DiRE: identifying distant regulatory elements of co-expressed genes (original) (raw)
Abstract
Regulation of gene expression in eukaryotic genomes is established through a complex cooperative activity of proximal promoters and distant regulatory elements (REs) such as enhancers, repressors and silencers. We have developed a web server named DiRE, based on the Enhancer Identification (EI) method, for predicting distant regulatory elements in higher eukaryotic genomes, namely for determining their chromosomal location and functional characteristics. The server uses gene co-expression data, comparative genomics and profiles of transcription factor binding sites (TFBSs) to determine TFBS-association signatures that can be used for discriminating specific regulatory functions. DiRE's unique feature is its ability to detect REs outside of proximal promoter regions, as it takes advantage of the full gene locus to conduct the search. DiRE can predict common REs for any set of input genes for which the user has prior knowledge of co-expression, co-function or other biologically meaningful grouping. The server predicts function-specific REs consisting of clusters of specifically-associated TFBSs and it also scores the association of individual transcription factors (TFs) with the biological function shared by the group of input genes. Its integration with the Array2BIO server allows users to start their analysis with raw microarray expression data. The DiRE web server is freely available at http://dire.dcode.org.
INTRODUCTION
High-quality sequencing of eukaryotic genomes provides the framework for understanding the mechanisms that underlie biological functions (1–3). In the case of the human genome, the complete genomic sequence has revealed fewer protein coding genes than expected (2,4), yet the complex nature of their regulation beyond proximal promoters remains poorly understood. The explosion in available genomic data raised the hopes for deciphering the ‘regulatory codes’ that govern gene expression specific to various developmental conditions (5,6). Considerable effort was put into finding regulatory elements in simpler organisms, such as yeast (7–9) and Drosophila (10–13), for which in silico predictions are easier to validate experimentally. Most of the prediction methods are based on local enrichment in binding sites for specific transcription factors (TFs), but it is additional information, such as sequence conservation across taxa (14), nucleosome occupancy (15) or binding competition between factors (16), that enables predictions to obtain remarkable accuracy. A significant effort was also put into predicting regulatory elements in mammalian genomes (17–19), with several computational tools being developed (20–28) for the purpose of predicting the locations of transcription factor binding sites (TFBS) and regulatory elements (REs). Central to most of these tools is the concept of proximal promoter, which is a natural extension from simpler organisms where promoter-based regulation plays the most important role. Some tools also provide the possibility to analyse sequences of up to 10 kb preceding (24) or surrounding the transcription start site (23,25), as well as any other sequences of interest (21,29), which have made possible the investigation of REs located further away from proximal promoters. In complex organisms, gene regulation is established through a cooperative activity of distant REs such as enhancers, repressors, silencers, etc. and proximal promoters (defined as 1.5 kb regions upstream of the transcription start site in this case). Recent experimental evidence indicates that distant regulatory elements can play an important role in gene regulation (30–33), but we can only speculate on their sequence signatures and on the extent to which these elements populate eukaryotic genomes. We have recently developed a new method for inferring positional and functional information on distant REs from the analysis of either microarray gene expression or co-regulation data (34). This method, called Enhancer Identification (EI), is one of the first in attempting to computationally predict distant REs in vertebrates directly from a list of co-regulated genes, irrespective of the absolute distance from REs to the genes they regulate. As described previously, in a study of 79 groups of tissue-specific genes, only 23% of candidate regulatory elements were found in promoter regions and over half of the remaining elements resided either in intronic or intergenic regions (34). The EI method combines gene co-expression data with conservation of regulatory signals across genomes and takes into account the combinatorial co-occurrence of TFBSs, which is known to enhance the prediction power of computational methods (35,36). As a result, EI can predict REs using a profile of evolutionarily conserved TFBSs, which can also be used as signatures of particular biological functions. By overlapping genome-wide predictions with a set of enhancers validated in vivo in transgenic mice, this method was previously shown to have 28% sensitivity and 50% precision (34). Here we present a generalized version of the method implemented as a web server, named DiRE, which provides computational means to investigate regulatory features of any user-submitted dataset of genes. Depending on the co-expression behaviour (e.g. up- or down-regulation) of the input genes, the DiRE server will predict function-specific (e.g. time, tissue) REs that can act as enhancers or repressors and the key regulatory TFs that potentially mediate their effects. A convenient feature of the DiRE server is its integration with the Array2BIO server (37), which provides users with the ability of using raw Affymetrix microarray expression data to start the investigation of the common regulatory features of the genes of interest. In short, we present a unique tool to effectively translate functional information shared by a group of genes into proximal and distant gene regulatory information. Identification of candidate REs and their TF profiles can be used for prioritizing candidates for experimental validation and potentially for de novo detection of synonymous REs in loci of genes not included into the input dataset. Ultimately, the DiRE server will facilitate enhancing the functional annotation of the human and other genomes by providing candidate distant REs responsible for specific biological functions.
