Human Cytidine Deaminase APOBEC3H Restricts HIV-1 Replication (original) (raw)
Abstract
The human genome encodes seven APOBEC3 (A3) cytidine deaminases with potential antiretroviral activity: A3A, A3B, A3C, A3DE, A3F, A3G, and A3H. A3G was the first identified to block replication of human immunodeficiency virus type 1 (HIV-1) and many other retroviruses. A3F, A3B, and A3DE were shown later to have similar activities. HIV-1 produces a protein called Vif that is able to neutralize the antiretroviral activities of A3DE, A3F, and A3G, but not A3B. Only the antiretroviral activity of A3H remains to be defined due to its poor expression in cell culture. Here, we studied the mechanism impairing A3H expression. When primate A3H sequences were compared, a premature termination codon was identified on the fifth exon of the human and chimpanzee A3H genes, which significantly decreased their protein expression. It causes a 29-residue deletion from the C terminus, and this truncation did not reduce human A3H protein stability. However, the mRNA levels of the truncated gene were significantly decreased. Human A3H protein expression could be restored to a normal level either by repairing this truncation or through expression from a vector containing an intron from human cytomegalovirus. Once expression was optimized, human A3H could reduce HIV-1 infectivity up to 150-fold. Importantly, HIV-1 Vif failed to neutralize A3H activity. Nevertheless, extensive sequence analysis could not detect any significant levels of G-to-A mutation in the HIV-1 genome by human A3H. Thus, A3H inhibits HIV-1 replication potently by a cytidine deamination-independent mechanism, and optimizing A3H expression in vivo should represent a novel therapeutic strategy for HIV-1 treatment.
Human chromosome 22 contains a cluster of APOBEC3 (A3)2 genes encoding seven potential antiretroviral proteins: A3A, A3B, A3C, A3DE, A3F, A3G, and A3H. They are members of the cytidine deaminase family (APOBEC) that have one or two copies of the cytidine deaminase domain (CDD) (1). All the A3 proteins, with the exception of A3H, have been shown to inhibit the replication of various retroviruses. In particular, A3B, A3DE, A3F, and A3G block HIV-1 replication (2-8). Within these, A3G has the most powerful anti-HIV-1 activity (6). A3A and A3C block the replication of the endogenous retrotransposons with or without long terminal repeat, whereas A3A additionally blocks replication of the adenovirus-associated virus (9,10). Previously, it was reported that human A3H is poorly expressed in cell culture, and its antiviral activity is not yet determined (11).
Initially, it was thought that A3 proteins inhibit retroviral replication via a catalytic mechanism. They could deaminate deoxycytidine to form deoxyuridine on nascent viral cDNAs during viral reverse transcription, resulting in G-to-A mutations in the plus-strand DNA leading to abortive replication (12-15). Later, a noncatalytic mechanism was reported through functional analysis of A3G deaminase active site mutants (16). This cytidine deamination-independent mechanism has been observed in many different viruses by various A3 proteins. For example, the inhibition of adenovirus-associated virus, human T-cell lymphotrophic virus type 1, and retrotransposon replication by A3A, A3B, A3F, or A3G could occur in the absence of G-to-A mutation (9,17-20). Further investigations demonstrated that A3G could interrupt viral reverse transcription by reducing the efficiency of tRNALys3 priming to the viral RNA template, elongation, and DNA strand transfer (21-24). In addition, it could also block viral cDNA integration (24,25). Nevertheless, HIV-1 is able to elude this defense mechanism and cause disease in humans. HIV-1 produces the viral infectivity factor Vif, which binds to and mediates the destruction of A3DE, A3F, and A3G (3,7,26-28) via recruitment of the Cullin-5 ubiquitin-protein isopeptide ligase (29). As a consequence, these A3 proteins are polyubiquitylated and directed to 26 S proteasomes for degradation (8,26-28,30). Additionally, a degradation-independent mechanism was also reported to inactivate A3G protein (31).
In this work, we studied the gene expression and antiretroviral activity of human A3H. We confirmed previous observations that human A3H was poorly expressed in cell culture and demonstrated that its poor expression was due to a premature termination codon (PTC) on its fifth exon, which severely impaired its mRNA expression. Once its expression was optimized, human A3H indeed blocked replication of both HIV-1 and SIV. Moreover, its antiretroviral activity was not countered by HIV-1 Vif. Our results shed light on the unique feature of the human A3H gene for its anti-HIV potential, which can be explored for future anti-HIV therapy.
