Hematopoietic Fingerprints: an expression database of stem cells and their progeny (original) (raw)
. Author manuscript; available in PMC: 2008 Nov 15.
Published in final edited form as: Cell Stem Cell. 2007 Nov 15;1(5):578–591. doi: 10.1016/j.stem.2007.10.003
Summary
Hematopoietic stem cells (HSC) continuously regenerate the hematologic system, yet few genes regulating this process have been defined. To identify candidate factors involved in differentiation and self-renewal, we have generated an expression database of hematopoietic stem cells and their differentiated progeny, including erythrocytes, granulocytes, monocytes, NK cells, activated and naïve T-cells, and B-cells. Bioinformatic analysis revealed HSC were more transcriptionally active than their progeny and shared a common activation mechanism with T-cells. Each cell type also displayed unique biases in the regulation of particular genetic pathways, with Wnt signaling particularly enhanced in HSCs. We identified ∼100 to 400 genes uniquely expressed in each cell type, termed lineage “fingerprints.” In overexpression studies, two of these genes, Zfp105 from the NK cell lineage, and Ets2 from the monocyte lineage, were able to significantly influence differentiation toward their respective lineages, demonstrating the utility of the fingerprints for identifying genes that regulate differentiation.
Introduction
Hematopoiesis entails differentiation of the hematopoietic stem cell (HSC) through progenitor intermediates to terminally differentiated blood cells exhibiting vastly different morphologies and functions. The transcriptional control of HSC differentiation is still poorly understood, despite advances in mouse genetics that have elucidated the role of certain pivotal molecules within the developmental hierarchy. A few transcription factors have been shown to be essential for specific lineages; for example, Early B-cell factor-1 (ebf1) in B lymphocytes (Lin and Grosschedl, 1995), and gata2 in megakaryocytic differentiation (Ling et al., 2004; Orkin et al., 1998). However, the number of genes demonstrated to be critical for differentiation within most hematopoietic lineages is extremely small. The few global approaches that have been used to study regulation of hematopoietic cells have focused either on comparisons between HSCs and other stem cell types (Ivanova et al., 2002; Ramalho-Santos et al., 2002), or between HSCs and pools of their differentiated progeny (Toren et al., 2005), which limits the ability to identify candidate regulators due to the choice of the comparator populations.
We have taken a systematic approach to identify genes uniquely expressed in murine HSCs, as well as in their differentiated counterparts. Bioinformatics enabled investigation of lineage relationships, dominant genetic pathways, and chromatin status in HSCs versus differentiated cells. We also identified “fingerprints” for each cell type comprised of genes uniquely expressed therein, and we show that at least two of these fingerprint genes participate in regulation of cell-type identity. These studies have uncovered a number of novel genes as candidate regulators of HSC and their differentiated progeny, many of which will be of interest for therapeutic modulation of these populations.
Results
To identify genes uniquely expressed in HSCs and their differentiated progeny, we took a global gene expression profiling approach, determining in parallel the transcribed genome of known coding transcripts in HSCs as well as their major differentiated lineage, including natural killer (NK) cells, T-cells, B-cells, monocytes, neutrophils, and nucleated erythrocytes. Because we were particularly interested in potential similarities between HSCs and T-cells, we examined both activated and naïve CD4+ (helper) and CD8+ (cytotoxic) T-cell subsets. Each population was purified to at least 95% purity, and multiple parameters were standardized to reduce technical variation (see supplemental Table S1 and Methods). RNA from the samples was processed and hybridized to Affymetrix MOE430 2.0 microarrays, which have probe sets representing about 20,000 genes. This is the first study to interrogate a stem cell and multiple progeny cell types. The entire data set is available (Supplemental Table S2, in GEO (accession number GSE6506), and a gene-by-gene query can be performed (http://franklin.imgen.bcm.tmc.edu/loligag/).
Gene expression patterns reflect ontogeny
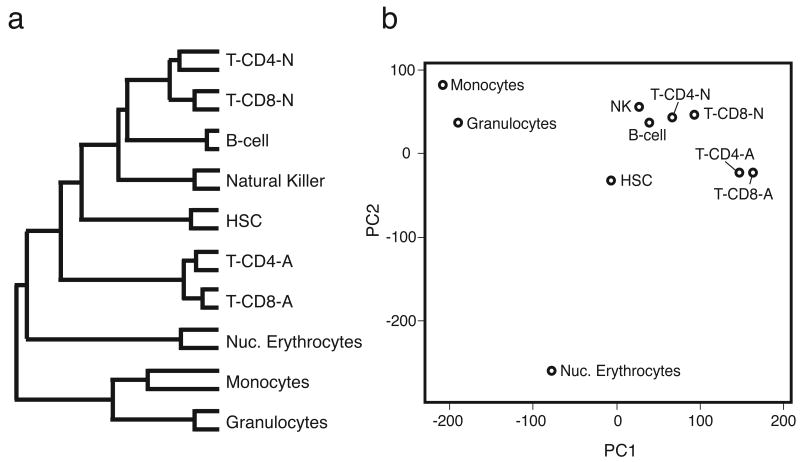
We first used the data set to investigate assumptions concerning relatedness of the hematopoietic cell types, as the similarity of gene expression profiles between different populations is thought to reflect their ontogeny relationships(Puthier et al., 2004), and may shed light on the debate regarding their developmental origins. We used cluster analysis to assess relative distance of the transcriptome of each cell type, resulting in a branched “family tree” based on the similarity between overall transcription patterns (Fig.1a). The HSC clustered with the lymphocytes, supporting a recent report that HSCs and lymphocytes share many similarities, and may indicate a conserved mechanism between HSCs and lymphocytes of long term quiescence interrupted by bursts of proliferative stimuli (Luckey et al., 2006). Also noteworthy is the very distinct nature of the activated T-cells (both CD4+ and CD8+).
Figure 1. Global transcription profile analysis reveals hematopoietic cell ontogeny.
Expression differences between microarrays were assessed on the basis of cluster analysis and principle components analysis (PCA). (a) A dendrogram displays distance in a branching fashion where the right-most branch point indicates the cell types with the highest degree of similarity, with replicates most similar. The HSC are most transcriptionally similar to lymphocytes, with activated T-cells quite distinct. (b) When relative distance is collapsed to two dimensions using PCA, the HSC resides at a midpoint between lymphocytes, myeloid cells, and erythrocytes.
We used principle components analysis (PCA) to further explore the cell-type relationships. When the relative expression distance between each averaged chip pair was examined with PCA in the two dimensions that contain the greatest variance within the data (first and second principle components; PC1, PC2), we found the cell types cluster on the basis of ontogeny, with the stem cell having a centralized position (Fig.1b). This analysis reveals the striking segregation of lymphoid, myeloid, and erythroid cells into three discrete clusters. Both cluster analysis and PCA indicate that erythrocytes are distant from myeloid cells (monocytes, granulocytes), supporting a recent observation that they arise independently of a common myeloid progenitor (Adolfsson et al., 2005).
Genetic fingerprints unique to each cell type
We next examined the data to determine whether there were genes exclusively expressed by particular cell types. Such a genetic “fingerprint” would be expected to contain some genes encoding proteins involved in unique cell functions, as well as regulatory proteins that may direct expression of other genes in the fingerprint.
