nitrogen source. For liquid growth A. fumigatus was cultured at 37°C up to the indicated time point either in AMM or in Aspergillus complete media (ACM) comprising 2% (wt/vol) glucose, 0.2% tryptone (wt/vol), 0.1% yeast extract (wt/vol) and 0.1% casamino acids (wt/vol). Media contained 10 µM FeSO4 and respectively for iron-depleted conditions, iron was omitted. For nitrogen starvation, 18-h AMM cultures were harvested and shifted for another 6 h into AMM lacking glutamine.
Growth conditions for conidiation of A. fumigatus ATCC46645
For synchronized asexual developmental A. fumigatus was grown in liquid ACM for 18 h (32). Then, mycelia were harvested by filtering and transferred to solid ACM, were conidiation is induced (32). Samples for RNA isolation were collected after 6, 12, 24, 48 and 72 h of growth on solid ACM.
Generation of an A. fumigatus cDNA library
Aspergillus fumigatus was cultured under various conditions to ensure expression of also growth-regulated ncRNAs. Total RNA was extracted from harvested mycelia of A. fumigatus by the TRI-zol method (Gibco BRL) (33). Subsequently, equal amounts of total RNAs were pooled and size-fractionated by denaturing 8% PAGE (7 M urea, 1× TBE buffer). RNAs in the size range between 15 and 500 nt were excised from the gel, passively eluted and ethanol-precipitated. RNAs were poly(C)-tailed employing poly(A) polymerase from yeast (USB). C-tailed RNAs were ligated to a 19-nt long 5′ linker by T4 RNA ligase, as described previously (29). RNAs from the library were subsequently converted into cDNAs by RT–PCR as described, employing complementary primers to 5′ linkers and the poly(C) tail (29), and cloned into pGEM-T vector (Promega).
Dot-blot hybridization
Aspergillus fumigatus library-derived cDNA clones were PCR-amplified using the primers M13 and M13 reverse. Two micro litres of diluted (1:20) and denatured (91°C, 2 min) PCR products were spotted onto a nylon membrane (Hybond N+, Amersham), cross linked employing the STRATAGENE UV crosslinker (120 mJ/cm2) and pre-hybridized for 2 h in 1 M sodium phosphate buffer (pH 6.2) with 7% SDS. Oligonucleotides, complementary to known and most abundant ncRNAs were [γ-32P]ATP end-labelled by T4 polynucleotide kinase. All six oligonucleotide probes were added to the hybridization tube and hybridization was carried out at 52°C in hybridization buffer (178 mM Na2HPO4, NaH2PO4, pH 6.2, 7% SDS) for 12 h. Blots were washed twice: at room temperature in 2× SSC buffer, 0.1% SDS for 10 min and subsequently at hybridization temperature in 0.1× SSC, 0.1% SDS for 10 min. Afterwards blots were rinsed in desalted water. Following stringency washes membranes were exposed to Kodak MS-1 film from 30 min to 2 days.
Sequence analysis of cDNA library
cDNA clones were sequenced using the M13 reverse primer and the BigDye terminator cycle sequencing reaction kit (PE Applied Biosystems). Sequencing reactions were run on an ABI Prism 3100 (Perkin Elmer) capillary sequencer. Subsequently, sequences were analysed with the LASERGENE sequence analysis program package (DNASTAR). In this analysis, cDNA sequences were compared to each other using the Lasergene Seqman II program package to identify identical sequences (DNASTAR). Following a BLASTN search against the GenBank database (NCBI, http://www.ncbi.nlm.nih.gov/BLAST) was performed using standard parameters (i.e. match/mismatch score: 1,-2 and linear gap costs) which were adjusted automatically for short input sequences. All RNA sequences, which were not annotated in the database and showed a perfect match within the A. fumigatus genome (34), were treated as potential candidates for novel ncRNAs.
Northern blot analysis
Total RNA was either size-separated on 1.2% agarose–2.2 M formaldehyde gels and blotted onto Hybond N membranes (Amersham) as described earlier (35), or size-fractionated on denaturing polyacrylamide gels (PAGE). For PAGE, 3–40 µg of total RNA isolated from different growth conditions (see above) was denatured for 1 min at 95°C, separated on a 8% denaturing polyacrylamide gel (7 M urea, 1× TBE buffer) and transferred onto a nylon membrane (Hybond N+, Amersham) using the Bio-Rad semi-dry blotting apparatus (Trans-blot SD; Bio-Rad).
After immobilizing of RNAs using the STRATAGENE UV crosslinker (120 mJ/cm2), nylon membranes were pre-incubated for 2 h in 1 M sodium phosphate buffer (pH 6.2) with 7% SDS. Oligonucleotides from 18 to 35 nt in size, complementary to potentially novel RNA species, were end-labelled with [γ-32P]ATP and T4 polynucleotide kinase. Depending on the _T_m of the respective oligonucleotides, hybridization was carried out from 42 to 58°C in hybridization buffer (178 mM Na2HPO4, NaH2PO4, pH 6.2, 7% SDS) for 12 h. Blots were washed twice, once at room temperature in 2× SSC buffer, 0.1% SDS for 10 min and subsequently at the respective hybridization temperature in 0.1× SSC, 0.1% SDS for 1–10 min. Membranes were exposed to Kodak MS-1 film from 15 min to 5 days.
Bioinformatical methods
To obtain secondary structure predictions and conservation of A. fumigatus ncRNAs, sequences were mapped to the genomes of related species via BLAST (36) search (E-value: 10–3). The homologous sequences were then aligned using a dynamic programming alignment algorithm as implemented in CLUSTAL W (37) and out of the multiple sequence alignment the secondary structure was predicted using the folding routines from the Vienna RNA package (38,39). Genomes of Aspergillus oryzae, Aspergillus nidulans and Aspergillus niger were downloaded from the NCBI database. Employing a BLASTN search in the JGI genome browser of A. niger (http://genome.jgi-psf.org/Aspni1/Aspni1.home.html) sequence conservation of snRNAs and novel ncRNA candidates was analysed in the genomes of A. niger, A. oryzae, A. fumigatus and A. nidulans (Supplementary Data, Figures 1 and 4). Since U3 snoRNA could not be found by means of a BLASTN search, a computationally more expensive strategy was employed. The semi-local variant of Gotoh's dynamic programming algorithm (40) was implemented in a memory-efficient scanning version in the C programming language (parameters: match +3, mismatch 1, gap opening 8 and gap extension 2).