EXPERIMENTAL PROCEDURES
_Plasmids_—HIV-1 proviral constructs pNL-Luc and pNL-LucΔVif and mammalian expression vectors for human A3G were described previously (3,8). The full-length macA3H cDNA clone was provided by M. Emerman (Fred Hutchinson Cancer Research Center), and the full-length agmA3H cDNA was amplified from COS-7 cells by RT-PCR. They were both cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen). The C-terminally truncated macA3H and agmA3H were created by PCR and cloned into the same vector. The full-length huA3H cDNA with a PTC and C-terminally truncated huA3H were cloned from human PBMCs by RT-PCR. The full-length clone was then used as template to repair the PTC using a QuikChange XL site-directed mutagenesis kit (Stratagene) to express full-length huA3H protein. cpzA3H cDNA was synthesized by overlapping 24 oligonucleotides with ∼50 bases, covering the entire gene with the PTC repaired. The C-terminally truncated cpzA3H gene was created by PCR. These human and chimpanzee A3H genes were cloned into the pcDNA3.1D/V5-His-TOPO vector. To create plasmids expressing A3H C-terminally fused with firefly luciferase (huA3H-FLuc and huA3H-L-FLuc), the firefly luciferase gene was amplified from pSP-luc+NF (Promega) by PCR and inserted into the pcDNA3-A3H expression vectors by NotI and XbaI digestion. This cloning did not interfere with the expression of the C-terminal V5 tag. To create plasmids expressing A3H N-terminally fused with Renilla luciferase (RLuc-huA3H and RLuc-huA3H-L), the Renilla luciferase gene was amplified from pRL-TK (Promega) by PCR with a FLAG-coding sequence embedded in the sense primer and cloned into the pcDNA3.1D/V5-His-TOPO vector. A3H genes were then inserted into this vector by NotI and XbaI digestion. To create plasmids expressing A3H C-terminally fused with a HA/FLAG tag, two oligonucleotides encoding the sense HA/FLAG gene with a 5′-NotI cohesive end or antisense HA/FLAG gene with a 5′-XbaI cohesive end were synthesized. After annealing, they were inserted into these A3H expression vectors by NotI and XbaI digestion. Various APOBEC genes were also cloned into another mammalian expression vector, VR (32). To create the VR-huA3H and VR-huA3H-L expression vectors, the human A3H and A3H-L genes were PCR-amplified with a forward primer, 5′-tgc_tctaga_ATGGCTCTGTTAACAGCCG-3′, and two different reverse primers, 5′-ga_agatct_ttaggcgtaatcgggcacgtcgtaggggta_ctcgag_GGACTTTATCC-3′ and 5′-ga_agatct_ttaggcgtaatcgggcacgtcgtaggggta_ctcgag_TCTTGAGTTGC-3′, respectively. The forward primer contains an XbaI site (lowercase, italicized), and the reverse primers contain BglII and XhoI sites (lowercase, italicized) and a HA tag-coding sequence (lowercase, underlined). These two fragments were then cloned into the VR-Cul5ΔNedd expression vector obtained from B. Liu and X. F. Yu (Johns Hopkins University) after removing the Cul5ΔNedd gene fragment by XbaI and BglII digestion. The human A3A, A3DE, A3F, and A3G and chimpanzee A3H genes were then cloned into this vector by replacing the human A3H gene by XbaI and XhoI digestion. The sequences of all these constructs were verified by nucleotide sequencing. SIV proviral clones SIVagm-Luc, SIVagm-LucΔVif, SIVmac-Luc, and SIVmac-LucΔVif were provided by N. Landau (New York University). pNL-A1, pNL-A1ΔVif, pNL-A1agmVif, and pNL-A1macVif were provided by K. Strebel (NIAID, National Institutes of Health).
_Viral Production and Infectivity Assay_—HIV-1 and SIV virions were produced from 293T cells by standard calcium phosphate transfection. Both SIVagm and SIVmac were pseudotyped with vesicular stomatitis virus glycoprotein because they did not encode a functional envelope protein. The production of HIV-1 was quantitated by p24Gag capture enzyme-linked immunosorbent assay and SIV by viral reverse transcriptase assay with a reaction mixture that contained 50 mm Tris-HCl, pH 8.0, 75 mm KCl, 2 mm dithiothreitol, 24 μg/ml poly(rA)/oligo(dT), 5 mm MgCl2, 1% Nonidet P-40, and 100 μCi/ml [3H]-dideoxythymidine 5′-triphosphate. Equal amounts of viruses were used to infect GHOST-R3/X4/R5 cells. Thirty-six hours later, cells were lysed in 25 mm Tris-HCl, pH 7.8, 2 mm dithiothreitol, 2 mm DCTA, 10% glycerol, and 1% Triton X-100. The cytosolic fraction was then used to determine the luciferase activity using a luciferase assay kit (Promega).
_Sequencing of Newly Synthesized Viral cDNA_—GHOST-R3/X4/R5 cells were infected with HIV-1 viruses produced from 293T cells in the presence of various A3 proteins after being treated with RQ1 RNase-free DNase (Promega) to remove any plasmid DNA contamination. Eight hours later, cellular DNAs were extracted by the DNeasy tissue kit (Qiagen). A 420-bp fragment was PCR-amplified by a previously described primer pair (8) and cloned into the pCR4-TOPO vector (Invitrogen). Multiple clones were selected and sequenced by the flanking T3 and T7 primers.
_Measurement of Human A3G and A3H mRNAs and A3H Genomic DNA by Real-time PCR_—293 cell lines stably transfected with the huA3H-FLuc or huA3H-L-FLuc expression vector were generated by G418 selection. Total cellular DNAs were extracted by the DNeasy tissue kit, and total cellular RNAs were extracted by TRIzol (Invitrogen) from these two cell lines. To measure A3H mRNA levels in these stable cell lines, 1 μg of total RNA was subjected to reverse transcription using Superscriptase II reverse transcriptase and oligo(dT)12-18 as a primer (Invitrogen). Synthesized cDNAs were then subjected to the real-time PCR using the TaqMan® Master Mix gene expression kit (Applied Biosystems). The forward primer was huA3H-183.txt-100F (5′-ctgacgccgcagaatgg-3′), the reverse primer was huA3H-183.txt-226R (5′-cttggtagcactgcgtttcg-3′), and the probe was huA3H-183.txt-139T (5′-6-carboxy-fluorescein-tttgaaaacaagaaaaagtgccatgcagaaa-6-carboxy-tetramethyl-rhodamine-3′). After initial incubation at 50 °C for 2 min and 95 °C for 10 min, 40 cycles of amplification were carried out for 15 s at 95 °C followed by 1 min at 60 °C. Reactions were analyzed using the 7900HT system (Applied Biosystems). To measure the integrated A3H DNA levels in these 293 cell lines, 1 μg of total cellular DNA was subjected to real-time PCR similarly. In addition, the levels of human A3G and A3H in PBMCs were also quantitated by the SYBR Green® PCR Master Mix kit (Applied Biosystems). The total RNA of human PBMCs was purchased from Clontech. After reverse transcription, transcripts were quantitated by this real-time PCR kit using A3H- or A3G-specific primers. The same primer pair was used for A3H amplification. The A3G primer pair was 5′-GAACCTTGGGTCAGAGGA-3′ and 5′-GACATCTTCCTTGATCAT-3′. In all these measurements, A3H and A3G mRNA levels were finally normalized to the levels of hypoxanthine-guanine phosphoribosyltransferase mRNA, which were determined by the SYBR Green® PCR Master Mix kit using primers 5′-AGATGGTCAAGGTCGCAAGC-3′ and 5′-GGACTCCAGATGTTTCCAAACTCAAC-3′. DNA levels were normalized to the amounts of input DNA.
