Eukaryotic Wobble Uridine Modifications Promote a Functionally Redundant Decoding System (original) (raw)
Abstract
The translational decoding properties of tRNAs are modulated by naturally occurring modifications of their nucleosides. Uridines located at the wobble position (nucleoside 34 [U34]) in eukaryotic cytoplasmic tRNAs often harbor a 5-methoxycarbonylmethyl (mcm5) or a 5-carbamoylmethyl (ncm5) side chain and sometimes an additional 2-thio (s2) or 2′-_O_-methyl group. Although a variety of models explaining the role of these modifications have been put forth, their in vivo functions have not been defined. In this study, we utilized recently characterized modification-deficient Saccharomyces cerevisiae cells to test the wobble rules in vivo. We show that mcm5 and ncm5 side chains promote decoding of G-ending codons and that concurrent mcm5 and s2 groups improve reading of both A- and G-ending codons. Moreover, the observation that the mcm5U34- and some ncm5U34-containing tRNAs efficiently read G-ending codons challenges the notion that eukaryotes do not use U-G wobbling.
The universal genetic code consists of 64 triplets of which 61 represent different amino acids and 3 signal translation termination (37). The deciphering of the code led to the realization that it is degenerate; that is, most amino acids are represented by more than one codon. The presence of isoaccepting tRNAs, different tRNAs charged with the same amino acid, helped to explain the translation of the code. However, the number of tRNA species is always fewer than the 61 sense codons, suggesting that some tRNAs decode more than one triplet. This ability is due to the fact that the first base of the anticodon (position 34 of the tRNA, also called the wobble nucleoside) may pair with more than one base in the third position of the codon (11). At the time Crick presented his wobble hypothesis, it was not known that cytoplasmic tRNA almost never harbors an unmodified uridine (U) as a wobble nucleoside. Revised wobble hypotheses have since emerged to explain some unexpected results regarding the decoding properties of tRNAs with modified wobble uridines (1, 31, 47, 53, 54). These hypotheses are based mainly on translational experiments performed in vitro and by analyses of the modifications' influence on the structure of nucleosides, nucleotides, or anticodons.
The xm5U type of modified wobble nucleosides, where x represents any of several different groups and m5 stands for a methylene carbon directly bonded to the C-5 atom of the uracil moiety, can be found in organisms from all three domains of life (42, 47). These nucleosides can also harbor an additional 2-thio (xm5s2U) or a 2′-_O_-methyl group (xm5Um). It is generally accepted that tRNA species containing xm5U34, xm5s2U34, or xm5Um34 residues do not read pyrimidine-ending codons (31, 53, 54). This is supported by the fact that these nucleosides are often found in tRNA species that decode in split codon boxes where the pyrimidine- and purine-ending codons are for different amino acids. The observation that a tRNA with an unmodified U34 can, under some circumstances, read codons ending with any nucleoside indicates that the xm5U derivatives may have evolved to restrict wobbling (31, 53). However, more recent genetic and structural data have suggested that the inability to pair with the pyrimidine-ending codons may not, at least for xm5s2U34-containing tRNAs, be a direct consequence of the modifications (20, 35). The models for how xm5U derivatives affect pairing with purine-ending codons are more diverse, but the modifications are believed to allow either efficient interaction with both A and G or efficient pairing with A while simultaneously reducing pairing with G (29, 31, 35, 47, 52-54).
The lack of defined mutants in eukaryotes has led to a poor understanding of the role of the xm5U derivatives found in this domain of life. However, in vitro translation systems have revealed that cytoplasmic tRNA species harboring xm5U derivatives preferentially read codons ending with A (19, 33, 43, 50). These data, in combination with the fact that C34-containing tRNAs are frequently found in eukaryotes, suggested that they may not use U-G wobbling (38, 47). This contrasts to xm5U-derivative-containing tRNA species in organelles and prokaryotes, which often read codons ending with G. This apparent discrepancy has been proposed to be a consequence of different compositions of the xm5 groups and/or differences in the ribosomes (47). In this study, we utilized recently characterized Saccharomyces cerevisiae mutants to analyze the in vivo decoding properties of tRNAs with or without the eukaryotic xm5 side chains 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5). We also investigated the role of the s2 group in tRNAs containing a wobble mcm5s2U residue. Our results show that many cytoplasmic tRNAs harboring an xm5U derivative can read G-ending codons and that this ability is enhanced by the presence of the modifications.
MATERIALS AND METHODS
Yeast strains, media, and genetic procedures.
The sources and genotypes of yeast strains used in this study are listed in Table 1. Yeast transformation (18), media, and genetic procedures have been described previously (7). The tuc1_Δ strain (UMY3165) was constructed as previously described (3). tRNA genes were deleted by a PCR-mediated strategy (6), replacing the tRNA gene with a selectable marker. Disruptions of tRNA genes were confirmed by PCR. The tV(CAC)H and tE(CUC)I genes were deleted in strain W303-1A. These strains were backcrossed once to W303-1B, confirming the viability of the deletion strain. The tS(CGA)C, tT(CGU)K, tQ(CUG)M, tV(CAC)D, tG(CCC)D, tG(CCC)O, tE(CUC)D, tR(CCU)J, t_P(AGG)C, and tP(AGG)N genes were independently deleted in either UMY2366 or UMY3104. UMY3104 is a diploid formed between W303-1A and W303-1B. The heterozygous strains were allowed to sporulate, and the viability of deletion strains was determined from tetrads. If a tRNA species was essential, a plasmid carrying the corresponding wild-type tRNA gene was introduced into the heterozygous strain before sporulation. The ade3::hisG allele in strains derived from UMY2366 was eliminated in subsequent crosses. For the tRNA species encoded by two genes, the double mutant was obtained in tetrads from a cross between strains with a deletion of the respective tRNA gene. The _elp3_Δ (UMY2843), _tuc1_Δ (UMY3165), and _trm9_Δ (UMY3297) mutants were crossed to the appropriate tRNA deletion strains and the double/triple mutants obtained from tetrads. In instances where the lack of modification influenced the viability of a strain lacking a tRNA, a plasmid carrying the corresponding wild-type tRNA gene was utilized to generate a rescued double/triple mutant. In standard S. cerevisiae laboratory strains, the number of genes encoding a tRNA species can vary (8). Southern blot analyses of the wild-type strains confirmed that the gene copy numbers of tP(AGG) and tV(CAC) are 2 and 2, respectively.
TABLE 1.
Yeast strains used in this study
| Yeast strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 | 16 |
| W303-1B | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 | 16 |
| UMY2843 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 elp3::kanMX4 | 32 |
| UMY3297 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 trm9::kanMX4 | 32 |
| UMY3165 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tuc1::TRP1 | This study |
| UMY2893 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 SUP4 | 22 |
| UMY2916 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 SUP4 elp3::kanMX4 | 22 |
| UMY3104 | MATa/_MAT_α ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 | This study |
| UMY2366 | MATa/_MAT_α ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 his3-11,15/his3-11,15 ade2-1/ade2-1 ade3::hisG/ade3::hisG can1-100/can1-100 | 24 |
| UMY3199 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tv(cac)d::TRP1 | This study |
| UMY3284 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tv(cac)h::TRP1 | This study |
| UMY3295 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tv(cac)d::TRP1 tv(cac)h::TRP1 | This study |
| UMY3296 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tv(cac)d::TRP1 tv(cac)h::TRP1 | This study |
| UMY3333 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tv(cac)d::TRP1 tv(cac)h::TRP1 elp3::kanMX4 p1725 | This study |
| UMY2406 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 ade3::hisG can1-100 ts(cga)c::TRP1 p1280 | This study |
| UMY3126 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 ts(cga)c::TRP1 p1280 | This study |
| UMY3127 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 ts(cga)c::TRP1 elp3::kanMX4 p1280 | This study |
| UMY3128 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tt(cgu)k::TRP1 p1244 | This study |
| UMY3129 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tt(cgu)k::TRP1 p1244 | This study |
| UMY3130 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tt(cgu)k::TRP1 elp3::kanMX4 p1244 | This study |
| UMY3132 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tq(cug)m::TRP1 p1605 | This study |
| UMY3133 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tq(cug)m::TRP1 p1605 | This study |
| UMY3134 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tq(cug)m::TRP1 elp3::kanMX4 p1605 | This study |
| UMY3345 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tq(cug)m::TRP1 trm9::kanMX4 p1605 | This study |
| UMY3347 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tq(cug)m::TRP1 tuc1::TRP1 p1605 | This study |
| UMY3322 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 | This study |
| UMY3270 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)d::TRP1 | This study |
| UMY3329 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 te(cuc)d::TRP1 p1723 | This study |
| UMY3348 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 te(cuc)d::TRP1 p1723 | This study |
| UMY3350 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 te(cuc)d::TRP1 elp3::kanMX4 p1723 | This study |
| UMY3354 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 te(cuc)d::TRP1 trm9::kanMX4 p1723 | This study |
| UMY3352 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 te(cuc)i::TRP1 te(cuc)d::TRP1 tuc1::TRP1 p1723 | This study |
| UMY3112 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 ade3::hisG can1-100 tr(ccu)j::TRP1 | This study |
| UMY3137 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tr(ccu)j::TRP1 | This study |
| UMY3136 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tr(ccu)j::TRP1 elp3::kanMX4 p1619 | This study |
| UMY3358 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tr(ccu)j::TRP1 trm9::kanMX4 | This study |
| UMY3223 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)d::TRP1 | This study |
| UMY3226 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)o::URA3 | This study |
| UMY3303 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)d::TRP1 tg(ccc)o::URA3 | This study |
| UMY3304 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)d::TRP1 tg(ccc)o::URA3 | This study |
| UMY3320 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)d::TRP1 tg(ccc)o::URA3 elp3::kanMX4 | This study |
| UMY3360 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tg(ccc)d::TRP1 tg(ccc)o::URA3 trm9::kanMX4 | This study |
| UMY3195 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tp(agg)c::TRP1 | This study |
| UMY3293 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tp(agg)n::TRP1 | This study |
| UMY3342 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tp(agg)c::TRP1 tp(agg)n::TRP1 | This study |
| UMY3343 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tp(agg)c::TRP1 tp(agg)n::TRP1 | This study |
| UMY3368 | _MAT_α ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 tp(agg)c::TRP1 tp(agg)n::TRP1 elp3::kanMX4 | This study |
Plasmid constructions.
