Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres (original) (raw)
Abstract
Point and regional centromeres specify a unique site on each chromosome for kinetochore assembly. The point centromere in budding yeast is a unique 150-bp DNA sequence, which supports a kinetochore with only one microtubule attachment. In contrast, regional centromeres are complex in architecture, can be up to 5 Mb in length, and typically support many kinetochore-microtubule attachments. We used quantitative fluorescence microscopy to count the number of core structural kinetochore protein complexes at the regional centromeres in fission yeast and Candida albicans. We find that the number of CENP-A nucleosomes at these centromeres reflects the number of kinetochore-microtubule attachments instead of their length. The numbers of kinetochore protein complexes per microtubule attachment are nearly identical to the numbers in a budding yeast kinetochore. These findings reveal that kinetochores with multiple microtubule attachments are mainly built by repeating a conserved structural subunit that is equivalent to a single microtubule attachment site.
Introduction
The kinetochore is a complex protein machine assembled at the centromere on each chromosome. Its primary function is to attach centromeric chromatin with the plus ends of one or more spindle microtubules in an end-on manner. Structure of the kinetochore is built from >40 different proteins, many of which are assembled into protein subcomplexes. Most of the kinetochore proteins are conserved in all eukaryotes (Meraldi et al., 2006). Biochemical and structural studies, as well as epistasis analyses, have produced much information about the structural and functional hierarchy among various protein complexes within the kinetochore (Westermann et al., 2007; Cheeseman and Desai, 2008). An important question regarding the architecture of a kinetochore with many microtubule attachments is whether it is built by repeating a structural subunit (“the repeat subunit” model; Zinkowski et al., 1991) or if it lacks an ordered structure (Dong et al., 2007). Accurate counts of the number of copies of structural kinetochore proteins for such a kinetochore can provide key information for understanding the overall organization of the kinetochore and the architecture of microtubule attachment sites.
The copy number for each of the eight structural proteins and protein complexes per kinetochore in budding yeast has been accurately determined using quantitative fluorescence microscopy (Joglekar et al., 2006). This study found that the centromeric nucleosome containing two molecules of ScCse4p (HsCENP-A) is linked to the microtubule plus end by a protein linkage that consists of 1–2 copies of Mif2p (HsCENP-C), 2–3 copies of the COMA complex (CENP-ANAC/CENP-ACAD CENP-A nucleosome-associated complex and nucleosome-associated distal complex), 5 copies of Spc105p (HsKNL-1/Blinkin), 6–7 copies of the MIND complex (HsMis12 complex), 8 copies of the NDC80 complex (HsNDC80/Hec1), and 16–20 copies of the Dam1–DASH complex. In budding yeast, the centromeric DNA is wrapped around one CENP-A nucleosome, which in turn supports one microtubule attachment (Furuyama and Biggins, 2007). In contrast, regional centromeres in higher eukaryotes span over large stretches of DNA and typically support multiple microtubule attachments. These characteristics of regional centromeres raise the possibility that the protein architecture of the associated kinetochore may also be distinctly different from the protein architecture of the budding yeast kinetochore that is built on a point centromere.
There are two critical aspects regarding the protein architecture of kinetochores and the chromatin architecture of regional centromeres that must be characterized to understand their overall organization. The first key question is the number of CENP-A nucleosomes in a regional centromere and its relationship with centromere length and the number of microtubule attachments. Chromatin immunoprecipitation assays on the regional centromeres in fission yeast and in Candida albicans (a multimorphic yeast) show that CENP-A is distributed over a 10–12-kb region of the fission yeast centromere (Partridge et al., 2000) and a 3–4-kb region of the C. albicans centromere (Sanyal and Carbon, 2002; Mishra et al., 2007). Biochemical assays in HeLa cells also estimate a full occupancy of the centromeric DNA with 15,000 CENP-A nucleosomes in 2.5 megabases of DNA (Black et al., 2007), although this number is likely lower, given that CENP-A nucleosomes are interspersed with canonical nucleosomes containing the conventional histone H3 (Blower et al., 2002; Sullivan and Karpen, 2004). The relationship of the number of CENP-A nucleosomes with the number of microtubule attachments is especially important for understanding the architecture of the centromeric chromatin that immediately surrounds a microtubule attachment at the regional centromere (Yeh et al., 2008). The second key question is how the numbers of structural kinetochore proteins at a regional centromere relate to the number of microtubule attachments. In this study, we address both these issues for regional centromeres in fission yeast (Schizosaccharomyces pombe) and in the multimorphic yeast C. albicans.