_Immunoprecipitation Assay_—To determine A3H and Vif interaction, 293T cells were transfected with vectors expressing Vif and HA/FLAG-tagged A3H proteins and lysed with radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 5 μg/ml aprotinin, and 5 μg/ml leupeptin). The cytosolic fraction was precleared with Sepharose 4B beads and then rocked with anti-FLAG antibody M2-conjugated beads (Sigma) for 4 h at 4 °C. After extensive washing with phosphate-buffered saline, bead-associated proteins were detected by Western blotting using a polyclonal anti-HIV-1 Vif antibody (catalog no. 2221).
Detection of Protein Expression_—Protein expression was directly determined by Western blotting. HIV-1 Gag and Vif proteins were detected by antibodies from the National Institutes of Health AIDS Research and Reference Reagent Program (catalog nos. 3537, 6459, and 2221). Commercial antibodies were horseradish peroxidase-conjugated anti-V5 antibody (Invitrogen), horseradish peroxidase-conjugated anti-HA antibody (Roche Applied Science), anti-actin polyclonal antibody (C-11; Santa Cruz Biotechnology), and horseradish peroxidase-conjugated anti-rabbit or mouse IgG secondary antibodies (Pierce). Detection of the horseradish peroxidase-conjugated antibody was performed using the SuperSignal West Pico chemiluminescence substrate kit (Pierce). In addition, the expression of various A3H proteins was also determined by luciferase assay. The A3H-Luc fusion expression vectors were cotransfected into 293T cells with the Vif expression vector or its control plus the pRL-TK vector expressing_Renilla luciferase. Twenty-four hours later, cells were lysed, and cytosolic fractions were used to determine the cellular luciferase activities using the Dual-Luciferase assay kit (Promega). Luciferase activity was then determined by a Veritas™ microplate luminometer (Turner Biosystems). The firefly luciferase activity is presented as a relative value after being normalized to the Renilla luciferase activity. The RLuc-A3H series reporter assays were performed similarly. In some experiments, cells were treated with 40 mm NH4Cl (J. T. Baker Inc.), 0.4 μm bafilomycin (Sigma), 20 μm MG132 (Sigma), and 100 nm wortmannin (Sigma) for 16 h before the detection of protein expression.
Pulse-Chase Radiolabeling_—293T cells were transfected with the VR vector expressing the huA3H or huA3H-L protein. The transfected cells were preincubated for 1 h in labeling medium (Dulbecco's modified Eagle's medium without methionine and cysteine (Cellgro) plus 10% dialyzed fetal bovine serum). Each sample was subsequently pulse-labeled for 30 min with 250 μCi of EasyTag Express35S protein labeling mixture (PerkinElmer Life Sciences). The initial pulse-labeled (t = 0) samples were harvested. The remaining radiolabeled samples were incubated with normal Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and harvested at various time points. The harvested cell pellets were lysed in radioimmune precipitation assay buffer for 15 min on ice and clarified at 14,000 ×_g at 4 °C for 10 min. Human A3H proteins were then immunoprecipitated by anti-HA antibody-conjugated beads (Roche Applied Science). After washing, the samples were analyzed by SDS-PAGE. The gels were scanned by Typhoon 9200, and the images were quantified using Image-Quant TL software (Amersham Biosciences).
RESULTS
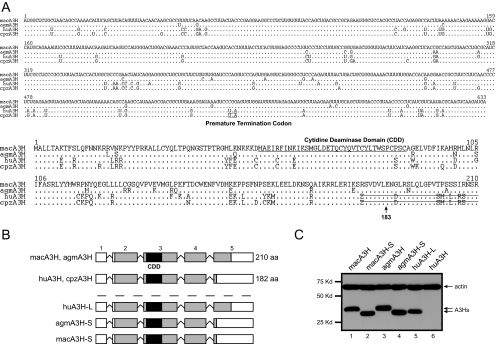
_A 29-Amino Acid C-terminal Deletion Disrupts Human but Not Simian A3H Gene Expression_—To study the antiretroviral mechanism of human A3H, the mechanism impairing its gene expression was investigated. First, primate A3H RNA and protein sequences were compared (Fig. 1_A_). huA3H, cpzA3H, and macA3H sequences were downloaded from the Ensembl Genome Browser, and their Ensembl IDs are ENSG00000100298, ENSPTRG00000014390, and ENSMMUG00000001016, respectively. The agmA3H gene was cloned from COS-7 cells by RT-PCR and sequenced. The Ensembl Database shows that primate A3H proteins are transcribed from five exons. When their RNA sequences were aligned, a PTC was found in the huA3H and cpzA3H mRNA genes at the last exon, which causes a 29-amino acid deletion. Second, to understand the functional consequence of this deletion, wild-type huA3H, which has a 29-amino acid C-terminal deletion, as well as the wild-type macA3H and agmA3H genes were cloned into the pcDNA3 vector. Three additional A3H mutants were created: 1) huA3H-L, which expresses the full-length 210-amino acid human A3H protein, and 2) agmA3H-S and 3) macA3H-S, which express the truncated 182-amino acid simian proteins (Fig. 1_B_). When their expressions were compared, macA3H, macA3H-S, agmA3H, agmA3H-S, and huA3H-L were expressed equally well, whereas the expression of wild-type huA3H was almost undetectable (Fig. 1_C_). These results confirm the previous observation that the wild-type human A3H gene, which has a 29-amino acid deletion, is poorly expressed (11). In addition, they demonstrate that although this deletion impairs human A3H gene expression, it has no effect on simian A3H genes.
FIGURE 1.
Primate A3H proteins. A, RNA and amino acid sequence alignment of primate A3H proteins. The macA3H sequence is listed on the top.Dots indicate identical sequence, and residues different from this sequence are directly indicated. The PTC in human and chimpanzee RNAs is_underlined_ and indicated. The CDD is underlined in the amino acid sequence, and the truncated sequences are marked by strikethrough. B, schematic representation of the primate A3H exon structure. Three mutant A3H genes including huA3H-L, agmA3H-S, and macA3H-S were generated.aa, amino acids. C, expression of primate A3H genes. The cDNAs of these genes were cloned into pcDNA3.1 with a V5 tag at their 3′-end, and they were transiently expressed in 293T cells. Protein expression was determined by Western blotting with antibodies against V5 and cellular protein actin.