RESULTS AND DISCUSSION
cDNA library construction and analysis
Aspergillus fumigatus was grown under seven different culture conditions (see Materials and methods section). Liquid complete medium (ACM) is the optimal growth condition resulting in maximal vegetative proliferation. In comparison, minimal medium demands expression of an increased number of anabolic pathways. Starvation for iron (AMM-Fe) or nitrogen (AMM-N) was found to induce virulence traits (41,42). In contrast to liquid cultures (vegetative growth), plate cultures induce conidiation, i.e. the formation of infectious propagules (43). These different growth conditions are reported to induce different mRNA transcriptomes and thus, in addition, are likely to also induce the expression of differentially expressed ncRNA species. Total RNA from each culture condition was isolated separately and converted into cDNA as previously described (29).
Subsequently, we screened about 7200 cDNA clones for identification of novel ncRNA species by dot-blot hybridization employing labelled oligonucleotides directed against the most abundant known ncRNAs, as described previously (44). This approach yielded 3120 cDNA sequences, which were subjected to further bioinformatical analysis (see Materials and methods section). Thereby, 58.2% of cDNA sequences corresponded to nuclear or mitochondrial rRNA fragments. tRNAs or tRNA cleavage fragments (see below) represented about 23.0% of cDNA clones (Figure 1A). About 4.9% of cDNA sequences corresponded to three different snRNAs (U1, U5 and U6 snRNA), which had escaped annotation in the current release of the A. fumigatus genome (27). Candidates for small nucleolar RNA (snoRNA) made up 11.3% of all cDNA clones. Due to the lack of conserved sequence or structure motifs the remaining 1.2% of cDNA clones could not be assigned to any class of known ncRNAs and thus might represent entirely novel ncRNAs in A. fumigatus (Figure 1A).
Figure 1.
(A) Distribution of 3120 cDNA clones from the A. fumigatus expression library encoding ncRNA candidates. The abundance of different ncRNA species is shown as percentage of total clones. (B) Numbers of ncRNA candidates derived from the A. fumigatus cDNA library are indicated in brackets.
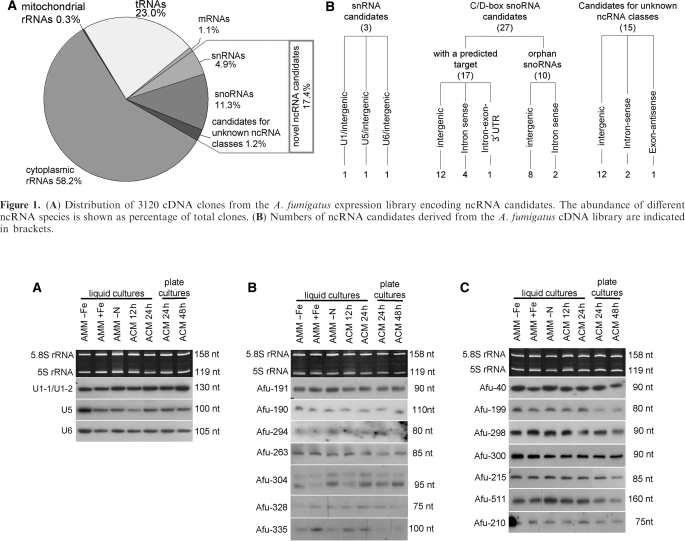
Subsequently, we confirmed expression and sizes of ncRNAs by northern blot analysis. To that aim, total RNA isolated from different growth conditions or developmental stages of A. fumigatus, was employed in northern blot analysis to investigate expression levels of ncRNAs (Figures 2 and 3). In our final analysis, we only listed those novel ncRNAs for which unambiguous northern blot signals were obtained, with the notable exception of some snoRNA species (Table 1, see below), which were computationally confirmed by the presence of canonical sequence and structure motifs.
Figure 2.
Northern blot analysis of A. fumigatus snRNAs and selected snoRNA candidates. Designation of clones is indicated on the left. Sizes of ncRNAs, as estimated by comparison with an internal RNA marker, are indicated on the right. Total RNA was isolated from A. fumigatus mycelia grown under seven different conditions and at different time points: AMM, minimal medium; ACM, complete medium; AMM-Fe, iron starvation; AMM-N, nitrogen starvation. Liquid cultures resemble vegetative growth and plate cultures induce conidiogenesis. In-gel ethidium bromide-stained 5.8 S rRNA and 5 S rRNA serve as loading controls. (A) U1-1/U1-2, U5 and U6 snRNAs, respectively. (B) C/D box snoRNAs with predicted targets (C) C/D box snoRNAs without predicted targets.
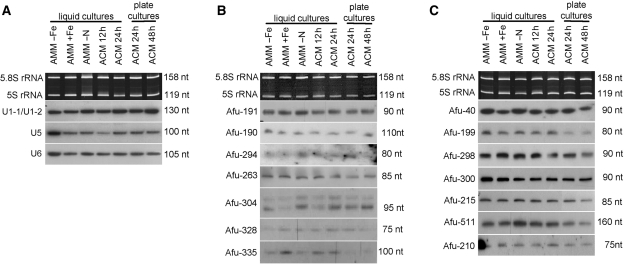
Figure 3.
Northern blot analysis of ncRNA candidates from unknown ncRNA classes. Designation of clones is indicated on the left. Sizes of ncRNAs, as estimated by comparison with an internal RNA marker, are indicated on the right. Growth conditions and loading controls (5.8 S rRNA and 5 S rRNA) as described in Figure 2. As an additional loading control, U1 snRNA was included in northern blot analysis.
Table 1.
