The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene (original) (raw)
Abstract
MicroRNAs (miRNAs) are noncoding RNAs that base pair imperfectly to homologous regions in target mRNAs and negatively influence the synthesis of the corresponding proteins. Repression is mediated by a number of mechanisms, one of which is the direct inhibition of protein synthesis. Surprisingly, previous studies have suggested that two mutually exclusive mechanisms exist, one acting at the initiation phase of protein synthesis and the other at a postinitiation event. Here, we resolve this apparent dichotomy by demonstrating that the promoter used to transcribe the mRNA influences the type of miRNA-mediated translational repression. Transcripts derived from the SV40 promoter that contain let-7 target sites in their 3′ UTRs are repressed at the initiation stage of translation, whereas essentially identical mRNAs derived from the TK promoter are repressed at a postinitiation step. We also show that there is a miR-34 target site within the 3′ UTR of c-myc mRNA and that promoter dependency is also true for this endogenous 3′ UTR. Overall, these data establish a link between the nuclear history of an mRNA and the mechanism of miRNA-mediated translational regulation in the cytoplasm.
Keywords: c-myc, protein synthesis, miRNA
MicroRNAs (miRNAs) are noncoding 21- to 25-nt RNA molecules that base pair imperfectly to target mRNAs (generally in the 3′ UTR) and repress the synthesis of the corresponding proteins (1). More than 800 (2) individual miRNAs have been identified in humans, which are estimated to regulate 74–92% of mRNAs (3). Malfunction of miRNA regulation is associated with human diseases, including cancer, diabetes and viral infection (4).
MicroRNA-mediated repression of gene expression appears to involve a number of posttranscriptional events. It has been shown that miRNA target mRNAs are subject to deadenylation and destabilization in addition to translational repression (5–7). However, miRNA-mediated translational repression can also occur on similar target mRNAs that lack a poly(A) tail in the absence of mRNA destabilization, so translation inhibition does not depend on deadenylation (5). Moreover, inhibition of translation is probably sufficient to account for the majority of the repression of gene expression observed in mRNAs that harbor miRNA binding sites (5, 7, 8), and it was recently shown that miRNA repression occurs before mRNA destabilization (8).
It is not yet fully understood how miRNAs repress mRNA translation. The hypothesis that miRNAs inhibit the translation of target mRNAs at the initiation stage of protein synthesis is supported by the observation that mRNAs targeted by miRNAs are found in translationally inactive subpolysomal particles (9, 10) and that miRNA-mediated translational repression depends on a 5′ cap structure and a poly(A) tail (11, 12, 8, 13). However, other studies suggest that miRNA-mediated translational repression occurs at a later stage of protein synthesis (14–17). For example, the C. elegans lin-14 and lin-28 mRNAs remain associated with the translationally active polysome fraction during miRNA-mediated translation inhibition (14, 15), and similar observations have been made in mammalian cell systems, using artificial mRNAs with miRNA binding sites in their 3′ UTRs (16, 17). Moreover, it has been shown that many miRNAs cosediment with polysomes (18, 19, 20). Therefore, the available evidence suggests two distinct and mutually exclusive mechanisms for miRNA-mediated translational repression.
For endogenous mRNAs, this apparent difference in the mechanism of translational inhibition by miRNAs could perhaps be explained by evolutionary variation (7). However, it is difficult to reconcile the differences in mechanism that are suggested by studies using reporter mRNAs. In all cases, very similar experimental systems (9, 16, 17) have been used to study miRNA-mediated translational repression, and, although the methods used to analyze the distribution of mRNA between the subpolysomes and polysomes are not identical, there are no obvious differences that could give rise to these mutually exclusive mechanisms of miRNA-mediated repression (7).
Here, we present data to resolve this dichotomy. We demonstrate that both mechanisms can operate on virtually identical mRNAs but that the promoter used to drive the transcription of the miRNA target mRNA dictates whether the translational inhibition occurs at or after initiation.
Results
The Polysomal Location of mRNAs That Are miRNA-Repressed Is Determined by the Promoter.