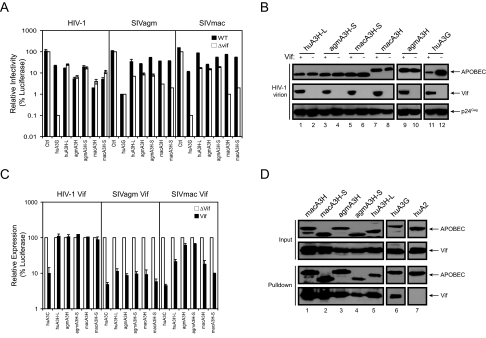
_Human A3H Antiretroviral Activity and Susceptibility to Vif_—Because we were able to express the human A3H gene, we next determined its antiretroviral activity against HIV-1 and two different SIV strains, SIVagm and SIVmac. To determine whether the C-terminal 29 residues affect A3H antiviral activity, the other simian A3H proteins were also included. In addition, huA3G was used as a positive control. As presented inFig. 2_A_, huA3G had the most powerful activity and reduced _vif_-defective HIV-1, SIVagm, and SIVmac infectivity by 1000-, 100-, and 1000-fold, respectively. As reported before, huA3G also decreased the infectivity of wild-type SIVagm, but not HIV-1 and SIVmac, confirming that SIVagm Vif could not neutralize huA3G (33). huA3H-L reduced_vif_-defective HIV-1, SIVagm, and SIVmac infectivity by 8-, 12.5-, and 10-fold, respectively, and it also reduced the wild-type HIV-1 infectivity. This result demonstrates that although it is lower than A3G, huA3H-L has antiretroviral activity, and this activity could be neutralized only by SIV Vif, but not by HIV-1 Vif. As reported previously, wild-type macA3H reduced the infectivity of HIV-1 and SIVagm by 25-fold, and its activity was only neutralized by Vif from SIVagm, but not from HIV-1 (11). In addition, wild-type macA3H reduced the _vif_-defective SIVmac infectivity by 50-fold, and it was neutralized by SIVmac Vif. Wild-type agmA3H reduced the_vif_-defective HIV-1, SIVagm, and SIVmac infectivity by 16-, 10-, and 7-fold, respectively, and it was neutralized by Vif from SIVagm, but not from HIV-1 and SIVmac. The truncated simian proteins agmA3H-S and macA3H-S had almost the same levels of antiretroviral activity as their wild-type proteins (only slight lower in the case of HIV-1), and their sensitivity to different Vif proteins was completely the same. Thus, human A3H has antiretroviral activity, which is not neutralized by HIV-1 Vif. In addition, the C-terminal 29 residues of simian A3H proteins should not have influence on their antiretroviral activity and sensitivity to Vif.
FIGURE 2.
Primate A3H antiretroviral activity. A, anti-HIV-1 and SIV activity. The pcDNA3 vectors expressing huA3H-L, agmA3H, agmA3H-S, macA3H, macA3H-S, or huA3G were cotransfected with wild-type (WT) or_vif_-deficient HIV-1 or SIV luciferase reporter proviral clone into 293T cells at a 5:1 ratio. Equal amounts of viruses were used to infect GHOST-R3/X4/R5 cells. Viral infectivity is presented as a relative value, where the infectivity of _vif_-deficient virus produced in the presence of pcDNA3.1 as a control (Ctrl) is set as 100. B, package of A3H proteins in HIV-1 virions. Wild-type or _vif_-deficient HIV-1 virions were produced from 293T cells in the presence of huA3H-L, agmA3H-S, macA3H-S, agmA3H, macA3H, or huA3G. Virions were purified through a 20% sucrose cushion via ultracentrifugation. Viral pellets were then lysed and analyzed by Western blotting with antibodies against V5 and HIV-1 Vif and p24Gag. C, susceptibility to Vif. Primate A3H-FLuc fusion proteins were coexpressed with HIV-1 Vif, SIVagm Vif, or SIVmac Vif. Cells were then lysed, and cellular luciferase activity was determined. Data are presented as a relative value, where its parallel control transfected with a Vif deletion vector (pNL-A1ΔVif) is set as 100. D, interactions of HIV-1 Vif with primate A3H proteins. A3 proteins were C-terminally fused to a HA/FLAG tag and then coexpressed in 293T cells with HIV-1 Vif. Proteins were pulled down by Sepharose beads conjugated with anti-FLAG antibody and determined by Western blotting with anti-HA antibody for A3 proteins and anti-Vif antibody. Error bars in A and C represent S.D. in at least three independent experiments. huA2, human APOBEC2.
To further confirm these observations, we determined how these primate A3H proteins are packaged into HIV virions. Wild-type or _vif_-deficient HIV-1 virions were produced from 293T cells in the presence of huA3H-L, agmA3H-S, macA3H-S, agmA3H, macA3H, or huA3G. As reported, huA3G was efficiently encapsidated by the _vif_-deficient virus, but not by the wild-type virus (Fig. 2_B_, lanes 11 and 12). All primate A3H proteins tested here were efficiently encapsidated by both wild-type and_vif_-deficient viruses (Fig. 2_B_, lanes 1-10). Thus, these results confirmed their anti-HIV activity and insensitivity to HIV-1 Vif.
To further compare the susceptibility of these A3H proteins with different Vif proteins, a FLuc-based Vif reporter assay was utilized (3). Primate A3H-Luc fusion proteins were coexpressed with HIV-1 or SIV Vif proteins, and cellular luciferase activities were determined. Human A3C was included as a positive control. As presented in Fig. 2_C_, HIV-1 Vif, SIVagm Vif, and SIVmac Vif significantly reduced huA3C expression (10-15-fold); HIV-1 Vif had little activity against any of the primate A3H proteins; SIVagm Vif could effectively reduce all these A3H protein expressions; and SIVmac Vif more effectively reduced human and rhesus monkey A3H protein expressions. These results are very consistent with the infectivity data in Fig. 2_A_, which further confirmed that HIV-1 Vif cannot neutralize huA3H-L.