Fission yeast and C. albicans both offer important advantages for counting the number of kinetochore proteins using the fluorescence ratio method that we developed for studying the budding yeast kinetochore. The most critical advantage is the ability to replace the endogenous copy of a gene with a GFP-tagged version through homologous recombination. This technique ensures that the GFP-tagged protein is expressed under the control of the endogenous promoter. Because of the use of the same GFP allele (EGFP) and similar growing conditions, GFP fluorescence can be expected to be equivalent in all three fungi. Geometry of the mitotic spindle in these two fungi is also well suited for accurate quantification of the fluorescence signal from GFP-tagged kinetochore proteins. Fission yeast is a haploid organism with three chromosomes. Before entry into mitosis, the three pairs of sister kinetochores on the replicated chromosomes remain bound to the spindle pole body, forming a tight cluster (Fig. 1, bottom). Microtubules are nucleated from the nuclear side of the spindle pole body after the onset of mitosis, and each kinetochore establishes two to four microtubule attachments (Ding et al., 1993). The regional centromeres in fission yeast span a 40–100-kb-long region. Each centromere consists of a central core element that is flanked by two sets of inverted repeats known as inner and outer repeats. C. albicans is diploid, with eight chromosomes. Spindle morphology in cells expressing Tub1p-GFP appears similar to the budding yeast spindle visualized by labeling Tub1p-GFP (Maddox et al., 2000; Finley and Berman, 2005). C. albicans centromeres span over a 4–18-kb-long DNA sequence (Sanyal et al., 2004; Mishra et al., 2007). In this paper, we characterize the protein architecture of the kinetochores built on regional centromeres in fission yeast and C. albicans by determining the numbers of structural kinetochore protein complexes per centromere using quantitative in vivo fluorescence microscopy.
Figure 1.
Pseudo-colored images for a comparison of kinetochore protein fluorescence in metaphase budding yeast and G2/M fission yeast cells. Images were obtained under identical imaging and image acquisition conditions. Each of the two spots in a metaphase budding yeast cell contains 16 kinetochores and 16 microtubule attachment sites. In G2/M fission yeast, there are six kinetochores in the single spot with a mean of ∼3 microtubule attachment sites per kinetochore and, therefore, 18 attachment sites in total. It should be noted that microtubule attachments are established only after G2/M transition. The intensity bar is color coded to represent the ratio (indicated by the numbers on the right) with the ScCse4p-GFP intensity in the top panel. Bar, 1 μm.
Results and discussion
Fission yeast and C. albicans strains were constructed by tagging the C terminus of the genomic copy of kinetochore proteins of interest with GFP. The fluorescence standard for our ratio method of fluorescence signal measurement is the mean signal from separated anaphase kinetochore clusters in budding yeast cells expressing ScCse4p-GFP. Such a kinetochore cluster contains 16 kinetochores. There are two molecules of ScCse4p per kinetochore and, thus, a total of 32 GFP molecules within each. The mean fluorescence signal from other GFP-tagged kinetochore proteins in fission yeast or C. albicans can be converted into the corresponding number of molecules from the ratio of its mean fluorescence signal to that of ScCse4p-GFP. In each experiment, fission yeast or C. albicans cells were mixed with budding yeast cells expressing ScCse4p-GFP, and the mean fluorescence signal for a kinetochore cluster was obtained for both strains (see Materials and methods). For fission yeast, fluorescence signal measurements were obtained for cells in G2/M and anaphase, whereas fluorescence signal was determined only for anaphase C. albicans cells.
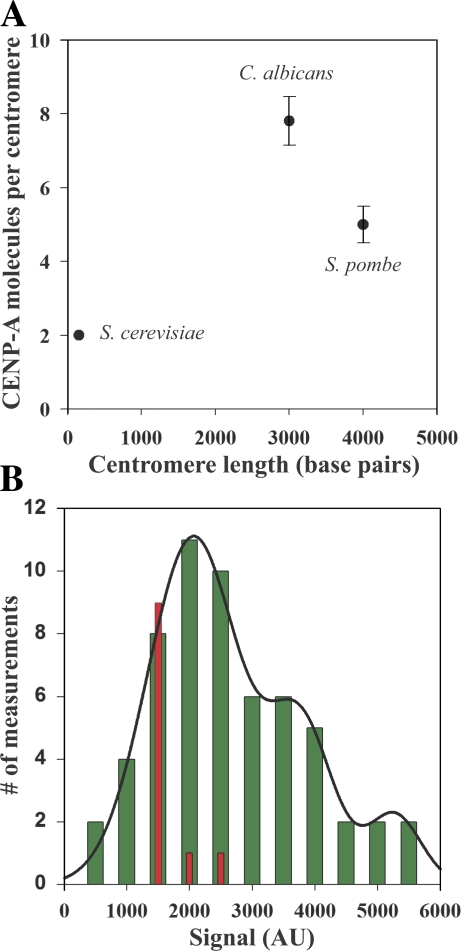
In fission yeast, we measured 5 ± 1 SpCnp1p (CENP-A) molecules per centromere on average (Fig. 2 A). This number translates into two to three CENP-A containing nucleosomes per centromere, and it is similar to the number of microtubule attachments per centromere (two to four) in fission yeast (Ding et al., 1993). Because C. albicans is diploid, we constructed four different strains that are homozygous, heterozygous, and hemizygous for CaCse4p-GFP. By quantifying the fluorescence signal from C. albicans cells homozygous for CaCse4p-GFP, we found that the C. albicans centromere contains 8 ± 1 molecules of CaCse4p (Fig. 2 A). This number translates into four CaCse4p-containing nucleosomes per centromere (Bloom et al., 1984; Saunders et al., 1990; Blower et al., 2002; Bouck and Bloom, 2005, 2007; Furuyama and Biggins, 2007).