Using the DiRE web server to predict distant regulatory elements
Data input
The DiRE server, located at http://dire.dcode.org, has a simple and intuitive interface, where users can input a list of co-regulated and a list of background (or control) genes. The list of genes for which users have prior knowledge of co-regulation is usually relatively small, so that users can paste this list of records into the main window of the DiRE server, with one record per line. The accuracy of the underlying EI method was tested on 79 diverse groups of human genes co-expressed in different tissues, with the number of genes per group ranging from around 200 to 300 (9). A set of genes ranging in size from a hundred to a thousand genes is not uncommon to microarray gene expression studies and will constitute an appropriate input gene set for this tool. We would strongly recommend using at least 50 genes as input to avoid overfitting the classifier by training it on a small set of genes. This user supplied list needs to match any one of the following recognized types of data (to be selected from a pull-down menu under the main window): GenBank nucleotide or protein accession numbers, official gene symbols, accession numbers from the UCSC known genes annotation or chromosomal coordinates. For the purpose of this analysis, DiRE explores not only the genomic region covered by gene transcripts, but also the flanking intergenic regions. Additionally, genes need to belong to one of the species for which precomputed alignments and TFBS content exist, which currently include human, mouse and rat (this list will be expanded in the future). In case of genes supplied in the form of genomic coordinates, users need to verify that these coordinates match the corresponding genome assembly.
In many cases the knowledge of co-regulation for a set of genes is inferred from microarray gene expression experiments. Users of Affymetrix gene expression microarrays have the option of starting a DiRE investigation with the raw gene expression data through the Array2BIO server located at http://array2bio.dcode.org (37). Array2BIO has a newly implemented feature that allows users to forward the list of co-regulated genes directly to the DiRE server, without additional data manipulation. Additionally, Array2BIO allows grouping genes into different Gene Ontology and KEGG functional categories. Each of these gene groupings can be automatically submitted to DiRE as well.
Users also need to input a list of genes that should serve for computing the background distribution of TFBS clusters. Alternatively, different static lists of up to 7500 background genes randomly selected from either the human, mouse or rat genome are provided for the convenience of users. These lists will remain the same, so that results can be reproduced and compared across different runs. However, if some genes are present in both the signal and the background lists of genes, they will be eliminated from the background set. Users may opt to provide their own list of background genes, which could be especially helpful if data for contrasting expression exists, such as generated from microarray gene expression experiments. In this case, the user-provided list of genes has to be formatted similarly to the list of co-regulated genes. For meaningful results, the list of background genes needs to contain at least a few thousands genes, in order to avoid biased representation of random expectations.