DNA manipulations, plasmid preparations, and bacterial transformations were performed according to standard protocols. Genes were PCR amplified using Pwo DNA polymerase (Roche Applied Science). Plasmid pRS316-ADE3-tS(CGA)C (p1280) was constructed by cloning a BglII/SacI fragment from pRS316-tS(CGA)C (sup61+) (23) into the BamHI/SacI sites of pDD44 (kindly provided by S. Åström). Plasmid p1244 is a derivative of a YCp50 library plasmid (41) that contains the tT(CGU)K gene. A SphI fragment was removed from the original plasmid, leaving a fragment that contains the tT(CGU)K gene and approximately 800 and 1,200 bp of upstream and downstream sequence, respectively. The tV(CAC)D, tQ(CUG)M, tE(CUC)D, and tR(CCU)J genes were separately PCR amplified from W303-1B and cloned into pRS316 (44) utilizing restriction sites present within the oligonucleotides. This generated the plasmids pRS316-tV(CAC)D (p1725), pRS316-tQ(CUG)M (p1605), pRS316-tE(CUC)D (p1723), and pRS316-tR(CCU)J (p1619). The oligonucleotides used were the following: for tV(CAC)D, 5′-CTCTGGATCCACGTACTTCCTGTAAGGAGC-3′ and 5′-CTCTCTCGAGGGAATGTACTTTCTGTAGGGC-3′; for tQ(CUG)M, 5′-GAACGGATCCTATCAGGCTCTCAGAAGGC-3′ and 5′-GAACCTCGAGAATATGACCAATCGGCGTGTG-3′; for tE(CUC)D, 5′-TTGAATTCTGGCCTGTATTTTCTATATTCC-3′ and 5′-TTCTCGAGCCCCCTAGAAGCATAGTTTTGT-3′; for tR(CCU)J, 5′-CATAGGATCCTCGAGGAGAACTTCTAGTA-3′ and 5′-CCACGAATTCGGAGCAATATCGTACGCCAC-3′. The tS(UGA)P gene was PCR amplified from strain W303-1B, followed by addition of an A overhang by Taq DNA polymerase (Roche Applied Science). This fragment was cloned into the pGEMR-T Easy Vector (Promega) generating p1456. The oligonucleotides used were 5′-TCTCGAATTCAAGTTCAGTATGCCATAGGTGC-3′ and 5′-CTCATCTAGACAGAATGAGCGGATGTTTCA-3′. The high-copy-number LEU2 plasmid pRS425-tS(UGA)P (p1477) was constructed by cloning an ApaI/SpeI fragment from p1456 into the corresponding sites of pRS425 (10). Plasmid pRS425-tT(UGU)G1 (p1779) was constructed by cloning an XhoI/SpeI fragment PCR amplified from W303-1B into the corresponding sites of pRS425. The oligonucleotides used were 5′-TTCTCGAGGCTAATTGGCGATCGTTTA-3′ and 5′-AAACTAGTGAAAGCAAATATCTGGCCTTC-3′. The pRS425-tQ(UUG) (pABY1499) and pRS425-tE(UUC) (pABY1479) plasmids have previously been described (32).
RNA analyses.
Single tRNA species were prepared and analyzed as previously described (4, 22). Steady-state tRNA levels were determined as described previously by using total RNA isolated by the hot phenol method (2, 24). Aminoacylation levels were determined from exponentially growing cells at 30°C. Cells were harvested by pouring the cultures into tubes containing an equivalent volume of ice, followed by centrifugation. All subsequent steps were performed in the cold using prechilled reagents. Cells were washed with water and resuspended in 300 μl of buffer A (0.1 M NaAc [pH 4.5], 10 mM EDTA) followed by addition of ∼0.3 g of glass beads and 300 μl of buffer A-equilibrated phenol-chloroform-isoamylalcohol (25:24:1) solution. The mixture was vortexed for two times for 1 min each time with 1 min on ice between steps. Following centrifugation, the RNA in the aqueous layer was precipitated by addition of 3 volumes of ethanol and incubation at −20°C. RNA pellets were dissolved in 10 mM NaAc (pH 4.5)-1 mM EDTA buffer. Polyacrylamide gel electrophoresis was performed essentially as described previously (48) by applying 10 μg of total RNA on an 8% polyacrylamide-8 M urea-0.1 M NaAc (pH 5.0) gel. After electroblotting to Zeta-Probe membranes (Bio-Rad), 32P-labeled oligonucleotides complementary to the tRNA of interest were used as probes. Signals in Northern blotting experiments were detected and quantified by phosphorimaging using a Storm imaging system and ImageQuant software.
Nonsense suppression assay.
The LEU2 derivatives of pUKC815, pUKC817, pUKC818, and pUKC819 (45, 46) were separately transformed into wild-type (W303-1B), SUP4 (UMY2893), and _SUP4 elp3_Δ (UMY2916) cells. Six independent clones from each transformation were grown in synthetic complete (SC)-Leu medium to an optical density at 600 nm (OD600) of ∼0.85. Cells were harvested, and the β-galactosidase activity determined in protein extracts as previously described (7).
RESULTS
Lack of a wobble mcm5, ncm5, or s2 group does not influence the steady-state or aminoacylation level of tRNA.
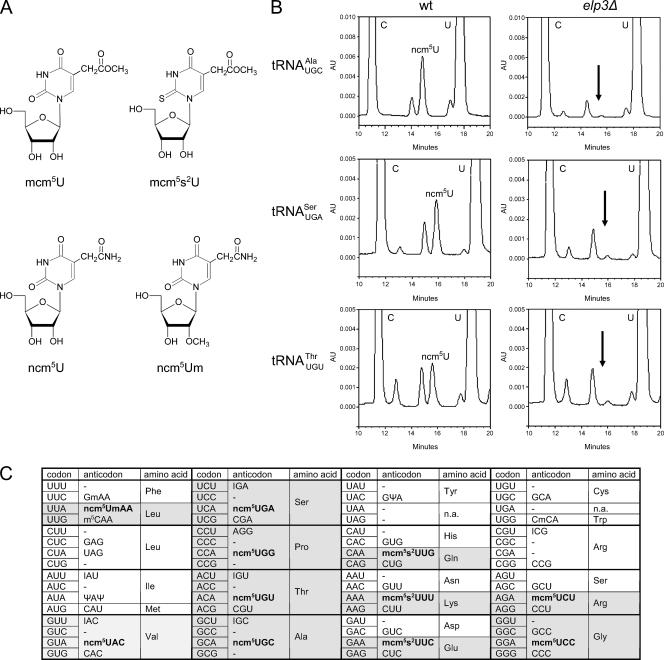
In S. cerevisiae, approximately 274 nuclear-encoded tRNA genes code for the 42 different cytoplasmic tRNA species that are responsible for the decoding of the 61 sense codons (21, 39). The number of genes coding for individual tRNA species varies between 1 and 16, with a good correlation between gene copy number and intracellular tRNA levels (39). The identity of the nucleoside at position 34 is known for 10 of the 13 S. cerevisiae tRNA species that in the primary transcript contain a U at this position (25, 32). Of these, one contains an unmodified U (tRNAUAGLeu), and one contains a pseudouridine (Ψ; tRNAΨAΨIle) residue. The remaining eight tRNA species contain ncm5U, ncm5Um, mcm5U, or mcm5s2U (Fig. 1A). Since the nature of the wobble nucleoside in tRNAUGCAla, tRNAUGASer, and tRNAUGUThrwas not clear, we purified these tRNA species from a wild-type strain. The purified tRNAs were degraded to nucleosides, and their composition analyzed by high-performance liquid chromatography (HPLC). Based on retention time and UV absorption spectra, all three tRNA species were found to contain ncm5U (Fig. 1B and data not shown). Thus, the eleven tRNA species that contain a wobble mcm5 or ncm5 side chain are as follows: tRNAmcm5UCUArg, tRNAmcm5UCCGly, tRNAmcm5s2UUULys, tRNAmcm5s2UUGGln, tRNAmcm5s2UUCGlu, tRNAncm5UACVal, tRNAncm5UGASer, tRNAncm5UGGPro, tRNAncm5UGUThr, tRNAncm5UGCAla, and tRNAncm5UmAALeu(Fig. 1C).