Figure 2.
The number of CENP-A molecules does not scale with the centromere DNA length. (A) The number of CENP-A molecules in the three fungi plotted as a function of centromere length. The number of CENP-A nucleosomes per centromere reflects the number of microtubule attachments per centromere. Error bars represent SD. (B) Signal measurement from clusters of one or more sister kinetochores in metaphase-arrested cells expressing SpNdc80p-GFP cells and a cold-sensitive nda3cs (β-tubulin) allele (green bars). A multipeak normal curve fit to the histogram (solid line) predicts peaks at 2,000, 3,800, and 5,600 intensity counts. Measurement of fluorescence signal from single lagging kinetochores in anaphase wild-type fission yeast expressing SpNuf2p-GFP yields a mean signal value of ∼1,000 counts (red bars). This confirms that the unit of minimum fluorescence signal in the nda3cs experiment corresponds to on pair of sister kinetochores. AU, arbitrary units.
We verified that the fluorescence signal measured from the single spot seen in fission yeast cells in G2/M corresponds to the cumulative signal from three pairs of sister kinetochores. A fission yeast strain expressing Ndc80-GFP and containing a cold-sensitive allele of β-tubulin (nda3cs) was used for this purpose. When these cells are maintained at the restrictive temperature, all microtubules depolymerize. Cells arrest in mitosis with replicated chromosomes scattered in the nucleus (Grishchuk and McIntosh, 2006), which makes it possible to image centromeres of individual chromosomes. By measuring the fluorescence signal from all the fluorescent spots imaged in such cells, we found that the minimum unit of fluorescence corresponds to one pair of sister kinetochores (Fig. 2 B, green bars). Measurement of fluorescence signal from lagging kinetochores in early anaphase cells expressing SpNuf2p-GFP showed that this signal was equivalent to the fluorescence signal for a single kinetochore (Fig. 2 B, red bars).
We selected a representative kinetochore protein from each of the core structural protein complexes for quantification of their number at the fission yeast centromere (Fig. 3 and Table S1). We measured three to four molecules of SpMif2p (CENP-C) per kinetochore. We found 12 copies of the proteins SpFta3p and SpSim4p from the fission yeast Sim4 complex (Liu et al., 2005). These proteins represent members of the CENP-ANAC (CENP-H and CENP-K, respectively; Foltz et al., 2006; Okada et al., 2006). Other members from this complex, SpMal2p and SpFta2p, representing CENP-ACAD proteins (Foltz et al., 2006; Okada et al., 2006), were found to be present in a slightly lower number (nine per kinetochore). SpFta1p was present in a much smaller number, which is consistent with biochemical evidence (Liu et al., 2005). Members of the KMN complex were present in higher numbers (16 copies of SpMis12p, 12 copies of SpSpc7p, and 21 copies of SpNdc80p per kinetochore; Liu et al., 2005). We quantified the copy number for three different members of the Dam1–DASH complex, namely Dam1p, Dad1p, and Ask1p. Previous studies have shown that only SpDad1p is a constitutive kinetochore protein present at the centromeres before entry into mitosis. Other components of the Dam1–DASH complex are recruited at the kinetochore only after the nucleation of intranuclear microtubules at the onset of mitosis (Liu et al., 2005; Sanchez-Perez et al., 2005). Surprisingly, we found that there are only six or seven copies of the Dam1–DASH complex per kinetochore in anaphase. Measurements of SpDad1p indicate that the number of copies of SpDad1p in G2 was lower than the number of copies of the Dam1–DASH complex measured in anaphase/telophase cells (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200803027/DC1).
Figure 3.