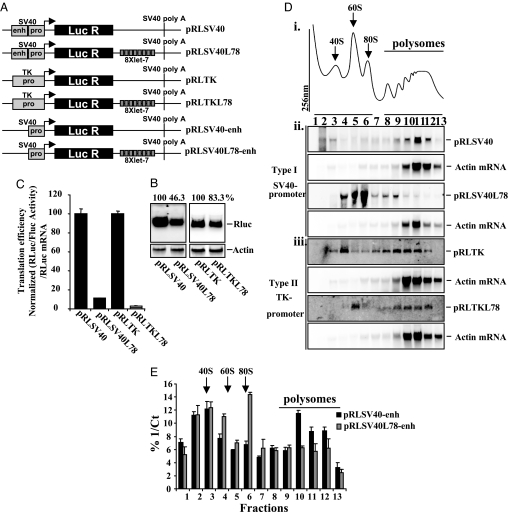
Sucrose density gradient analysis, allowing mRNAs to be separated based on the number of ribosomes associated, is the main experimental technique used to support either the initiation or postinitiation modes of miRNA repression (7). Discrepancies between the methodologies used may explain the disparate results obtained. First, differences in sucrose gradient composition could cause artifactual differences in these studies (9, 16, 17), but we found that these conditions did not affect the association of miRNA target mRNAs with ribosomes (data not shown). Second, plasmids containing different promoters were used in the studies in refs. 9, 16, and 17. To test whether the promoter influences the mechanism of miRNA-mediated repression, repeats of a let-7 miRNA target site (21) were introduced into the 3′UTR of a Renilla luciferase reporter gene under the control of either the SV40 or TK promoter (Fig. 1A). After transfection of these constructs into HeLa cells, Northern blot analysis showed that the let-7 target sites decreased Renilla luciferase mRNA levels by 54% and 17% when the mRNA was transcribed from the SV40 promoter and the TK promoter, respectively (Fig. 1B). Renilla luciferase activity was determined and normalized to the level of Renilla luciferase mRNA as a measure of translation efficiency. Let-7 target sites reduced the translational efficiency by 88% for the mRNA transcribed from the SV40 promoter and by 97% from the TK promoter (Fig. 1C). Virtually identical data were obtained by using transfection efficiency as a method of normalization [supporting information (SI) Fig. S1_A_]. A 2′-_O_-methyl oligonucleotide directed against let-7 restored translation efficiency, confirming that translational repression is let-7 dependent (Fig. S1Aiii). Sucrose density gradient analysis was performed on postnuclear lysates prepared from the transfected cells to determine the effect of let-7 target sites on the polysomal distribution of Renilla luciferase mRNA (Fig. 1Di and Fig. S1_B_). Northern blot analysis showed that the polysomal localization of actin mRNA did not change (Fig. 1D ii and iii and Fig. S1_C_). Renilla luciferase mRNA derived from the SV40 promoter was predominantly associated with the polysomes in the absence of let-7 target sites but sedimented primarily with the subpolysomes when let-7 target sites were present (Fig. 1Dii and Fig. S1_C_). In contrast, both the Renilla luciferase mRNAs derived from the TK promoter were mainly associated with polysomes, regardless of the presence of let-7 target sites (Fig. 1Diii and Fig. S1_C_).
Fig. 1.
Translation repression of mRNAs that contain let-7 target sites. (A) Diagrammatic representation of the SV40 and TK reporter constructs. See SI Materials and Methods for details. (B) HeLa cells were transfected by using the constructs shown in A. Total mRNA was prepared, Northern blot analysis was performed, and the resultant membranes were probed with radiolabeled DNA derived from Renilla luciferase and actin mRNA. Quantification of the Renilla luciferase mRNA to actin mRNA levels is shown at the top and is expressed as a percentage of the control levels. Quantitative RT-PCR was also performed on the same samples (Fig. S1_Ai_). Experiments were performed on three independent occasions. (C) Lysates from cells transfected in B were assayed for luciferase activity, and Renilla luciferase levels were normalized to the transfection control, firefly luciferase. Values were normalized to Renilla luciferase mRNA levels (B) as a measure of translational efficiency. Experiments were performed in triplicate on three independent occasions. (D) HeLa cells (6 × 106) were transfected with constructs as indicated. Postnuclear lysates were prepared and subjected to sucrose density gradient centrifugation analysis. (Di) An example of a trace from one gradient is shown (additional traces can be found in Fig. S1_B_). (Dii and Diii) Northern blot analysis was performed on equal volumes of RNA and membranes were probed with radiolabeled DNA derived from Renilla luciferase. These same membranes were then reprobed for actin mRNA. Experiments were performed on three independent occasions. (E) The SV40 enhancer was removed from both the control construct (pRLSV40) and the construct containing 8X let-7 target sites (pRLSV40L78), creating two new vectors, pRLSV40-enh and pRLSV40L78-enh, respectively. After transfection of these constructs, postnuclear lysates were subjected to sucrose density gradient analysis. RNA was isolated from gradient fractions and real-time PCR was carried out to determine the relative amount of Renilla luciferase mRNA in each fraction and expressed as a percentage of the total value of 1/Ct per fraction. Real-time PCR was preformed as the mRNA levels were too low to detect by Northern blot analysis. Average values for three independent experiments are shown.