DiRE processing
Once the initial data submitted, the DiRE server will display a dynamically updated progress report. The report will highlight the main computational steps (parameter optimization, characterization of REs) which were previously described in great detail (34). In short, DiRE server first maps the input genes onto the reference genome, and then it selects a set of candidate REs from their loci based on the inter-species conservation pattern, which is available in the form of precomputed alignments if evolutionary conserved regions (ECR) (38). Also the locations of putative TFBS are precomputed and are determined independently for each genome using ∼400 families of vertebrate position weight matrices (PWM) available from the 10.2 version of the TRANSFAC Professional database (due to redundancy in TRANSFAC, 584 TRANSFAC PWMs effectively represent only ∼400 TFBSs). Using these readily available datasets, DiRE determines the TFBS content of these candidate REs. This step is followed by the maximization of the F t scoring function (34), which is established by assigning and varying TFBS weights. The optimization effectively increases the number of candidate REs recognized by the profile of TFBS in the loci of input genes while decreasing the number of such predictions in the loci of background genes. The optimization process is stopped once no positively scoring elements are found in at least 85% loci of background genes. Next, positively scoring elements in loci of signal genes are reported as candidate REs for driving the expression pattern specific to the initial set of genes. The run time depends on the total number of input and background genes; a typical analysis, with <500 co-regulated genes and 5000 background genes, is usually completed within 10 min. Using fewer background genes can decrease the running time significantly (to less than a minute), but this might result in unreliable predictions because of the background signal under-sampling.
DiRE output
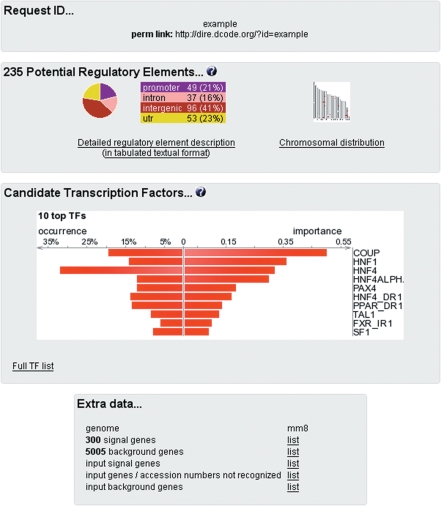
Users are provided with the results as exemplified in Figure 1. The Request ID provided at the top can be used for future data retrieval. A summary of the detected REs follows, both relative to the input genes (categorized as promoter, intronic, intergenic or UTR elements) and as a graphical representation of their chromosomal distribution. Also provided is a link to the detailed description of candidate REs. The candidate RE score, as described above and defined previously (34), is obtained for each RE by summing the assigned weights of all its constituent TFBSs (scores below 0.1 should generally indicate low confidence predictions). Candidate RE description also contains an annotation based on the element location relative to the features of the gene locus (UTR, intron, intergenic), the coordinates of the gene locus, the official symbol(s) of the gene and a list of positively-scoring TFBSs located in that element. Users have the option of exploring the conservation and the genomic landscape of each candidate element in the ECR Browser (38) by clicking on its coordinates in the ‘Regulatory element’ column. The next section of the output is dedicated to TFs found in candidate REs; the occurrence and importance measures (34) being reported for each TF. The occurrence indicates the fraction of REs containing a particular TF, while the importance is the product between the occurrence and the weight assigned to each TF after the optimization. Finally, the original list of genes used in the computation and their mapped location on the target genome is provided for convenience.
Figure 1.
Example of a summary output generated by the DiRE server, publicly available at http://dire.dcode.org/?id=example. The request ID is normally a 16-digit number that replaces ‘example’ in this figure. While visually comprehensive, the output provides easy access to the detailed description (chromosomal location, score, TF content) of candidate REs, to the graphical representation of their genomic distribution and to the list of most important TFs. The ‘occurrence’ represents the fraction of putative REs that contain a particular TFBS, while the ‘importance’ is defined as the product of the TF occurrence and its weight. For users’ convenience links to the original gene lists are provided, as well as to the results of their mapping onto the corresponding genome.
DISCUSSION
The DiRE web server is the latest addition to the Dcode.org set of comparative genomic tools, allowing researchers to computationally predict common regulatory features of co-regulated genes. While other tools (20–28) focus on detecting REs in promoter regions, DiRE predicts distant REs in vertebrate genomes independently of their position relative to the gene they regulate. It can predict either enhancer or repressor elements, depending on whether the genes of interest are up- or down-regulated, or general regulatory elements of any type if the input data originates from a particular biological group that does not necessarily involve expression data (such as a Gene Ontology or KEGG category, for example). The EI method underlying the DiRE server was previously used to predict REs for different sets of genes co-expressed in 79 human and 61 mouse tissues and the predictions were experimentally validated in transgenic mice (34). This provided the motivation for the generalization of the method and its implementation into a web server that would allow exploring other sets of co-regulated genes, thus contributing to enhancing the functional annotation of genomes.