To understand why HIV-1 Vif failed to neutralize huA3H-L, we determined the interactions of HIV-1 Vif with these different primate A3H proteins. We wanted to know whether there was a correlation between binding ability and susceptibility to Vif. These A3 proteins were C-terminally fused to a HA/FLAG tag and then coexpressed with HIV-1 Vif in 293T cells. When these fusion proteins were pulled down by Sepharose beads conjugated with anti-FLAG antibody, Vif was copurified with macA3H, macA3H-S, agmA3H, agmA3H-S, and huA3H-L with a similar efficiency as huA3G (Fig. 2_D_, lanes 1-6) and was not copurified with human APOBEC2 (lane 7). These results indicate that HIV-1 Vif binds to these A3H proteins, although it could not neutralize these proteins, suggesting that binding to Vif cannot be used as an indicator for the sensitivity to Vif-triggered degradation.
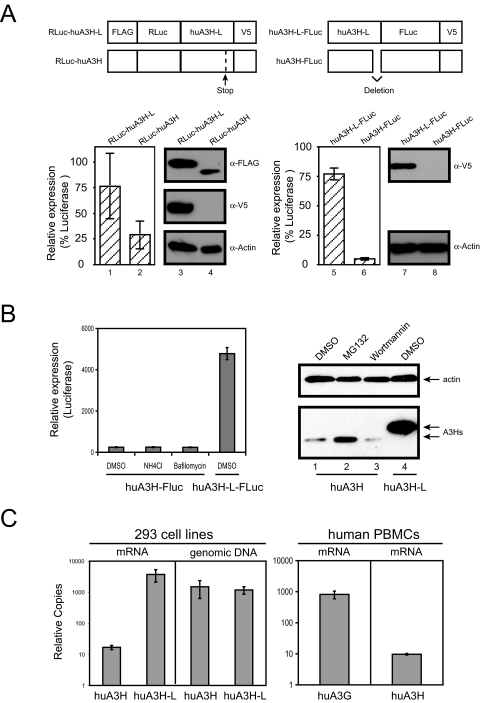
_Wild-type Human A3H Expression_—We next investigated the mechanism that leads to the poor expression of wild-type human A3H protein. We have already demonstrated that a PTC impairs the expression of human A3H (Fig. 1_C_). Two types of reporter systems were generated to address this question. First, the RLuc containing an N-terminal FLAG tag was fused to the N terminus of the full-length human A3H protein containing a PTC at amino acid 182 and a C-terminal V5 tag (RLuc-huA3H) (Fig. 3_A_, top left). This PTC was then repaired by site-directed mutagenesis to express the full-length A3H protein (RLuc-huA3H-L). As expected, Western blotting using anti-V5 antibody could detect the expression of only RLuc-huA3H-L, but not RLuc-huA3H (Fig. 3_A_, middle panel, lanes 3 and 4). However, if anti-FLAG antibody was used, the expression of both proteins could be detected, and the expression of the full-length protein was much higher compared with the truncated protein (Fig. 3_A_, top panel, lanes 3 and 4). Consistently, RLuc-huA3H-L produced 3-fold higher luciferase activity compared with RLuc-huA3H (Fig. 3_A_, lanes 1 and 2). Second, the firefly luciferase containing a C-terminal V5 tag was fused to the C terminus of the full-length human A3H protein (huA3H-L-FLuc) (Fig. 3_A_, top right). To generate a truncated version of this protein, the A3H-coding sequence from amino acids 183 to 210 was completely removed (huA3H-FLuc). Because this is an in-frame deletion, huA3H-FLuc should be detected by Western blotting with anti-V5 antibody. However, only huA3H-L-FLuc was detected (Fig. 3_A_, lanes 7 and 8). Consistently, huA3H-L-FLuc produced at least 20-fold higher luciferase activity compared with huA3H-FLuc (Fig. 3_A_, lanes 5 and 6). Thus, these results confirm that the C-terminal truncation causes the poor expression of human A3H.
FIGURE 3.
Human A3H expression. A, comparison of huA3H and huA3H-L expressions by luciferase reporter systems. Full-length and truncated human A3H proteins were fused to RLuc or FLuc at the N or C terminus, respectively. Proteins were expressed in 293T cells, and cellular luciferase activities were determined. The same cell lysates were also subject to Western blotting with antibodies against FLAG, V5, or actin. B, rescue of huA3H expression by various inhibitors. 293T cells were transfected with either FLuc fusion or non-fusion expression vectors and then treated with 40 mm NH4Cl, 0.4 μm bafilomycin, 20 μm MG132, or 100 nm wortmannin for 16 h. Human A3H protein expression was determined by measuring cellular luciferase activity or by Western blotting.DMSO, Me2SO. C, quantitation of A3H genes in vivo. Two 293 cell-derived cell lines stably expressing huA3H-FLuc or huA3H-L-FLuc were created by G418 selection. The levels of A3H mRNA and genomic DNA in these cell lines were determined by TaqMan® real-time PCR. In addition, the mRNA levels of A3G and A3H in human PBMCs were determined by SYBR Green® real-time PCR. mRNA levels were normalized to hypoxanthine-guanine phosphoribosyltransferase mRNA, and DNA levels were normalized to the amounts of DNA input. Error bars represent S.D. in at least three independent experiments.
To understand how huA3H protein expression is decreased, we first determined whether the low levels of A3H expression were due to protein degradation. The FLuc reporter constructs were expressed in 293T cells, which were then treated with lysosomal inhibitors NH4Cl and bafilomycin. None of them could increase the huA3H expression (Fig. 3_B_, left panel). Next, huA3H protein was expressed in 293T cells, which were then treated with the proteasomal inhibitor MG123 or the phosphatidylinositol 3-kinase inhibitor wortmannin as a control (Fig. 3_B_, right panel). As reported previously (11), MG132 only marginally increased huA3H expression, but the increased levels of expression were still much lower than the levels of huA3H-L expression. Thus, the low expression of huA3H should not be caused by protein degradation in cells.
We next determined the steady-state levels of A3H mRNA by real-time quantitative RT-PCR. We created two 293 cell-derived cell lines stably expressing the huA3H or huA3H-L gene by G418 selection. Although both cell lines had similar levels of A3H genomic DNA, the mRNA levels of huA3H-L were at least 100-fold higher than those of huA3H (Fig. 3_C_). In addition, we measured A3H mRNA levels directly in human PBMCs by RT-PCR. Because A3G is well expressed in vivo, its mRNA levels were also determined for comparison. It was found that huA3H mRNA levels were at least 80-fold lower than huA3G mRNA levels (Fig. 3_C_), providing more evidence for low huA3H mRNA levels_in vivo_. Thus, the low levels of A3H mRNA could be responsible for the reduced A3H protein expression.