Because the SV40 promoter/enhancer is considerably more efficient than the TK promoter, it is possible that these promoter effects could be due to mRNA abundance. To test this hypothesis, the SV40 enhancer was deleted from the SV40 promoter constructs (Fig. 1A). The resulting constructs were transfected into HeLa cells and expressed less mRNA than the corresponding TK promoter constructs (Fig. S1_Di_). After fractionation on sucrose density gradients (Fig. S1_Dii_), polysome analysis was performed, using qRT-PCR (because of low expression) to detect the Renilla luciferase mRNA (Fig. 1E) and distribution compared with actin mRNA (Fig. S1_Diii_). In the presence of let-7 target sites, a reduction in the translational efficiency similar to that obtained by using the vectors that contained the enhancer was observed, accompanied by an equivalent shift from the polysomes to subpolysomal fractions (Fig. 1A and Fig. S1_D iv_ and v). This suggests that the promoter-specific behavior of miRNA target mRNAs is not due to mRNA abundance or the presence of the SV40 enhancer.
There Are Two Distinct Mechanisms of miRNA-Mediated Translational Repression.
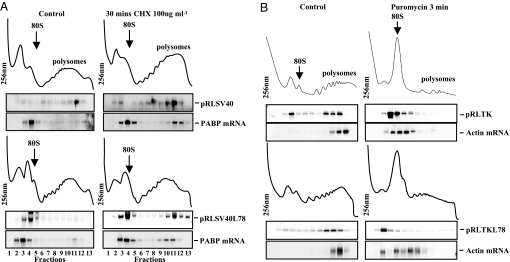
The behavior of the mRNAs derived from the SV40 promoter suggests that miRNAs may inhibit translation during initiation. However, there are three possible explanations for these data: a complete inhibition of initiation, a reduction in the rate of initiation, or a complete inhibition of translation elongation resulting in mRNAs harboring a single 80S ribosome. To distinguish between these three types of inhibition, low concentrations of cycloheximide (1,000-fold less than would cause a total elongation block) were used to reduce the rate of elongation rather than totally inhibit this process (ref. 22; Fig. S2_A_). An mRNA repressed at the initiation step would be driven into the polysomes if elongation is slowed down, whereas an mRNA repressed at elongation would be affected to a much lesser degree by further repression. Cells were transfected with pRLSV40 or pRLSV40L78 and then treated for 30 min with 100 ng/ml cycloheximide. After sucrose density gradient analysis and Northern blot analysis of RNA fractions, we observed an accumulation of the let-7 targeted Renilla luciferase mRNA in the polysomes (Fig. 2A and Fig. S2_B_), and a similar redistribution of poly(A) binding protein (PABP) mRNA, which is known to be repressed at the initiation stage (23) (Fig. 2A and Fig. S2_C_). These data strongly suggest that in the case of SV40-derived mRNAs, initiation and not elongation is the rate-limiting step of miRNA repression, and that miRNA-mediated repression reduces the frequency of initiation, rather than completely blocking this process. The polysomal distribution of the TK derived transcripts was also examined after cycloheximide treatment but no further movement was identified (data not shown).
Fig. 2.
Translational inhibitors confirm that miRNA-mediated repression can occur at both initiation and postinitiation stages of translation. (A) HeLa cells were transfected with pRLSV40 and pRLSV40L78 and treated with 100 ng/ml cycloheximide for 30 min. Cell lysates were prepared and subjected to sucrose density gradient analysis. RNA was isolated from gradient fractions, and Northern blot analysis performed. Membranes were probed with radiolabeled DNA derived from a fragment of Renilla luciferase, and the same membranes were then reprobed for PABP mRNA. The data shown are representative of three independent experiments. (B) HeLa cells were transfected with pRLTK or pRLTKL78, and cells were treated with 100 μg/ml puromycin for 3 min. Sucrose density gradient analysis and Northern blot analysis for Renilla luciferase and actin mRNA were performed as described above. Representative data from three independent experiments are shown.