When using the DiRE web server, users should keep in mind that the results depend on a series of precomputed datasets and future updates should positively impact the data processing of the DiRE server. Precomputed ECR Browser (39) alignments might be compromised by draft quality of genomes (human and mouse genomes are of highest available quality). Gene annotation is very important, because for the purpose of the EI method, the locus of a gene is defined as the sequence between its two neighbouring genes, which can be altered by adding new genes or eliminating incorrect annotations with direct implications on the predictions associated with that particular locus. The TRANSFAC database (40), which defines TFBS used by DiRE, is another important factor. A TF missing from TRANSFAC, a poorly defined TF binding specificity, different TFs with very similar binding specificities—all these and other factors might negatively impact the quality of DiRE predictions. One should thus expect that the constant update of the TRANSFAC database should result in improved DiRE predictions.
Despite these uncertainties, it is possible to demonstrate that the DiRE analysis can consistently lead to informative biological findings. For example, Figure 1, which illustrates the main DiRE output, represents the result of re-analysing the regulatory landscape of the top 300 genes highly expressed in the mouse liver (34,41). It was generated using mm8 mouse and hg18 human genome assemblies, corresponding gene annotation tracks, and the 10.2 version of the TRANSFAC Professional database. We compared these results to the original mouse liver analysis (34), which used previous genome assemblies, mm7 and hg17, accompanied by an earlier, version 9.4, release of the TRANSFAC database. Comparing the lists of top 10 TFs with the highest predicted importance in the liver gene regulation, we observed that Nr2f1a, Hnf1a, Hnf4a, Ppara and Nr1h4 TFs, known to play an important role in liver-specific gene activation, are shared by both the new and the previous analysis (Table 1). One factor, Srebf1 [Figure 4A in (34)], however, was missing from the top 10 TF list in the new analysis. However, the updated analysis picked up two other additional known liver regulatory factors, Nr5a1 (42) and Pax4 (43). There was only 1 TF, Tal1, in the top 10 TF list predicted by the new analysis, for which we cannot confirm its liver regulatory function, despite its known expression in liver (44).
Table 1.
List of TFs with the highest importance found in regulatory elements detected by the DiRE server to be associated with liver up-regulation of 300 mouse genes (Figure 1)
| TF name in TRANSFAC | Gene name | TF function | Reference |
|---|---|---|---|
| COUP | Nr2f1a | Essential for postnatal development and normal lymphopoiesis; required for hematopoietic development. | (47) |
| HNF1 | Hnf1a | Regulates proline metabolism in adult liver; | (48) |
| HNF4/ HNF4ALPHA | Hnf4a | Regulates genes involved in drug metabolism and detoxification as well as maintenance of liver function. | (49) |
| PAX4 | Pax4 | Promotes late-stage beta-cell differentiation and maturation. | (43) |
| PPAR | Ppara | Hepatic activation of Ppara underlies glucocorticoid-induced insulin resistance. | (50) |
| DR1 | Dr1 | Forms dimers with Hnf4a and Ppara. | |
| TAL1 | Tal1 | Development of murine primitive hematopoiesis. | (51) |
| FXR | Nr1h4 | Modulator of hepatic carbohydrate metabolism. | (52) |
| IR1 | Ir1 | Forms dimer with Nr1h4. | |
| SF1 | Nr5a1 | Regulates genes that are involved in sterol and steroid metabolism in gonads, adrenals, liver and other tissues. | (42) |
It is interesting to note that despite big differences in the computational approach, other tools produce results comparable to those of DiRE. Among the available tools, oPOSSUM (23) is probably the most similar to DiRE. It uses a fixed precomputed set of phylogenetically conserved TFBSs, employs matrices from the JASPAR database and can estimate the statistical significance of TFBS co-occurrence (only for groups of two or three TFBSs) in windows of up to 20 kb surrounding the transcription start sites. We provided the oPOSSUM Combination Site Analysis (CSA) with the initial set of 300 genes highly expressed in the mouse liver mentioned above to find shared groups of three TFBSs. It only used 193 of the input genes for the analysis, which identified Nr2f1 and Hnf1a as the most abundant TFs in the five top scoring groups of TFBSs (data not shown). These two TFs are also the top two scoring factors in the DiRE output. As expected, despite these similarities, there were discrepancies between the oPOSSUM and DiRE predictions that highlight differences in computational approaches employed by these tools, such as using matrices from the JASPAR database in the case of oPOSSUM as opposed to matrices from TRASNFAC in the case of DiRE. From the user's point of view, it might be practical to give priority to overlapping predictions, while treating predictions specific to a single tool with more caution.