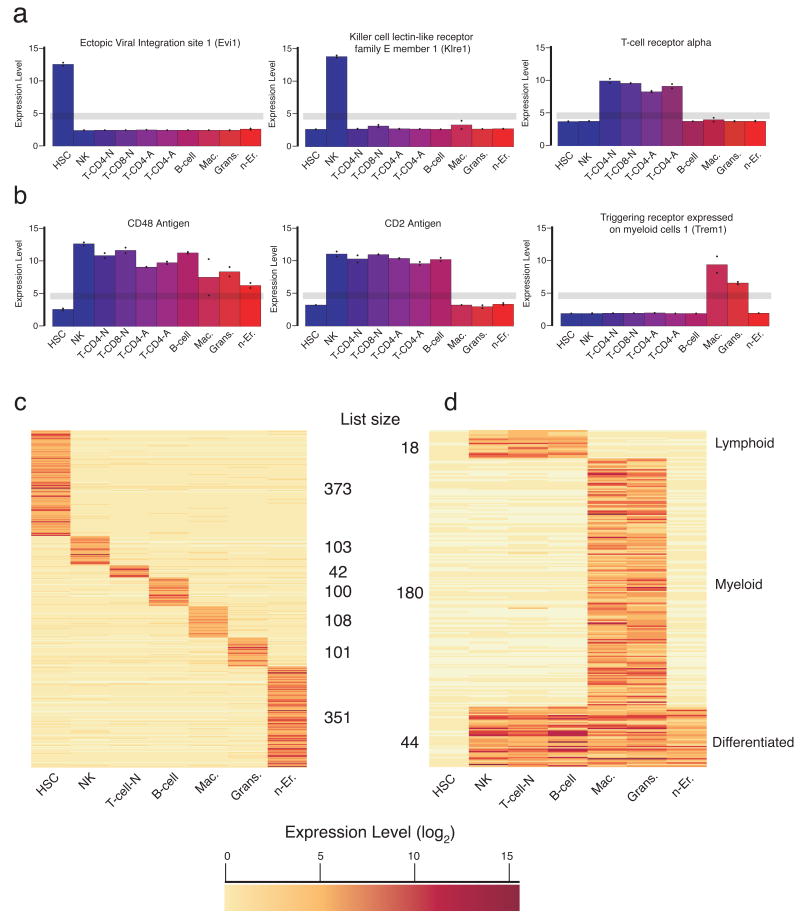
We defined fingerprint genes as those that were “on” in one cell type, and “off” in all other profiles by establishing expression thresholds above and below a conservative window selected by real-time PCR (Supplemental Fig. 1). An expression value of 5 or greater was considered “on”, and 4 or less was considered “off” (log(2) scale). Examples of uniquely expressed genes found in fingerprints are ecotropic viral integration site-1 (Evi1), a known proto-oncogene (Mucenski et al., 1988) and the most differentially expressed gene in HSC, killer cell lectin-like receptor family E member-1 (Klre1), an NK cell receptor (Wilhelm and Mager, 2003), and T cell receptor alpha on T-cells (Fig. 2a).
Figure 2. Unique genetic fingerprints for hematopoietic cell types.
The normalized (log2) expression intensity is shown. The grey line indicates the threshold window, above which a gene was considered to be expressed, and below which, non-expressed, for selection of uniquely expressed genes in the “fingerprints.” (a) Shown are three examples of genes expressed in a specific cell type, or (b) a group of cell types including all differentiated cells, lymphocytes, or myeloid cells. The fingerprints are summarized in a heat map (color indicates normalized expression level (log2); darker corresponds to higher expression): (c) cell-type specific fingerprints, and (d) shared fingerprints.
The filter was also applied to groups of cell-types to determine a “shared” fingerprint for all differentiated cells, as well as the lymphoid (NK, T-, and B-cells) and myeloid (monocytes and granulocytes) branches, which defined mutually expressed genes excluded from other cells (Fig. 2b). Strikingly, expression of the gene for the cell surface marker CD48 identifies all the differentiated cell types but not HSCs, consistent with recent reports (Kiel et al., 2005). Similarly, CD2 expression marks the lymphoid lineage, and Trem1 the myeloid lineage. For the purpose of identifying a T-cell fingerprint, we chose to combine both types of naïve T-cells and exclude the activated T-cells in order uncover genes responsible for T-cell differentiation and not T-cell activation.
Each cell-type fingerprint contained 40-100 genes, except the HSC and erythroid cell fingerprints, which both had ∼ 350 genes (Fig. 2c, Supplemental Table S3). The shared lymphoid fingerprint, representing genes shared by all lymphoid cells and excluded from stem cells, contained only 18 genes, reflecting their similarity to stem cells. The myeloid fingerprint contained 154 genes, and 36 genes were shared among all differentiated cells. Finally, there were ∼9000 genes expressed by all hematopoietic cells (Supplemental Table 3); many are likely ubiquitously expressed house-keeping genes, but some may be unique to the entire hematopoietic system, such as CD45.
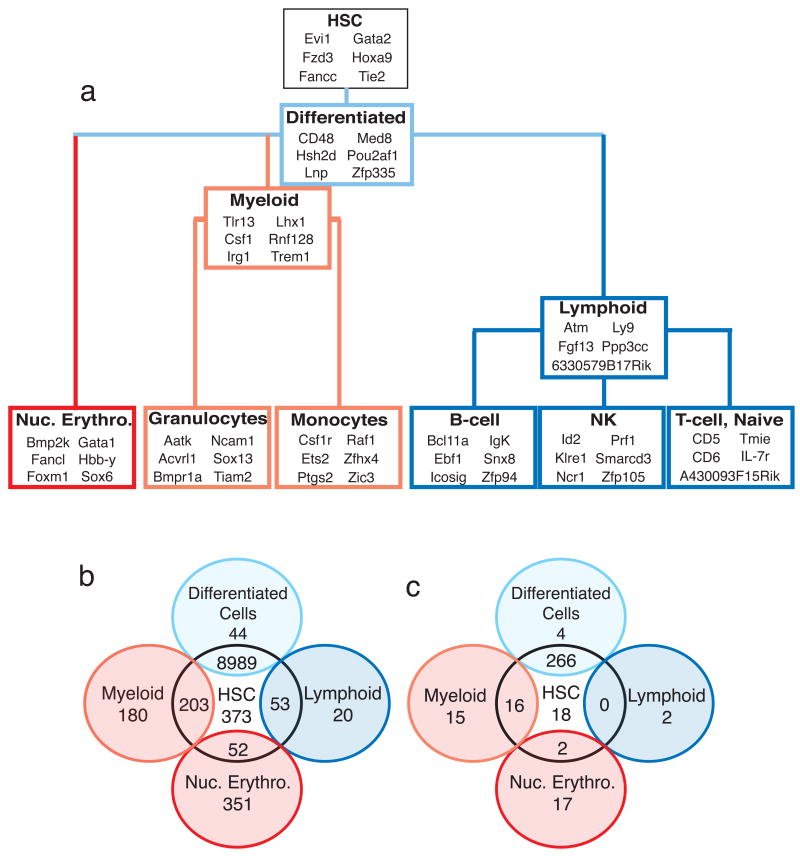
The fingerprints contain some genes known to be important for the function of certain cell types, such as Gata1 in the erythroid lineage (Evans et al., 1988; Wall et al., 1988) and the CSF1 receptor (Csf1r) in the monocyte lineage (Roussel and Sherr, 1989), validating the approach for identification of cell-type-specific genes (Fig. 3a), as well as many under-studied genes that may have regulatory role or serve as novel markers. Likewise, the shared fingerprint genes are either required for the function of all family cell types, such as Atm in the lymphoid fingerprint (Ito et al., 2004), or for differentiation of the cell-type family (Fig. 3a). We predict that common progenitors will express some of the shared fingerprint genes (Fig. 3a,b), which could be developed as tools for markers of those progenitors, or as regulators of their fate.