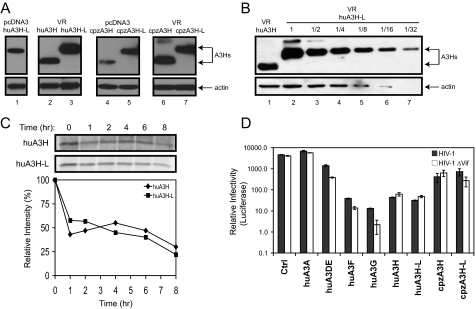
_Wild-type Human A3H Anti-HIV Activity_—After determining the low levels of the wild-type human A3H mRNA, an increase in A3H protein expression was attempted through a different expression vector system. The eukaryotic expression vector VR has been successfully employed for gene delivery in vivo (32). It contains a cytomegalovirus immediate early gene promoter, enhancer, and intron A. When the human A3H proteins were expressed from this system, the expression of wild-type human A3H was significantly increased (Fig. 4_A_, lane 2), which was comparable with full-length huA3H-L expressed from the pcDNA3 vector (lane 1). However, this level of expression was still lower than that of full-length protein expressed from the same VR vector (Fig. 4_A_, lane 3). To compare this difference, we made dilution series of the huA3H-L protein sample, which were then compared with the wild-type protein. As presented in Fig. 4_B_, huA3H-L proteins were expressed 2-4-fold higher than huA3H proteins.
FIGURE 4.
Wild-type human and chimpanzee A3H anti-HIV-1 activity. A, stable expression of the wild-type huA3H and cpzA3H genes from the VR vector. The huA3H, huA3H-L, cpzA3H, and cpzA3H-L genes were cloned into the VR vector containing a C-terminal HA tag. Their expressions were then compared with those from the pcDNA3.1 vector by Western blotting after transfection into 293T cells. B, comparison of huA3H and huA3H-L expressions from the VR vector. 293T cells were transfected with the VR vector expressing huA3H or huA3H-L, and cell lysates were prepared. The huA3H-L cell lysates were serially diluted, and the levels of huA3H-L in these samples were then compared with the levels of undiluted huA3H sample by Western blotting.C, pulse-chase radiolabeling of human A3H protein. 293T cells were transiently transfected with the VR vector expressing huA3H or huA3H-L protein. Cells were pulse-labeled for 30 min and then incubated for the indicated time points. After immunoprecipitation of lysates with anti-HA antibody, proteins were separated by SDS-PAGE, and the gels were subjected to autoradiography. The relative intensity of the bands was quantified, normalized to 100, and graphed. D, anti-HIV activity. The VR vectors expressing huA3A, huA3DE, huA3F, huA3G, huA3H, huA3H-L, cpzA3H, and cpzA3H-L were cotransfected with either wild-type or _vif_-deficient HIV-1 luciferase reporter proviral clone into 293T cells at a 1:9 ratio (0.6 μg of the A3H expression vector plus 5.4 μg of the HIV-1 proviral vector). HIV-1 infectivity was determined as before. Error bars represent S.D. in at least three independent experiments. Ctrl, control.
We have shown that the cpzA3H gene contains a similar PTC (Fig. 1_A_). To understand the influence of C-terminal deletion on cpzA3H expression, the full-length (cpzA3H-L) and truncated (cpzA3H) genes were cloned into the pcDNA3 or VR expression vector. It was found that this deletion also impaired cpzA3H gene expression in the pcDNA3 vector (Fig. 4_A_, lanes 4 and 5), and the expressional difference between the wild-type and full-length cpzA3H genes could be completely suppressed in the VR vector (Fig. 4_B_, lanes 6 and 7). Thus, a similar mechanism might be shared by human and chimpanzee. In addition, it implies that the cytomegalovirus intron A has an unknown function to increase A3H mRNA expression.
The expression of wild-type human A3H from the VR vector made it possible to measure its protein stability by pulse-chase radiolabeling. 293T cells were transfected with the VR vector expressing either wild-type or full-length A3H protein. After a 30-min pulse with 35S-labeled Met/Cys, cultures were chased for 8 h. A3H proteins were then immunoprecipitated from cell lysates harvested at different time points to determine how proteins were rapidly degraded. Both wild-type and full-length A3H proteins lost about half of the 35S label in the first hour, followed by very marginal loss over the next 7 h (Fig. 4_C_), indicating that they should have a similar stability_in vivo_. Thus, we finally proved that the poor expression of the wild-type human A3H protein is not due to protein instability.
Up-regulation of wild-type huA3H and cpzA3H expression also made it possible to test their anti-HIV-1 activity directly. As controls, human A3A, A3DE, A3F, and A3G were cloned into the VR vector. To avoid overexpression, the ratio between the HIV-1 proviral construct and VR expression vector was reduced to 9:1 for each transfection. Under this condition, the full-length and truncated huA3H proteins both reduced the infectivity of wild-type and_vif_-defective HIV-1 by 150-fold; the full-length and truncated cpzA3H proteins reduced the infectivity of wild-type and _vif_-defective HIV-1 by 10-fold; huA3A did not have any activity; and huA3DE, huA3F, and huA3G reduced the _vif_-defective HIV-1 infectivity by 10-, 400-, and 2000-fold, respectively (Fig. 4_D_). These results demonstrate that human A3H proteins have potent anti-HIV-1 activity and that chimpanzee A3H proteins are less effective in blocking HIV-1 replication. In addition, because huA3H and huA3H-L had equal anti-HIV-1 activity, although the expression of huA3H-L was higher, the wild-type human A3H protein might have higher antiviral activity than the full-length human A3H protein.
