APOBEC3G Is Degraded by the Proteasomal Pathway in a Vif-dependent Manner without Being Polyubiquitylated (original) (raw)
Abstract
Although the covalent attachment of a polyubiquitin is the prevailing paradigm for entry into proteasomes, accumulating evidence suggests that poorly defined ubiquitin-free pathways also degrade proteins. The cytidine deaminase APOBEC3G (A3G) potently inhibits human immunodeficiency virus type 1 replication by disrupting viral reverse transcription. However, human immunodeficiency virus type 1 produces a viral infectivity factor (Vif) to destroy this antiretroviral protein. It was shown that Vif binds to both A3G and a Cullin 5 ubiquitin-protein isopeptide ligase. It is currently accepted that this enzyme polyubiquitylates A3G on lysine residues, resulting in its degradation by proteasomes. Here, we find that A3G without ubiquitylation is still degraded by proteasomes in a Vif-dependent manner. We further show that Vif is polyubiquitylated and that this event could be critical for A3G proteasomal degradation. Thus, A3G is degraded by a novel pathway that might involve ubiquitylation of one protein and then targets a second binding partner for proteasomal entry and degradation. We propose that instead of triggering A3G polyubiquitylation, polyubiquitylated Vif might serve as a vehicle to transport A3G into proteasomes for degradation.
Eukaryotes employ proteasomes to destroy misfolded proteins and control levels of protein expression. The 26 S proteasome, composed of a 20 S proteolytic cylinder and two 19 S regulatory caps, specializes in recognition and proteolysis of proteins covalently derivatized with a polyubiquitin chain on certain lysine residues (1). The process of polyubiquitylation involves three enzymes: E1,2 E2, and E3 (2). E1 activates ubiquitin for transfer to E2. E2 interacts with a specific E3 partner, a multiprotein complex that binds directly to the substrate protein, to ensure the target specificity of ubiquitin conjugation. After a single ubiquitin is attached, additional ubiquitins can be linked to one of the seven lysines of ubiquitin to form a multiubiquitin chain, which serves as a signal for proteasomal degradation. Although this model has been verified for a great number of proteins, it also has been reported that some proteins can be degraded in proteasomes in the absence of polyubiquitylation (reviewed in Refs.3 and4).
On human chromosome 22, a cluster of genes encodes a family of seven APOBEC3 (apolipoprotein B mRNA-editingcatalytic polypeptide 3; A3) antiretroviral proteins: A3A, A3B, A3C, A3DE, A3F, A3G, and A3H. They are cytidine deaminases with one or two cytidine deaminase domains (reviewed in Ref.5). A3G potently blocks HIV-1 replication (6). However, HIV-1 produces a protein called Vif to counteract A3G by triggering its proteasomal degradation (7–9). Vif was shown to bridge A3G with a cellular E3 ligase (10). In fact, Vif has a BC-box motif (Ser144/Leu145/Gln146) that binds to elongin C (EloC) (11,12) and an HCCH motif (Cys114/Cys133) that binds to Cullin 5 (Cul5), the core subunits of a Cul5-based E3 ligase (13–15). As a consequence, it is believed that A3G is polyubiquitylated by this enzyme and directed to 26 S proteasomes for degradation. According to this model, A3G is the target of this Cul5 E3 ligase, and polyubiquitylation of A3G is required for proteasomal degradation. Here, we present evidence that the proteasomal degradation of A3G does not require polyubiquitylation of this protein.
EXPERIMENTAL PROCEDURES
Plasmids_—HIV-1 reporter constructs pNL-Luc and pNL-LucΔVif and pcDNA3.1 vectors expressing C-terminally V5-tagged human A3G, A3G-GST, and A3G-FLuc were described previously (16,17). Mammalian expression vector pEF-BOS-HA contains three HA coding sequences upstream of the multiple cloning site. The A3G gene was inserted into this vector by EcoRI and XbaI digestion to express the N-terminally 3× HA-tagged A3G (3× HA-A3G). To express a C-terminally 1× HA-tagged A3G (A3G-HA), the HA gene was fused by PCR to the 3′-end of A3G with an XhoI site between them. This fragment was then inserted into the pEF-BOS-pLinker vector following EcoRI and XbaI digestion. A3G lysine mutants were created by changing lysine to arginine of A3G in the pcDNA3.1 vector using a QuikChange XL site-directed mutagenesis kit (Stratagene). Lysine-free A3G and HIV-1_vif genes were created by a commercial resource (GENEART). The lysine-free vif gene was inserted into the pNL-A1 vector by BssHII and EcoRI digestion. The Vif expression vector pNL-A1 and its control, pNL-A1ΔVif, were provided by K. Strebel (National Institute of Allergy and Infectious Diseases). The expression vectors for Cul5, EloC, Cul5ΔNedd8, and Cul5ΔRbx1 were from L. Liu and X. Yu (Johns Hopkins University).