Figure 3. Examples of fingerprint genes and phenotype assessment.
(a) Selected genes from each of the cell-type and shared fingerprints are shown in context with their developmental relationships. (b) A Venn diagram summarizes the number of shared genes in the different fingerprints. (c) Venn diagram summarizes the number of knockout mice with hematopoietic phenotypes in each of the indicated gene groups. Not all cell types and intersections are shown.
To assess whether deficiencies of fingerprint genes result in hematopoietic defects, we examined phenotypes in the Jax Mouse Genome Informatics database (MGI; v.3.44) for all genes that had a reported knock-out allele. Remarkably, several fingerprint genes with available knock-outs show defects in either differentiation or functionality of a lineage (Fig. 3c, Table 1). A striking example from the B-cell fingerprint is the Ebf1 knockout, which exhibits a complete loss of B-cells (Lin and Grosschedl, 1995). Likewise, ablation of HoxA9, an HSC fingerprint gene, results in severely impaired HSCs (Lawrence et al., 2005). All fingerprints, with one exception, demonstrated a statistically significant enrichment for genes exhibiting a hematopoietic phenotype when knocked out (defined by MGI as either “hematopoietic system” or “immune system” phenotypes) (z-test, Supplemental Table S4). Only the 9000-gene common fingerprint was not found to be enriched for a hematopoietic phenotype, most likely due to the preponderance of house-keeping genes in this list.
HSC and T-cells share a similar transcriptional program of activation
Anecdotal observations have suggested that HSCs and T-cells share a number of markers as well as regulatory genes. These similarities could result from a similar lifestyle of long-term quiescence followed by expansion (Luckey et al., 2006), or could be related to the evolutionary history of the hematopoietic system; specifically, HSC transcription factors may have been adapted for lymphocyte development and maintained for T-cell differentiation (Rothenberg and Pant, 2004). Because we were interested in a potential relationship between HSC and T cells, and also the similarity between HSC activation and T-cell activation, we examined this in greater detail.
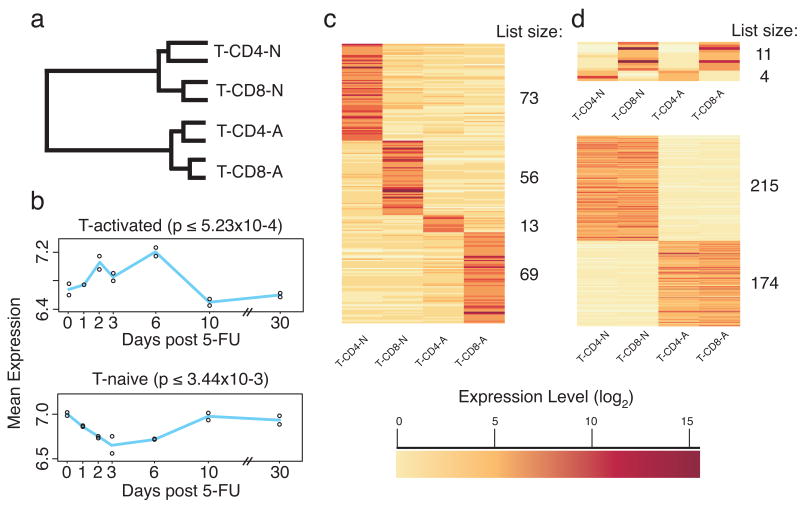
Cluster analysis was repeated with T-cell data alone. This analysis again showed the importance of activation for distinguishing T-cell sub-types compared to their CD4/CD8 status (Fig. 4a). We next compared the activated vs. naïve T-cell data to our previous study, in which we identified genes that change during HSC activation, as stimulated by 5-fluorouracil treatment (Venezia et al., 2004). A list of genes that differ between activated and naïve T-cells was generated from a pair-wise comparison. The average expression values of these genes in HSCs over the activation time-course were then plotted. On average, genes that were upregulated in activated T-cells were also upregulated in activated HSCs (Fig. 4b; genes increasing in expression on days 2-6 of HSC activation; one-way ANOVA p ≤ 5.23×10-4, alpha=0.05), whereas naïve T-cell genes have the reciprocal signature, being more highly expressed in quiescent HSCs (p ≤ 3.44×10-3, alpha=0.05). This analysis suggests that the mechanisms that regulate the activation state in HSCs and T-cells may be similar.
Figure 4. T-cell and HSC activation are transcriptionally similar.
T-cell data was analyzed independent of other cell types. (a) Cluster analysis indicates that activation state drives T-cell sub-set differences. (b) Genes involved in T-cell activation are up regulated during HSC activation. A pair-wise comparison between naïve and activated T-cells (including both CD4+ and CD8+ cells) was used to generate a list of genes up regulated in activated or naïve T-cells. The mean expression values of genes in these lists (Up-in-Naïve and Up-in-Activated) was plotted from the HSC activation time-course data. Genes up regulated during T-cell activation were upregulated during HSC activation (top panel), peaking at day 6 when HSCs are most highly replicating, mirroring genes in the HSC proliferation signature (P-sig). Genes upregulated in naïve T-cells were down regulated during HSC activation (bottom panel), as genes in the quiescence signature (Q-sig). The p-value is from a one-way ANOVA for significant changes across the 5-FU time course. (c) A T-cell fingerprint independent of the other cell types. (d) Shared fingerprints for CD4+, CD8+, and ‘naïve’ and ‘activated’ T-cells were also identified. Gene list sizes are indicated to the right side of each heat map (color indicates normalized expression level (log2); darker corresponds to higher expression).
We next sought to identify genes distinguishing the T-cell subsets, irrespective of their expression pattern in the other hematopoietic cells, using the same threshold criteria applied to the hematopoietic fingerprints. For each T-cell type, the number of genes uniquely expressed in each ranged from 13 (CD4-activated) to 73 (CD4-naïve) (Fig. 4c, Supplemental Table S5). This strategy also revealed that the number of genes shared between activated and naïve CD4+ T-cells, but excluded from their CD8+ counterparts, was only four; included is Zbtb7b, a gene that permits selection of CD4+ T-cells in the thymus (Sun et al., 2005). Activated and naïve CD8+ cells shared eleven genes, including the α and β chains of CD8 (Fig. 4d). However, the CD4+ and CD8+ naïve T-cells shared 215 genes with each other, while the activated cells shared 174, emphasizing the dramatic changes helper and effector T-cells undergo during activation (Fig. 4a, Table S4). As expected, activated T-cells upregulated markers such as Il2ra (CD25) (Crabtree, 1989) and Icos (Flesch, 2002). This list includes a substantial number of under-characterized genes, such as two Riken clones confirmed by RT-PCR (3110082i17Rik and 4933403f05Rik, Fig. S4) named Heist (for Higher Expression in Stimulated T-cells) -1 and -2.