Candidates for snRNAs or snoRNAs
| Name | Copies | cDNA (nt) | Northern blot (nt) | Location | Putative target | Accession number |
|---|---|---|---|---|---|---|
| snRNAs | ||||||
| U1-1 | 157 | 132 | 130 | Intergenic; Afu1g06980/Afu1g07000 | AM921915 | |
| U1-2 | 67 | 132 | 130 | Intergenic; Afu4g12490/Afu4g12500 | AM921916 | |
| U5 | 19 | 99 | 100 | Intergenic; Afu6g12670/Afu6g12680 | AM921917 | |
| U6 | 3 | 50 | 105 | Intergenic; Afu4g12500/Afu4g12520 Intergenic; Afu2g10150/Afu2g10160 | AM921918 | |
| C/D box snoRNAs with predicted target | ||||||
| Afu-34 | 18 | 84 | n.d. | Intergenic; Afu2g15970/Afu2g15980 | Am1131 and Gm2506 in 26S | AM921919 |
| Afu-191 | 11 | 92 | 90 | Intergenic; Afu1g10270/Afu1g10280 | Gm75 in 5.8S and Am32 in U2 snRNA | AM921920 |
| Afu-190 | 8 | 107 | 110 | Intergenic; Afu4g11320/Afu4g11330 | Gm557 in 18S | AM921921 |
| Afu-198 | 8 | 130 | n.d. | Intergenic; Afu1g02700/Afu1g02680 | Cm2856 and Um 2859 in 26S | AM921922 |
| Afu-294 | 7 | 85 | 80 | Intergenic; Afu1g09750/Afu1g09760 | Cm1851 in 26S; Am43 in 5.8S | AM921923 |
| Afu-264 | 4 | 103 | n.d. | Intergenic; Afu7g05290/Afu7g05300 | Gm2770 and Gm2773 in 26S; | AM921924 |
| Afu-277 | 4 | 100 | n.d. | Intergenic; Afu1g09740/Afu1g09760 | Um2706 in 26S; Am97 in 18S; Cm47 in 5.8S | AM921925 |
| Afu-263 | 3 | 96 | 85 | Intron1 - Exon2 -3′UTR (sense); | Am815 and Gm906 in 26S | AM921926 |
| Afu-188 | 2 | 84 | n.d. | Intergenic; Afu4g11310/Afu4g11320 | Am25 and Um26 in 18S | AM921927 |
| Afu-200 | 2 | 91 | n.d. | Intron 3 (sense); Afu1g12390 | Am415 and Gm1422 in 18S | AM921928 |
| Afu-304 | 2 | 106 | 95 | Intron 2 (sense); Afu1g09800 | Cm2925 in 26S | AM921929 |
| Afu-380 | 2 | 87 | n.d. | Intergenic; Afu4g06780/Afu4g06770 | Um2701 in 26S | AM921930 |
| Afu-513 | 2 | 129 | n.d. | Intergenic; Afu4g11310/Afu4g11320 | Gm1338 and Gm3719 in 26S | AM921931 |
| Afu-328 | 1 | 75 | 75 | Intron 2 (sense); Afu6g04570 | Gm2792 in 26S | AM921932 |
| Afu-335 | 1 | 97 | 100 | Intron 2 (sense); Afu1g04840 | Cm2316 in 26S | AM921933 |
| Afu-438 | 1 | 83 | n.d. | Intergenic; Afu2g15980/Afu2g15970 | Am2877 in 26S | AM921934 |
| Afu-455 | 1 | 92 | n.d. | Intergenic; Afu1g09760/Afu1g09740 | Gm1122 in 18S | AM921935 |
| C/D box snoRNAs without predicted target (orphan snoRNAs) | ||||||
| Afu-40 | 123 | 90 | 90 | Intergenic; Afu4g11310/Afu4g11320 | AM921936 | |
| Afu-514 | 74 | 105 | n.d. | Intergenic; Afu6g03830/Afu6g03840 | AM921937 | |
| Afu-515 | 40 | 89 | n.d. | Intergenic; Afu4g11310/Afu4g11320 | AM921938 | |
| Afu-199 | 26 | 79 | 80 | Intron 1 (sense); Afu7g02320 | AM921939 | |
| Afu-298 | 3 | 93 | 90 | Intergenic; Afu1g05080/Afu1g05100 | AM921940 | |
| Afu-300 | 3 | 88 | 90 | Intergenic; Afu4g11310/Afu4g11320 | AM921941 | |
| Afu-215 | 2 | 24 | 85 | Intergenic; Afu5g12870/Afu5g12880 | AM921942 | |
| Afu-511 | 2 | 164 | 160 | Intergenic; Afu1g03400/Afu1g03410 | AM921943 | |
| Afu-210 | 1 | 24 | 75 | Intron 2 (sense); Afu2g03610 | AM921944 | |
| Afu-265 | 1 | 23 | n.d. | Intergenic; Afu3g02340/Afu3g02370 | AM921945 |
snRNA candidates
Four different cDNA clones representing putative snRNA candidates were annotated as U1-1, U1-2, U5 and U6 snRNAs, respectively, by comparison to the RNA family database of alignments and Covariance Models (45). Sequences of U1-1 and U1-2 snRNAs differ by 1 nt, only (A or T at position 128, respectively), and exhibit different genomic locations, implying that they represent two distinct isoforms of U1 snRNA (Table 1). A comparison of the predicted secondary structures of A. fumigatus snRNAs U1-1, U1-2, U5 and U6 snRNAs and their homologues with the corresponding Saccharomyces cerevisae homologues shows highly conserved regions in the loops of the hairpins. Compared to the consensus secondary structure as reported by the Rfam database (45) the Aspergillus snRNA structures show the same stem–loop distribution and arrangement (Supplementary Data Figure 1A–D).
Except for nuclear encoded 26S, 18S, 5.8S rRNAs and tRNAs, U1-1 snRNA appeared as the most abundant clone in the library (Table 1). Three identical truncated cDNA clones of U6 snRNA, mapping to two different loci within the A. fumigatus genome, were also identified in our screen (Table 1).
Expression of A. fumigatus U1, U5 and U6 snRNAs could be confirmed by northern blot analysis (Figure 2A). Sizes, as estimated by comparison to an internal RNA marker, were determined as 130 nt, 100 nt or 105 nt, respectively (Figure 2A). The absence of U2 and U4 snRNAs in our screen might be explained by RNA modifications or structural constraints, impeding reverse transcription into cDNAs.
Therefore, we employed, as a bioinformatical approach, a BLASTN search of all known U2 and U4 snRNAs as annotated in the Rfam database, including all yeast snRNA genes. Indeed, we were able to predict U2 and U4 homologues within the A. fumigatus genome (Supplementary Data Figure 1B and D). However, A. fumigatus U2 and U4 snRNAs appeared to be less conserved on the sequence level than expected and could only be partially aligned. Manually extending the alignment yielded a set of nearly perfect U2 and U4 snRNA sequences in A. fumigatus and related species (e.g. A. niger). In addition, we were able to compute the interaction structure of the U4–U6 complex (Supplementary Data Figure 1D) using the RNAalifold program (47).
A comparison of the predicted consensus secondary structures to annotated secondary structures in the Rfam database and in (48) shows that these computational candidates match perfectly, although they show high variance in their primary sequence (Supplementary Data, Figure 1A–C). This is consistently observed for the evolution of ncRNAs, since these genes are mainly conserved on the secondary structure level. Table 3 indicates levels of conservation within all identified snRNA candidates in A. fumigatus. In addition to their bioinformatical identification, expression of U2 and U4 snRNAs was verified by northern blot analysis (Supplementary Data, Figure 2A). It is noteworthy, however, that sizes of U2 and U4 snRNAs, as estimated by northern blot analysis, differ from their bioinformatical predicted sizes due to the fact that precise 5′- and 3′ termini cannot be obtained by computational analysis.