FIG. 1.
Eleven S. cerevisiae tRNA species contain an xm5U derivative at the wobble position. (A) Structures of mcm5U, mcm5s2U, ncm5U, and ncm5Um. (B) The indicated tRNA species was isolated from either wild-type (UMY2893) or _elp3_Δ (UMY2916) cells, and their nucleoside compositions were analyzed by HPLC. The part of the chromatogram between 10 and 20 min is shown in each case. The arrow indicates expected retention time of ncm5U (right panels). The small peak at this position represents an unrelated compound with an UV absorption spectrum different from that of ncm5U. (C) The genetic code and distribution of cytoplasmic S. cerevisiae tRNAs. The anticodon sequences of the 42 different tRNA species (1 initiator and 41 elongator tRNAs) are indicated (21, 25, 32, 39). For anticodons with an uncharacterized RNA sequence, the primary sequence is shown. The initiator and elongator tRNAMet species have identical anticodon sequences. The wobble rules suggest that an inosine (I34) residue allows paring with U, C, and sometimes A. A tRNA with a G or its 2′-_O_-methyl derivative (Gm) at the wobble position should read U- and C-ending codons. Presence of a C34 residue or its 5-methyl (m5C) or 2′-_O_-methyl (Cm) variant should only allow pairing with G. The pseudouridine (Ψ)-containing tRNAIle is presumably unable to pair with the methionine AUG codon. The anticodons containing an xm5U derivative are shown in bold. In tRNAA66Pro, the A34 residue is most likely modified to I34 (see text). AU, absorbance units; wt, wild type.
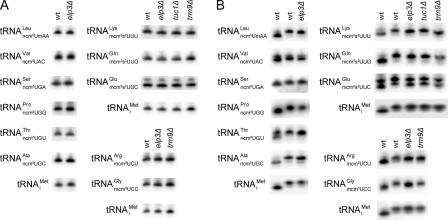
We previously showed that the six subunits in the elongator complex (Elp1p to Elp6p) are all required for the formation of mcm5 and ncm5 side chains at the wobble position (22). In an elp3 mutant, the formation of the side chain at position 5 of the wobble uridine is abolished in all 11 tRNA species that normally contain such groups (22, 32) (Fig. 1B). The TUC1 and TUC2 genes (previously known as NCS6 and NCS2) were recently shown to be required for formation of the s2 group in mcm5s2U34-containing tRNA species (3, 14). The formation of s2 and mcm5 groups occurs independently of each other (3, 14, 22), which means that _elp3_Δ and _tuc1_Δ mutants can be used to dissect their individual functions. Although the wobble mcm5/ncm5 and s2 groups are likely to influence the decoding properties of tRNA, it was conceivable that their absence could generate tRNAs that are destabilized and/or inefficiently aminoacylated (17, 25). To address this concern, we determined the steady-state and in vivo aminoacylation levels for each of the affected tRNA species in wild-type, _elp3_Δ, and _tuc1_Δ cells. The analyses revealed that the absence of an mcm5, ncm5, or s2 group at the wobble position does not reduce abundance or aminoacylation of any of the hypomodified tRNA species (Fig. 2 and Tables 2 and 3). These data suggest that _elp3_Δ and _tuc1_Δ mutants can be utilized to study the decoding properties of tRNA species lacking a wobble mcm5, ncm5, or s2 group.
FIG. 2.
Hypomodification induced by _elp3_Δ, _tuc1_Δ, or _trm9_Δ alleles does not reduce abundance or aminoacylation of any of the affected tRNA species. Northern blot analyses of total RNA isolated under either basic (A) or acidic (B) conditions from wild-type (W303-1B), _elp3_Δ (UMY2843), _tuc1_Δ (UMY3165), or _trm9_Δ (UMY3297) cells. After polyacrylamide gel electrophoresis and transfer under appropriate conditions, blots were probed for the U34-containing tRNA of interest using 32P-labeled oligonucleotides. To control for loading and indirect effects on aminoacylation, the blots were also probed for the C34-containing initiator methionine tRNA (tRNAiMet). (A) Steady-state tRNA levels. (B) Aminoacylation levels. The first lane in each individual blot in panel B represents a deacylated wild-type RNA sample. The symbols for the U34-containing tRNAs indicate the anticodon sequence of wild-type cells. The _elp3_Δ strain lacks the mcm5 and ncm5 side chains whereas the _tuc1_Δ strain lacks the s2 groups. Cells deleted of TRM9 lack the esterified methyl group of mcm5 side chains. wt, wild type.
TABLE 2.
Steady-state levels of tRNAs with hypomodified wobble uridines
| tRNA speciesa | Relative tRNA levelb | ||
|---|---|---|---|
| _elp3_Δ | _tuc1_Δ | _trm9_Δ | |
| tRNAncm5UmAALeu | 0.99 ± 0.13 | ND | ND |
| tRNAncm5UACVal | 0.83 ± 0.04 | ND | ND |
| tRNAncm5UGASer | 0.97 ± 0.11 | ND | ND |
| tRNAncm5UGGPro | 0.99 ± 0.20 | ND | ND |
| tRNAncm5UGUThr | 0.85 ± 0.11 | ND | ND |
| tRNAncm5UGCAla | 0.93 ± 0.12 | ND | ND |
| tRNAmcm5UCUArg | 0.95 ± 0.10 | ND | 0.79 ± 0.12 |
| tRNAmcm5UCCGly | 1.09 ± 0.07 | ND | 0.87 ± 0.08 |
| tRNAmcm5s2UUGGln | 0.98 ± 0.09 | 1.04 ± 0.12 | 0.90 ± 0.13 |
| tRNAmcm5s2UUULys | 0.96 ± 0.27 | 1.02 ± 0.14 | 0.88 ± 0.27 |
| tRNAmcm5s2UUCGlu | 1.02 ± 0.13 | 1.06 ± 0.17 | 0.80 ± 0.15 |
TABLE 3.
In vivo aminoacylation levels of tRNAs with hypomodified wobble uridines
| tRNA speciesa | In vivo aminoacylation level (%)c | |||||||
|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||||||
| wt | _elp3_Δ | _tuc1_Δ | _trm9_Δ | wt | _elp3_Δ | _tuc1_Δ | _trm9_Δ | |
| tRNAncm5UmAALeu | 83 | 87 | ND | ND | 58 | 67 | ND | ND |
| tRNAncm5UACVal | 88 | 92 | ND | ND | 86 | 86 | ND | ND |
| tRNAncm5UGASer | 61 | 61 | ND | ND | 82 | 76 | ND | ND |
| tRNAncm5UGGPro | 69 | 76 | ND | ND | 46 | 52 | ND | ND |
| tRNAncm5UGUThr | 80 | 82 | ND | ND | 67 | 68 | ND | ND |
| tRNAncm5UGCAla | 80 | 79 | ND | ND | 53 | 62 | ND | ND |
| tRNAmcm5UCUArg | 67 | 68 | ND | 65 | 39 | 45 | ND | 50 |
| tRNAmcm5UCCGly | 84 | 87 | ND | 92 | 75 | 73 | ND | 80 |
| tRNAmcm5s2UUGGln | 83 | 81 | 82 | 80 | 60 | 62 | 59 | 65 |
| tRNAmcm5s2UUULys | 79 | 81 | 86 | 84 | 61 | 56 | 63 | 61 |
| tRNAmcm5s2UUCGlub | 85 | 83 | 85 | 83 | 94 | 92 | 91 | 94 |
| tRNAiMet | 72 | 71 | 71 | 68 | 58 | 59 | 56 | 54 |
A wobble mcm5 side chain improves decoding of G-ending codons.