Lineages enriched for key pathways
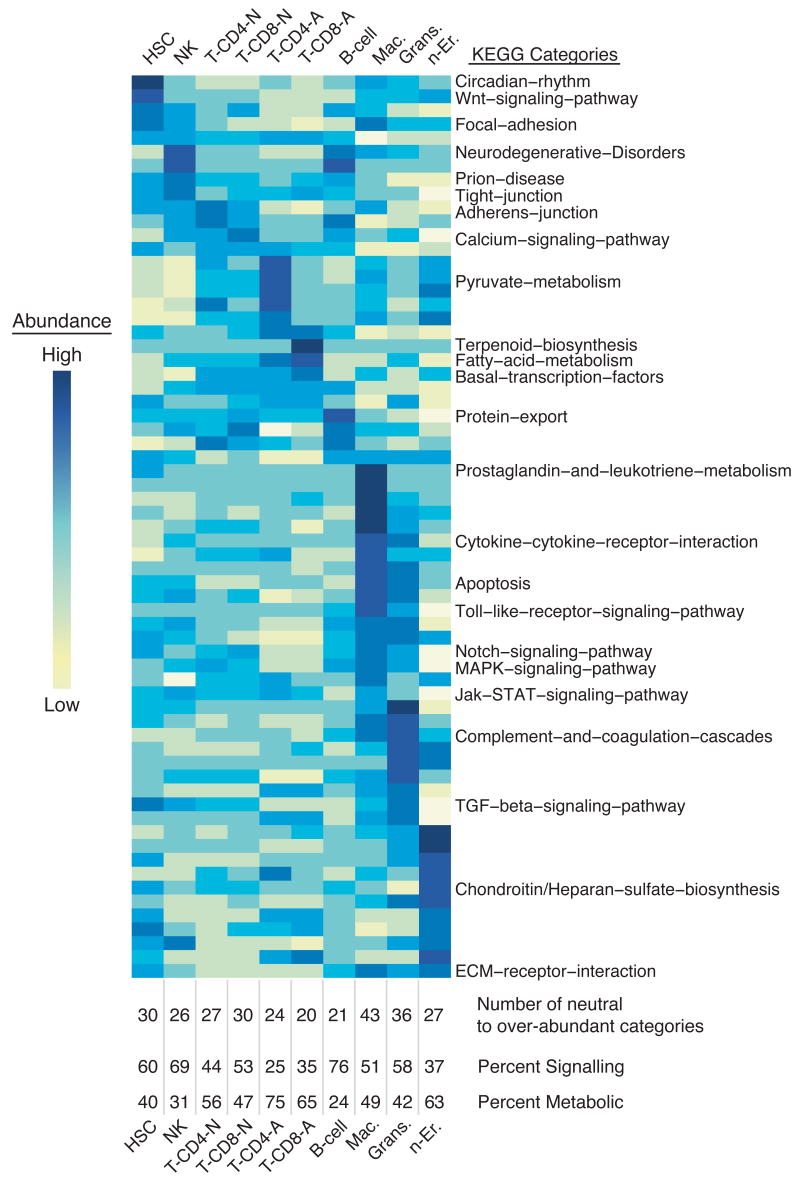
To investigate whether certain signaling or metabolic pathways are likely to be operating in specific cell types, we utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG) to group genes into specific pathways (Ogata et al., 1999). The mean expression value for all arrayed genes in each pathway was determined for every cell type, and categories with significant variation (i.e. difference between the cell types; 65 categories; ANOVA p-value ≤ 0.05, alpha=0.05) were displayed (Fig. 5, Supplemental Fig. 2). Notably, genes involved in Wnt signaling were over-represented in HSCs, consistent with a putative role for this pathway in cell fate decisions (Kirstetter et al., 2006; Reya et al., 2003; Scheller et al., 2006). In addition, we find prostaglandin and leukotriene metabolism enriched in myeloid cells, and Notch signaling enriched in HSCs, activated CD8+ T-cells, and monocytes.
Figure 5. Molecular pathway analysis of hematopoietic cells using KEGG.
KEGG was used to identify which cell types exhibit an over- or under-abundance of components of a molecular pathways, with significant differences between cell types determined by an ANOVA (one way, α = 0.05). For pathways containing genes found on the array, the mean expression value for all genes within a pathway for each cell type is displayed as a function of color (yellow corresponds to low average expression, dark blue to high). Pathways are ordered by high expression within a cell type from left to right. Below the heat map, the number of neutral to over-abundant categories is denoted, and the percent of those pathways that are signaling and metabolic is indicated. This method identified differences in KEGG pathway activity between the cell types; pathways equally active in all cell types will not appear significant.
The KEGG database contains six categories with some redundancy; therefore we chose to classify the 65 significant pathways as either metabolism or signaling and assess each cell type for abundance within these two pathways. Myeloid cells displayed the greatest abundance of both metabolic and signaling pathways. T-cells showed a pattern of increasing metabolic pathways and decreasing signaling pathways upon activation. B-cells and NK cells showed a greater abundance of signaling pathways, perhaps indicative of their role as primary sensors of the immune system. Nucleated erythrocytes displayed lower abundance of signaling pathways and a greater proportion of metabolic pathways, as would be expected during the final stages erythrocyte differentiation. HSCs showed the third highest abundance of active pathways with signaling pathways represented most, consistent with a state of differentiation readiness.
Bioinformatic analysis indicates HSC have an open chromatin state
Stem cells have been speculated to maintain an open chromatin state in order to facilitate rapid differentiation (Akashi et al., 2003; Hu et al., 1997). This is consistent with the idea of stem cell ‘priming’ where progenitors may express low levels of genes necessary for multiple differentiation programs, most of which become repressed as lineage choices become restricted during differentiation (Hu et al., 1997; Jimenez et al., 1992). This suggests a major role for chromatin remodeling during differentiation.