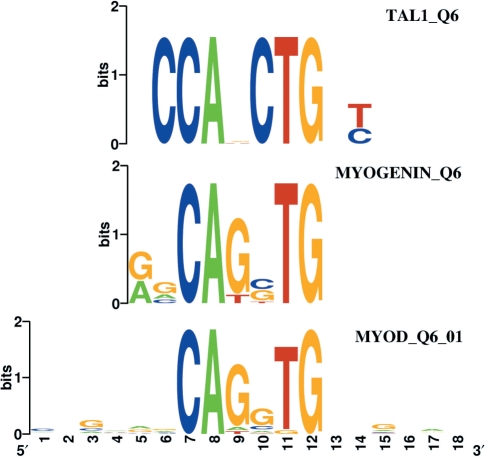
We also show here that the DiRE server produces results consistent with known biological facts. We compared the gene expression profile of mouse embryonic fibroblasts (MEFs) derived from MyoD−/−/Myf5−/− mice after transcriptional induction of the wild type MyoD TF to that of MEFs in which an unacetylable MyoD version was transcriptionally induced (45). The experiment effectively contrasts expression profiles of two cell lines that differ only by the impact of functional and non-functional versions of the MyoD TF, respectively. Therefore, the difference in gene expression levels should only be due to the deviant behaviour of the mutant MyoD, given the same double knockout background for the two MEF cell lines. We used the dataset of genes differentially expressed in these two experiments to determine whether DiRE can trace back the difference in expression to the disruption of the MyoD regulatory pathway or not. Using the Gene Expression Omnibus (46) with the accession number GDS2854, we found 135 genes with significantly (at the 0.005 level) lower expression in MEFs expressing the mutant MyoD 24 h following its transcriptional induction. By optimizing the weights of ∼400 TF families and selecting candidate regulatory elements in loci of the co-regulated genes, DiRE determined MyoD to be second only to Tal1 in the list of the predicted most important TFs. However, the binding site for MyoD is present in more gene loci than that for Tal1 (Table 2). Interestingly, the three most frequent predicted TFBSs, those for MyoD, Tal1 and Myogenin, all share a palindromic CAgcTG core (Figure 2). This might indicate that some of the predicted Tal1 and Myogenin binding sites might be in fact MyoD binding sites and the DiRE server was not capable of effectively distinguishing them given the mapping of such highly similar binding sites. We found that REs containing at least one of the three TFs sharing the same binding core motif are present in 54 out of the 117 loci (46.1%) found by DiRE to contain positively scoring REs. Assuming that MyoD actually binds to all predicted TFBSs containing this core motif, one can speculate that these down-regulated genes are being directly regulated by MyoD, while the remaining genes represent secondary effects—genes located downstream in the regulatory pathway of MyoD. It is interesting to note that MyoD was not found to be positively associated with genes down-regulated at the 6- and 12-h time points. This indicates that regulatory effects of TFs might be detectable only at certain time points, as cell expression profiles are highly dynamic. This is reinforced by the fact that we found only six genes to be common among the sets of genes down-regulated at 6-, 12- and 24-h time points.
Table 2.