_Antibodies_—The following antibodies were obtained through the AIDS Research and Reference Reagent Program: a polyclonal anti-Vif antiserum (catalog no. 2221) from D. Gabuzda (Dana-Farber Cancer Institute), two monoclonal anti-Vif antibodies (clones 319 and 564) from M. Malim (King's College London), and a monoclonal anti-p24Gag antibody (catalog no. 3537) from B. Chesebro and K. Wehrly (National Institute of Allergy and Infectious Diseases). Other antibodies used included horseradish peroxidase-conjugated anti-V5 antibody (Invitrogen), horseradish peroxidase-conjugated anti-HA antibody (Roche Applied Science), polyclonal anti-actin antibody (clone C-11; Santa Cruz Biotechnology), and horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Pierce).
_Viral Production and Infectivity Assay_—HIV-1 virions were produced from 293T cells by standard calcium phosphate transfection. Typically, 25 μg of total plasmid DNA were used for each transfection in a 100-mm culture dish with 40% cell confluence. The ratio between these proviral constructs and the APOBEC expression vectors was normally kept at 1:4. The production of HIV-1 was quantitated by p24Gag capture enzyme-linked immunosorbent assay. Equal amounts of viruses were used to infect GHOST-R3/X4/R5 cells. Thirty-six hours later, cells were lysed in buffer composed of 25 mm Tris-HCl, pH 7.8, 2 mm dithiothreitol, 2 mm 1,2-diaminocyclohexane-_N,N,N_′,_N_′-tetraacetic acid, 10% glycerol, and 1% Triton X-100. After removing the nuclei, the cytosolic fraction was used to determine luciferase activity using a luciferase assay kit (Promega).
_Luciferase Reporter Assay for Vif-triggered A3G Degradation_— The A3G-FLuc fusion expression vector was cotransfected into 293T cells with a Vif expression vector plus the pRL-TK vector expressing the Renilla luciferase to serve as an internal control. Twenty-four hours later, cells were lysed, and cytosolic fractions were used to determine the cellular luciferase activities using the Dual-Luciferase assay kit (Promega). The activities are presented as a relative value after normalization to the internal controls.
_In Vivo Polyubiquitylation Assay_—293T cells were cotransfected with the HA-tagged A3G expression vector, Vif expression vector pNL-A1, and FLAG-tagged Ub48A expression vector at a 1:1:1 ratio by a normal calcium phosphate method. Thirty-six hours later, cells were harvested and lysed. Polyubiquitylated proteins were affinity-selected by anti-FLAG antibody M2-conjugated beads (Sigma) and detected by Western blotting with anti-HA antibody.
_Pulse-Chase Radiolabeling_—293T cells were transfected or cotransfected with the Vif and/or A3G-GST expression vectors. The transfected cells were preincubated for 1 h in labeling medium (Dulbecco's modified Eagle's medium without methionine and cysteine (Cellgro) plus 10% dialyzed fetal bovine serum). Each sample was pulse-labeled for 30 min with 250 μCi of EasyTag Expre35S35S Protein Labeling Mix (PerkinElmer Life Sciences). The initial pulse-labeled (t = 0) samples were harvested. The remaining radiolabeled samples were incubated with normal Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and harvested at the indicated times. The cell pellets were lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 5 μg/ml aprotinin, and 5 μg/ml leupeptin) for 15 min on ice and clarified at 14,000 × g for 10 min at 4 °C. After a brief incubation with anti-Vif monoclonal antibodies (catalog no. 564), proteins were pulled down by protein A-conjugated beads (Amersham Biosciences) or glutathione-Sepharose beads (GE Healthcare). After washing, the samples were analyzed by SDS-PAGE. The gels were scanned by Typhoon 9200, and the images were quantified using Image-Quant TL software (Amersham Biosciences).
RESULTS
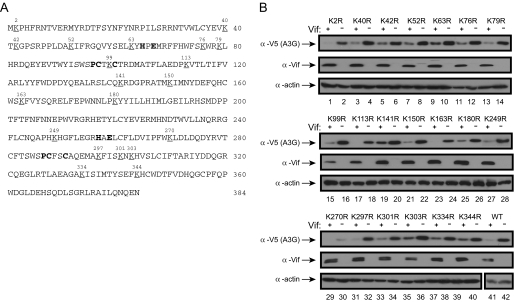
_Mutation of Each Individual Lysine in A3G Does Not Reduce Its Sensitivity to HIV-1 Vif_—As introduced, the polyubiquitylation of A3G was thought to be essential for proteasomal turnover. Accordingly, we tried to map the specific site for A3G polyubiquitylation. Our rationale was that if such a site exists, we should be able to block A3G polyubiquitylation by removal of this site and generate an A3G protein insensitive to Vif. Because polyubiquitylation usually occurs on lysine residues, we first mutated each lysine in A3G to arginine to see if the protein becomes stable in the presence of HIV-1 Vif. Human A3G has 20 lysines (Fig. 1_A_), and we generated 20 A3G lysine mutants in which each lysine (K) was changed to arginine (R): K2R, K40R, K42R, K52R, K63R, K76R, K79R, K99R, K113R, K141R, K150R, K163R, K180R, K249R, K270R, K297R, K301R, K303R, K334R, and K344R. Almost all mutants were well expressed, and the expression of K270R was relatively low (Fig. 1_B_, lane 30). Nonetheless, the expression of each Lys-to-Arg mutant was significantly reduced in the presence of HIV-1 Vif, and the reduction was similar to that of wild-type protein (Fig. 1_B_, lanes 41 and 42). Thus, none of these mutants became resistant to the Vif-triggered degradation, suggesting that there might not be a specific polyubiquitylation site in A3G responsible for proteasomal turnover.