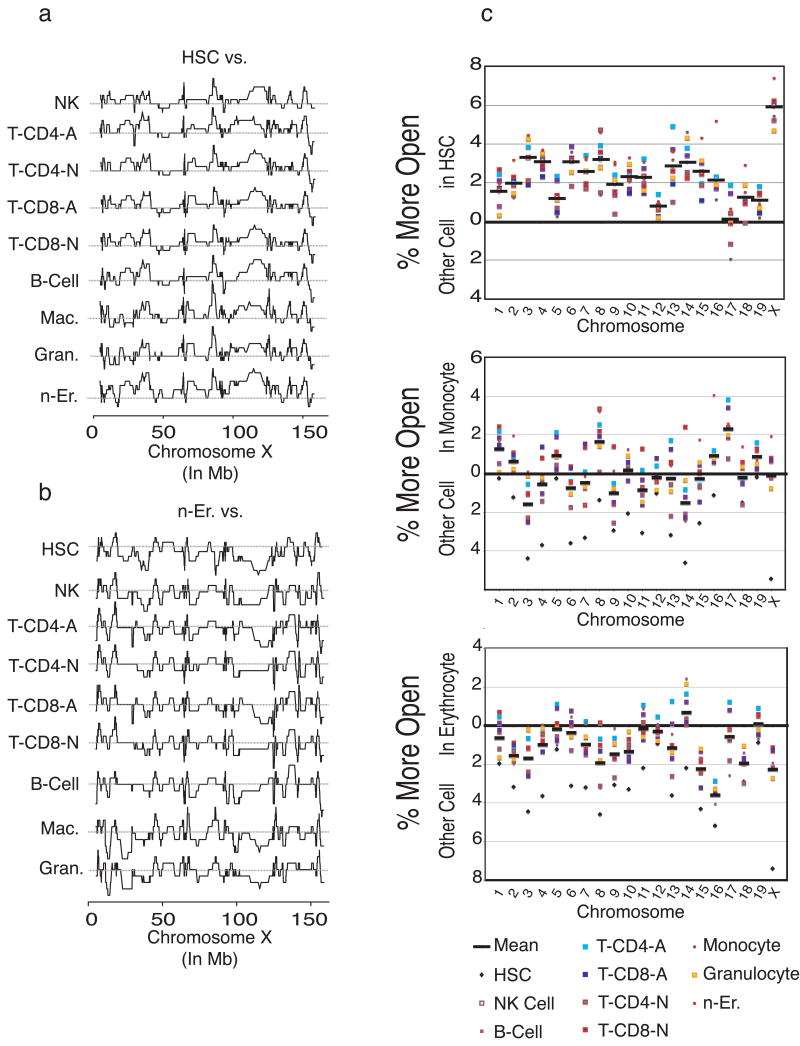
We probed chromatin status using high-level analysis of our gene expression data, reasoning that “open” chromatin would translate into measurable expression by cohorts of chromosomally adjacent genes. We generated a map, for each cell type, of all transcribed genes (using 4.5 as the expression threshold), and identified genes within windows of at least 20 adjacent co-expressed probe sets (representing 14 genes on average). We then subtracted each cell type's chromosomal expression map from the chromosomal expression maps of each other population to observe differences in chromosomal transcribed regions, as illustrated in Fig. 6a. [The complete set of comparative analyses, including 2000 maps, is downloadable http://franklin.imgen.bcm.tmc.edu/loligag/]. To assess this novel method we examined two loci known to be regulated via chromatin remodeling: the interleukin 4/interleukin-13 locus on chromosome 11 (Rogan et al., 2004) and the globin locus on chromosome 7 (Li et al., 2002). Both loci were found to have a higher density of transcription within the anticipated cell type (activated T-cells and nucleated erythrocytes, respectively; Supplemental Figure 5). When HSCs were compared to all other cell types across the X-chromosome, the HSCs exhibited many more regions of open chromatin, evidenced by peaks on the plots. The reverse is true for the erythrocytes (Fig. 6b), with mostly valleys. By calculating the area under the curve, the percentage of open chromatin for each cell type was determined and displayed graphically for each chromosome (Fig. 6c; the entire set, including tabular representation is available at http://franklin.imgen.bcm.tmc.edu/loligag/). On all chromosomes except 17, the chromatin is highly accessible in HSC, relative to the other cell types. Remarkably, the X-chromosome displays an extreme degree of openness. The same analysis with monocytes shows a variable degree of chromatin accessibility, while erythrocytes were largely closed except for chromosome 14. The marked openness of chromosome 14 in erythrocytes may indicate the presence of a number of linked genes involved in erythrocyte generation.
Figure 6. Chromosomal expression density indicates a more open chromatin state for HSC compared to other cell types.
(a) Chromosomal expression maps were plotted as described in the text, and then subtracted against each other. On the X-chromosome, HSCs exhibit many more regions where multiple adjacent genes are expressed, seen as the curve above the grey line (representing 0, or no difference between the cell types). (b) The converse is true for erythrocytes on the X-chromosome. (c) On a per chromosome basis, differences in expression density between cell types were determined, where a positive value (% more open) represents a greater number of expressed genes and open chromatin for the indicated cell type compared to each other cell type. Results indicate more open chromatin for HSCs and a closed chromatin state for n-Er. Chromosomes 17 and 8 are relatively more open in monocytes, whereas there are more highly expressed regions on chromosome 14 in erythroid cells.
Fingerprint transcription factors can bias differentiation to their lineage
To test whether the lineage fingerprints contain genes involved in lineage specification, we attempted to bias hematopoietic differentiation by over expression of putative regulatory fingerprint transcription factors. We chose three transcription factors: Sox13, Ets2, and Zfp105 from the granulocyte, monocyte, and NK cell fingerprints respectively. Ets2 is member of the ETS family of transcription factors and is implicated in acute myeloid leukemia (Aperlo et al., 1996); Zfp105 has so far only been implicated in spermatogenesis (Przyborski et al., 1998).
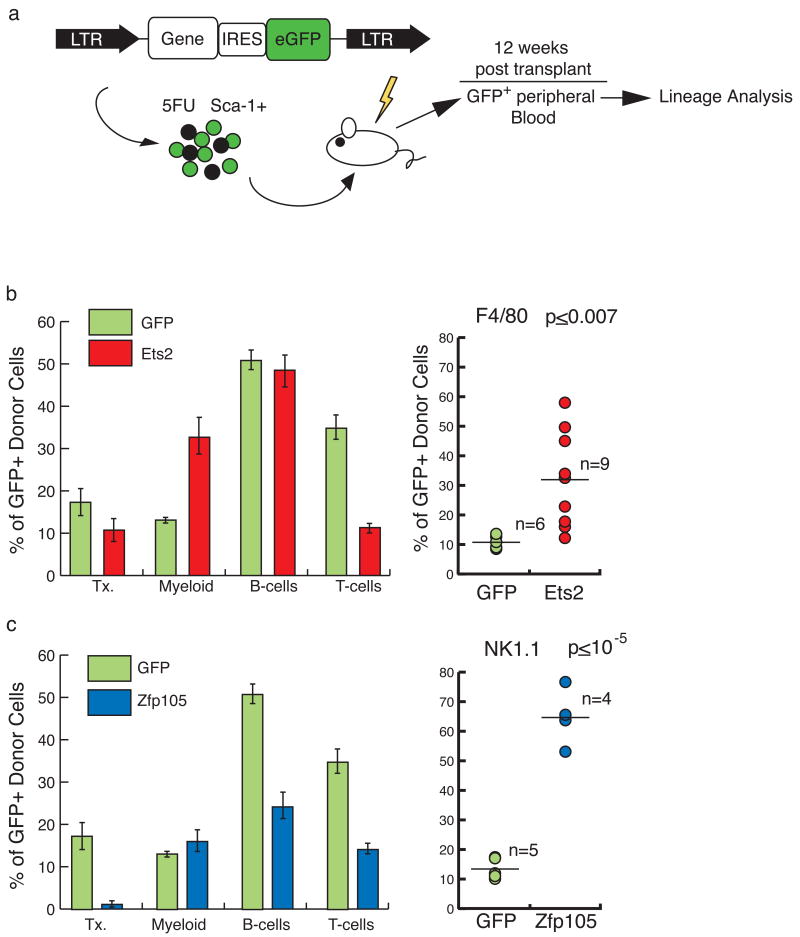
MSCV-based retroviral vectors containing each of the genes coupled to eGFP via an IRES were used to transduce bone marrow progenitors, which were then transplanted into lethally irradiated recipients (Fig. 7a). Peripheral blood of the recipients was analyzed 12 weeks after transplantation for the proportion of transduced GFP+ cells in different blood lineages, compared to controls (transduced with GFP-vector alone). Sox13 overexpression had no discernable effect on lineage distribution (data not shown). However, Ets2 over expression resulted in an increase in myeloid cells, and a decline in T-cells (Fig. 7b). When the F4/80 marker was used to specifically track monocytes, a ∼4-fold increase in monocytes derived from _Ets2_-transduced cells was evident in the peripheral blood, relative to controls (two sample T-test assuming equal variances, two-sided p≤ 0.007, alpha=0.05), indicating that over expression of Ets2 can bias differentiation toward the monocyte lineage. Even more striking were the results of over-expression of Zfp105. Both T- and B-cells were significantly reduced in the Zfp105 over expressing cells relative to controls (Fig. 7c). NK cells, enumerated using the NK1.1 marker, showed a greater than 5-fold increase (two sample T-test assuming equal variances, two-sided p≤ 10-6, alpha=0.05). Interestingly, the total percentage of _Zfp105_-transduced cells was quite low in these mice (∼2% at 12 weeks, compared to ∼18% in GFP-alone), suggesting that over-expression of a powerful differentiation factor is incompatible with maintenance of stem cell function. These data do not exclude the possibility that over-expression of Ets2 and Zfp105 exert their effects on cell-type representation in part by increasing cell survival or proliferation; the mechanism through which these genes act will require more in-depth analysis.