The list of 10 most important TFs determined by DiRE to be associated with down-regulation of 135 genes at 24 h after transcriptional induction of unacetylable MyoD mutant in double knockout mutant mouse fibroblast cells (DiRE ID: myod)
| TF | Importance | DiRE rank | Occurrence in REs (%) | Occurrence in gene loci (%) |
|---|---|---|---|---|
| MYOD | 0.483 | 2 | 27.91 | 36.75 |
| TAL1 | 1.013 | 1 | 29.07 | 34.19 |
| MYOGENIN | 0.225 | 5 | 20.93 | 27.35 |
| YY1 | 0.341 | 4 | 20.35 | 24.79 |
| NFKAPPAB50 | 0.344 | 3 | 13.37 | 16.24 |
| MOVOB | 0.140 | 10 | 9.3 | 13.68 |
| LFA1 | 0.166 | 7 | 9.3 | 11.97 |
| FOX | 0.171 | 6 | 8.14 | 9.40 |
| PBX | 0.160 | 8 | 6.4 | 9.40 |
| PU1 | 0.159 | 9 | 8.14 | 9.40 |
Figure 2.
Logos of the binding sites of the three most frequent TFs associated with the MyoD knockout. Note the common CAgcTG core binding motif. Logos were created with WebLogo (53).
In conclusion, the DiRE web server is capable of predicting distant REs and candidate regulatory TFs for a set of vertebrate input genes. This tool can help narrowing down and prioritizing genomic regions and TFs as candidates for further experimental validation. This should ultimately lead to enhancing the functional annotation of genomes and better understanding of mechanism of gene regulation in higher eukaryotes.
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. We thank Gabriela Loots, Kannan Tharakaraman, Leelavati Narlikar and two anonymous reviewers for critically reading the manuscript and for helpful suggestions. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Collins FS, Lander ES, Rogers J, Waterston RH, Abdellah Z, Ahmadi A, Ahmed S, Aimable M, Ainscough R, Almeida J, et al. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 5.Michelson AM. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc. Natl Acad. Sci. USA. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston M. The yeast genome: on the road to the Golden Age. Curr. Opin. Genet. Dev. 2000;10:617–623. doi: 10.1016/s0959-437x(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 7.Vilo J, Brazma A, Jonassen I, Robinson A, Ukkonen E. Mining for putative regulatory elements in the yeast genome using gene expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:384–394. [PubMed] [Google Scholar]
- 8.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 9.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajewsky N, Vergassola M, Gaul U, Siggia ED. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics. 2002;3:30. doi: 10.1186/1471-2105-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebeiz M, Reeves NL, Posakony JW. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl Acad. Sci. USA. 2002;99:9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazina AG, Papatsenko DA. Statistical extraction of Drosophila cis-regulatory modules using exhaustive assessment of local word frequency. BMC Bioinformatics. 2003;4:65. doi: 10.1186/1471-2105-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halfon MS, Grad Y, Church GM, Michelson AM. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 2002;12:1019–1028. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad YH, Roth FP, Halfon MS, Church GM. Prediction of similarly acting cis-regulatory modules by subsequence profiling and comparative genomics in Drosophila melanogaster and D.pseudoobscura. Bioinformatics. 2004;20:2738–2750. doi: 10.1093/bioinformatics/bth320. [DOI] [PubMed] [Google Scholar]
- 15.Narlikar L, Gordân R, Hartemink AJ. A nucleosome-guided map of transcription factor binding sites in yeast. PLoS Comput. Biol. 2007;3:e215. doi: 10.1371/journal.pcbi.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 17.Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O’Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- 19.Jegga AG, Sherwood SP, Carman JW, Pinski AT, Phillips JL, Pestian JP, Aronow BJ. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 2002;12:1408–1417. doi: 10.1101/gr.255002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Wu J, Sun Z. An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Res. 2003;31:1995–2005. doi: 10.1093/nar/gkg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkema WB, Johansson O, Lagergren J, Wasserman WW. MSCAN: identification of functional clusters of transcription factor binding sites. Nucleic Acids Res. 2004;32:W195–W198. doi: 10.1093/nar/gkh387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung TH, Kwan YL, Hamady M, Liu X. Unraveling transcriptional control and cis-regulatory codes using the software suite GeneACT. Genome Biol. 2006;7:R97. doi: 10.1186/gb-2006-7-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Defrance M, Touzet H. Predicting transcription factor binding sites using local over-representation and comparative genomics. BMC Bioinformatics. 