Table 3.
Conservation level of snRNA candidates in Aspergillus species
| snRNAs | Location in A.fumigatus | Conserved in related species (% sequence identity) |
|---|---|---|
| U1-1 | chr1:1991385-1991564 | A.nidulans (78), A.oryzae (94), A.niger (90) |
| U1-2 | chr4:3279813-3279991 | A.nidulans (88), A.oryzae (91), A.niger (93) |
| U2-1 | chr3:3242037-3242312 | A.nidulans (76), A.oryzae (85), A.niger (75) |
| U2-2 | chr1:3223904-3224180 | A.nidulans (88), A.oryzae (84), A.niger (74) |
| U4 | chr1:1274827-1275331 | A.nidulans (49), A.oryzae (53), A.niger (53) |
| U5 | chr6:3202420-3202638 | A.nidulans (44), A.oryzae (60), A.niger (64) |
| U6 | chr1:1651441-1651760 | A.nidulans (42), A.oryzae (39), A.niger (43) |
| chr2:2598365-2598684 | A.nidulans (50), A.oryzae (54), A.niger (52) | |
| chr4:3281001-3281320 | A.nidulans (42), A.oryzae (49), A.niger (67) |
snoRNA candidates
Two classes of small nucleolar ncRNAs (snoRNAs) have been detected in eukaryal as well as in archaeal species (49–52): C/D box snoRNAs, which guide 2′-_O_-methylation of ribosomal, spliceosomal and tRNAs (the latter in Archaea, only), and H/ACA snoRNAs which guide pseudouridylation in these RNA species (53). In our screen, we identified 27 candidates for C/D box snoRNAs based on conserved sequence or structural motifs by employing the SnoReport program (54).
All C/D box snoRNAs from our screen contained bona fide sequence motifs of canonical snoRNAs, namely C, D′, C′ and D boxes, respectively (1,55). We noticed, however, that all C/D box snoRNAs from A. fumigatus lacked the canonical terminal stem structure, in contrast to mammalian C/D snoRNAs. This is in agreement with previous observations on archaeal C/D snoRNAs (56) or C/D snoRNAs from the slime mold Dyctiostelium discoideum, which are also devoid of a terminal stem (57).
Surprisingly, the abundant U3 C/D snoRNA was missing among these candidates. A standard BLASTN search also failed to identify U3 snoRNA candidates within the A. fumigatus genome. Thus, we applied a computationally more sophisticated method, employing the semi-local variant of the Gotoh dynamic programming algorithm (parameters: match +3, mismatch –1, gap opening –8 and gap extension –2; see Materials and methods section). By this method, we were able to also identify a U3 snoRNA candidate and verify its expression by northern blot analysis (Supplementary Data, Figure 2B and C).
Most C/D snoRNAs identified in our screen, with some exceptions (Table 1) are within the size range of a canonical C/D snoRNAs (i.e. from 80 to 100 nt; Table 1). Identification of some larger C/D box snoRNAs in our library is in agreement with previous findings in Oryza sativa (58) or Trypanosoma brucei (59) (Table 1). From the 27 C/D snoRNA candidates, six are intron-derived (Table 1, Figure 1B) as observed for most mammalian and plant snoRNAs (53). One C/D box snoRNA candidate, Afu-263, extends beyond a predicted intron–exon border within the 3′-untranslated region (UTR; Figure 1B) of a hypothetical protein, encoded by the Afu3g14080 gene. However, this might be due to a wrong annotation of Afu3g14080 as a protein-coding gene, consistent with its unusually small transcript length of 96 nt. Comparison of Afu-263 with the Rfam database reveals homology to S. cerevisiae snR60. In addition, Afu-263 is also conserved in the genome of the related fungus A.oryzae.
The remaining 20 candidates map to intergenic regions. Twelve snoRNA candidates from this group are present as singletons, whereas the remaining eight candidates are distributed in two clusters, comprised of three and five snoRNA genes, respectively. A similar clustered gene organization has been previously observed in S. cerevisiae (60–63). The first snoRNA cluster is located on chromosome 1 between protein-coding genes Afu1g09740 and Afu1g09760 and comprises the C/D box snoRNAs Afu-455, Afu-277 and Afu-294. The second cluster maps to chromosome 4 between the protein-coding genes Afu4g11310 and Afu4g11320 and contains five C/D box snoRNAs, Afu-40, Afu-188, Afu-300, Afu-513 and Afu-515. Database searches with genomes from the related moulds A. nidulans and A. oryzae revealed their conservation in both filamentous fungi. SnoRNA clusters have been detected in a multitude of different eukaryal organisms, including S. cerevisiae, which may suggest an ancient origin of this gene arrangement (62,63).
By employing the Snoscan Server 1.0 program (64) we identified putative targets for 17 out of 27 C/D box snoRNAs (Table 1 and Figure 1B). As a probabilistic search model for filamentous fungi is currently not available, the search model for target evaluation was adjusted to S. cerevisiae. As potential target sequences we considered the previously identified canonical targets, i.e. 26 S, 18 S, 5.8 S rRNAs and all snRNAs [identified by our screen (i.e. U1, U5 or U6 snRNA, respectively)]. Most of the C/D box snoRNAs were predicted to guide methylation of 26 S rRNA, and, to a lesser extent, also 18 S and 5.8 S rRNAs. Several snoRNA candidates are predicted to guide two different rRNA modifications, while C/D box snoRNA Afu-277 is predicted to guide methylation of three RNAs, i.e. 26 S rRNA, 18 S rRNAs and 5.8 S rRNA, respectively (Table 1). No C/D box snoRNAs targeting U1, U5 or U6 snRNAs were found in our screen.
For the remaining 10 C/D box snoRNA candidates, no rRNA or snRNA targets could be identified (Table 1 and Figure 1B). Hence, these were termed ‘orphan snoRNAs’ in agreement with earlier findings of similar snoRNAs in other species including mouse (1,44). Some orphan snoRNAs were represented by numerous cDNA clones (Table 1), indicating their high abundance in A. fumigatus. Interestingly, MBII-52 an abundant orphan snoRNA from mouse brain, was proposed to target the serotonin receptor 2C mRNA, (65). Whether some of these orphan snoRNAs from A. fumigatus might fulfil similar functions in mRNA targeting needs to be determined.