Figure 7. Retroviral transduction of fingerprint genes Ets2 and Zfp105 biases cell fate.
(a) HSCs were transduced using a MSCV-based vector that generates a bicistronic mRNA containing eGFP linked to either Ets2 or Zfp105. The control vector contained eGFP alone. Transduced cells were transplanted into recipients, and eGFP-positive blood was analyzed 12 weeks later and compared to the eGFP-vector control. (b) The overall proportion of transduced cells (transduction, Tx.), and the proportions of transduced myeloid, and lymphoid cells were determined (left; error bars represent standard error; GFP n=6; Ets2 n=9). Compared to vector control, Ets2 enhanced myeloid differentiation to the detriment of T-cell contribution. F4/80-positive transduced monocytes in the blood were elevated (P≤0.007). (c) Zfp105 transplants exhibited a depression in B- and T-cells, and low transduction (left; GFP n=5; Zfp105 n=4). NK1.1-positive GFP+NK cells in peripheral blood were elevated ∼5-fold (p≤10-5) [P-values are based on a two sample T-test assuming equal variances, alpha=0.05.] * Indicates a P-value ≤ 0.05.
Discussion
We have taken a systematic approach to identifying the molecular components of the hematopoietic system, including stem cells and their differentiated counterparts. The unique cellular fingerprints enabled us to identify genes exclusively expressed in specific cell types, or shared by related populations. These fingerprints contain some known genes, and ablation of many of the genes results in a phenotype consistent with their expression pattern (Table 1).
The fingerprints also contain many unstudied genes, some of which may play a functional role, and others which may regulate other genes in that fingerprint, thus participating in cell type specification. The proportion of genes currently identified as unnamed Riken clones or other expressed sequence tags (ESTs) ranged from 14% in HSCs to 35% in naïve T-cells, with an average of 25% in the lineage-specific fingerprint genes. These data open many new avenues for study in all of the populations we have characterized, and we predict that modulation of at least some of these unknown genes would impact hematopoiesis.
Some of these fingerprint genes may be used as markers to uniquely identify and purify particular populations. While markers are available for many cell types, these have been developed empirically over the years and in many cases are not exclusively expressed on the population of interest, complicating data analysis. For example, an unexploited cell surface marker whose expression is common to the differentiated cells is CD82. In contrast is the HSC-specific Tie2, shown to be important in maintaining HSC quiescence (Arai et al., 2004). All of the fingerprints contain additional cell surface molecules that could be used to develop new cell-identification strategies.
Most exciting is the identification in the fingerprints of potential new regulators of HSCs and of hematopoietic differentiation. Each fingerprint contains transcription factors that are candidates for playing a regulatory role in that population. As shown above, overexpression of two of these, Ets2 and Zfp105, unique to monocytes and NK cells respectively, resulted in significantly biased differentiation toward their respective lineages. Again, each fingerprint contains a number of other candidate regulators that could be similarly exploited. Novel regulators that are truly unique to specific lineages could be a powerful tool to modulate production of those cell types for therapeutic purposes. For example, inhibition of gene products specific to activated T-cells may be useful in autoimmune disease; similarly, inhibition of products specific to B-cells could be applied to inhibit production of B-cells in B-cell lymphoma, as has been shown to be clinically successful in the application of anti-CD20 antibody (Press et al., 1987).
Analysis of the fingerprints using KEGG (Fig. 5) showed distinct pathways enriched in different cell types. HSCs were notably enriched in the Wnt signaling pathway, which has previously been implicated in self-renewal (Reya et al., 2003), but not explored in-depth. We found members of both canonical and non-canonical Wnt pathway activation in stem cells, including unique expression in HSC of multiple Wnt receptor genes (Fzd3, 4, 6, 7), as well as the tyrosine kinase Ryk, thought to be involved in non-canonical Wnt signal transduction (Cadigan and Liu, 2006). Other members of the Wnt signal-transduction pathway, such as disheveled 1 and β-catenin were present in all cell types examined.
Also emerging from these data is the observation that a number of cell-type-specific genes, are associated with oncogenic transformation when aberrantly expressed in other cell types. For example, Pbx1, a shared myeloid fingerprint, is vital to myeloid development and differentiation (Piccaluga et al., 2006), but is commonly found overexpressed in B-cell leukemia. Likewise, Evi-1, an HSC-fingerprint gene, is frequently involved in human myeloid leukemias (Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2003).
HSCs had the largest fingerprint, with over 10% of the genes being proven or putative transcription factors. This suggests that quiescent HSC are poised for action, and ready to differentiate. This is consistent with the idea of stem cell priming (Hu et al., 1997; Jimenez et al., 1992), where many genes in stem cells are accessible, and in some cases already expressing low-levels of differentiation markers (Akashi et al., 2003). The observation that ES cells show similarly “poised” patterns of regulation at the chromatin level (Boyer et al., 2005) suggests that even disparate kinds of stem cells may use similar strategies to effect rapid differentiation. Our chromatin state analysis also supports this idea, by showing that in HSC, nearly every chromosome is more “open”, as measured by co-expressed adjacent genes. The X chromosome exhibited an extreme degree of openness, consistent with a previous observation that there are a large number of active genes on the X chromosome in HSC (Forsberg et al., 2005). Genetic linkage analysis has also implicated the X chromosome in regulation of HSC (Henckaerts et al., 2002). The quantitative trait locus (QTL) scpro11, is found within an open region of this chromosome, providing evidence for the importance of an X chromosome with open chromatin for stem cell function (Geiger et al., 2001). Equally striking was the degree to which chromosome 17, which also contains a QTL determining HSC frequency (Morrison et al., 2002), appeared most closed in HSC. Previous work from our lab showed that several genes on chromosome 17 become up regulated when HSC are stimulated to proliferate (Venezia et al., 2004), suggesting a role in maintenance of quiescence.
Other investigators have used expression profiling to identify HSC-enriched genes (Akashi et al., 2003; Ramalho-Santos et al., 2002). We found that the majority of these were found in our data to be expressed in HSC (∼94% within Ramalho-Santos et al. and 78 % within Akashi, et al.) suggesting high similarity between the analyzed cell populations; however, few of these were found to be specific to HSC according to our fingerprint analysis. Nevertheless, eleven of our HSC fingerprint genes were found within one or the other HSC-enriched lists (Ghr, Ryk, Gab1, Yes1, Yap1, Tead2, Gsta4, Meis1, Gata-2, TCF-3, and Nr4a2). The lack of overlap between our HSC fingerprint list and other HSC-enriched gene lists likely reflects the different comparator populations used to generate the lists. The Ramahlo-Santos and Akashi studies respectively used ES and neural stem cells or hematopoietic progenitors as their comparators, whereas we used differentiated hematopoietic cells. Further comparison of our expression data to that obtained from other studies of differentiated hematopoietic lineages may yield additional insights.