_Levels of G-to-A Mutation in the HIV Genome_—Having detected the potent anti-HIV activity, we next determined whether human A3H could introduce G-to-A hypermutations to the HIV-1 genome. Wild-type or_vif_-deficient HIV-1 was produced in the presence of huA3H or huA3H-L proteins expressed from the VR vector. These viruses were used to infect GHOST cells, and newly synthesized viral cDNAs were sequenced. For comparison, huA3C, huA3G, and four different simian A3H proteins were also included in this investigation. As summarized in Fig. 5, viral genomes produced in the absence of APOBEC protein or in the presence of huA3C that does not block HIV replication contained only a small number of random mutations, including one G-to-A mutation in each case. In contrast, human A3G caused significant high levels of G-to-A hypermutations in the _vif_-deficient HIV-1 genome (51 per 4486). As reported, macA3H-L could introduce certain levels of G-to-A mutations in both the wild-type and _vif_-defective HIV genomes (11), and similar levels of G-to-A mutation were also detected in the case of macA3H, agmA3H-L, and agm-A3H. No G-to-A mutation was detected in the case of huA3H, and only one G-to-A mutation was detected in huA3H-L when similar amount of nucleotides were sequenced. This single G-to-A mutation should represent a random event because three other mutations were also detected in the huA3H-L-derived sample. Thus, although human A3H potently inhibited HIV-1 replication, it did not introduce G-to-A mutation, suggesting that huA3H may employ a cytidine deamination-independent mechanism for this inhibition.
FIGURE 5.
G-to-A hypermutations in the HIV-1 genome. Wild-type (WT) or _vif_-deficient HIV-1 was produced in the presence of various A3 proteins. huA3H, hua3H-L, and huA3G were expressed from the VR vector. The other proteins were expressed from pcDNA3.1, which was also used as the control vector (Ctrl). These viruses were used to infect GHOST-R3/X4/R5 cells. 8 h later, cellular DNAs were extracted from infected cells, and viral DNAs from nucleotides 5707 to 6066 were amplified by PCR. After cloning into TA cloning vector, viral DNAs were sequenced. The type of mutations is summarized in tabular forms, where the original HIV-1 sequence is given at left, and the new sequence or positions from the mutated cytosine are given across the top. N at the lower right of each box indicates the total numbers of bases sequenced.
DISCUSSION
In this study, we explored the mechanism impairing human A3H gene expression and studied its antiretroviral activity in comparison with other primate homologues. We confirmed previous observations that human A3H protein is poorly expressed in cell culture and demonstrated that this is caused by a C-terminal deletion that impairs its mRNA expression. Once its expression is optimized, human A3H exhibited potent antiretroviral activity against HIV-1. Importantly, HIV-1 Vif cannot neutralize this protein.
The poor expression of the human A3H gene is caused by a PTC, and a similar PTC also decreases chimpanzee A3H expression. The presence of this PTC, which causes a 29-amino acid C-terminal deletion, did not reduce protein stability. The expression of truncated protein was not rescued by treatment with lysosomal inhibitors and was increased marginally by treatment with proteasomal inhibitor (Fig. 3_B_). Importantly, the truncated protein was as stable as the full-length protein (Fig. 4_C_). Interestingly, a significant decrease in A3H mRNA levels was detected in cells expressing this truncated protein. Thus, the mRNA of the truncated gene could be very unstable, which decreases the steady-state levels of mRNA.
Human A3H potently inhibits HIV-1 replication once expressed. Including A3H, five of seven human A3 proteins have been shown to block HIV-1 replication. The activity of A3H was lower than that of A3G but was comparable with that of A3F (Fig. 4_D_). Notably, A3B, A3DE, A3F, and A3G all have two CDDs, whereas A3H has only one. In the process of blocking HIV-1 replication, the functions of these two CDDs are divided into a catalytically active domain for cytidine deamination and a RNA-binding domain for substrate recognition (34). How a single CDD achieves these different functions remains unclear. In addition, our results indicate that human A3H antiviral activity is governed by a cytidine deamination-independent mechanism. After extensive sequence analysis, only background levels of G-to-A mutation were detected in the HIV-1 genome in the presence of human A3H (Fig. 5). Previously, a similar observation was made in the case of huA3A, when it was shown to inhibit the replication of adenovirus-associated virus and retrotransposons, including MusD, IAP, and LINE-1 (9,18). Because both A3A and A3H have a single CDD, they may share a very similar antiviral mechanism.
Unlike A3DE, A3F, and A3G, human A3H is insensitive to HIV-1 Vif. Interestingly, HIV-1 Vif still binds to human A3H (Fig. 2_D_), suggesting that this protein is not a suitable substrate for a HIV-1 Vif-triggered proteasomal degradation pathway. Human A3B is also resistant to HIV-1 Vif, and it is poorly expressed in vivo (4). However, because A3B could be easily expressed in cell culture, a much more complicated mechanism is involved for A3B gene regulation. In addition, the A3B gene is completely deleted in certain human populations (35). It is therefore apparent that human A3H is distinguishable from all the other anti-HIV proteins in the A3 subfamily, and optimizing its expression in vivo may become a novel strategy for anti-HIV therapy. Compared with using a viral protein as a drug target, targeting A3H should have a much smaller likelihood of inducing drug-resistant strains. Thus, our results shed light on the unique candidacy of human A3H for the future of HIV-1 therapeutics.
Acknowledgments
We thank W. J. Esselman and P. Venta for comments on this manuscript and X. F. Yu for discussion. We thank Y. Lee from the Bieniasz laboratory for protocol and M. Emerman, N. Landau, K. Strebel, B. Liu, and X. F. Yu for generous reagent support. We also thank the National Institutes of Health AIDS Reagent Repository for antibodies contributed by M. Malim, D. Gabuzda, B. Chesebro, and K. Wehrly.
*
This work was supported by National Institutes of Health Grant AI063944 (to Y.-H. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
2
The abbreviations used are: A3, APOBEC3; APOBEC, apolipoprotein B mRNA-editing catalytic polypeptide; CDD, cytidine deaminase domain; HIV-1, human immunodeficiency virus type 1; SIV, simian immunodeficiency virus; PTC, premature termination codon; Vif, viral infectivity factor; hu, human; cpz, chimpanzee; mac, rhesus macaque; agm, African green monkey; FLuc, firefly luciferase; RLuc, Renilla luciferase; RT, reverse transcription; HA, hemagglutinin; PBMCs, peripheral blood mononuclear cells; DCTA, 1,2-diaminocyclohexane-_N,N,N_′,_N_′-tetraacetic acid.