To investigate the mechanism of miRNA-mediated repression on mRNAs derived from the TK promoter, HeLa cells were transfected with pRLTK and pRLTKL78 and treated with puromycin, which causes premature polypeptide chain termination and ribosome release (Fig. 2B and Fig. S2_D_). Sucrose density gradient analysis followed by northern blot analysis revealed that puromycin treatment resulted in a shift of the control Renilla luciferase, let-7 targeted Renilla luciferase mRNAs, and actin mRNAs from the polysomes to the subpolysomes. These data indicate that ribosomes can translocate on miRNA-repressed mRNAs; consequently, miRNAs do not completely block elongation (Fig. 2B, S2D), in agreement with observations in refs. 16 and 17.
Taken together, these data show that, although the degree of let-7-mediated translational repression is similar for mRNAs derived from either promoter, the mechanism of translational repression depends on the promoter. We have termed these type I and type II repression, where type I (SV40) repression is mediated at the initiation stage of translation, and type II (TK) is a postinitiation event.
The Promoter Can Alter the Type of miRNA-Mediated Repression of an Endogenous 3′ UTR.
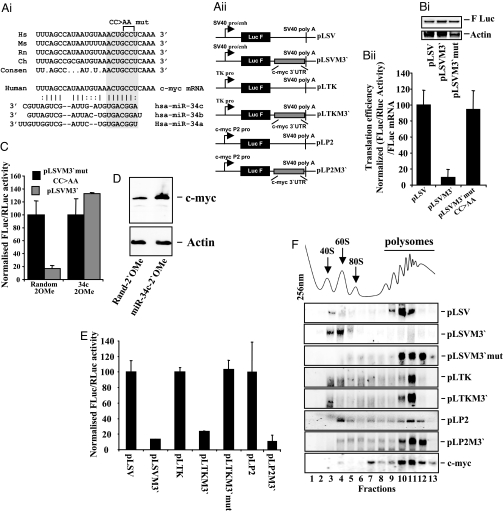
We showed that c-myc mRNA is almost exclusively associated with polysomes; however, the translation of this mRNA is partially repressed (24), suggesting that it may be subject to type II miRNA-mediated repression. To test this hypothesis, the c-myc 3′ UTR was examined for potential miRNA target sites and a conserved target site for the miR-34 family [a, b, c; known to be expressed in HeLa cells (25)] was identified (Fig. 3Ai). Two point mutations were introduced in the c-myc 3′UTR to disrupt the miR-34a-c target site within the seeding sequence, and wild-type and mutant c-myc 3′UTR sequences were inserted into the vector pLSV (Fig. 3Aii). These constructs were transfected into HeLa cells, and firefly luciferase activities were determined and normalized to the levels of firefly luciferase mRNA, determined by Northern blot analysis, as a measure of translational efficiency (Fig. 3B). The c-myc 3′UTR reduced translational efficiency by ≈90%, and mutation of the miR-34a-c site completely restored translation rates to control levels (Fig. 3Bii). These data were also normalized by using transfection efficiency as an additional control and virtually identical data were obtained (Fig. S3_A_). To confirm the presence of a miRNA target sequence in the c-myc 3′ UTR, 2′-_O_-methyl oligonucleotides directed against miR-34c or a control sequence were included in transfections (Fig. 3C). In the presence of the 2′-_O_-methyl oligonucleotide directed against miR-34c the repression of luciferase expression was relieved (Fig. 3C). In addition, the levels of c-Myc protein were determined by Western blot analysis, and increased twofold without a change in mRNA level in the presence of the 2′-_O_-methyl oligonucleotide directed against miR-34c, consistent with relief of miRNA-mediated translational repression (Fig. 3D and Fig. S3_B_). Overall, these data confirm the presence of a functional miR-34c target site in the 3′ UTR of c-myc (Fig. 3 A–D).