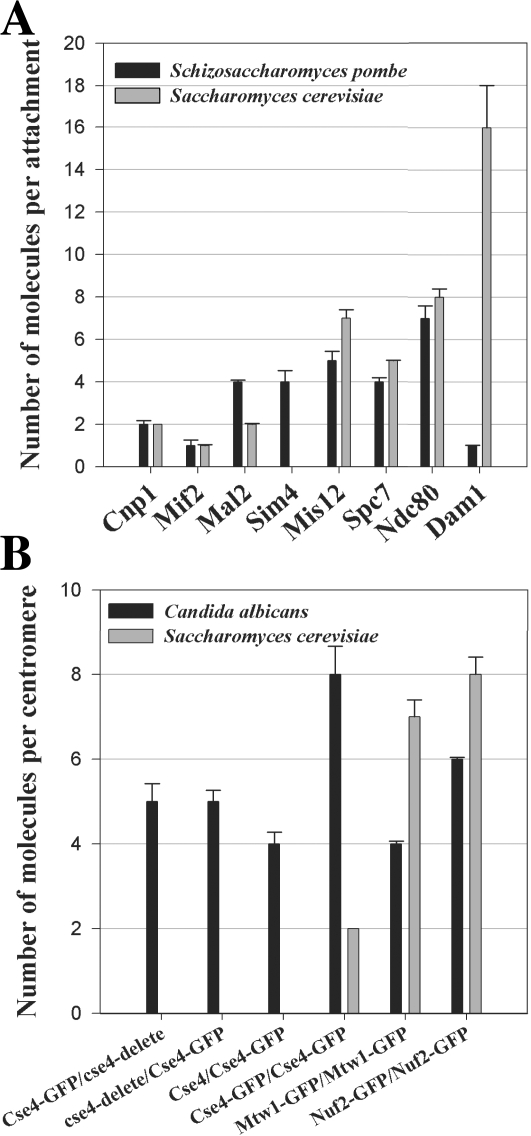
Comparison of kinetochore protein numbers in the three fungi. (A) The number of core structural kinetochore proteins per microtubule attachment in fission yeast is very similar to the number of corresponding proteins at a budding yeast kinetochore. The Dam1–DASH complex, which is nonessential in fission yeast, is the sole exception (black bars, mean protein numbers in fission yeast; gray bars, mean numbers in budding yeast). (B) Although each C. albicans centromere contains approximately four CENP-A nucleosomes on average, the mean number of outer kinetochore proteins, such as Mtw1 and Nuf2, is sufficient to build only one microtubule attachment site (black bars, mean protein numbers in C. albicans; gray bars, mean numbers in budding yeast). Error bars represent SD from a minimum of two experiments.
Because there are three microtubule attachments per fission yeast centromere on average, we converted the copy number of kinetochore proteins per centromere into the mean number of molecules per microtubule attachment by distributing the total kinetochore protein count per centromere equally among the three microtubule attachments. We found that, with one exception, the copy numbers of structural kinetochore proteins per microtubule attachment are very similar to the protein numbers measured at the budding yeast kinetochore, which has only one microtubule attachment (Fig. 3 A). The exception is the Dam1–DASH complex. We measured six to seven copies of Dam1–DASH complex per fission yeast kinetochore. This number is much smaller than the 16–23 copies necessary to form rings around the attached microtubules (Miranda et al., 2005; Westermann et al., 2005). The Dam1–DASH complex is not essential in fission yeast (Sanchez-Perez et al., 2005). It is, however, synthetically lethal with the plus-end microtubule depolymerases Klp5 and 6 and the microtubule-associated protein Dis1p. We verified that the number of Dam1–DASH subunits per centromere does not show a significant change in strains that have either Klp5 or 6 knocked out (unpublished data). Thus, the concentration of the Dam1–DASH complex at the kinetochores is not suppressed by these proteins in wild-type cells. The low number of Dam1–DASH complex per centromere could be indicative of altered protein architecture of the microtubule attachment site at the fission yeast kinetochore. This is unlikely, however, because the numbers for the rest of the structural kinetochore proteins per microtubule attachment are nearly identical in both budding yeast and fission yeast.
The NDC80 complex is most proximal to the microtubule plus end within the kinetochore because members of this protein complex, Nuf2p and Ndc80p, can directly bind microtubules (Cheeseman et al., 2006). Therefore, we decided to count the number of copies of the NDC80 complex at the centromere in C. albicans to understand the protein architecture of the kinetochore. The number of NDC80 complex molecules was 7–8 times the number of CENP-A nucleosomes in budding as well as in fission yeast. Because we measured four CENP-A nucleosomes per centromere in C. albicans, the expected number of NDC80 molecules would be ∼32 if all four CENP-A nucleosomes support a microtubule attachment site and if each microtubule attachment site in C. albicans recruits seven to eight copies of the NDC80 complex. Unexpectedly, our measurements yielded only six molecules of CaNuf2p per centromere (Fig. 3 B). Similarly, we measured only four molecules of CaMtw1p (Mis12 complex) per centromere (Fig. 3 B). These numbers suggest that the C. albicans centromere likely supports only one kinetochore microtubule attachment site, although it recruits a mean of four CENP-A. Interestingly, the number of CENP-A molecules per centromere reduces to five in hemizygotic C. albicans strains (CSE4:GFP/cse4Δ), which is sufficient to form only two CENP-A nucleosomes (Fig. 3 B). The copy number for the NDC80 complex in this hemizygotic strain did not change (unpublished data). Together with the number of NDC80 complex copies per centromere, these data indicate that the C. albicans centromere likely bears only one microtubule attachment site, although it normally contains four CENP-A nucleosomes.