Expression analysis of snoRNAs by northern blot analysis revealed that most snoRNAs are equally expressed under most growth conditions with some exceptions, (Figure 2B and C). Down-regulation during iron starvation, as observed for Afu-328, might implicate iron-related functions; iron starvation, for example, down-regulates transcription of genes encoding iron-containing proteins or iron-consuming pathways in A. nidulans (66). Likewise, down-regulation of expression during conidiogenesis, as observed for Afu-335, Afu-199 or Afu-511, suggests their developmental regulation.
In contrast to C/D box snoRNAs, we were so far unable to experimentally identify any representatives for H/ACA box snoRNAs. Therefore, we applied the snoReport program (54) to chromosome 1 of the A. fumigatus genome and were indeed able to bioinformatically identify candidates for H/ACA snoRNAs. The majority of those candidates show a canonical H/ACA secondary structure and both of the sequence motifs H and ACA (for examples see Supplementary Data, Figure 3A–E).
Candidates for entirely novel ncRNAs
Due to the lack of conserved sequence motifs 1.2% of cDNA clones, amounting to 15 different cDNA sequences, could not be assigned to any class of known ncRNAs and thus might encode entirely novel ncRNAs in A. fumigatus. Northern blot analysis verified expression of all 15 ncRNA candidates (Figure 3) and the genomic location could be determined for all 15 candidates (Table 2).
Table 2.
Candidates for entirely novel ncRNAs
| Name | Copies | cDNA (nt) | Northern blot (nt) | Location | Accession number |
|---|---|---|---|---|---|
| Afu-182 | 17 | 219 | 300 | Intergenic; Afu4g07680/Afu4g07690 | AM921946 |
| Afu-202 | 5 | 266 | 270 | Intergenic; Afu1g10420/Afu1g10430 | AM921947 |
| Afu-67 | 2 | 21 | 600/190 | Intergenic; Afu4g01630/Afu4g02610 | AM921948 |
| Afu-254 | 2 | 163 | 90/420 | Intergenic; Afu7g04110/Afu7g04120 | AM921949 |
| Afu-203 | 1 | 111 | 110 | Intron 1 (sense); Afu3g14240 | AM921950 |
| Afu-222 | 1 | 22 | 535/250 | Intergenic; Afu4g12410/Afu4g12420 | AM921951 |
| Afu-262 | 1 | 25 | 225 | Intergenic; Afu1g10370/Afu1g10380 | AM921952 |
| Afu-309 | 1 | 318 | 320 | Intergenic; Afu4g10430/Afu4g10420 | AM921953 |
| Afu-318 | 1 | 24 | 70 | Intergenic; Afu7g01590/Afu7g01600 | AM921954 |
| Afu-322 | 1 | 46 | 75/35 | Intergenic; Afu2g13250/Afu2g13240 | AM921955 |
| Afu-336 | 1 | 28 | 130 | Intergenic; Afu1g09740/Afu1g09760 | AM921956 |
| Afu-364 | 1 | 18 | 100 | Intergenic; Afu1g13820/Afu1g13830; | AM921957 |
| Intergenic; Afu3g03090/Afu3g03130; | |||||
| Exon4-Intron4; Afu3g02730; | |||||
| Intergenic; Afu6g11780/Afu6g11790 | |||||
| Afu-448 | 1 | 29 | 315 | Intergenic; Afu1g11550/Afu1g11540 | AM921958 |
| Afu-465 | 1 | 17 | 270 | Intron 2 (sense); Afu4g08930 | AM921959 |
| Afu-484 | 1 | 41 | 600 | Exon2 (antisense); Afu3g07170 | AM921960 |
Expression of ncRNA candidates was verified by northern blot analysis, pointing towards a significant abundance within A. fumigatus. Interestingly, many of these candidates, appear differentially expressed under growth conditions tested, which might indicate their involvement in regulatory processes. In particular, expression of Afu-336, Afu-364 and Afu-448 appears to be down-regulated following growth on plates for 48 h, indicating developmental regulation. In several cases, northern blot analysis revealed signals of larger sizes compared to sizes as determined by cDNA cloning. This might be explained by cDNA sequences representing partial sequence fragments of full-length ncRNAs. In addition, for some cDNA clones several bands could be observed, the larger ones potentially reflecting precursor forms of mature ncRNA candidates.
No obvious conserved sequence or structure motifs could be identified among these candidates. However, six candidates turned out to be highly conserved in related Aspergillus species. In particular, Afu-182, Afu-202, Afu-309 and Afu-336 are conserved in A. nidulans, A. oryzae and A. niger (Table 4) over their entire length from 220 to 350 nt. Hence, consensus secondary structures could be predicted employing the JGI genome browser for A. niger (http://genome.jgi-psf.org; Supplementary Data Figure 4A–D). One conserved candidate is located in the vicinity of a known ncRNA gene: Afu-67 is present in two copies within the A. fumigatus genome and is located 550-nt upstream of the rRNA operon. None of the remaining novel ncRNA candidates could be assigned to any known ncRNA gene or function.
Table 4.
Conservation, annotation and secondary structure prediction of novel ncRNA candidates
| Name | Location in A. fumigatus | Conservation | Structure conservation | Annotation |
|---|---|---|---|---|
| Afu-182 | chr4:1997138-1996922 | A.niger, A.oryzae, A.nidulans | + | − |
| Afu-202 | chr1:2714678-2714414 | A.niger, A.oryzae, A.nidulans | + | − |
| Afu-67 | chr4:444973-444954 | – | − | 550 nt upstream of rRNA operon |
| chr4:706179-706160 | – | − | ||
| Afu-254 | chr7:927348-927204 | – | − | − |
| Afu-203 | chr3:3785105-3785214 | – | − | − |
| Afu-222 | chr4:3252985-3252964 | – | − | − |
| Afu-262 | chr1:2675372-2675395 | – | − | − |
| Afu-309 | chr4:2732230-2732547 | A.niger, A.oryzae | + | − |
| Afu-318 | chr7:415029-415006 | – | − | − |
| Afu-322 | chr2:3404938-3404893 | – | − | − |
| Afu-336 | chr1:2523503-2523530 | A.niger, A.oryzae, A.nidulans | + | − |
| Afu-364 | chr1:3693906-3693889 | A.niger | + | |
| chr3:710476-710493 | A.niger | + | ||
| chr3:836808-836825 | A.niger | + | ||
| chr6:2935513 2935530 | A.niger | + | ||
| Afu-448 | chr1:3048137-3048109 | A.niger, A.oryzae | − | − |
| Afu-465 | chr4:2316203-2316188 | – | − | − |
| Afu-484 | chr3:1802200-1802240 | – | − | − |
A large number of known ncRNAs function as anti-sense RNAs targeting mRNAs (67). Thus, it would be desirable to identify potential targets for novel ncRNAs in A. fumigatus. However, in the absence of any hints on the location of anti-sense boxes, this is currently a difficult task. As an exception, Afu-484 ncRNA is transcribed in anti-sense orientation to Exon 2 of the PIGC mRNA and thus might regulate the translation or stability of the mRNA. Interestingly, under conditions where A. fumigatus undergoes conidiation (ACM plate cultures at 48 hr) expression of Afu-484 decreases while expression of the corresponding mRNA increases, consistent with a regulation of stability of the PIGC mRNA by Afu-484 (Supplementary Data, Figure 4E).