This resource is the first broad molecular portrait of any developmental system including a stem/progenitor cell and most major progeny. The database is the first step toward a comprehensive transcriptional profile of the entire hematopoietic system, and in the future may be refined with the addition of other cell types (e.g. megakaryocytes, and progenitors), as well as the use of platforms that enable analysis of the complete coding and non-coding genomes. Even the current data set can be more deeply mined as some of the poorly characterized transcripts represented on the arrays are better annotated, or using different analysis strategies. Nevertheless, the data and analyses presented offer a resource for further investigation into the regulation of HSC and their differentiation. The wealth of genes identified as candidates for functional and regulatory roles in hematopoietic cells will enable new lines of investigation and therapeutic approaches to modulate the activity of these cell types.
Experimental Procedures
Mice and Cell Purification
All cell types were purified according to published protocols using a combination of magnetic and flow cytometric cell sorting to achieve at least 95% purity. Details are given in supplemental Table S1. C57Bl/6-CD45.1 mice housed in a specific pathogen free barrier and fed autoclaved acidified water and mouse chow ad libitum were used. Cells were purified from 8-12 week-old female mice; each population was purified on two separate occasions from pools of tissue from at least 4 mice (biological replicates). RNA from all samples were processed together and were amplified from approximately the same number of cells prior to hybridization to Affymetrix MOE430 2.0 microarrays, which include ∼45,000 probe sets representing about two-thirds of the coding genome. Nucleated erythrocytes from WBM were Ter-119+, CD3-,CD4-,CD8-,Mac-1-,Gr-1-, and B220-. Granulocytes (from WBM) were Gr-1+, clone 7/4+ (Cedarlane Labs), CD2-, CD5-, B220-, F4/80- (eBiosciences), ICAM-1-, Ter-119-. LT-HSCs were isolated as shown previously (Camargo et al., 2006). Briefly, WBM was stained with Hoescht 33342 and the Sca-1+ cells were enriched by magnetic separation, followed by flow cytometry for side-population (SP) and Sca-1+, c-Kit+, and Lin- (Mac-1, Gr-1, Ter119, B220, CD4, CD8 (eBiosciences). Naïve T-cells were freshly isolated from spleen as CD4+, CD25-, CD69- or CD8+, CD25-, and CD69-. Activated T-cells were isolated by enriched naïve T-cells with Concanavalin A (1 μg/ml, Sigma) for eight to eleven hours followed by sorting for CD25+ and CD69+. B-cells were CD19+ and 33D1- splenocytes. Monocytes were isolated from PB based on Forward and Side Scatter properties as well as being Mac-1+. NK cells from spleen were Nk1.1+ CD3- cells.
RNA purification and hybridization to microarrays
RNA was isolated from 1 × 105 cells (except for HSCs which was from 2.5-5 × 104 cells), using the RNAqeuous kit (Ambion, Austin, TX, USA), treated with DNAseI, and precipitated with phenol:chloroform:isoamyl alcohol. The RNA was linearly amplified as previously reported (Venezia et al., 2004). Briefly, two rounds of T7-based in vitro transcription using the MessageAmp kit (Ambion) was undertaken and the RNA was labeled with biotin-conjugated UTP and CTP (Enzo Biotech). Amplified biotinylated RNA (20 ug) was diluted in fragmentation buffer, incubated at 94° C for 25 minutes, and stored at −80° C. A sample was run on a 4% agarose gel to confirm an RNA fragment length of approximately 50 bp. The labeled RNA was hybridized to MOE430.2 chips according to standard protocols. For signal amplification, the chips were washed and counterstained with PE-conjugated strepavidin and a biotinylated anti-streptavidin antibody. The raw image and intensity files were generated using GCOS 1.0 software (Affymetrix). All microarrays used in this study had to pass several quality control tests including a scale factor < 5, a 5′ to 3′ probe ratio < 20, and replicate correlation coefficient > 0.96.
Microarray Analysis
Normalization and model-based expression measurements were performed with GC-RMA. GC-RMA, as well as additional analytical and annotation packages, are available as part of the open-source Bioconductor project (www.bioconductor.org) within the statistical programming language R (http://cran.r-project.org/) (Team”, 2006). Cluster analysis and Principle Components Analysis were performed with the R base package. Lineage fingerprints were established by setting on/off expression thresholds (Off <4, On >5) and using Boolean logic to find uniquely expressed genes in each cell type and each grouping. When selecting a T-cell fingerprint, activated T-cells were removed from the data set and naïve T-cells (both CD4+ and CD8+) were averaged together. The FDR q-value for each list ranged between 0.00185 and 0.08933.
RT-PCR analysis
RNA was isolated using the RNAqeuous kit (Ambion) from FACS-sorted cells isolated independently from the samples used for array analysis. For qRT-PCR, RNA was reverse transcribed with random hexamer primers using Super Script II (Invitrogen). cDNA input was standardized and qRT-PCRs were performed with Taqman master mix, 18s-rRNA probe (VIC-MGB), and a gene specific probe (FAM-MGB, Applied Biosystems) for 55 cycles using a AbiPrism 7900HT (Applied Biosystems). For standard RT-PCR used to confirm T-cell genes, RNA was prepared as above with the addition of DNAse I treament before RT followed by phenol:chloroform extraction. cDNA input was standardized and PCR was performed for 40 cycles.
The reader should note that some genes (e.g. Evi1) are represented by multiple probe sets, which in some cases give differing results, typically appearing with a differential expression profile for one set, and appearing as “off” in all cell types with other probe sets. This is due to variations in the efficacy of Affymetrix probe sets, and the 5′ bias of some probe sets. False positives are much less likely than false negatives, so we assume the positive probe-set is correct, as borne out by the analysis shown in Figure S1 with Evi1. The reader is urged to verify expression profiles of particular interest with qRT-PCR and to be aware of the limitations of Affymetrix probe sets (Allison et al., 2006).
Knockout Analysis
All knockout data was obtained via the 3.44 MGI database (http://www.informatics.jax.org/). The phenotypes are classified into a hierarchical tree with the terminal categories being very precise descriptions and the initial categories being very broad descriptions. To obtain all knockouts within ‘hematopoietic system phenotype’ and ‘immune system phenotype’ grouped into a single list (‘hematopoietic knockout data’). All genes on the array that have a reported knockout were identified. The frequency of fingerprint genes that have a hematopoietic phenotype was compared to the frequency of hematopoietic knockouts found by chance within all knock-out data. A Z-statistic was used to determine the significance of enrichment fingerprint genes found within the knockouts with a reported hematopoietic phenotype and reported in Supplemental Table S4. A Z-statistic above 3 S.D. was considered significant for this enrichment.
KEGG analysis
KEGG is a suite of gene databases systematically organized into metabolic and signaling pathways (http://www.genome.ad.jp/kegg/) (Ogata et al., 1999). The mean expression value for each KEGG pathway was obtained by taking the mean of the expression values of each gene on the microarray for a given KEGG pathway. The mean expression value of all KEGG pathways for each hematopoietic cell type was obtained using all the genes on the array (not limited to the fingerprints). An ANOVA to identify pathways with a significant variation between the cell types (ANOVA p-value ≤ .05) was performed and 65 significant pathways were identified. The pathways were then ranked by maximal abundance for each cell type and a heat map was generated using the centered data, enabling identification of differences in KEGG pathway activity between each of the cell types. KEGG pathways equally active in all cell types will not appear significant.