FIGURE 1.
Sensitivity of A3G single lysine mutants to HIV-1 Vif. A, human A3G amino acid sequence. It has 20 lysines that are underlined and numbered. B, expression profile of A3G lysine mutants in the presence of HIV-1 Vif. Twenty A3G single lysine mutants were created by mutating each lysine (K) to arginine (R). The lysine mutants were coexpressed with HIV-1 Vif in 293T cells, and their expressions were determined by Western blotting. Actin blots served as controls for loading. WT, wild-type.
_Lys-free A3G Is Also Destabilized by Vif_—Because we were unable to identify a single lysine in A3G that converts it to Vif resistance, we mutated all lysine residues and created a Lys-free protein, A3G20K/R. We assumed that this protein would not be polyubiquitylated and therefore be much more stable.
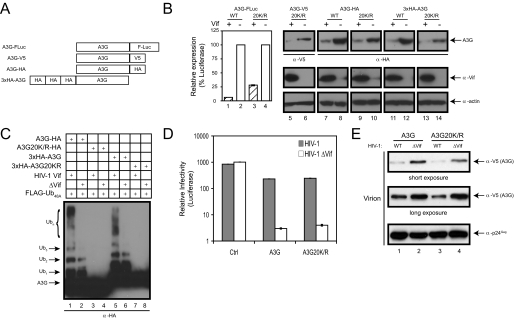
To determine A3G expression, we created four different expression constructs: A3G-FLuc, A3G-V5, and A3G-HA (FLuc, V5, and HA tags fused to the C terminus of A3G) and 3× HA-A3G (three HA repeats fused in tandem to the N terminus of A3G) (Fig. 2_A_). The V5 tag contains one lysine, the FLuc tag contains multiple lysines, and the HA tag contains no lysines. Therefore, the HA-tagged proteins served as controls for introduction of artificial polyubiquitylation sites. Unexpectedly, the Lys-free A3G protein was still sensitive to Vif, as the expressions of A3G20K/R-FLuc, A3G20K/R-V5, A3G20K/R-HA, and 3× HA-A3G20K/R were all significantly decreased in the presence of Vif (Fig. 2_B_). Thus, removal of all lysines did not convert A3G to Vif resistance.
FIGURE 2.
Sensitivity of Lys-free A3G mutants to HIV-1 Vif. A, schematic description of A3G expression constructs. The constructs express wild-type or lysine-free A3G proteins either C-terminally fused with FLuc, V5, or HA or N-terminally fused with 3× HA. B, expression of Lys-free A3G in the presence of HIV-1 Vif. The four A3G expression constructs were transfected alone or coexpressed with HIV-1 Vif, and their levels were determined either by measuring luciferase activity or by Western blotting.WT, wild-type. C, polyubiquitylation of Lys-free A3G in vivo. HA-tagged wild-type or Lys-free A3G was coexpressed with HIV-1 Vif and a FLAG-tagged ubiquitin mutant (Ub48A). Polyubiquitylated proteins were isolated via anti-FLAG-Sepharose beads and visualized by Western blotting using anti-HA antibody. D, anti-HIV-1 activity of Lys-free A3G. Wild-type and _vif_-deficient HIV-1 luciferase reporter viruses were produced in the presence of the wild-type or Lys-free A3G expression vector, and viral infectivity was determined. The pcDNA3.1 empty vector was used as a control (Ctrl). Error bars represent S.D. in three independent experiments. E, virion packaging of Lys-free A3G. Wild-type and _vif_-deficient HIV-1 were produced in the presence of wild-type or Lys-free A3G, and virions were purified by ultracentrifugation. Virion-associated proteins were determined by Western blotting using antibodies for V5 or p24Gag.