Two S. cerevisiae tRNA species, tRNAmcm5UCUArgand tRNAmcm5UCCGlycontain mcm5U at the wobble position (Fig. 1C). The tRNAmcm5UCUArgspecies decodes in the split codon box AGN, where the AGA and AGG codons are for arginine, whereas tRNAmcm5UCCGlydecodes in the glycine family codon box GGN. A C34-containing tRNA species complementary to the G-ending codon is present in both the AGN and GGN box (tRNACCUArgand tRNACCCGly) (Fig. 1C), indicating that tRNAmcm5UCUArgand tRNAmcm5UCCGlydo not have to read these codons. Consistent with this suggestion, the tRNAmcm5UCUArgspecies was able to read the AGA but not the AGG codon in an in vitro translation system (50).
We previously showed that lack of the mcm5 group in the _SUP4_-encoded ochre suppressor tRNA, where the primary anticodon sequence is UUA, abolished suppression of the ade2-1 (UAA) and can1-100 (UAA) nonsense alleles in vivo (22). These results implied that the function of the wobble modification in tRNAmcm5UCUArgand tRNAmcm5UCCGlymay be to improve reading of their respective A-ending codons. To test this hypothesis, we used a strategy that involved reducing the copy number of the genes coding for tRNAmcm5UCCGlyand combined that with an _elp3_Δ allele. We deleted two of the three genes coding for tRNAmcm5UCCGly, which resulted in a strain with no apparent growth defect but with about a twofold decrease in the tRNAmcm5UCCGlylevel (data not shown). Introduction of the _elp3_Δ allele into this strain did not generate a synergistic growth defect (data not shown), indicating that the wobble mcm5 group does not affect the ability of a wild-type tRNA, i.e., a nonsuppressor tRNA, to read the cognate A-ending codon.
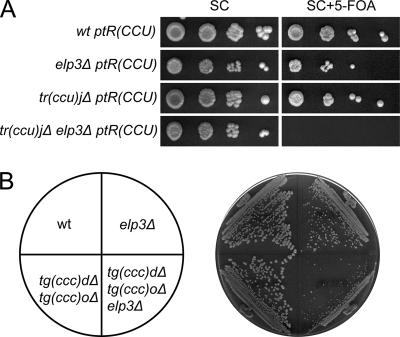
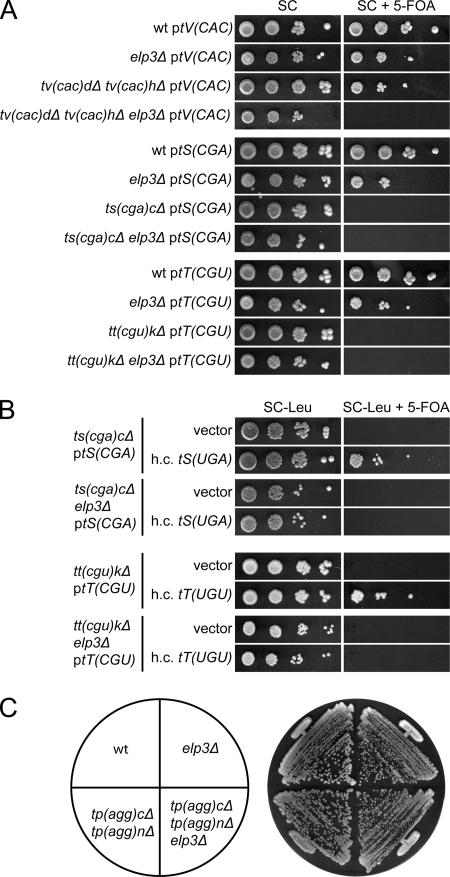
The viability of a strain with a deletion of the single copy of tRNACCUArg(27) [Fig. 3A, tR(CCU)_J_] or the two tRNACCCGlygenes [Fig. 3B, tG(CCC)D and tG(CCC)_O_] shows that tRNAmcm5UCUArgand tRNAmcm5UCCGlyare capable of pairing with their corresponding G-ending codons. We will hereafter use the systematic tRNA gene names in figures since they indicate both the one-letter amino acid abbreviation and the primary sequence of the anticodon. To further assess the role of the mcm5 group, we introduced an _elp3_Δ allele into a strain lacking either tRNACCUArgor tRNACCCGly. Interestingly, synergistic growth defects were observed in both cases, suggesting that the presence of an mcm5 side chain promotes reading of G-ending codons (Fig. 3A and B).
FIG. 3.
The wobble mcm5 side chain in tRNAmcm5UCUArgand tRNAmcm5UCCGlyimproves reading of G-ending codons. (A) Wild-type (W303-1B), _elp3_Δ (UMY2843), tr(ccu)_j_Δ (UMY3137), and _elp3_Δ tr(ccu)_j_Δ (UMY3136) cells carrying the low-copy-number URA3 plasmid pRS316-tR(CCU)J were grown in liquid SC medium for 24 h. The cells were serially diluted, spotted onto SC plates and SC plates containing 5-fluoroorotic acid (5-FOA), and incubated for 3 days at 30°C. On 5-FOA-containing medium, only the cells that lost the URA3 plasmid were able to grow (5). The tR(CCU)J gene codes for tRNACCUArg. (B) The wild-type (W303-1B), _elp3_Δ (UMY2843), tg(ccc)_d_Δ tg(ccc)_o_Δ (UMY3304), and tg(ccc)_d_Δ tg(ccc)_o_Δ _elp3_Δ (UMY3320) strains were streaked on a YEPD (yeast extract, peptone, dextrose) plate and incubated at 30°C for 2 days. The tG(CCC) genes code for tRNACCCGly. wt, wild type.
The observation that the wobble mcm5 group in wild-type tRNAs seems to preferentially affect decoding of G-ending codons prompted us to reexamine the effect of the modification on the _SUP4_-encoded suppressor tRNA. This was accomplished by utilizing a plasmid-borne nonsense suppression assay, which is based on a low-copy-number plasmid carrying a PGK1-LacZ fusion (45, 46). The three nonsense suppression reporter constructs have one of the stop codons (UAA, UAG, or UGA) placed in frame at the junction of the PGK1 and LacZ sequences. Thus, nonsense suppression can be quantified by determining the β-galactosidase activity in cells harboring a nonsense reporter construct and then comparing that to the activity in cells containing the control construct, which lacks a premature stop codon (45, 46). Analyses of wild-type, SUP4, and _SUP4 elp3_Δ strains transformed with the respective plasmid revealed that the presence of the SUP4 allele caused read-through of the UAA and, to a lesser extent, the UAG codon (Table 4). As predicted from our previous results (22), lack of the mcm5 group decreased suppression of the UAA codon. Interestingly, the influence of the mcm5 group was even larger on the UAG codon (Table 4). These results validate our genetic approach and suggest that the function of an mcm5U34 residue is not primarily to improve reading of the A-ending codons but to improve pairing with the codon ending with G.
TABLE 4.
The wobble mcm5 group in an ochre suppressor tRNA promotes reading of UAA and UAG stop codons
| Termination codon | % Read-througha | Reduction (_n_-fold) in read-through (value for SUP4 /value for SUP4 elp3) | ||
|---|---|---|---|---|
| wt | SUP4 | _SUP4 elp3_Δ | ||
| UAA | 0.10 ± 0.02 | 29.49 ± 9.03 | 11.31 ± 1.63 | 2.6 |
| UAG | 0.12 ± 0.03 | 1.39 ± 0.24 | 0.31 ± 0.04 | 4.5 |
| UGA | 0.10 ± 0.02 | 0.09 ± 0.01 | 0.09 ± 0.02 | 1.0 |
Influence of mcm5 and s2 groups on the decoding properties of tRNA.
The three mcm5s2U-containing tRNA species, tRNAmcm5s2UUGGln, tRNAmcm5s2UUULys, and tRNAmcm5s2UUCGlu, decode in split codon boxes where the pyrimidine-ending codons are for another amino acid (Fig. 1C). In each of these codon boxes a C34-containing tRNA species complementary to the G-ending codon is present (tRNACUGGln, tRNACUULys, and tRNACUCGlu), which is consistent with the suggestion that mcm5s2U34-containing tRNA species preferentially decode triplets ending with A (33, 43, 54).