The number of microtubule attachments per C. albicans centromere has not been determined. To experimentally verify that there is only one microtubule attachment site per C. albicans centromere, we devised a strategy for estimating the number of microtubules in the C. albicans spindle by quantifying the fluorescence signal from individual cytoplasmic microtubules and the cumulative signal from all the microtubules in a metaphase half-spindle in cells expressing Tub1p-GFP (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200803027/DC1). This method takes the ratio of the mean fluorescence signal from single cytoplasmic microtubules to the maximum fluorescence signal from a spindle half as an estimate of the maximum number of microtubules in a half-spindle. For budding yeast, this method estimates 24 ± 5 microtubules per half-spindle. This measurement confirms the known number of microtubules in each budding yeast half-spindle (16 kinetochore microtubule and 4 microtubules from each spindle pole to construct a central spindle with 8 interpolar microtubules; Winey et al., 1995). Similar measurements on the C. albicans spindle showed that there are 18 ± 3 microtubules in each half-spindle. This number is sufficient to form a mean of only one microtubule attachment per kinetochore on each of the sixteen chromosomes, with two additional interpolar microtubules. This data further supports the prediction that the regional centromere in C. albicans has only one microtubule attachment.
The excess of three CENP-A nucleosomes over the one needed for a single microtubule attachment at the C. albicans centromere prompted us to test the role of CENP-A in controlling the number of microtubule attachment sites at the kinetochore. We examined a fission yeast strain with a four- to fivefold constitutive overexpression of CENP-A (Takahashi et al., 2000), which leads to a similar increase in the number of CENP-A molecules at the centromere (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200803027/DC1). Overexpression of mutant alleles of CENP-A does not mislocalize other kinetochore proteins in budding yeast (Collins et al., 2004), although it has been shown to mislocalize CENP-C in vertebrate cells (Van Hooser et al., 2001). A 30-fold overexpression of Drosophila melanogaster CENP-A homologue leads to the recruitment of kinetochore proteins at ectopic sites and massive chromosome missegregation (Heun et al., 2006). We counted the copy numbers of kinetochore proteins in the fission yeast strain with constitutive CENP-A overexpression and compared them with the wild-type copy numbers. We found that the copy numbers of structural kinetochore proteins remained comparable to wild-type protein numbers (Fig. S2). These results show that although CENP-A is a necessary factor for recruiting kinetochore components at the centromere, its fivefold overexpression does not affect the numbers of kinetochore proteins recruited to the centromere.
We verified that the structural kinetochore protein complexes are stably associated with the kinetochore by measuring the fluorescence recovery after photobleaching of GFP-tagged kinetochore proteins. None of the GFP-tagged kinetochore proteins tested (SpCnp1p, SpMis12p, SpSpc7p, and SpNdc80p) showed significant recovery in G2, when there are no microtubule attachments. Interestingly, we found the highest recovery percentage for the outermost kinetochore protein complex NDC80, which exhibited only 10% recovery over 5 min (10.8 ± 5.5%; mean ± SD; n = 5). SpCnp1p showed no detectable recovery (1.6 ± 1.7%; n = 5; measured in the strain with fivefold overexpression). Microtubule attachment is likely to suppress this turnover further to undetectable levels, as found previously for kinetochores in metaphase budding yeast cells (Joglekar et al., 2006). These results show that the core protein architecture of the kinetochore-microtubule attachment site in fission yeast is stable in G2/M.
Accurate quantification of the number of copies of structural kinetochore protein complexes at regional centromeres elucidates two key aspects of the molecular architecture of the kinetochore. First, the number of CENP-A molecules at the regional centromeres in fungi does not scale with the length of the centromeric DNA. Based on the exact correspondence between the number of CENP-A nucleosomes and the number of microtubule attachment sites in budding yeast and fission yeast, we hypothesize that each microtubule attachment site is normally built around one CENP-A nucleosome. In budding yeast, the pericentric chromatin region ∼25 kb long was recently shown to form a loop with the CENP-A nucleosome and the microtubule attachment site residing at the apex of the loop (Yeh et al., 2008). The existence of one CENP-A nucleosome per microtubule attachment raises the possibility that similar chromatin architecture may also be present within regional centromeres. The number of CENP-A nucleosomes alone is not sufficient to decide the number of microtubule attachments. This conclusion is based on the observation that the number of CENP-A nucleosomes can be larger than the number of microtubule attachment sites in these fungi. Wild-type C. albicans centromeres have CENP-A nucleosomes in excess of the number of microtubule attachments. Similarly, a fission yeast strain that has a four- to fivefold overexpression of CENP-A does not recruit any more kinetochore proteins than wild-type centromeres. Experiments in vertebrate cells similarly show that the CENP-A levels must be depleted by >90% before a noticeable decrease was observed in the levels of outer kinetochore proteins (Liu et al., 2006). Thus, although a CENP-A nucleosome is necessary for building a microtubule attachment site, it is not sufficient. Factors other than CENP-A, and possibly the architecture of centromeric chromatin, play a role in regulating the recruitment of kinetochore proteins and the assembly of microtubule attachment sites (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007). The second key finding of this study is that the molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromere. The number of copies of kinetochore proteins and protein complexes per microtubule attachment at the fission yeast kinetochore is nearly identical to corresponding numbers at the budding yeast kinetochore that has only one microtubule attachment. Although high resolution electron tomography has failed to find any visual evidence of structural subunits within the vertebrate kinetochore (Dong et al., 2007), our data demonstrate that each microtubule attachment site at a kinetochore with many microtubule attachments is built from a specific number of copies of each of the structural protein complexes.