Because it was reported that polyubiquitylation might also take place on non-lysine residues such as cysteine, serine, and threonine or through the N terminus (18–20), we wanted to confirm that Lys-free mutants were, indeed, not polyubiquitylated_in vivo_. Ubiquitin conjugates commonly represent only a small fraction of the steady-state population, and inhibition of chain elongation is often required for such conjugates to accumulate to detectable levels. The 76-amino acid ubiquitin has seven lysines (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) that could participate in multi-ubiquitin chain elongation. Lys48-linked polyubiquitylation generally associates with proteasomal degradation, whereas Lys63-linked polyubiquitylation signifies lysosomal degradation. Two ubiquitin lysine mutants, Ub48A and Ub63A, bearing a single Lys-to-Ala mutation at position 48 or 63, respectively, should interrupt Lys48- or Lys63-linked ubiquitin chain initiation and specifically block these degradations. To detect Lys48-linked polyubiquitylation, the Ub48A mutant was coexpressed with A3G in the presence of HIV-1 Vif. We detected many polyubiquitylated intermediates of wild-type A3G (A3G-HA and 3× HA-A3G) in the presence of Vif, suggesting that wild-type A3G is efficiently polyubiquitylated in vivo (Fig. 2_C_, lanes 1 and 5). In contrast, no such intermediate was detected using the Lys-free protein (A3G20K/R-HA and 3× HA-A3G20K/R) (Fig. 2_C_, lanes 3 and 7). These results confirm that Lys-free A3G was not polyubiquitylated and also exclude lysine-independent polyubiquitylation of A3G.
To ensure that mutagenesis manipulations did not change A3G conformation and alter its function, the anti-HIV activity and virion incorporation of the Lys-free A3G were compared with the wild-type protein. A3G20K/R reduced the infectivity of the _vif_-deficient HIV-1 to a level similar to the wild-type protein (Fig. 2_D_), and Lys-free A3G was also efficiently packaged into_vif_-deficient HIV-1 virions (Fig. 2_E_). Thus, Lys-free A3G retains its antiretroviral activity.
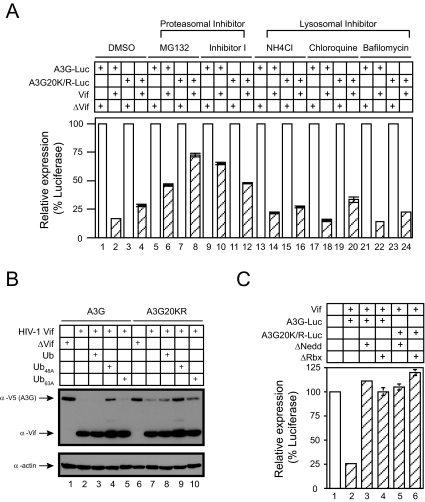
_Lys-free A3G Degradation Requires Proteasome, Polyubiquitylation System, and Cul5 E3 Ligase_—To further understand Lys-free A3G degradation, we first tested proteasome-mediated degradation. We attempted to block A3G degradation using either proteasomal inhibitors (MG132 and proteasome inhibitor I) or lysosomal inhibitors (NH4Cl, chloroquine, and bafilomycin). None of the lysosomal inhibitors blocked wild-type or Lys-free A3G protein degradation (Fig. 3_A_, bars 13–24), suggesting that lysosomes are not responsible for A3G degradation. Instead, degradation of both proteins was effectively, albeit not completely, blocked by MG132 or proteasome inhibitor I (Fig. 3_A_, bars 5–12). Thus, like the wild-type protein, Lys-free A3G is also degraded in proteasomes even though it is not polyubiquitylated.
FIGURE 3.
Lys-free A3G is degraded in proteasomes. A, requirement of a functional proteasomal system for degradation. Wild-type or Lys-free A3G-FLuc reporter constructs were cotransfected with HIV-1 Vif expression vector pNL-A1 or its control, pNL-A1ΔVif. Cells were treated with proteasomal inhibitor MG132 (20 μm) or proteasome inhibitor I (10 μm) and lysosomal inhibitor NH4Cl (20 mm), chloroquine (100 nm), or bafilomycin (200 nm). Cellular luciferase activities were then determined.B, requirement of a functional polyubiquitylation system for degradation. Wild-type or Lys-free A3G proteins were coexpressed with HIV-1 Vif in the presence of wild-type ubiquitin or mutant ubiquitins (Ub48A and Ub63A). A3G and Vif expressions were determined by Western blotting. C, requirement of the Cul5 E3 ligase for degradation. Wild-type or Lys-free A3G-FLuc fusion proteins were coexpressed with Vif proteins in the presence of Cul5ΔNedd8 (Δ_Nedd_) or Cul5ΔRbx1 (Δ_Rbx_). Cellular luciferase activities were then determined as before. Error bars in_A_ and C represent S.D. of at least three independent experiments.
We then determined whether a functional polyubiquitylation system is required for degradation. As introduced before, Ub48A and Ub63A mutants specifically interfere with proteasomal or lysosomal degradation by blocking Lys48-or Lys63-linked polyubiquitylation, respectively. Our results indicate that in the presence of HIV-1 Vif, the degradation of wild-type and Lys-free A3G was blocked by Ub48A (Fig. 3_B_, lanes 4 and 9). In contrast, wild-type Ub and Ub63A had no effect on degradation (Fig. 3_B_, lanes 3, 5, 8, and 10). Thus, these results not only confirm the proteasomal degradation but also suggest that a functional polyubiquitylation system, specifically Lys48-linked polyubiquitylation, is required for Lys-free A3G degradation.