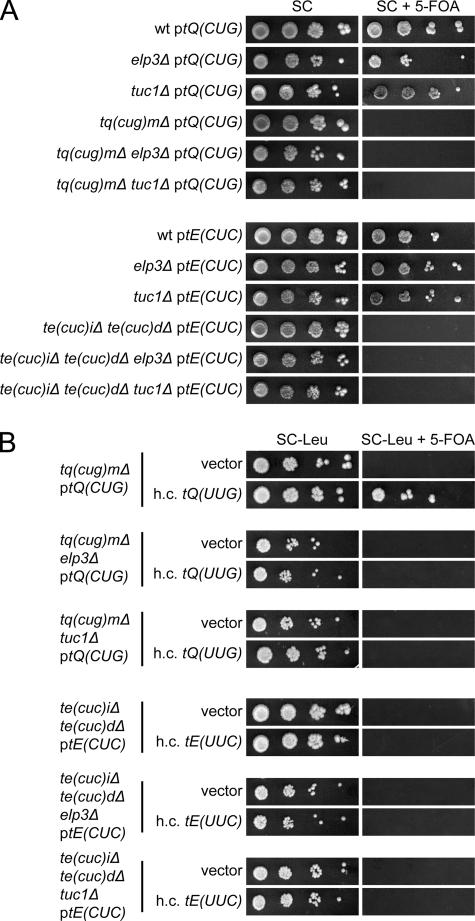
To examine the role of mcm5s2U34 residues, strains with deletions of the single copy tRNACUGGln[tQ(CUG)_M_] or the two tRNACUCGlu[tE(CUC)I and tE(CUC)_D_] genes were constructed. These strains were nonviable unless rescued by a wild-type copy of the respective tRNA gene on a plasmid (49) (Fig. 4A), suggesting that tRNAmcm5s2UUGGlnand tRNAmcm5s2UUCGluare unable to read the codons ending with G. The evidence presented above that the mcm5 group in tRNAmcm5UCUArgand tRNAmcm5UCCGlypromoted reading of the AGG and GGG codons implied that the inability of mcm5s2U-containing tRNA species to read G-ending codons might be caused by a restricting effect of the s2 group. However, introduction of a _tuc1_Δ allele, which abolishes formation of the s2 but not the mcm5 group (3), did not suppress the inviability of a strain lacking tRNACUGGlnor tRNACUCGlu(Fig. 4A). Moreover, introduction of an _elp3_Δ allele into a strain lacking either the tRNACUGGlnor the tRNACUCGluspecies did not overcome the requirement for the rescuing plasmid (Fig. 4A). Thus, neither the mcm5 nor the s2 group in mcm5s2U-containing tRNA species is solely responsible for the inability to read G-ending codons.
FIG. 4.
Neither the mcm5 nor the s2 group restricts tRNAmcm5s2UUGGlnand tRNAmcm5s2UUCGluto A-ending codons. (A) The appropriate strains (W303-1B, UMY2843, UMY3165, UMY3133, UMY3134, UMY3347, UMY3348, UMY3350, and UMY3352) carrying the indicated low-copy-number URA3 plasmid were grown in liquid SC medium for 24 h. The cells were serially diluted, spotted onto SC plates and SC plates containing 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 3 days. The tQ(CUG) and tE(CUC) genes code for tRNACUGGlnand tRNACUCGlu, respectively. (B) The tq(cug)Δ, te(cuc)Δ, and their _elp3_Δ or _tuc1_Δ derivatives from panel A were transformed with an empty high-copy-number (h.c.) LEU2 vector (10) or the same plasmid harboring a gene coding for the relevant U34-containing tRNA [tQ(UUG) or tE(UUC)]. The transformants were grown in SC-Leu medium for 24 h, serially diluted, spotted onto SC-Leu plates and SC-Leu plates containing 5-FOA, and incubated at 30°C for 3 days. wt, wild type.
We recently showed that the phenotypes of a strain lacking wobble mcm5/ncm5 and/or s2 groups can be suppressed by increased dosage of the genes coding for the tRNALys and tRNAGln species that normally contain mcm5s2U (3, 14). This prompted us to investigate whether the lethality of a strain lacking tRNACUGGlnor tRNACUCGlucould be suppressed by increased expression of the relevant U34-containing tRNA species. Increased expression of the tRNAGlu species did not suppress the inviability of a strain lacking tRNACUCGlu, regardless of whether the tRNA contained mcm5s2U34, mcm5U34, or s2U34 (Fig. 4B). However, overexpression of tRNAmcm5s2UUGGlnsuppressed the need for tRNACUGGlnin an otherwise wild-type background (Fig. 4B). This suppression was not observed in an _elp3_Δ or _tuc1_Δ background (Fig. 4B), suggesting that the mcm5 and s2 groups cooperatively improve pairing with G.
The wobble ncm5 side chain in tRNA_ncm5UAC_ Val is not required for efficient decoding of the A-ending codon.
An ncm5U34 residue is present in tRNAncm5UACVal, tRNAncm5UGASer, tRNAncm5UGGPro, tRNAncm5UGUThr, and tRNAncm5UGCAla, which all decode in family codon boxes (Fig. 1C). To assess the influence of a wobble ncm5 group on the ability to read the A-ending codon, we reduced the copy number of the genes coding for tRNAncm5UACValor tRNAncm5UGASerand combined that with an _elp3_Δ allele. The strains with a deletion of one of the two genes coding for tRNAncm5UACValor two of the three genes for tRNAncm5UGASershowed no apparent growth defect although the steady-state level was reduced to about 50% (tRNAncm5UACVal) or 30% (tRNAncm5UGASer) of that in wild type (data not shown). Introduction of the _elp3_Δ allele into any of these strains did not generate a synergistic growth defect (data not shown), indicating that an ncm5U34 residue does not improve reading of A-ending codons. However, both the valine (GUN) and serine (UCN) family boxes harbor a tRNA species with an inosine (I) at the wobble position (tRNAIACValand tRNAIGASer) (Fig. 1C). Since I34-containing tRNAs may read not only codons ending with U and C but also those ending with A (11), it seemed possible that a synergistic growth defect could be masked by the ability of tRNAIACValor tRNAIGASerto read its respective A-ending codon (GUA and UCA). We cannot exclude this possibility for the UCA codon since a strain lacking the three genes coding for tRNAncm5UGASeris viable (data not shown). In contrast, a strain with deletions of both tRNAncm5UACValgenes is inviable (data not shown), showing that tRNAIACValcannot efficiently decode the GUA codon. This observation in combination with the above-mentioned lack of synergistic growth defect suggests that the ncm5 side chain in tRNAncm5UACValdoes not improve reading of the cognate A-ending codon.
A wobble ncm5 side chain influences the ability to read G-ending codons.
Two of the five family codon boxes where ncm5U34-containing tRNAs decode lack a tRNA with an anticodon complementary to the G-ending codon (proline CCN and alanine GCN boxes) (Fig. 1C), showing that an ncm5U-containing tRNA is able to read such codons. However, a C34-containing tRNA species with an anticodon complementary to the G-ending codon (tRNACGASer, tRNACGUThr, and tRNACACVal) is present in the remaining three codon boxes (Fig. 1C). It was previously shown that the single-copy genes coding for tRNACGASer[tS(CGA)_C_] and tRNACGUThr[tT(CGU)_K_] are essential for viability (9, 15). In contrast, a strain with deletions of the two genes coding for tRNACACVal[tV(CAC)D and tV(CAC)_H_] is viable although with a growth defect (Fig. 5A). These data suggested that the ncm5U34-containing tRNAncm5UGASerand tRNAncm5UGUThrdo not efficiently read the UCG and ACG codons, whereas tRNAncm5UACValcan read the GUG codon.
FIG. 5.
A wobble ncm5 side chain improves reading of codons ending with G but does not restrict the tRNA to purine-ending codons. (A) The appropriate strains (W303-1B, UMY2843, UMY3296, UMY3333, UMY3126, UMY3127, UMY3129, and UMY3130) carrying the indicated low-copy-number URA3 plasmid were grown in liquid SC medium for 24 h. The cells were serially diluted, spotted onto SC plates and SC plates containing 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 3 days. The tV(CAC), tS(CGA), and tT(CGU) genes code for tRNACACVal, tRNACGASer, and tRNACGUThr, respectively. (B) The ts(cga)_c_Δ, tt(cgu)_k_Δ, and their _elp3_Δ derivatives from panel A were transformed with an empty high-copy-number (h.c.) LEU2 vector (10) or the same plasmid harboring a gene coding for the relevant U34-containing tRNA [tS(UGA) or tT(UGU)]. The transformants were grown in SC-Leu medium for 24 h, serially diluted, spotted onto SC-Leu plates and SC-Leu plates containing 5-FOA, and incubated at 30°C for 3 days. (C) The wild-type (W303-1B), _elp3_Δ (UMY2843), tp(agg)cΔ tp(agg)_n_Δ (UMY3343), and tp(agg)cΔ tp(agg)_n_Δ _elp3_Δ (UMY3368) strains were streaked on a YEPD (yeast extract, peptone, dextrose) plate and incubated at 30°C for 2 days. The tP(AGG) genes code for tRNAAGGPro. wt, wild type.