Chromosomal Analysis
Affymetrix probes were assigned a value of 1 if their expression value was equal to or greater than 4.5 and 0 if they were below 4.5. Each probe was then plotted to its chromosomal position and a chromosomal expression map was created for each chromosome for each cell type by an R function we developed. Briefly, this function uses a sliding and overlapping window, which covers 20 probe sets (representing 14 genes on average) at one time and overlaps with the previous nineteen, that takes the sum of 1's within the window for the y-axis and the mean chromosomal position for the x-axis and uses these values to plot the expression map. For comparison between cell types, the expression map of one cell type was directly subtracted from all the other cell types in turn. To quantitate openness of chromatin we calculated the area under the curves and divided that by the possible area under the curve (if all y-values were 20) to calculate a percentage of more open chromatin as shown by the chromosomal expression maps.
Retroviral HSC transduction
Clones for Ets2, Zfp105, and Sox13 were TA-cloned into a pENTR/D-TOPO vector and the Gateway system was used to recombine Ets2, Zfp105, and Sox13 into a murine stem cell virus (MSCV) vector containing attR recombination sites. Vectors were packaged using 293T cells by cotransfection with pCL-Eco (Naviaux et al., 1996). CD45.2 mice were treated with 5-Fluorouracil (150 mg/kg American Pharmaceutical Partners) 6 days before harvesting WBM. WBM was enriched for Sca-1+ cells using magnetic enrichment (AutoMACS, Miltenyi) and adjusted to a concentration of 5×105 cells/ml in transduction medium, containing Stempro 34 (Gibco), nutrient supplement, penicillin/streptomycin, L-Glutamine (2mM), mSCF (10ng/ml, R&D Systems), mTPO (100ng/ml, R&D Systems), and polybrene (4ug/ml, Sigma). Thawed virus was applied at an MOI of 1 and the suspension was spin-infected at 250g at room temperature for 2 hours (Kotani et al., 1994). After transduction, the cells were incubated at 37°C for 3 hours and transplanted into lethally irradiated CD45.1 mice. Engraftment, transduction and lineage analysis was performed on peripheral blood 12 weeks after transplantation by FACS (FACSaria, BD). The lineage analysis method is shown in Supplemental Fig. 3.
Supplementary Material
01
Acknowledgments
We thank all Goodell lab members for helpful comments. The work was supported by DK63588 and DK 58192 and MAG was a Scholar of the Leukemia and Lymphoma Society. SC was supported by T32 AG000183. KL, DW, and MT were partially supported by T32 DK064717. SBB was supported by T32 AI07495. We thank Dr. Zhou Songyang at BCM for providing the MSCV-IRES-GFP backbone and the pCL-Eco packaging vectors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Aperlo C, Pognonec P, Stanley ER, Boulukos KE. Constitutive c-ets2 expression in M1D+ myeloblast leukemic cells induces their differentiation to macrophages. Mol Cell Biol. 1996;16:6851–6858. doi: 10.1128/mcb.16.12.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB, Verhoef G, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Liu YI. Wnt signaling: complexity at the surface. J Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Chambers SM, Drew E, McNagny KM, Goodell MA. Hematopoietic stem cells do not engraft with absolute efficiencies. Blood. 2006;107:501–507. doi: 10.1182/blood-2005-02-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesch IE. Inducible costimulator (ICOS) J Biol Regul Homeost Agents. 2002;16:214–216. [PubMed] [Google Scholar]
- Forsberg EC, Prohaska SS, Katzman S, Heffner GC, Stuart JM, Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H, True JM, de Haan G, Van Zant G. Age- and stage-specific regulation patterns in the hematopoietic stem cell hierarchy. Blood. 2001;98:2966–2972. doi: 10.1182/blood.v98.10.2966. [DOI] [PubMed] [Google Scholar]
- Henckaerts E, Geiger H, Langer JC, Rebollo P, Van Zant G, Snoeck HW. Genetically determined variation in the number of phenotypically defined hematopoietic progenitor and stem cells and in their response to early-acting cytokines. Blood. 2002;99:3947–3954. doi: 10.1182/blood.v99.11.3947. [DOI] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Griffiths SD, Ford AM, Greaves MF, Enver T. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci U S A. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- Kotani H, Newton PB, 3rd, Zhang S, Chiang YL, Otto E, Weaver L, Blaese RM, Anderson WF, McGarrity GJ. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Qian D, Jerabek L, Thiel BA, Park IK, Ford PS, Kiel MJ, Schork NJ, Weissman IL, Clarke MF. A genetic determinant that specifically regulates the frequency of hematopoietic stem cells. J Immunol. 2002;168:635–642. doi: 10.4049/jimmunol.168.2.635. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, Taylor BA, Ihle JN, Hartley JW, Morse HC, 3rd, Jenkins NA, Copeland NG. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol Cell Biol. 1988;8:301–308. doi: 10.1128/mcb.8.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Transcription factor GATA-1 in megakaryocyte development. Stem Cells. 1998;16 2:79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- Piccaluga PP, Malagola M, Rondoni M, Ottaviani E, Testoni N, Laterza C, Visani G, Pileri SA, Martinelli G, Baccarani M. Poor outcome of adult acute lymphoblastic leukemia patients carrying the (1;19)(q23;p13) translocation. Leuk Lymphoma. 2006;47:469–472. doi: 10.1080/10428190500331261. [DOI] [PubMed] [Google Scholar]
- Press OW, Appelbaum F, Ledbetter JA, Martin PJ, Zarling J, Kidd P, Thomas ED. Monoclonal antibody 1F5 (anti-CD20) serotherapy of human B cell lymphomas. Blood. 1987;69:584–591. [PubMed] [Google Scholar]
- Przyborski SA, Knowles BB, Handel MA, Gurwitch SA, Ackerman SL. Differential expression of the zinc finger gene Zfp105 during spermatogenesis. Mamm Genome. 1998;9:758–762. doi: 10.1007/s003359900859. [DOI] [PubMed] [Google Scholar]
- Puthier D, Joly F, Irla M, Saade M, Victorero G, Loriod B, Nguyen C. A general survey of thymocyte differentiation by transcriptional analysis of knockout mouse models. J Immunol. 2004;173:6109–6118. doi: 10.4049/jimmunol.173.10.6109. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, Staynov DZ. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci U S A. 2004;101:2446–2451. doi: 10.1073/pnas.0308327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Pant R. Origins of lymphocyte developmental programs: transcription factor evidence. Semin Immunol. 2004;16:227–238. doi: 10.1016/j.smim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Roussel MF, Sherr CJ. Mouse NIH 3T3 cells expressing human colony-stimulating factor 1 (CSF-1) receptors overgrow in serum-free medium containing human CSF-1 as their only growth factor. Proc Natl Acad Sci U S A. 1989;86:7924–7927. doi: 10.1073/pnas.86.20.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Team”, R.D.C. R: A Language and Environment for Statistical Computing. 2006. [Google Scholar]
- Toren A, Bielorai B, Jacob-Hirsch J, Fisher T, Kreiser D, Moran O, Zeligson S, Givol D, Yitzhaky A, Itskovitz-Eldor J, et al. CD133-positive hematopoietic stem cell “stemness” genes contain many genes mutated or abnormally expressed in leukemia. Stem Cells. 2005;23:1142–1153. doi: 10.1634/stemcells.2004-0317. [DOI] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L, deBoer E, Grosveld F. The human beta-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 1988;2:1089–1100. doi: 10.1101/gad.2.9.1089. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Mager DL. Identification of a new murine lectin-like gene in close proximity to CD94. Immunogenetics. 2003;55:53–56. doi: 10.1007/s00251-003-0540-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01