The requirement for polyubiquitylation implies the involvement of E3 ligases. To test whether the same Cul5 E3 ligase was involved in A3G degradation, we disrupted its assembly with two Cul5 mutants: Cul5ΔNedd8 defective for Nedd8 modification and Cul5ΔRbx1 defective for Rbx1 binding (10). In the presence of these two mutants, we completely blocked the degradation of wild-type A3G triggered by Vif (Fig. 3_C_, bars 3 and 4), as well as Lys-free A3G degradation (bars 5 and 6). Thus, the Cul5 E3 ligase is required for degradation of Lys-free A3G.
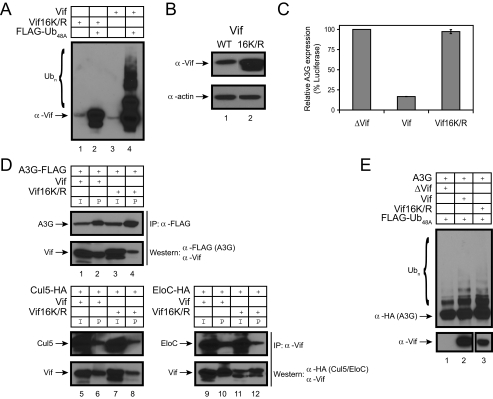
_Role of Vif Polyubiquitylation in A3G Degradation_—These results suggest that there is another Cul5 E3 ligase substrate that plays a critical role in A3G proteasomal degradation. HIV-1 Vif has 190 amino acids, including 16 lysines. It was reported that HIV-1 Vif itself is unstable and rapidly degraded in proteasomes (21,22). Moreover, it is polyubiquitylated by the Cul5 E3 ligase (11). We confirmed that HIV-1 Vif was polyubiquitylated in vivo (Fig. 4_A_, lane 4). In contrast, Lys-free HIV-1 Vif (Vif16K/R) ws not polyubiquitylated (Fig. 4_A_, lane 2). Lys-free Vif was more abundant than the wild-type protein when expressed in 293T cells (Fig. 4_B_), suggesting that it could be more stable. Importantly, Lys-free Vif was inactive in triggering wild-type A3G protein degradation (Fig. 4_C_).
FIGURE 4.
Role of Vif polyubiquitylation in A3G degradation. A, polyubiquitylation of HIV-1 Vif protein in vivo. Wild-type or Lys-free (Vif16K/R) HIV-1 Vif proteins were coexpressed with FLAG-tagged Ub48A. Proteins were isolated on anti-FLAG beads, and polyubiquitylated proteins were visualized by Western blotting using anti-Vif polyclonal antiserum. B, expression of Lys-free Vif. Equal amounts of expression vectors for wild-type (WT) or Lys-free Vif were transfected into 293T cells, and protein expression was determined by Western blotting. C, activity of Lys-free Vif. Wild-type or Lys-free Vif proteins were coexpressed with A3G protein fused with FLuc in 293T cells, and the cellular luciferase activity was determined as before. Error bars represent S.D. in at least three independent experiments. D, interaction of HIV-1 Vif with A3G and Cul5 E3 ligase. Vif proteins were coexpressed with FLAG-tagged A3G or HA-tagged Cul5 or EloC. To detect the interaction between Vif and A3G, proteins were pulled down by anti-FLAG beads and analyzed by Western blotting using anti-FLAG and anti-Vif antibodies. To detect the interaction between Vif and Cul5 or EloC, proteins were immunoprecipitated (IP) by anti-Vif polyclonal antibody and analyzed by Western blotting using anti-Vif and anti-HA antibodies. I, input;P, pull down. E, A3G polyubiquitylation by Lys-free Vif. Wild-type A3G was coexpressed with wild-type or Lys-free Vif in the presence of the FLAG-tagged Ub48A expression vector. Proteins were isolated on anti-FLAG beads, and polyubiquitylated proteins were visualized by Western blotting using anti-Vif polyclonal antiserum.
The inability of Lys-free Vif to trigger A3G degradation could be due to a conformational change in the Lys-free protein resulting in loss of function. We performed the following experiments to rule out this possibility. First, we determined the binding of Lys-free Vif to A3G and Cul5 E3 ligase. Although the binding of Lys-free Vif to A3G seemed to be reduced compared with the wild-type protein (Fig. 4_D_, lanes 2 and 4), the Lys-free mutant protein still bound to Cul5 and EloC as efficiently as wild-type Vif (lanes 5–12). Second, we determined the ability of Lys-free Vif to trigger A3G polyubiquitylation. Indeed, this activity was retained by Lys-free Vif (Fig. 4_E_,lane 3). Finally, even though Lys-free Vif interacted with the E3 ligase and triggered A3G polyubiquitylation, a conformational change could result in cellular mislocalization and a “functional” null phenotype for A3G degradation. As reported previously for wild-type Vif (23–26), we detected Lys-free Vif in the nucleus and cytoplasm (data not shown). Lys-free Vif retains those functions associated with unmodified Vif. Thus, polyubiquitylation of Vif could be very critical for A3G degradation in proteasomes.