To investigate the role of the ncm5 group in tRNAncm5UGASer, tRNAncm5UGUThr, and tRNAncm5UACVal, we constructed strains lacking the C34-containing tRNACGASer, tRNACGUThr, or tRNACACValthat harbored a wild-type copy of the respective C34-containing tRNA gene on a plasmid. Introduction of the _elp3_Δ allele into a strain with a deletion of the tRNACACValgenes generated a requirement for the plasmid (Fig. 5A), showing that the ncm5 group in tRNAncm5UACValis important for reading the GUG codons. Lack of tRNACGASeror tRNACGUThrwas lethal both in an ELP3+ and an _elp3_Δ background (Fig. 5A), suggesting that neither the modified nor the unmodified tRNAncm5UGASerand tRNAncm5UGUThrcan efficiently read their respective G-ending codons. However, we previously showed that increased dosage of a gene coding for tRNAncm5UGASersuppresses the lethality of a strain deficient for tRNACGASer(24) (Fig. 5B). Similarly, increased dosage of a tRNAncm5UGUThrgene suppressed the lethality of a strain lacking tRNACGUThr(Fig. 5B). These dosage suppressions were not observed in an _elp3_Δ background (Fig. 5B), suggesting that a wobble ncm5 group improves reading of G-ending codons also for these tRNAs. Based on these results, we conclude that the presence of the ncm5 side chain improves reading of G-ending codons but that features other than the modification status of the wobble uridine determine the efficiency.
The wobble ncm5 side chain in tRNAncm5UGGPro does not prevent reading of pyrimidine-ending codons.
A tRNA with an unmodified U34 residue may, under some circumstances, pair with a codon ending with any nucleoside (31, 53). Interestingly, the only yeast tRNA species (tRNAUAGLeu) shown to have an unmodified wobble uridine (40) was able to read all four CUN codons in vitro (19, 51). Accordingly, a strain with a deletion of the gene encoding the other tRNA species in this box (tRNAGAGLeu) (Fig. 1C) is viable (data not shown). In an attempt to address whether this indiscriminate decoding is a unique feature of tRNAUAGLeuor whether it would be a feature of other tRNAs harboring an unmodified U34, we utilized the distribution of tRNA species in the proline family codon box (Fig. 1C). In this box, only two tRNA species are present, one contains ncm5U (tRNAncm5UGGPro), and the other has in the primary sequence an A at the wobble position. That an unmodified A34 is almost never found in tRNA and that I34 is present in the corresponding tRNA species in higher eukaryotes suggest that the A34 residue in the yeast species is also deaminated to I34 (28). Unexpectedly, a strain with deletions of the two tRNAAGGProgenes [tP(AGG)C and tP(AGG)_N_] was viable with no apparent growth defect (Fig. 5C), indicating that the ncm5U34-containing tRNAncm5UGGProcan read all four codons. Moreover, introduction of an _elp3_Δ allele did not generate a synergistic growth defect (Fig. 5C), suggesting that the ncm5 group in tRNAncm5UGGProhas no influence on the ability to read the pyrimidine-ending codons.
The esterified methyl group of mcm5 side chains has a modest effect on decoding.
The biosynthesis of mcm5 and ncm5 side chains is likely to involve many steps and gene products (22). The last step in the formation of mcm5 side chains is dependent on the Trm9 protein, which in vitro catalyzes formation of the methylester using _S_-adenosyl-methionine as the donor and tRNAs with a cm5 group at U34 as substrates (26). Although, a _trm9_Δ strain was shown to lack methyl-esterified nucleosides, the identity of the hypomodified wobble nucleoside in the mutant was not clear (26). Analysis of the tRNAmcm5UCUArgand tRNAmcm5s2UUCGluspecies isolated from a _trm9_Δ strain revealed that they did not contain the expected Trm9p substrates, cm5U and cm5s2U, but that they contained ncm5U and ncm5s2U (data not shown). It is not clear whether these nucleosides represent intermediates in the mcm5 biosynthesis pathway or whether they reflect shunting of cm5-modified nucleosides to the pathway responsible for ncm5 formation. In analogy with the elp3 mutant, no obvious reduction in abundance or aminoacylation level was observed in a _trm9_Δ strain (Fig. 2 and Tables 2 and 3).
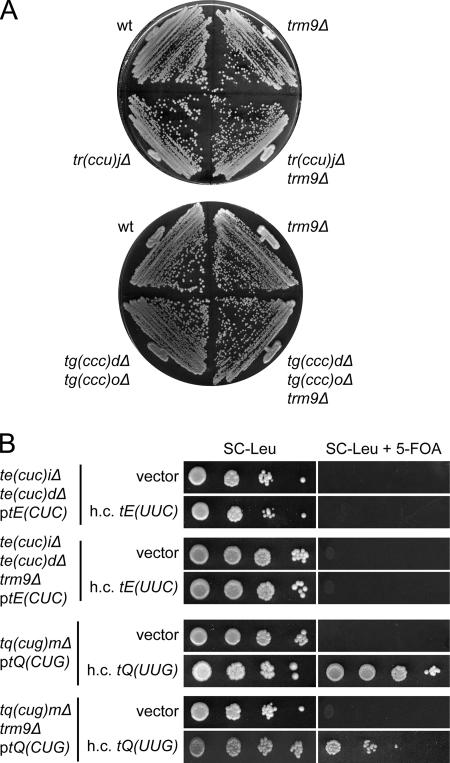
A strain with a deletion of the TRM9 gene does not show the same severe growth defect as _elp3_Δ or _tuc1_Δ mutants (Fig. 6 and data not shown). Furthermore, introduction of a _trm9_Δ allele does not prevent an ochre suppressor tRNA, which normally contains mcm5U, from reading ochre stop codons (32). Consistent with this, we saw no or small synergistic effects when a _trm9_Δ allele was introduced into a strain containing only one tRNAmcm5UCCGlygene or a strain deleted of the C34-containing tRNACCUArgor tRNACCCGlygene (Fig. 6A and data not shown). Nevertheless, introduction of a _trm9_Δ allele into the _tuc1_Δ strain generated a small synergistic growth defect (data not shown), suggesting that the methyl ester is important for tRNA species that also contain an s2 group at the wobble uridine. Although increased dosage of a tRNAmcm5s2UUGGlngene suppressed the need for the C34-containing tRNACUGGlnin a _trm9_Δ background, the strain grew at a slower rate than the corresponding TRM9+ strain, implying that the ncm5s2U34-containing tRNAGln is less capable of reading the CAG codon (Fig. 6B). We conclude that the lack of the esterified methyl group of mcm5 side chains has a modest effect on the decoding properties of tRNA.
FIG. 6.
Influence of the esterified methyl group of mcm5 side-chains on the decoding properties of tRNA. (A) The indicated strains (W303-1B, UMY3297, UMY3137, UMY3358, UMY3304, and UMY3360) were streaked on a YEPD (yeast extract, peptone, dextrose) plate and incubated at 30°C for 2 days. (B) The indicated strains (UMY3348, UMY3354, UMY3134, or UMY3345) were transformed with an empty high-copy-number (h.c.) LEU2 vector (10) or the same plasmid harboring a gene coding for the relevant U34-containing tRNA [tE(UUC) or tQ(UUG)]. The transformants were grown in SC-Leu medium for 24 h, serially diluted, spotted onto SC-Leu plates and SC-Leu plates containing 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 2 days. wt, wild type.