2006;7:396. doi: 10.1186/1471-2105-7-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson IJ, Gottgens B. TFBScluster web server for the identification of mammalian composite regulatory elements. Nucleic Acids Res. 2006;34:W524–W528. doi: 10.1093/nar/gkl041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donaldson IJ, Gottgens B. CoMoDis: composite motif discovery in mammalian genomes. Nucleic Acids Res. 2007;35:e1. doi: 10.1093/nar/gkl839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh LN, Wang LS, Hannenhalli S. TREMOR- -a tool for retrieving transcriptional modules by incorporating motif covariance. Nucleic Acids Res. 2007;35:7360–7371. doi: 10.1093/nar/gkm885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 30.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 31.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 33.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 34.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terai G, Takagi T. Predicting rules on organization of cis-regulatory elements, taking the order of elements into account. Bioinformatics. 2004;20:1119–1128. doi: 10.1093/bioinformatics/bth049. [DOI] [PubMed] [Google Scholar]
- 36.Thompson W, Palumbo MJ, Wasserman WW, Liu JS, Lawrence CE. Decoding human regulatory circuits. Genome Res. 2004;14:1967–1974. doi: 10.1101/gr.2589004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loots GG, Chain PS, Mabery S, Rasley A, Garcia E, Ovcharenko I. Array2BIO: from microarray expression data to functional annotation of co-regulated genes. BMC Bioinformatics. 2006;7:307. doi: 10.1186/1471-2105-7-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo MW, Postlethwait J, Lee WC, Lou SW, Chan WK, Chung BC. Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family. Biochem. J. 2005;389:19–26. doi: 10.1042/BJ20050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang DQ, Cao LZ, Chou W, Shun L, Farag C, Atkinson MA, Li SW, Chang LJ, Yang LJ. Role of Pax4 in Pdx1-VP16-mediated liver-to-endocrine pancreas transdifferentiation. Lab. Invest. 2006;86:829–841. doi: 10.1038/labinvest.3700434. [DOI] [PubMed] [Google Scholar]
- 44.Pulford K, Lecointe N, Leroy-Viard K, Jones M, Mathieu-Mahul D, Mason DY. Expression of TAL-1 proteins in human tissues. Blood. 1995;85:675–684. [PubMed] [Google Scholar]
- 45.Di Padova M, Caretti G, Zhao P, Hoffman EP, Sartorelli V. MyoD acetylation influences temporal patterns of skeletal muscle gene expression. J. Biol. Chem. 2007;282:37650–37659. doi: 10.1074/jbc.M707309200. [DOI] [PubMed] [Google Scholar]
- 46.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles- -database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 48.Kamiya A, Inoue Y, Kodama T, Gonzalez FJ. Hepatocyte nuclear factors 1alpha and 4alpha control expression of proline oxidase in adult liver. FEBS Lett. 2004;578:63–68. doi: 10.1016/j.febslet.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 49.Bell AW, Michalopoulos GK. Phenobarbital regulates nuclear expression of HNF-4alpha in mouse and rat hepatocytes independent of CAR and PXR. Hepatology. 2006;44:186–194. doi: 10.1002/hep.21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernal-Mizrachi C, Weng S, Feng C, Finck BN, Knutsen RH, Leone TC, Coleman T, Mecham RP, Kelly DP, Semenkovich CF. Dexamethasone induction of hypertension and diabetes is PPAR-alpha dependent in LDL receptor-null mice. Nat. Med. 2003;9:1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- 51.Brunet de la Grange P, Armstrong F, Duval V, Rouyez MC, Goardon N, Romeo PH, Pflumio F. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood. 2006;108:2998–3004. doi: 10.1182/blood-2006-05-022988. [DOI] [PubMed] [Google Scholar]
- 52.Duran-Sandoval D, Cariou B, Percevault F, Hennuyer N, Grefhorst A, van Dijk TH, Gonzalez FJ, Fruchart JC, Kuipers F, Staels B. The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J. Biol. Chem. 2005;280:29971–29979. doi: 10.1074/jbc.M501931200. [DOI] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]