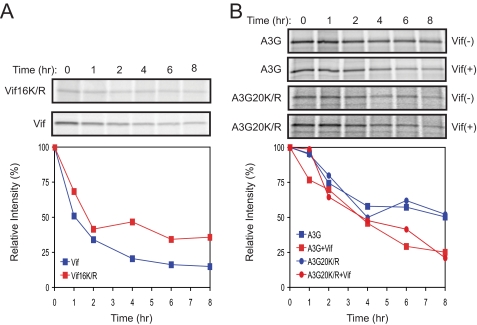
_Half-life of Lys-free Vif and A3G_— Having determined the role of polyubiquitylation in the degradation of Vif and A3G, we determined the stability of these proteins in the presence and absence of this post-translational modification. Pulse-chase radiolabeling studies were performed to measure half-life (_t_½). We first compared the _t_½ of Lys-free Vif with its wild-type protein. 293T cells were transfected with vector expressing either wild-type or mutant protein. After incubation in Met/Cys-free medium followed by a 30-min pulse, cultures were chased for 8 h. Vif was immunoprecipitated from cell lysates harvested at the indicated times. The_t_½ of wild-type Vif was ∼1 h, whereas that of the Lys-free Vif (Vif16K/R) was extended to at least 2 h (Fig. 5_A_). Next, we compared the stability of Lys-free A3G with its wild-type protein. The wild-type or mutant A3G-GST protein was expressed alone or coexpressed with Vif in 293T cells. After pulse-chase labeling, A3G was isolated by absorption to glutathione beads. In the absence of Vif, the _t_½ values of A3G and Lys-free A3G were similar (∼8 h). However, in the presence of Vif, the _t_½ of both proteins decreased at a similar rate, with a _t_½ that was ∼4 h (Fig. 5_B_). These findings demonstrate that polyubiquitylation destabilizes Vif, but it does not decrease the stability of A3G, which is consistent with our previous results.
FIGURE 5.
Half-life of Lys-free Vif and A3G. A, pulse-chase radiolabeling of Vif. 293T cells were transiently transfected with the wild-type or Lys-free Vif expression vector. Cells were pulse-labeled for 30 min and then incubated for the indicated time points. After immunoprecipitation of lysates with anti-Vif antibody, proteins were separated by SDS-PAGE, and the gels were subjected to autoradiography. The relative intensity of the bands was quantified, normalized to 100, and graphed.B, pulse-chase radiolabeling of A3G. GST-tagged wild-type or Lys-free A3G was transiently expressed in 293T cells in the presence or absence of Vif. Cells were pulse-chased as above, and proteins were isolated on glutathione-Sepharose beads. Labeled A3G proteins were analyzed and quantified as in A.
DISCUSSION
In this study, we have demonstrated that HIV-1 Vif triggers the proteasomal turnover of A3G but that this event does not require A3G polyubiquitylation. Instead, A3G degradation requires polyubiquitylation of Vif. Disruption of Vif polyubiquitylation completely abolishes A3G proteasomal turnover. Thus, the polyubiquitylation of Vif might contribute significantly to A3G proteasomal degradation.
Since the discovery of A3G inhibitory activity to HIV-1, efforts have been made to understand how A3G blocks retroviral replication and why A3G is so vulnerable to HIV-1 Vif. Soon after the finding that A3G induced C-to-U hypermutations on the viral genome, it was found that Vif triggers A3G polyubiquitylation and proteasomal turnover (7–9). Although the interaction between Vif and A3G was demonstrated, it was not understood how Vif could initiate this degradation until it was found that Vif also interacts with Cul5 E3 ligase (10). Because this Cul5 E3 ligase polyubiquitylates A3G and is required for A3G degradation, it has been assumed that the polyubiquitylation of A3G is necessary for its proteasomal entry and turnover. A well accepted model was therefore generated in which Vif acts as an adaptor protein that bridges A3G with Cul5 E3 ligase to trigger polyubiquitylation of A3G. However, the demonstration of such ubiquitin-protein conjugates does not eliminate the possibility that A3G can also be degraded in the absence of prior ubiquitylation.