The results presented in this study provide compelling evidence that the complex kinetochores at the regional centromeres in fission yeast and C. albicans are built by repeating a structural subunit, which corresponds to a single microtubule attachment site. Furthermore, the protein architecture of this microtubule attachment site is conserved between point and regional centromeres. Despite more complex centromere and kinetochore architectures in metazoa, most of the structural kinetochore proteins are conserved in all eukaryotes, from yeast to humans. The remarkable conservation of kinetochore protein architecture, despite highly divergent centromeric architecture in the three fungal systems investigated in this study, leads us to believe that the subunit structure of individual microtubule attachment sites is likely conserved in higher eukaryotes.
Materials and methods
Strains and media
All the strains (listed in Table I) were grown in YPD at 32°C (with the exception of the budding yeast strain expressing ScCse4p-GFP, which was grown at 25°C). YE5S, YPD, and YPD supplemented with 50 μg/ml of uridine (Sigma-Aldrich) were used to grow fission yeast, budding yeast, and C. albicans, respectively. GFP fusions were made by PCR amplification of a GFP–KANr cassette (from pFA6a–GFP(S65T) KANr MX6) flanked with 60 bp of homology to the site of integration at the 3′ end of the gene. The fission yeast strains with constitutive Cnp1p and Cnp1p-GFP overexpression were supplied by K. Takahashi and M. Yanagida (National BioResource Project, Okinawa Institute of Science and Technology, Okinawa, Japan).
Table I. List of strains used in this study.
| Genotype | Source |
|---|---|
| KBY7006: S. cerevisiae 473a CSE4-GFP:KAN | KB |
| YWY277: S. pombe h− Cnp1-GFP-KanMX6, ade6-m12/0, leu1-32, ura4D | XH |
| XHE255: S. pombe native promoter Cnp1-6GFP(lys1+) h− | XH |
| XL403: S. pombe NDC80:GFP | XH |
| XL101: S. Pombe MAL2:GFP | XH |
| XL174: S. pombe h−, mif2-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL103: S. pombe h−, spc7-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL358: S. pombe h+, mis12-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL017: S. pombe h−, ask1-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL101: S. pombe h−, mal2-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL099: S. pombe h−, sim4-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL059: S. pombe h−, fta1-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL069: S. pombe h−, fta2-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL067: S. pombe h−, fta3-GFP-KanMx6, ade6-m210, leu1-32, ura4D | XH |
| XL097: S. pombe h−, Dam1-GFP-KanMX6, ade6-m210, leu1-32, ura4D | XH |
| XL127: S. pombe h+, Dad1-GFP-KanMX6, ade6-m210, leu1-32, ura4D, DAD1 | XH |
| XL071: S. pombe h−, Dad2-GFP-KanMX6, ade6-m210, leu1-32, ura4D | XH |
| XL332: S. pombe h−, Nuf2-GFP-KanMX6, ade6-m210, leu1-32, ura4D | XH |
| XL462: S. pombe ask1-GFP-KANMX6, klp5∷ura4, leu1-32, ura4-D18, ade6- | XH |
| XL464: S. pombe ask1-GFP-KANMX6, klp6∷ura4, leu1-32, ura4-D18 | XH |
| XL468: S. pombe ndc80-GFP-KANMX6, nda3cs, ade6-m210, leu1-32, ura4- | XH |
| XL487: S. pombe h−, fta3-GFP-KANMX6, ade6-m210, leu1-32, ura4D, native promoter-cnp1+-3xHA6xHis[lys1+] | XH |
| XL489: S. pombe h−, mal2-GFP-KANMX6, ade6-m210, leu1-32, ura4D, native promoter-cnp1+-3xHA6xHis[lys1+] | XH |
| XL491: S. pombe h−, mis12-GFP-KANMX6, ade6-m210, leu1-32, ura4D, native promoter-cnp1+-3xHA6xHis[lys1+] | XH |
| XL493: S. pombe h−, ndc80-GFP-KANMX6, ade6-m210, leu1-32, ura4D, native promoter-cnp1+-3xHA6xHis[lys1+] | XH |
| XL 497: S. pombe mif2-GFP-KANMX6, ade6-m210, leu1-32, ura4D, native promoter cnp1+-3xHA-6xHis [lys1+] | XH |
| S. pombe XL 506: mis6GFP[leu+], native promoter-cnp1+-3xHA-6xHis [lys1+] | XH |
| CAJS1-1: C. albicans arg4∷hisG/ arg4∷hisG his1∷hisG/ his1∷hisG ura3Δ∷λimm434/ ura3Δ∷λimm434 CSE4:GFP:CSE4/cse4Δ∷hisG∷URA3∷hisG | JB |
| 10118: C. albicans ura3Δ∷λimm434/ura3Δimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG cse4∷dpl200-URA3/CSE4:GFP:CSE4 | JB |