Fig. 3.
c-myc mRNA translation is repressed by miR-34c at a postinitiation stage. (Ai) Comparison of a section of the c-myc 3′ UTRs from human (Hs), mouse (Ms), rat (Rn), and chicken (Ch), showing that the putative miR-34a-c target site is conserved. (Aii) Schematic representation of plasmids containing the c-myc 3′UTR and the SV40, TK and endogenous P2 c-myc promoters. See SI Materials and Methods for details. (Bi) The constructs pLSV, pLSVM3′ and pLSVM3′mut were transfected into HeLa cells. Total RNA was isolated and Northern blot analysis was performed by using a radiolabeled probe directed against firefly luciferase. All experiments were performed in triplicate on three independent occasions. (Bii) After the transfections in Bi, firefly luciferase expression was assayed by using the Dual-Luciferase assay system and normalized to the transfection control, Renilla luciferase. Values were normalized to firefly luciferase mRNA levels (Bi) as a measure of translational efficiency. All experiments were performed in triplicate on three independent occasions. (C) The constructs pLSVM3′ and pLSVM3′mut were transfected into HeLa cells with either a 2′-_O_-methyl oligonucleotide directed against miR-34c or a control oligonucleotide. Firefly luciferase activity was determined and expressed relative to the transfection control Renilla luciferase. All experiments were performed in triplicate on three independent occasions. (D) Western analysis was performed on cells transfected with 2′-_O_-methyl oligonucleotides directed against miR-34c or a control oligonucleotide, and membranes were immunoblotted for c-Myc and actin protein levels. (E) The constructs shown in Aii were transfected into HeLa cells. Firefly luciferase activity was determined and expressed relative to the transfection control Renilla luciferase. All experiments were performed in triplicate on three independent occasions. (F) The constructs shown in Aii were transfected into HeLa cells and postnuclear lysates were subjected to sucrose density gradient analysis. RNA was isolated and Northern blot analysis performed to detect firefly luciferase mRNA, and the same membranes were reprobed for actin mRNA (Fig. S3_D_). Data are representative of three independent experiments.
To investigate whether the mechanism of miRNA-mediated repression via a target site in an endogenous 3′UTR also depends on the promoter, the SV40 promoter in the constructs pLSV and pLSVM3′ was replaced with the TK promoter (Fig. 3Aii). The presence of the c-myc 3′UTR reduced luciferase expression significantly whether transcription was driven by the SV40 or the TK promoter (Fig. 3E), whereas a mutation in the seeding sequence of miR34a-c restored firefly luciferase synthesis (Fig. 3E). Sucrose density gradient analysis was performed on HeLa cells transfected with the c-myc 3′UTR constructs (Fig. 3F and Fig. S3_C_). When transcription was driven by the SV40 promoter, the c-myc 3′UTR caused firefly luciferase mRNA to accumulate in subpolysomal fractions, whereas mRNAs containing the mutant 3′ UTR associated with polysomes (Fig. 3F). In contrast, when transcription was controlled by the TK promoter, both the control and c-myc 3′UTR-containing mRNAs were predominantly associated with polysomes (Fig. 3F). There was no significant change in the distribution of actin mRNA (Fig. S3 D and E). qRT-PCR showed there was no destabilization of the mRNA derived from the vectors that contained the TK promoter (Fig. S3_F_). These data show that the mechanism of miRNA-mediated repression is determined by the promoter in an identical manner for mRNAs harbouring artificial or endogenous miRNA target sites.
To establish whether the endogenous c-myc promoter conferred type I or type II repression on mRNAs bearing the c-myc 3′UTR, the c-myc P2 promoter (26) and corresponding 5′ UTR were placed upstream of the firefly luciferase coding region (Fig. 3Aii), and insertion of the c-myc 3′UTR into this construct substantially reduced firefly luciferase expression compared with the control construct, in the absence of any change in the levels of mRNA (Fig. 3E and Fig. S3_F_). Sucrose density gradient analysis indicated that the control firefly luciferase mRNA was distributed across the gradient, possibly because of the 5′ UTR present in these constructs, whereas firefly luciferase mRNA bearing the c-myc 3′ UTR cosedimented almost exclusively with the polysomes (Fig. 3F and Fig. S3_Eiii_). Endogenous c-myc mRNA was also mainly associated with the polysomes (Fig. 3F and Fig. S3_Eiii_). Therefore, a combination of the c-myc promoter and 3′UTR inhibits the translation of an mRNA postinitiation. Taken together with the data in Fig. 3D, these data indicate that miRNA-mediated translational repression of c-myc mRNA occurs by a type II mechanism.