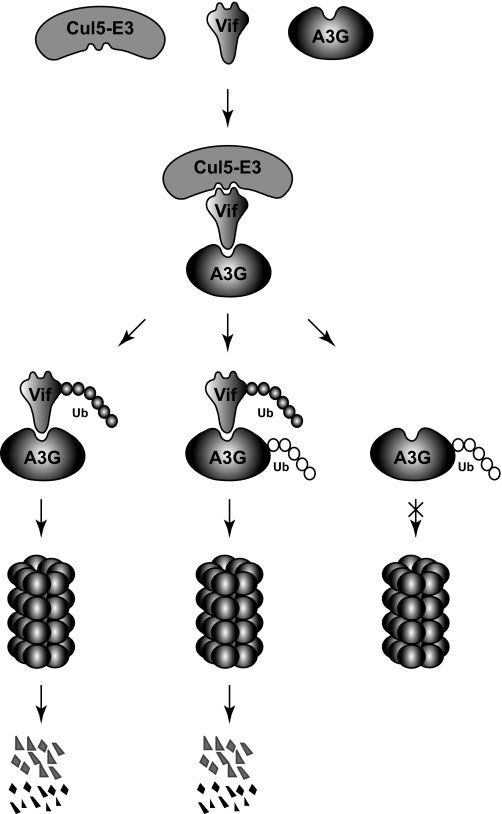
As mentioned previously, it was reported that Vif is also polyubiquitylated and subject to proteasomal degradation (21,22). Unfortunately, the function of this polyubiquitylation has attracted little attention even though this reaction is also catalyzed by the same Cul5 E3 ligase (11). Here, by creating Lys-free Vif and A3G, we have demonstrated that Vif polyubiquitylation could be critical for A3G proteasomal turnover. Indeed, our pulse-chase experiments demonstrated that A3G polyubiquitylation does not lead to more rapid degradation, which is reminiscent of a finding from the yeast Met4 transcription factor (27). It was reported that even though Met4 was polyubiquitylated via a Lys48-linked ubiquitin chain, it was still quite stable. Thus, we propose a possible new model explaining Vif destabilization of A3G (Fig. 6). In this model, although Vif can trigger A3G polyubiquitylation via Cul5 E3 ligase, it is more important that Vif itself is polyubiquitylated by this enzyme. The polyubiquitylated Vif then functions as a vehicle to transport A3G to proteasomes for degradation. In the absence of this vehicle, A3G does not have access to proteasomes even though it is polyubiquitylated.
FIGURE 6.
Possible model of how Vif targets A3G for proteasomal degradation. Vif interacts with both A3G and a Cul5 E3 ligase. These interactions result in the polyubiquitylation of both Vif and A3G proteins by this enzyme. However, only the polyubiquitin chain of Vif can direct the Vif and A3G complex to proteasomes for degradation.
A small number of proteins have been identified as substrates for the proteasome yet do not require ubiquitin for their degradation, indicating the complexities of the cellular proteolytic machinery. In most of these cases, degradation does not require a functional ubiquitylation system. Examples include ornithine decarboxylase (28), the cyclin-dependent kinase inhibitor p21Cip/Kip (29), retinoblastoma protein (30), p53 (31), and thymidylate synthase (32). These proteins could be targeted to the proteasome by a proteasome-recognizable signal other than ubiquitin or by binding to a proteasome subunit directly. For example, when a 35–40-amino acid peptide from the C-terminal extension of ornithine decarboxylase is linked to a variety of stable proteins, these proteins become vulnerable to proteasomal degradation. Also, both p21 and retinoblastoma protein interact with the C8 (α7) subunit of the 20 S core particle of the proteasome.
In addition, although polyubiquitylation is not required for proteasomal turnover, the protein degradation process may still require a functional ubiquitylation system. For example, the unassembled T-cell receptor α subunits are degraded in proteasomes after dislocation from the endoplasmic reticulum membrane. When polyubiquitylation of T-cell receptor α was blocked by mutating all lysines, it was still degraded by proteasomes. This degradation could be blocked by Ub48R or by mutating E1 in a temperature-sensitive cell line (ts20) (33), suggesting that a functional ubiquitin pathway is necessary for this process. It was speculated that an unknown polyubiquitylated factor might target T-cell receptor α for dislocation from the endoplasmic reticulum and proteasomal degradation. This case is quite similar to what we have found for A3G proteasomal turnover. Here, we have documented the involvement of polyubiquitylated Vif in A3G degradation. This is the first demonstration that a protein can be targeted to proteasomes by another protein via a direct interaction. The precise mechanism for this indirect proteasomal targeting becomes an interesting subject for future studies. Understanding this novel mechanism should lead to new strategies to block Vif-triggered A3G degradation, which could serve as an important reference for HIV drug development.
Acknowledgments
We thank Ronald Patterson, Ian York, Patrick Venta, and Walter J. Esselman for critical reading of this manuscript. We thank K. Strebel, B. Liu, and X. F. Yu for reagents. We thank the National Institutes of Health AIDS Reagent Repository for antibodies contributed by M. Malim, D. Gabuzda, B. Chesebro, and K. Wehrly.