References
- 1.Holmes, R. K., Malim, M. H., and Bishop, K. N. (2007) Trends Biochem. Sci. 32 118-128 [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., Holmes, R. K., Sheehy, A. M., Davidson, N. O., Cho, S. J., and Malim, M. H. (2004) Curr. Biol. 14 1392-1396 [DOI] [PubMed] [Google Scholar]
- 3.Dang, Y., Wang, X., Esselman, W. J., and Zheng, Y.-H. (2006) J. Virol. 80 10522-10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doehle, B. P., Schafer, A., and Cullen, B. R. (2005) Virology 339 281-288 [DOI] [PubMed] [Google Scholar]
- 5.Liddament, M. T., Brown, W. L., Schumacher, A. J., and Harris, R. S. (2004) Curr. Biol. 14 1385-1391 [DOI] [PubMed] [Google Scholar]
- 6.Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002) Nature 418 646-650 [DOI] [PubMed] [Google Scholar]
- 7.Wiegand, H. L., Doehle, B. P., Bogerd, H. P., and Cullen, B. R. (2004) EMBO J. 23 2451-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng, Y.-H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T., and Peterlin, B. M. (2004) J. Virol. 78 6073-6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., Lilley, C. E., Yu, Q., Lee, D. V., Chou, J., Narvaiza, I., Landau, N. R., and Weitzman, M. D. (2006) Curr. Biol. 16 480-485 [DOI] [PubMed] [Google Scholar]
- 10.Dutko, J. A., Schafer, A., Kenny, A. E., Cullen, B. R., and Curcio, M. J. (2005) Curr. Biol. 15 661-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OhAinle, M., Kerns, J. A., Malik, H. S., and Emerman, M. (2006) J. Virol. 80 3853-3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S., and Malim, M. H. (2003) Cell 113 803-809 [DOI] [PubMed] [Google Scholar]
- 13.Lecossier, D., Bouchonnet, F., Clavel, F., and Hance, A. J. (2003) Science 300 1112. [DOI] [PubMed] [Google Scholar]
- 14.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003) Nature 424 99-103 [DOI] [PubMed] [Google Scholar]
- 15.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C., and Gao, L. (2003) Nature 424 94-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman, E. N., Holmes, R. K., Craig, H. M., Klein, K. C., Lingappa, J. R., Malim, M. H., and Sheehy, A. M. (2005) Curr. Biol. 15 166-170 [DOI] [PubMed] [Google Scholar]
- 17.Bogerd, H. P., Wiegand, H. L., Doehle, B. P., Lueders, K. K., and Cullen, B. R. (2006) Nucleic Acids Res. 34 89-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogerd, H. P., Wiegand, H. L., Hulme, A. E., Garcia-Perez, J. L., O'Shea, K. S., Moran, J. V., and Cullen, B. R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8780-8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasada, A., Takaori-Kondo, A., Shirakawa, K., Kobayashi, M., Abudu, A., Hishizawa, M., Imada, K., Tanaka, Y., and Uchiyama, T. (2005) Retrovirology 2 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenglein, M. D., and Harris, R. S. (2006) J. Biol. Chem. 281 16837-16841 [DOI] [PubMed] [Google Scholar]
- 21.Guo, F., Cen, S., Niu, M., Saadatmand, J., and Kleiman, L. (2006) J. Virol. 80 11710-11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, F., Cen, S., Niu, M., Yang, Y., Gorelick, R. J., and Kleiman, L. (2007) J. Virol. 81 11322-11331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwatani, Y., Chan, D. S., Wang, F., Maynard, K. S., Sugiura, W., Gronenborn, A. M., Rouzina, I., Williams, M. C., Musier-Forsyth, K., and Levin, J. G. (2007) Nucleic Acids Res. 35 7096-7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mbisa, J. L., Barr, R., Thomas, J. A., Vandegraaff, N., Dorweiler, I. J., Svarovskaia, E. S., Brown, W. L., Mansky, L. M., Gorelick, R. J., Harris, R. S., Engelman, A., and Pathak, V. K. (2007) J. Virol. 81 7099-7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, K., Wang, T., Liu, B., Tian, C., Xiao, Z., Kappes, J., and Yu, X. F. (2007) J. Virol. 81 7238-7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marin, M., Rose, K. M., Kozak, S. L., and Kabat, D. (2003) Nat. Med. 9 1398-1403 [DOI] [PubMed] [Google Scholar]
- 27.Sheehy, A. M., Gaddis, N. C., and Malim, M. H. (2003) Nat. Med. 9 1404-1407 [DOI] [PubMed] [Google Scholar]
- 28.Stopak, K., de Noronha, C., Yonemoto, W., and Greene, W. C. (2003) Mol. Cell 12 591-601 [DOI] [PubMed] [Google Scholar]
- 29.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X. F. (2003) Science 302 1056-1060 [DOI] [PubMed] [Google Scholar]
- 30.Liu, B., Sarkis, P. T., Luo, K., Yu, Y., and Yu, X. F. (2005) J. Virol. 79 9579-9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opi, S., Kao, S., Goila-Gaur, R., Khan, M. A., Miyagi, E., Takeuchi, H., and Strebel, K. (2007) J. Virol. 81 8236-8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartikka, J., Sawdey, M., Cornefert-Jensen, F., Margalith, M., Barnhart, K., Nolasco, M., Vahlsing, H. L., Meek, J., Marquet, M., Hobart, P., Norman, J., and Manthorpe, M. (1996) Hum. Gene Ther. 7 1205-1217 [DOI] [PubMed] [Google Scholar]
- 33.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H., and Landau, N. R. (2003) Cell 114 21-31 [DOI] [PubMed] [Google Scholar]
- 34.Navarro, F., Bollman, B., Chen, H., Konig, R., Yu, Q., Chiles, K., and Landau, N. R. (2005) Virology 333 374-386 [DOI] [PubMed] [Google Scholar]
- 35.Kidd, J. M., Newman, T. L., Tuzun, E., Kaul, R., and Eichler, E. E. (2007) PLoS Genet. 3 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]