Discussion
Our data demonstrate that there are two distinct types of miRNA-mediated translational repression. In our experimental system, type I repression is observed for miRNA targeted mRNAs that originate from the SV40 promoter. In addition, we have shown that miRNA- targeted mRNAs transcribed from the CMV promoter undergo type I repression, in agreement with ref. 9 (Fig. S4). Type I repression is characterized by increased association of the target mRNA with the subpolysomes on miRNA binding, and appears to be due to inhibition of translation during initiation (Figs. 1Dii and 3F). These observations are in agreement with other studies that have described inhibition of translation initiation by miRNAs, using artificial constructs that contain miRNA target sites (9). Our data also show that miRNAs do not completely inhibit translation initiation on their target mRNAs, because low concentrations of cycloheximide can restore the association of these target mRNAs with the polysomes (Fig. 2A). Based on this observation, we propose that miRNAs reduce the rate of initiation of protein synthesis on a target mRNA in the type I mechanism. Recently, it has been shown that the endogenous CAT1 mRNA is repressed by miR-122 at the initiation phase of protein synthesis, indicating that the type I mechanism of miRNA-mediated repression also occurs on endogenous mRNAs (10).
Type II repression occurs on miRNA targeted mRNAs that are transcribed from the TK promoter and is characterized by translational inhibition despite the continued association of the target mRNA with the polysomes (Fig. 1Diii). These data are in agreement with those obtained by using artificial constructs that contained the TK promoter and either CXCR4 target sites or let-7 target sites (16, 17). Importantly, we show that this type of repression also occurs on an endogenous mRNA. We have identified a miR-34c binding site in the c-myc 3′UTR that causes translational repression of the target mRNA. mRNAs derived from the endogenous c-myc P2 promoter are translationally repressed by the c-myc 3′UTR but have increased association with polysomes (Fig. 3F), suggesting that miR-34c can repress the translation of c-myc mRNAs through a type II mechanism. In addition, polysomes formed on mRNAs subject to type II miRNA-mediated repression can be dissociated by using puromycin, indicating that ribosomes are actively translocating along these mRNAs (Fig. 2B), in agreement with the studies in refs. 16 and 17. The puromycin sensitivity also shows that we are not observing pseudopolysomes (27). We cannot rule out the possibility that initiation is inhibited during type II repression, but the polysomal distribution of these mRNAs shows that inhibition at the postinitiation stage predominates.
There are several possible explanations why the promoter of a miRNA target gene may influence the mechanism of miRNA-mediated translational repression. It is unlikely that different promoter efficiencies result in the preferential use of a particular mechanism, because we have shown that the mechanism of repression does not change when transcription from the SV40 promoter is reduced to below that from the TK promoter (Fig. 1E and Fig. S1_Di_).
There is no apparent correlation between the mechanism of miRNA-mediated repression and the 5′ ends of the mRNAs used in this study. The mRNAs transcribed from the TK promoter begin with an adenine, as do CAT-1 and c-myc mRNAs (28), whereas three possible mRNAs can be derived from the SV40 promoter that start with either guanine or adenine (29). To determine whether there were differences in the 5′ start sites of the mRNAs derived from the SV40 promoter that could lead to alternative mechanisms of miRNA-mediated repression, RACE was performed to map the 5′ ends of the transcripts present in either the polysomal or subpolysomal fractions. All three possible 5′ ends were present in both pools (data not shown). Furthermore, there is no obvious similarity between the 5′ UTRs either in the constructs used here or those found in endogenous mRNAs and the type of repression observed. Because both the CAT-1 and c-myc mRNAs contain IRESs (30, 31) yet exhibit different mechanisms of miRNA repression, it is unlikely that internal translation initiation influences the mode of miRNA-mediated inhibition. Finally, we have examined whether the method of transfection influences the degree of miRNA-mediated repression but find no difference (Fig. S5).
Therefore, our data suggest that an intrinsic property of the promoter determines the mechanism of miRNA-mediated translational repression. Studies using mRNAs transfected directly into the cytoplasm indicate that under these conditions miRNAs target the initiation phase of protein synthesis (11), reminiscent of type I repression. Therefore, a possible explanation for the difference between the types of miRNA-mediated repression is that a nuclear event linked to the promoter, such as cotranscriptional loading of factors onto the nascent mRNA, identifies miRNA target mRNAs for type II repression. Target mRNAs that do not experience this nuclear event would be subject to type I repression by default. The reason for two distinct mechanisms is unclear at present, but it is likely that these mechanisms reflect different modes of regulation of these mRNAs.