tRNA cleavage: a novel mechanism to regulate protein synthesis?
tRNAs corresponded to 23% of all cDNA clones from the expression library (Figure 1A). All nuclear encoded tRNAs except cytoplasmic tRNAMet, tRNATrp and tRNAAsp were identified. This suggests a reasonable good coverage of the cDNA library for ncRNA species, all the more so, since tRNAs, because of their highly stable tertiary structure and their base modifications, are generally refractory to reverse transcription and cDNA cloning (44).
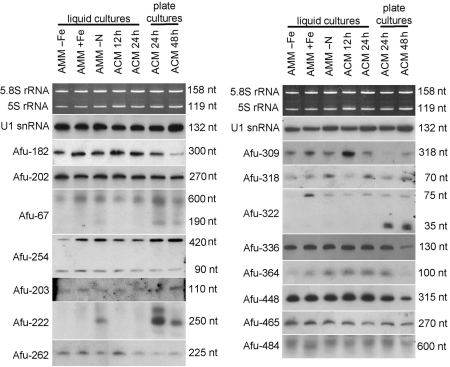
From mitochondrial tRNAs, only two different cDNA clones were obtained encoding tRNASer and tRNALys. Both tRNA species are encoded by the mitochondrial genome of A. fumigatus (34). Interestingly, in contrast to nuclear encoded tRNAs, expression of the two mitochondrial tRNA species was down-regulated by 2.1 and 3.7-fold, respectively, in media lacking iron (Figure 4). Within mitochondria, several enzymes containing iron-sulphur clusters are present; also, the final step of haeme biosynthesis, the incorporation of iron into protoporphyrinogen IX, takes place (68). Hence, these cell organelles are the primary consumers of cellular iron. It is tempting to speculate that down-regulation of mitochondrial tRNA levels represents a mechanism for decreasing mitochondrial activity during iron starvation due to the strong iron-dependence of metabolism within this cellular compartment.
Figure 4.
Northern blot analysis of selected nuclear and mitochondrial encoded tRNAs. Total RNA was isolated from A. fumigatus mycelia grown under iron-depleted (AMM-Fe) or iron-repleted conditions. Loading controls (5.8 S rRNA, 5 S rRNA and U1 snRNA) as described in Figure 3.
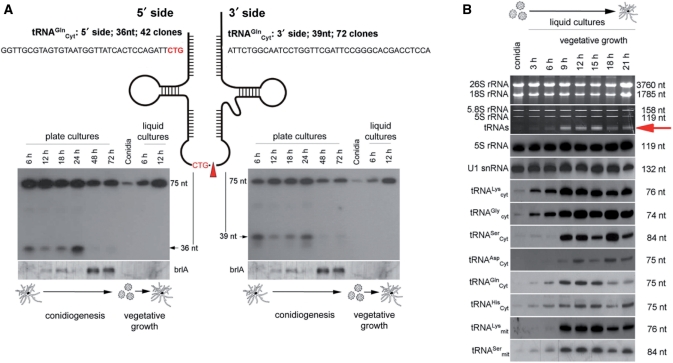
Interestingly, from cytoplasmic tRNAs, 16 were also represented as partial sequences in the cDNA library by several identical clones, corresponding either to the 5′- or 3′-halves of tRNAs (Figure 5A). The majority of tRNA 5′-halves contained the anti-codon sequence at their 3′-ends, whereas sequences of cDNAs encoding the tRNA 3′-halves started right after the anti-codon (Figure 5A). This strongly suggests an endonucleolytic cleavage of tRNAs within their anti-codon loop, at a position 3′ adjacent to the anti-codon. For cytoplasmic tRNAGln and tRNAHis we confirmed these stable cleavage intermediates by northern blot analysis employing oligonucleotides directed against the 5′- and the 3′-halves of the tRNAs (Figure 5A, and data not shown). Thereby, the estimated sizes of the northern blot signals from the 5′- and the 3′-halves of tRNAs are in agreement with the lengths of the corresponding cloned cDNAs. In addition, for cytoplasmic tRNAGln, tRNAGly and tRNAHis we verified cleavage sites 3′ adjacent to the anti-codon by primer extension (data not shown).
Figure 5.
(A) Upper: alignment of cDNA sequences from the cDNA library, representing 5′ and 3′-halves of cytoplasmic tRNAGln. The anti-codon is boxed in red. Northern blot analysis of cytoplasmic tRNAGln (bottom) employing oligonucleotide probes directed against 5′- and 3′-halves of tRNAGln. Northern blot signals correspond to full-length tRNAGln (75 nt) or 5′ or 3′ cleavage products (36 nt or 39 nt), respectively. Lower: total RNAs were isolated from A. fumigatus conidia and from mycelia undergoing conidiogenesis (solid ACM, 6 h, 12 h, 18 h, 24 h, 48 h and 72 h), germination (liquid ACM 6 h) and vegetative growth (liquid ACM 12 h). Conidiogenesis is indicated by expression of the brlA gene. The proposed site of endonucleolytic cleavage within the anti-codon loop is indicated by a red triangle; (B) Comparison of expression levels of most abundant ncRNA species by in-gel ethidium bromide staining or by northern blot analysis from germination to hyphal growth; expression levels of the entire tRNA fraction is indicated by a red arrow; total RNA was isolated from A. fumigatus conidia, germinating conidia (3 h and 6 h) and hyphae (9–21 h), respectively.