DISCUSSION
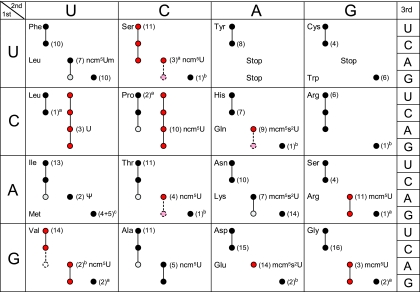
Analyses of the distribution of cytoplasmic tRNA species in eukaryotes have shown that they normally contain both U34- and C34-containing isoacceptors, suggesting that A- and G-ending codons are decoded by distinct tRNAs (13, 34, 38, 39). This observation suggested that eukaryotic organisms may not use U-G wobbling, since this would make the C34-containing tRNA species functionally redundant (38). Further support for this view comes from the fact that the U34 residues are normally modified to an xm5U derivative, which might restrict wobbling, and from the observation that tRNA species harboring such derivatives were unable to efficiently read G-ending codons in vitro (19, 33, 43, 50, 54). Here, we provide a comprehensive analysis of the decoding properties of tRNA species harboring an xm5U derivative. In contrast to previous suggestions, we show that many eukaryotic U34-containing tRNA species read G-ending codons (Fig. 7) and that the presence of an xm5U derivative promotes this ability. These results also show that the ability of an xm5U34-containing tRNA to read the G-ending codon cannot be predicted from whether the codon box includes a C34-containing tRNA.
FIG. 7.
The genetic code and decoding abilities of individual tRNA species. Codons read by a tRNA are indicated by circles and connecting lines. Red and pink circles represent tRNA species for which the decoding properties were investigated in this article. Pink circles connected with a dashed line indicate that the tRNA species reads the codon only when it is overexpressed. The empty dashed circle for tRNAIACValis shown only to indicate that this inosine-containing tRNA species does not efficiently read the GUA codon. The nucleoside at the wobble position is given for the 13 wobble uridine-containing tRNA species. Black and gray circles represent decoding abilities predicted by the wobble hypothesis, the revised wobble rules, and the distribution of tRNA species. A gray circle indicates that the tRNA species is less likely to read the codon. The number of genes coding for a tRNA species is indicated next to the circle for the complementary codon. The following qualifications apply: a superscript a indicates that the gene(s) encoding the tRNA is nonessential; a superscript b indicates that the gene(s) encoding the tRNA is essential; where two values are given (superscript c) four genes code for tRNAiMet, and five code for tRNAmMet.
An mcm5U34 residue promotes decoding of G-ending codons.
Based on an in vitro translation system, it was suggested that the yeast tRNAmcm5UCUArgspecies is able to decode the AGA but not the AGG codon (50). These results are consistent with the notion that the mcm5 group may prevent U-G wobbling. However, the observation that strains lacking the C34-containing tRNACCUArgor tRNACCCGlyspecies are viable (27) (Fig. 3A) shows that the mcm5U34-containing tRNAmcm5UCUArgand tRNAmcm5UCCGlyare able to read the AGG and GGG codons in vivo. In fact, we found that the wobble mcm5 group in tRNAmcm5UCUArgand tRNAmcm5UCCGlypromotes reading of their respective G-ending codons (Fig. 3). A possible explanation for the contradictory results in vivo and in vitro is that the mcm5 group at U34 may not significantly influence intrinsic differences in the efficiency by which the tRNA reads the A- or G-ending codon, i.e., it may only be possible to detect reading of the complementary codon in vitro. It cannot, however, be excluded that the inability of tRNAmcm5UCUArgto read the AGG codon in vitro was caused by the fact that that Escherichia coli and not S. cerevisiae ribosomes were used in the translation system (50).
We did not detect a decrease in the ability of the hypomodified tRNAmcm5UCCGlyto read the GGA codon. This result contrasted our previous finding that the mcm5 group in an ochre suppressor tRNA was required for suppression of the ade2-1 and can1-100 alleles (22). By utilizing a nonsense suppression reporter system, we confirmed that the modification in the suppressor tRNA improved reading of UAA codons (Table 4). However, the relative influence of the modification was even larger on the UAG codon, providing further support for the notion that an mcm5U34 residue preferentially improves reading of the G-ending codon. It remains to be determined if the effect observed on the UAA codon is applicable to A-ending sense codons or if it is caused by the fact that the modification is present in an atypical context, i.e., in an altered anticodon.
An mcm5s2U34 residue promotes decoding of A- and G-ending codons.
The presence of an mcm5s2U34 residue was originally proposed to allow the tRNA to efficiently read the cognate A-ending codon and simultaneously reduce the ability to pair with the G-ending codon (43, 54). However, more recent data have suggested that a mcm5s2U-containing tRNA may read both A- and G-ending codons (35). We have found here that neither tRNAmcm5s2UUGGlnnor tRNAmcm5s2UUCGlucan efficiently read its respective G-ending codon in vivo (Fig. 4A). However, tRNAmcm5s2UUGGlnbut not tRNAmcm5s2UUCGlucan read the codon if its expression is increased (Fig. 4B). This ability of tRNAmcm5s2UUGGlnrequired both the mcm5 and s2 groups, indicating that they cooperatively improve pairing with the CAG codon (Fig. 4B).
We previously showed that the growth defects of strains lacking mcm5/ncm5 and/or s2 groups are suppressed by increased expression of tRNAmcm5s2UUGGlnand tRNAmcm5s2UUULys(3, 14). These data suggested that the lack of a wobble mcm5 and/or s2 group in tRNAmcm5s2UUGGlnand tRNAmcm5s2UUULyscaused a reduced functionality and that this defect can be counteracted by increasing the tRNA levels. However, these studies did not determine if it was the A- or G-ending Gln and Lys codons that are poorly translated in the modification-deficient cells. The finding that the hypomodified tRNAmcm5s2UUGGlncannot read the CAG codon even when it is overexpressed indicates that the phenotypes are caused by poor translation of the A-ending codons (Fig. 4B). This also suggests that the relevant function of an mcm5s2U34 residue is to improve reading of the codon ending with A. These results are consistent with a role for the modifications in promoting an appropriate anticodon conformation (12), which would improve pairing with both the A- and G-ending codon.
An ncm5U34 residue promotes decoding of G-ending codons.
An ncm5U34 residue is found in tRNAs species decoding in codon boxes where all four codons specify the same amino acid (Fig. 7). Our results suggest that the function of the ncm5 modification is to improve reading of G-ending codons and that any influence on reading of the A-ending codons is small (Fig. 5 and data not shown). However, the evidence that tRNAncm5UGASerand tRNAncm5UGUThrare only able to read their respective G-ending codons when they are overexpressed suggests that features other than the modification status of the wobble uridine determine the decoding efficiency. Consistent with this hypothesis, the hypomodified forms of tRNAncm5UGGProand tRNAncm5UGCAlamust be able to read their respective G-ending codons efficiently since the major growth defect of an _elp3_Δ mutant is not caused by a reduced functionality of these tRNA species (14), even though there are no C34-containing tRNA species for the proline CCG and alanine GCG codons (Fig. 7).
The observation that tRNAncm5UGGProcan read the four CCN codons in vivo (Fig. 5C) implied that the ncm5U34 residue might extend the wobble capacity of the tRNA, analogous to the modified wobble nucleoside uridine-5-oxyacetic acid (cmo5U) in bacteria. In fact, the Salmonella enterica tRNAcmo5UGGProspecies has been shown to read all four proline codons, and the modification promoted decoding of the CCU and CCC codons (36). However, the ability of yeast tRNAncm5UGGProto read the CCU and CCC codons is independent of the ncm5 group (Fig. 5C), implying that pairing with these codons involves a two-out-of-three interaction (30). The observation that the presence of a wobble ncm5 group does not prevent pairing with pyrimidine-ending codons provides a possible explanation to the wobble 2′-_O_-methyl group found in the only ncm5-containing tRNA that decodes in a split codon box (Fig. 7). The tRNAncm5UmAALeuspecies was shown in an in vitro translation system to preferentially read the UUA codon, which is presumably due to the influence of the 2′-_O_-methyl group on the conformation of the wobble nucleoside and/or the anticodon (19, 31). Moreover, the observation that the tRNAs that normally harbor an mcm5 group contain an ncm5 group in a _trm9_Δ strain may provide a clue to the specific function of the esterified methyl group of mcm5 side chains. In four of the five codon boxes where mcm5-containing tRNA species decode, the pyrimidine-ending codons code for another amino acid (Fig. 7). It is therefore feasible that a mcm5 group, in contrast to an ncm5 group, would prevent misreading of codons ending with U or C and thereby improve the fidelity of translation. On the other hand, the minor growth defect of a _trm9_Δ mutant (Fig. 6 and data not shown) suggests that any influence on translational fidelity is relatively small.
Concluding remarks.