Materials and Methods
Vectors and Constructs.
The vectors used were based on pRLSV40 and pGL3 (Promega). Eight copies of the let-7 target site (5′-AACTATACAACGTCTACCTCA-3′) (21) were cloned into the 3′UTR (XbaI site) of pRLSV40, creating the construct pRLSV40L78. The SV40 promoter/enhancer elements from these constructs were replaced with the TK promoter element, giving the constructs pRLTK and pRLTKL78. The vector pLSVM3′ was created by cloning the c-myc 3′ UTR into pLSV and was modified by replacing the SV40 promoter with either the TK promoter or the c-myc P2 promoter to create vectors pLTK, pLTKM3′, pLP2, and pLP2M3′.
Sucrose Density Gradient Centrifugation and RNA Detection.
Sucrose density gradient centrifugation was used to separate ribosomes into polysomal and subpolysomal forms. Cells (6 × 106) were incubated with 0.1 mg/ml cycloheximide for 3 min at 37°C, washed in PBS containing 0.1 mg/ml cycloheximide, and lysed in lysis buffer [15 mM Tris·HCl (pH 7.4), 15 mM MgCl2, 0.15M NaCl, 1% Triton X-100, 0.1 mg/ml cycloheximide, and 1 mg/ml heparin). Please see SI Materials and Methods for details.
RNA Analysis.
Northern blot analysis of RNA isolated from sucrose density gradients was performed as described in ref. 24. Radiolabeled DNA hybridization probes were generated by using the RadPrime kit according to the manufacturer's instructions (Invitrogen). Quantification of Northern blot analysis was preformed on QuantityOne HD analysis software from Bio-Rad after scanning on Bio-Rad molecular Imager FX. Real-time PCR was carried out by using the Stratagene MX3005P QPCR system. Please see SI Materials and Methods for details.
Cell Culture and Transient Transfections.
HeLa cells were cultured in DMEM containing 10% FCS in a humidified atmosphere containing 5% CO2. For DNA transfections, FuGENE 6 (Roche) was used, following the supplier's instructions. For a 10-cm plate of cells containing ≈2 × 106 cells, a total of 10 μg of DNA was transfected. HeLa cells were transfected with 2′-_O_-methyl oligonucleotides, using lipofectamine 2000 according to manufacturer's instructions (Invitrogen).
Cells were harvested after 48 h, and the activities of firefly and Renilla luciferases in lysates were measured by using a dual luciferase reporter assay system (Promega). Light emission was measured over 10 s, using an OPTOCOMP I luminometer.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Rachel Edwards for making the plasmid construct pRLSV40L78 in the laboratory of Drs. C. H. de Moor and Catherine Jopling for critical reading of this manuscript. This work was supported by grants from the Biotechnology and Biological Sciences Research Council (to Y.W.K., I.G.C., T.L.H., P.G.G., and H.C.D.), the Wellcome Trust (to K.A.S., M.S., H.A.M.), and the Leukaemia Research Fund (K.H.). M.B. holds a Biotechnology and Biological Sciences Research Council David Phillips Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNA and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 2.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 3.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;367:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 8.Mathonnet G, et al. MicroRNA inhibition of translation tnitiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 9.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of mircoRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 13.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21(15):1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 15.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 16.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 17.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 20.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walden WE, Godefroy-Colburn T, Thach RE. The role of mRNA competition in regulating translation. I. Demonstration of competition in vivo. J Biol Chem. 1981;256:11739–11746. [PubMed] [Google Scholar]
- 23.de Melo Neto OP, Standart N, Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushell M, et al. Polypyrimidine tract binding protein regulates gene expression during apoptosis via modulation of IRES-mediation translation of mRNAs including those encoding members of Notch signalling pathway and chromatin remodelling enzymes. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 27.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 28.Watt R, et al. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983;303:725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- 29.Haegeman G, Fiers W. Characterization of the 5′-terminal cap structures of early simian virus 40 mRNA. J Virol. 1980;35:955–961. doi: 10.1128/jvi.35.3.955-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez J, et al. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information