We next investigated whether cleavage of tRNAs was developmentally regulated in A. fumigatus. To synchronize conidiation (43), mycelia from 18-h liquid cultures were transferred to plates and RNA was isolated at different time points (Figure 5A). Indeed, northern blot analysis of tRNAGln revealed cleavage products of the expected sizes (see above) from 6 to 24-h after the transfer from liquid to plate culturing. In comparison to full-length tRNAs, tRNA-halves are expected to undergo rapid degradation by exonucleases. Thus, the high abundance of these tRNA cleavage products, as determined by northern blot analysis, is unexpected and therefore might even reflect a lower estimate of tRNA cleavage at these time points, only. At the 48- and 72-h time points (where conidia production is maximal) tRNAGln levels were significantly decreased and in conidia largely decreased. The absence of 5′- or 3′-halves of tRNAGln at these time points probably reflects an increase in exonuclease activity, thus rapidly degrading the unstable tRNA intermediates during conidiogenesis (Figure 5A). As a control for conidiation, the expression of the conidiation-specific transcription factor brlA was measured (32). Consistently, brlA transcripts were detected in plate cultures with a maximum at 48 to 72 h, but not in liquid cultures and in conidia (Figure 5A).
We then analysed whether tRNA cleavage during conidiogenesis is only restricted to tRNAs or also involves other abundant ncRNA species. To that aim, we investigated expression levels of various house keeping ncRNAs during A. fumigatus development (Figure 5B). Northern blot analysis and in-gel ethidium bromide staining of total RNA (24) revealed that all large and small ribosomal RNAs (i.e. 26 S, 18 S, 5.8 S and 5 S rRNAs, respectively), as well as spliceosomal U1 snRNA showed comparable abundance at all developmental stages. In contrast, levels of all selected tRNAs, either from the nuclear or mitochondrial genome, were strongly reduced in conidia, compared to hyphae, as shown by northern blot analysis (Figure 5B).
Levels of all tRNAs steadily increase during germination (Figure 5A: right lanes 6 and 12 h time points; Figure 5B: 3-, 6- and 9-h time points). Under these conditions, cleavage products are not detectable, consistent with suppression of conidiogenesis in liquid culture. Thereby, the 3-, 6- and 9-h time points resemble conidial swelling, conidial germ tube formation and hyphal growth, respectively (Figure 5B). Maximal tRNA levels are observed during hyphal growth from 9 to 15 h. The significant lower abundance of tRNAs in conidia, compared to other ncRNAs, can readily be detected by in-gel ethidium bromide staining of total RNA (indicated by a red arrow, Figure 5B).
We currently envision two alternative models for a decrease of total tRNA levels during conidiogenesis: (i) a decrease in the de novo synthesis of tRNAs or (i) an increase in tRNA cleavage. At this point, we favour the second model, which is consistent with the observed tRNA halves derived from cleavage within the anti-codon loop of tRNAs (see above). What would be the function of tRNA degradation in fungi during conidiogenesis? Since filamentous fungi usually start their asexual life cycle as metabolically inactive asexual spores, the conidia (69), this requires stalling of protein synthesis (25).
Several different mechanisms for stalling of protein synthesis have been identified. For example, bacteria react to nutritional stress, like amino acid deprivation, by the so-called stringent response, which primarily results in inhibition of RNA synthesis (70). The effector of the stringent control is guanosine 3′,5′-bisdiphosphate (ppGpp), synthesized by the ribosome-associated RelA protein. Upon amino acid deprivation, uncharged tRNAs bind to the ribosome, which triggers increased ppGpp synthesis by RelA, resulting in changes in gene expression including inhibition of promoters for ribosomal and most tRNA operons and stimulation of promoters for many amino acid biosynthesis operons (71). As an alternative mechanism, in the single-cellular organism Tetrahymena, it has recently been reported that amino acid deprivation triggers endonucleolytic cleavage within several positions in the anti-codon loop of functional tRNAs (72). This suggests that anti-codon loop cleavage reduces the accumulation of uncharged tRNAs as a part of a specific response induced by starvation (72).
Inhibition of protein synthesis by a similar mechanism, i.e. tRNA cleavage, is also found for Escherichia coli cells infected by the phage T4. Here, it was demonstrated that an _E. coli_-encoded anti-codon-specific endonuclease, termed ACNase, cleaves tRNALys within its anti-codon loop as a ‘suicide-response’ to T4 phage infection. This lesion could deplete the infected cell of functional tRNALys, inhibit translation of late T4 proteins and, consequently, contain the infection (73). Interestingly, the T4 phage employs an RNA repair mechanism to offset the damage, namely by religation of both tRNA halves through polynucleotide kinase and RNA ligase (73).
It should be noted that the tRNA cleavage mechanism, proposed for A. fumigatus, differs from the one described for E. coli: here the ACNase cleaves tRNALys, only, at a position 5′ to the wobble base of the anti-codon, while the proposed endonucleolytic cleavage in A. fumigatus occurs at all tRNAs investigated but 3′ adjacent to the anti-codon. Hence, endonucleases involved in this process might differ significantly from each other between the two species, consistent with the lack of ACN homologues found in A. fumigatus employing a BlastP database search (data not shown).
By employing this mechanism, resuming protein synthesis in A. fumigatus, would initially require de novo transcription of tRNAs, only, followed by rapid protein synthesis which would ensure a fast response to growth stimuli. Accordingly, the initiation of the fungal germination process is reported to be associated with the onset of protein synthesis (25).
CONCLUSION
In this study, we have analysed the small transcriptome of the filamentous fungus A. fumigatus by generating a specialized cDNA library from RNAs sized from 20 to 500 nt. We were able to identify 30 ncRNAs from known classes such as snRNAs and snoRNAs and thus complement the current annotation of only protein-coding genes within the A. fumigatus genome (27,34). From the class of snRNAs, we experimentally identified U1, U5 and U6 snRNA while U2, U3 and U4 snRNAs were found by bioinformatical in silico analysis. From the class of snoRNAs we experimentally detected 27 representatives, all belonging to the class of C/D box snoRNAs. Employing the SnoReport program we bioinformatically identified H/ACA snoRNA candidates in the A. fumigatus genome exhibiting canonical H and ACA sequence box motifs as well as a canonical double stem–loop structure; we also found 15 candidates of ncRNAs, which could not be assigned to any known ncRNA class. Some of these ncRNA species are developmentally regulated implying a possible function in A. fumigatus development. Four of these novel candidates are conserved on the sequence and structure level in at least two of the other three available Aspergillus species.
For other eukaryal organisms, such as fission yeast Schizosaccharomyces pombe (74,75) or the unicellular green algae Chlamydomonas reinhardtii (18,76), the presence of siRNAs, or miRNAs has been reported. We were unable to detect any small ncRNA species in A. fumigatus with signatures of precursors or mature forms of these abundant ncRNAs.