The in vivo roles of modified wobble nucleosides are poorly understood. By utilizing genetic approaches, we have discovered several important features of eukaryotic xm5U34 derivatives: (i) the presence of an mcm5U34, ncm5U34, or mcm5s2U34 residue improves the ability of tRNA to read G-ending codons; (ii) an mcm5s2U34 residue enhances the ability to decode the A-ending codon; (iii) an ncm5U34 residue does not restrict tRNA to purine-ending codons; and (iv) the importance of a wobble modification depends on its context, which presumably includes structural attributes of the tRNA as well as properties of the codon-anticodon interaction. Although these general features are likely to be conserved in eukaryotes, the apparent requirement for the modifications in any given organism is likely to be influenced by variations in tRNA sequences, intracellular tRNA levels, and distribution of tRNA species.
Acknowledgments
We thank S. Åström and M. Tuite for plasmids, and K. Jacobsson for HPLC analysis of modified nucleosides. We thank S. Tuck, T. Hagervall, and S. Ghosh for editorial comments.
This work was supported by grants from the Swedish Cancer Foundation (Projects 680 to G.R.B. and 3516-B05-12XAB to A.S.B.), Swedish Research Council (Projects BU-2930 to G.R.B. and 621-2006-4269 to A.S.B.), and Margareta Dannbergs Foundation (Project 223-302-06 to A.S.B.).
Footnotes
▿
Published ahead of print on 10 March 2008.
REFERENCES
- 1.Agris, P. F. 1991. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 731345-1349. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 3.Björk, G. R., B. Huang, O. P. Persson, and A. S. Byström. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 131245-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björk, G. R., K. Jacobsson, K. Nilsson, M. J. O. Johansson, A. S. Byström, and O. P. Persson. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197345-346. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D., D. Dawon, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Byström, A. S., U. Pawel-Rammingen von, and S. U. Åström. 1993. Genetic systems in yeast for analysis of initiator/elongator tRNA specificity, p. 35-45. In K. H. Nierhaus (ed.), The translational apparatus. Plenum Press, New York, NY.
- 9.Chakshusmathi, G., S. D. Kim, D. A. Rubinson, and S. L. Wolin. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 226562-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 11.Crick, F. H. 1966. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19548-555. [DOI] [PubMed] [Google Scholar]
- 12.Durant, P. C., A. C. Bajji, M. Sundaram, R. K. Kumar, and D. R. Davis. 2005. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 448078-8089. [DOI] [PubMed] [Google Scholar]
- 13.Duret, L. 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16287-289. [DOI] [PubMed] [Google Scholar]
- 14.Esberg, A., B. Huang, M. J. O. Johansson, and A. S. Byström. 2006. Elevated levels of two tRNA species bypass the requirement for Elongator complex in transcription and exocytosis. Mol. Cell 24139-148. [DOI] [PubMed] [Google Scholar]
- 15.Etcheverry, T., M. Salvato, and C. Guthrie. 1982. Recessive lethality of yeast strains carrying the SUP61 suppressor results from loss of a transfer RNA with a unique decoding function. J. Mol. Biol. 158599-618. [DOI] [PubMed] [Google Scholar]
- 16.Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner, and L. S. Symington. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 172764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giege, R., M. Sissler, and C. Florentz. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 265017-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasser, A. L., C. el Adlouni, G. Keith, E. Sochacka, A. Malkiewicz, M. Santos, M. F. Tuite, and J. Desgres. 1992. Presence and coding properties of 2′-_O_-methyl-5-carbamoylmethyluridine (ncm5Um) in the wobble position of the anticodon of tRNA(Leu) (U*AA) from brewer's yeast. FEBS Lett. 314381-385. [DOI] [PubMed] [Google Scholar]
- 20.Hagervall, T. G., S. C. Pomerantz, and J. A. McCloskey. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 28433-42. [DOI] [PubMed] [Google Scholar]
- 21.Hani, J., and H. Feldmann. 1998. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 26689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, B., M. J. O. Johansson, and A. S. Byström. 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson, M. J. O., and A. S. Byström. 2002. Dual function of the tRNA(m5U54)methyltransferase in tRNA maturation. RNA 8324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson, M. J. O., and A. S. Byström. 2004. The Saccharomyces cerevisiae TAN1 gene is required for _N_4-acetylcytidine formation in tRNA. RNA 10712-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson, M. J. O., and A. S. Byström. 2005. Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae, p. 87-120. In H. Grosjean (ed.), Fine-tuning of RNA functions by modification and editing. Springer-Verlag, New York, NY.
- 26.Kalhor, H. R., and S. Clarke. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 239283-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawakami, K., B. K. Shafer, D. J. Garfinkel, J. N. Strathern, and Y. Nakamura. 1992. Ty element-induced temperature-sensitive mutations of Saccharomyces cerevisiae. Genetics 131821-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keith, G., J. Desgres, P. Pochart, T. Heyman, K. C. Kuo, and C. W. Gehrke. 1990. Eukaryotic tRNAs(Pro): primary structure of the anticodon loop; presence of 5-carbamoylmethyluridine or inosine as the first nucleoside of the anticodon. Biochim. Biophys. Acta 1049255-260. [DOI] [PubMed] [Google Scholar]
- 29.Kruger, M. K., S. Pedersen, T. G. Hagervall, and M. A. Sorensen. 1998. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284621-631. [DOI] [PubMed] [Google Scholar]
- 30.Lagerkvist, U. 1978. “Two out of three”: an alternative method for codon reading. Proc. Natl. Acad. Sci. USA 751759-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim, V. I. 1994. Analysis of action of wobble nucleoside modifications on codon-anticodon pairing within the ribosome. J. Mol. Biol. 2408-19. [DOI] [PubMed] [Google Scholar]
- 32.Lu, J., B. Huang, A. Esberg, M. J. O. Johansson, and A. S. Byström. 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 111648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lustig, F., P. Elias, T. Axberg, T. Samuelsson, I. Tittawella, and U. Lagerkvist. 1981. Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem. 2562635-2643. [PubMed] [Google Scholar]
- 34.Marck, C., and H. Grosjean. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 81189-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, F. V., V. Ramakrishnan, A. Malkiewicz, and P. F. Agris. 2004. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 111186-1191. [DOI] [PubMed] [Google Scholar]
- 36.Näsvall, S. J., P. Chen, and G. R. Björk. 2004. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 101662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nirenberg, M. 2004. Historical review: deciphering the genetic code-a personal account. Trends Biochem. Sci. 2946-54. [DOI] [PubMed] [Google Scholar]
- 38.Percudani, R. 2001. Restricted wobble rules for eukaryotic genomes. Trends Genet. 17133-135. [DOI] [PubMed] [Google Scholar]
- 39.Percudani, R., A. Pavesi, and S. Ottonello. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268322-330. [DOI] [PubMed] [Google Scholar]
- 40.Randerath, E., R. C. Gupta, L. L. Chia, S. H. Chang, and K. Randerath. 1979. Yeast tRNA Leu UAG. Purification, properties and determination of the nucleotide sequence by radioactive derivative methods. Eur. J. Biochem. 9379-94. [DOI] [PubMed] [Google Scholar]
- 41.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60237-243. [DOI] [PubMed] [Google Scholar]
- 42.Rozenski, J., P. F. Crain, and J. A. McCloskey. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekiya, T., K. Takeishi, and T. Ukita. 1969. Specificity of yeast glutamic acid transfer RNA for codon recognition. Biochim. Biophys. Acta 182411-426. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stansfield, I., Akhmaloka, and M. F. Tuite. 1995. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 27417-426. [DOI] [PubMed] [Google Scholar]
- 46.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 144365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takai, K., and S. Yokoyama. 2003. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 316383-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varshney, U., C. P. Lee, and U. L. RajBhandary. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 26624712-24718. [PubMed] [Google Scholar]
- 49.Weiss, W. A., and E. C. Friedberg. 1986. Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J. Mol. Biol. 192725-735. [DOI] [PubMed] [Google Scholar]
- 50.Weissenbach, J., and G. Dirheimer. 1978. Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim. Biophys. Acta 518530-534. [DOI] [PubMed] [Google Scholar]
- 51.Weissenbach, J., G. Dirheimer, R. Falcoff, J. Sanceau, and E. Falcoff. 1977. Yeast tRNALeu (anticodon UAG) translates all six leucine codons in extracts from interferon treated cells. FEBS Lett. 8271-76. [DOI] [PubMed] [Google Scholar]
- 52.Yarian, C., H. Townsend, W. Czestkowski, E. Sochacka, A. J. Malkiewicz, R. Guenther, A. Miskiewicz, and P. F. Agris. 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 27716391-16395. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama, S., and S. Nishimura. 1995. Modified nucleosides and codon recognition, p. 207-223. In D. Söll and U. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC.
- 54.Yokoyama, S., T. Watanabe, K. Murao, H. Ishikura, Z. Yamaizumi, S. Nishimura, and T. Miyazawa. 1985. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. USA 824905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]