| 8675: C. albicans ura3Δ∷λimm434/ura3Δimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG CSE4/CSE4:GFP:CSE4 | JB |
| 8676: C. albicans ura3Δ∷λimm434/ura3Δimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG CSE4/CSE4:GFP:CSE4 | JB |
| 10116: C. albicans ura3Δ∷_imm434/ura3Δimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG CSE4:GFP:CSE4/CSE4:GFP:CSE4 | JB |
| 10418: C. albicans ura3∷imm434/ura3∷imm434, Nuf2-GFP-URA3/Nuf2-GFP-NAT | JB |
| 10702 ura3Δ∷λimm434/ura3Δ∷λimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG MTW1-GFP-URA3/MTW1-GFP-NAT1 | JB |
| 10635 ura3∷imm434/ura3∷imm434, Nuf2-GFP:NAT1/Nuf2-GFP/NAT1, CSE4/cse4∷URA3 | JB |
| YMG5629: C. albicans ura3Δ∷λimm434/ura3Δimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG TUB1/TUB1:GFP-URA3 | JB |
Microscopy
An inverted microscope (TE-2000U; Nikon) with a 100× 1.4 NA differential interference contrast objective (Nikon) was used for imaging cells at 25°C. Cells suspended in filter sterile SD complete media were immobilized on coverslips coated with concanavaline A (Sigma-Aldrich) for imaging. A standard HQ EGFP long pass filter set (Chroma Technology Corp.) was used for fluorescence imaging. Images were acquired with a cooled charge-coupled device camera (Orca II ER; Hamamatsu Photonics) with 2 × 2 binning (1 pixel, ∼133 nm). The microscope shutters and the camera were operated by Metamorph 6.1 (MDS Analytical Technologies). A stack of 13 images was obtained for each chosen field (300 × 300 camera pixels in the center of the field to minimize excitation intensity irregularities), with an exposure time of 400 ms and a 200-nm axial separation between successive images in the stack.
Data analysis
Image analysis was performed with a custom-written graphical user interface in MATLAB (MathWorks) in the in-focus image plane for each kinetochore cluster (this plane also contains the pixel with maximum intensity value). Measurements of fluorescence signal from kinetochore clusters in fission yeast cells in G2/M were performed by placing a 6 × 6 pixel box on the signal region. It was placed so that the central 2 × 2 pixel region in the box had the maximum cumulative signal. For fission yeast and C. albicans cells in anaphase, a 5 × 5 pixel box was used to measure the fluorescence signal.
The number of pixels for signal measurement for GFP-tagged kinetochore proteins was determined by fitting 1-D Gaussian curves to line scans through kinetochore clusters in the in-focus plane (the maximum intensity plane), with the standard deviation of the Gaussian as a free parameter. Fluorescence signal was measured over an area corresponding to the 4× (SD) around the maxima. For fission yeast cells expressing Ndc80p-GFP, this fitting procedure yielded a mean spot size of 600 ± 60 nm for anaphase cells and 700 ± 44 nm for G2 cells corresponding to an area defined by a 5 × 5 and 6 × 6 pixel square, respectively, in the acquired images. Measurements from C. albicans cells expressing Cse4p-GFP in anaphase prompted the use of a 5 × 5 pixel square for signal measurement. Background correction was applied by measuring the background from a larger square region (8 × 8 pixels for fission yeast and 7 × 7 pixels for budding yeast and C. albicans) that is concentric with the signal region, following the scheme detailed in Hoffman et al. (2001).
Online supplemental material
Fig. S1 shows quantification of the number of microtubules in the budding yeast and C. albicans spindles. Fig. S2 shows that a four- to fivefold excess of CENP-A nucleosomes does not alter the number of kinetochore proteins at the centromere in fission yeast. Table S1 contains a summary of fluorescence signal measurements for fission yeast. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803027/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
The authors would like to thank Dr. Mary Baum and Nathaniel H.B. Thaler for critical reading of the manuscript and technical assistance and the National BioResource Project (Japan), Kohta Takahashi, and Mitsuhiro Yanagida for providing fission yeast strains.