*
This work was supported, in whole or in part, by National Institutes of Health Grant AI063344 (to Y.-H. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
2
The abbreviations used are: E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; HIV-1, human immunodeficiency virus type 1; A3, APOBEC3; Vif, viral infectivity factor; FLuc, firefly luciferase; Cul5, Cullin 5; EloC, elongin C; HA, hemagglutinin; GST, glutathione _S_-transferase; Ub, ubiquitin.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 2.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 3.Hoyt, M. A., and Coffino, A. (2007) Protein Degradation 4 107-121 [Google Scholar]
- 4.Verma, R., and Deshaies, R. J. (2000) Cell 101 341-344 [DOI] [PubMed] [Google Scholar]
- 5.Holmes, R. K., Malim, M. H., and Bishop, K. N. (2007) Trends Biochem. Sci. 32 118-128 [DOI] [PubMed] [Google Scholar]
- 6.Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002) Nature 418 646-650 [DOI] [PubMed] [Google Scholar]
- 7.Marin, M., Rose, K. M., Kozak, S. L., and Kabat, D. (2003) Nat. Med. 9 1398-1403 [DOI] [PubMed] [Google Scholar]
- 8.Sheehy, A. M., Gaddis, N. C., and Malim, M. H. (2003) Nat. Med. 9 1404-1407 [DOI] [PubMed] [Google Scholar]
- 9.Stopak, K., de Noronha, C., Yonemoto, W., and Greene, W. C. (2003) Mol. Cell 12 591-601 [DOI] [PubMed] [Google Scholar]
- 10.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X. F. (2003) Science 302 1056-1060 [DOI] [PubMed] [Google Scholar]
- 11.Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M., and Gabuzda, D. (2004) Genes Dev. 18 2861-2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu, Y., Xiao, Z., Ehrlich, E. S., Yu, X., and Yu, X. F. (2004) Genes Dev. 18 2867-2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo, K., Xiao, Z., Ehrlich, E., Yu, Y., Liu, B., Zheng, S., and Yu, X. F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11444-11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehle, A., Thomas, E. R., Rajendran, K. S., and Gabuzda, D. (2006) J. Biol. Chem. 281 17259-17265 [DOI] [PubMed] [Google Scholar]
- 15.Xiao, Z., Ehrlich, E., Yu, Y., Luo, K., Wang, T., Tian, C., and Yu, X. F. (2006) Virology 349 290-299 [DOI] [PubMed] [Google Scholar]
- 16.Dang, Y., Wang, X., Esselman, W. J., and Zheng, Y.-H. (2006) J. Virol. 80 10522-10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng, Y.-H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T., and Peterlin, B. M. (2004) J. Virol. 78 6073-6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadwell, K., and Coscoy, L. (2005) Science 309 127-130 [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover, A., and Ben-Saadon, R. (2004) Trends Cell Biol. 14 103-106 [DOI] [PubMed] [Google Scholar]
- 20.Wang, X., Herr, R. A., Chua, W. J., Lybarger, L., Wiertz, E. J., and Hansen, T. H. (2007) J. Cell Biol. 177 613-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akari, H., Fujita, M., Kao, S., Khan, M. A., Shehu-Xhilaga, M., Adachi, A., and Strebel, K. (2004) J. Biol. Chem. 279 12355-12362 [DOI] [PubMed] [Google Scholar]
- 22.Mehle, A., Strack, B., Ancuta, P., Zhang, C., McPike, M., and Gabuzda, D. (2004) J. Biol. Chem. 279 7792-7798 [DOI] [PubMed] [Google Scholar]
- 23.Goncalves, J., Shi, B., Yang, X., and Gabuzda, D. (1995) J. Virol. 69 7196-7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, S., Miyagi, E., Khan, M. A., Takeuchi, H., Opi, S., Goila-Gaur, R., and Strebel, K. (2004) Retrovirology 1 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madani, N., Millette, R., Platt, E. J., Marin, M., Kozak, S. L., Bloch, D. B., and Kabat, D. (2002) J. Virol. 76 11133-11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon, J. H., Fouchier, R. A., Southerling, T. E., Guerra, C. B., Grant, C. K., and Malim, M. H. (1997) J. Virol. 71 5259-5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flick, K., Ouni, I., Wohlschlegel, J. A., Capati, C., McDonald, W. H., Yates, J. R., and Kaiser, P. (2004) Nat. Cell Biol. 6 634-641 [DOI] [PubMed] [Google Scholar]
- 28.Murakami, Y., Matsufuji, S., Kameji, T., Hayashi, S., Igarashi, K., Tamura, T., Tanaka, K., and Ichihara, A. (1992) Nature 360 597-599 [DOI] [PubMed] [Google Scholar]
- 29.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5 403-410 [DOI] [PubMed] [Google Scholar]
- 30.Kalejta, R. F., and Shenk, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3263-3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asher, G., Lotem, J., Sachs, L., Kahana, C., and Shaul, Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13125-13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena, M. M., Xing, Y. Y., Koli, S., and Berger, F. G. (2006) Biochem. J. 394 355-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, H., and Kopito, R. R. (1999) J. Biol. Chem. 274 36852-36858 [DOI] [PubMed] [Google Scholar]