Surprisingly, we also observed ncRNA species corresponding to the 5′- or 3′ half of tRNAs, which are likely generated by endonucleolytic cleavage within the anti-codon loop during conidiogenesis. This led to a model, in which the requirement for stalling protein synthesis in conidia is achieved by cleavage and subsequent degradation of tRNAs in A. fumigatus. To verify this model, future experiments have to address the identification of the tRNA cleavage activity in A. fumigatus. It is tempting to speculate whether similar mechanisms to regulate protein synthesis are also employed in resting states of other, multi-cellular, organisms, such as plants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Data]
ACKNOWLEDGEMENTS
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung (FWF grant P171370) and an Austrian genome research (Gen-Au grant D 110420-011-013) to A.H. and a FWF grant (P18606) to H.H. We thank Norbert Polacek for helpful discussions and reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Gen-Au grant D 110420-011-013.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 2.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 4.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′ and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick JS. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 6.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 9.Fragnet L, Kut E, Rasschaert D. Comparative functional study of the viral telomerase RNA based on natural mutations. J. Biol. Chem. 2005;280:23502–23515. doi: 10.1074/jbc.M501163200. [DOI] [PubMed] [Google Scholar]
- 10.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 11.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 12.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 13.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 15.Pauler FM, Barlow DP. Imprinting mechanisms—it only takes two. Genes Dev. 2006;20:1203–1206. doi: 10.1101/gad.1437306. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 17.Bagnyukova TV, Pogribny IP, Chekhun VF. MicroRNAs in normal and cancer cells: a new class of gene expression regulators. Exp. Oncol. 2006;28:263–269. [PubMed] [Google Scholar]
- 18.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinas A, Reimegard J, Wagner EG, Nellen W, Ambros VR, Soderbom F. The small RNA repertoire of Dictyostelium discoideum and its regulation by components of the RNAi pathway. Nucleic Acids Res. 2007;35:6714–26. doi: 10.1093/nar/gkm707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 21.Steigele S, Huber W, Stocsits C, Stadler PF, Nieselt K. Comparative analysis of structured RNAs in S. cerevisiae indicates a multitude of different functions. BMC Biol. 2007;5:25. doi: 10.1186/1741-7007-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latge JP. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denning DW. Invasive Aspergillosis. Clin. Infect. Dis. 1998;26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 24.Furebring M, Oberg G, Sjolin J. Side-effects of amphotericin B lipid complex (Abelcet) in the Scandinavian population. Bone Marrow Transpl. 2000;25:341–343. doi: 10.1038/sj.bmt.1702156. [DOI] [PubMed] [Google Scholar]
- 25.Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–656. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.d’Enfert C. Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet. Biol. 1997;21:163–172. [Google Scholar]
- 27.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 28.Huttenhofer A, Cavaille J, Bachellerie JP. Experimental RNomics: a global approach to identifying small nuclear RNAs and their targets in different model organisms. Methods Mol. Biol. 2004;265:409–428. doi: 10.1385/1-59259-775-0:409. [DOI] [PubMed] [Google Scholar]
- 29.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hearn VM, Mackenzie DW. Mycelial antigens from two strains of Aspergillus fumigatus: an analysis by two-dimensional immunoelectrophoresis. Mykosen. 1980;23:549–562. [PubMed] [Google Scholar]
- 31.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 32.Mah JH, Yu JH. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell. 2006;5:1585–1595. doi: 10.1128/EC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 34.Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, Bornberg-Bauer E, Robson GD, Oliver SG, Denning DW, et al. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 2004;32:D401–D405. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook JEFF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte Chemie. 1994;125:167–188. [Google Scholar]
- 39.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 40.Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 41.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN, Jr, Haynes K, Haas H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 2004;200:1213–1219. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krappmann S, Braus GH. Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med. Mycol. 2005;43(Suppl. 1):S31–S40. doi: 10.1080/13693780400024271. [DOI] [PubMed] [Google Scholar]
- 43.Yu JH, Mah JH, Seo JA. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell. 2006;5:1577–1584. doi: 10.1128/EC.00193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofacker IL. RNA consensus structure prediction with RNAalifold. Methods Mol. Biol. 2007;395:527–544. doi: 10.1007/978-1-59745-514-5_33. [DOI] [PubMed] [Google Scholar]
- 47.Mitrovich QM, Guthrie C. Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA. 2007;13:2066–2080. doi: 10.1261/rna.766607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 49.Gaspin C, Cavaille J, Erauso G, Bachellerie JP. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 50.Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 51.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A, et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl Acad. Sci. USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 53.Hertel J, Hofacker IL, Stadler PF. SnoReport: Computational identification of snoRNAs with unknown targets. Bioinformatics. 2007;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 54.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 55.Omer AD, Zago M, Chang A, Dennis PP. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA. 2006;12:1708–1720. doi: 10.1261/rna.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aspegren A, Hinas A, Larsson P, Larsson A, Soderbom F. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res. 2004;32:4646–4656. doi: 10.1093/nar/gkh804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CL, Liang D, Zhou H, Zhuo M, Chen YQ, Qu LH. The high diversity of snoRNAs in plants: identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003;31:2601–2613. doi: 10.1093/nar/gkg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galardi S, Fatica A, Bachi A, Scaloni A, Presutti C, Bozzoni I. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 2002;22:6663–6668. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qu LH, Henras A, Lu YJ, Zhou H, Zhou WX, Zhu YQ, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie JP. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell. Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 61.Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA. 2005;11:1303–1316. doi: 10.1261/rna.2380905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang D, Zhou H, Zhang P, Chen YQ, Chen X, Chen CL, Qu LH. A novel gene organization: intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res. 2002;30:3262–3272. doi: 10.1093/nar/gkf426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 64.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thön M, Kniemeyer O, Abt B, Seeber B, Werner ER, et al. Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron. EMBO J. 2007;26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huttenhofer A, Schattner P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nat. Rev. Genet. 2006;7:475–482. doi: 10.1038/nrg1855. [DOI] [PubMed] [Google Scholar]
- 67.Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu. Rev. Cell Dev. Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 68.Bainbridge BW. Macromolecular composition and nuclear division during spore germination in Aspergillus nidulans. J. Gen. Microbiol. 1971;66:319–325. doi: 10.1099/00221287-66-3-319. [DOI] [PubMed] [Google Scholar]
- 69.Cashel M, Gentry D, Hernandez VJ, Vinella D. Am. Soc. Microbiol. 1996. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. [Google Scholar]
- 70.van Delden C, Comte R, Bally AM. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 72.Kaufmann G. Anticodon nucleases. Trends Biochem. Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- 73.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 74.Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 75.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]