A.P. Joglekar holds a Career award at the Scientific Interface from the Burroughs-Wellcome Fund. K. Finley was supported by Biotechnology training grant 5T32GM008347. This work was supported by National Institutes of Health grant GM32238 to K.S. Bloom, National Institutes of Health/National Institute of General Medical Sciences grant GM068676 to X. He, National Institutes of Health grant AI062427 to J. Berman, and National Institutes of Health grant GM24364 to E.D. Salmon.
References
- Black, B.E., L.E. Jansen, P.S. Maddox, D.R. Foltz, A.B. Desai, J.V. Shah, and D.W. Cleveland. 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 25:309–322. [DOI] [PubMed] [Google Scholar]
- Bloom, K.S., E. Amaya, J. Carbon, L. Clarke, A. Hill, and E. Yeh. 1984. Chromatin conformation of yeast centromeres. J. Cell Biol. 99:1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., B.A. Sullivan, and G.H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck, D.C., and K.S. Bloom. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 102:5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck, D.C., and K. Bloom. 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort, R., B. Li, L. Florens, S.K. Swanson, M.P. Washburn, and J.L. Gerton. 2007. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 26:853–865. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., and A. Desai. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., J.S. Chappie, E.M. Wilson-Kubalek, and A. Desai. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. [DOI] [PubMed] [Google Scholar]
- Collins, K.A., S. Furuyama, and S. Biggins. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14:1968–1972. [DOI] [PubMed] [Google Scholar]
- Ding, R., K.L. McDonald, and J.R. McIntosh. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., K.J. Vanden Beldt, X. Meng, A. Khodjakov, and B.F. McEwen. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, K.R., and J. Berman. 2005. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot. Cell. 4:1697–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz, D.R., L.E. Jansen, B.E. Black, A.O. Bailey, J.R. Yates III, and D.W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Furuyama, S., and S. Biggins. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 104:14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, E.L., and J.R. McIntosh. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25:4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun, P., S. Erhardt, M.D. Blower, S. Weiss, A.D. Skora, and G.H. Karpen. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 10:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, D.B., C.G. Pearson, T.J. Yen, B.J. Howell, and E.D. Salmon. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., D.C. Bouck, J.N. Molk, K.S. Bloom, and E.D. Salmon. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.T., J.B. Rattner, S.A. Jablonski, and T.J. Yen. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., I. McLeod, S. Anderson, J.R. Yates III, and X. He. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P.S., K.S. Bloom, and E.D. Salmon. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., A.D. McAinsh, E. Rheinbay, and P.K. Sorger. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, J.J., P. De Wulf, P.K. Sorger, and S.C. Harrison. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143. [DOI] [PubMed] [Google Scholar]
- Mishra, P.K., M. Baum, and J. Carbon. 2007. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol. Genet. Genomics. 278:455–465. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., H. Xiao, J. Wisniewski, M.M. Smith, and C. Wu. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- Okada, M., I.M. Cheeseman, T. Hori, K. Okawa, I.X. McLeod, J.R. Yates III, A. Desai, and T. Fukagawa. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Partridge, J.F., B. Borgstrom, and R.C. Allshire. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez, I., S.J. Renwick, K. Crawley, I. Karig, V. Buck, J.C. Meadows, A. Franco-Sanchez, U. Fleig, T. Toda, and J.B. Millar. 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24:2931–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, K., and J. Carbon. 2002. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. USA. 99:12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, K., M. Baum, and J. Carbon. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA. 101:11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, M.J., E. Yeh, M. Grunstein, and K. Bloom. 1990. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol. 10:5721–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K. Rogers, S. Weitze, L. Morey, M. Fitzgerald-Hayes, and R.E. Baker. 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA. 104:10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., and G.H. Karpen. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., E.S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A.A., I.I. Ouspenski, H.C. Gregson, D.A. Starr, T.J. Yen, M.L. Goldberg, K. Yokomori, W.C. Earnshaw, K.F. Sullivan, and B.R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529–3542. [DOI] [PubMed] [Google Scholar]
- Westermann, S., A. Avila-Sakar, H.W. Wang, H. Niederstrasser, J. Wong, D.G. Drubin, E. Nogales, and G. Barnes. 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290. [DOI] [PubMed] [Google Scholar]
- Westermann, S., D.G. Drubin, and G. Barnes. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591. [DOI] [PubMed] [Google Scholar]
- Winey, M., C.L. Mamay, E.T. O'Toole, D.N. Mastronarde, T.H. Giddings Jr., K.L. McDonald, and J.R. McIntosh. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., J. Haase, L.V. Paliulis, A. Joglekar, L. Bond, D. Bouck, E.D. Salmon, and K.S. Bloom. 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski, R.P., J. Meyne, and B.R. Brinkley. 1991. The centromere–kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]