Identification of cis-acting sites for condensin loading onto budding yeast chromosomes (original) (raw)
Abstract
Eukaryotic chromosomes reach their stable rod-shaped appearance in mitosis in a reaction dependent on the evolutionarily conserved condensin complex. Little is known about how and where condensin associates with chromosomes. Here, we analyze condensin binding to budding yeast chromosomes using high-resolution oligonucleotide tiling arrays. Condensin-binding sites coincide with those of the loading factor Scc2/4 of the related cohesin complex. The sites map to tRNA and other genes bound by the RNA polymerase III transcription factor TFIIIC, and ribosomal protein and SNR genes. An ectopic B-box element, recognized by TFIIIC, constitutes a minimal condensin-binding site, and TFIIIC and the Scc2/4 complex promote functional condensin association with chromosomes. A similar pattern of condensin binding is conserved along fission yeast chromosomes. This reveals that TFIIIC-binding sites, including tRNA genes, constitute a hitherto unknown chromosomal feature with important implications for chromosome architecture during both interphase and mitosis.
Keywords: Chromosome condensation, condensin, Scc2/4, tRNA genes, TFIIIC
Mitotic chromosome structure and stability are determined by the chromosomal condensin complex. Condensin is one of three essential SMC (structural maintenance of chromosomes) subunit-containing protein complexes that exist in all eukaryotes and share important roles in chromosome biology (Nasmyth and Haering 2005; Hirano 2006). The other SMC complexes are cohesin, required for sister chromatid cohesion and DNA repair by homologous recombination, and the Smc5/Smc6 complex, with a less well understood role in preventing DNA damage and facilitating DNA repair. Condensin’s most apparent role relates to mitotic chromosome architecture and to sister chromatid resolution during anaphase (Hirano and Mitchison 1994; Saka et al. 1994; Strunnikov et al. 1995; Bhat et al. 1996; Hudson et al. 2003; Ono et al. 2003). In addition, condensin fulfills important roles during interphase. These include transcriptional silencing at the budding yeast mating type and the Drosophila Fab-7 loci, as well as DNA replication checkpoint signaling in fission yeast (Lupo et al. 2001; Aono et al. 2002; Bhalla et al. 2002). How and where condensin binds to interphase and mitotic chromosomes to carry out these functions are poorly understood.
Condensin is built on a dimer of Smc2 and Smc4, two long coiled-coil proteins that interact with each other at both ends. On one side lies a dimerization interface known as the hinge, and on the other end lie interacting ATPase head domains. Studies using recombinant human condensin subunits have shown that the kleisin subunit CAP-H (Brn1 in budding yeast) directly binds the Smc head domains (Onn et al. 2007). The kleisin subunit in turn recruits the remaining two HEAT repeat-containing non-Smc subunits, CAP-D2 and CAP-G (Ycs4 and Ycg1 in budding yeast), to the complex. Human cells contain two isoforms of the complex, condensin I and condensin II, that share the Smc2 and Smc4 subunits but contain alternate non-Smc subunits (Ono et al. 2003, 2004; Hirota et al. 2004). While condensin II is nuclear throughout the cell cycle, condensin I is excluded from the interphase nucleus and associates with chromosomes only in mitosis. Detailed biochemical characterization of the vertebrate condensin I complex has shown that it binds to DNA in vitro by wrapping ∼190 bp of DNA in an ATP hydrolysis-dependent reaction (Bazett-Jones et al. 2002). The resulting supercoiling activity of condensin is stimulated by mitotic phosphorylation of its CAP-D2 subunit, thus correlating this activity to mitotic chromosome condensation (Kimura et al. 1998). In organisms from yeast to human, condensin is present at a stoichiometry of one molecule per 5–10 kb of DNA (MacCallum et al. 2002; Wang et al. 2005). The relationship between condensin’s in vitro activities, and its function on interphase and mitotic chromosomes in vivo, is not yet understood.
Chromosome binding of the related cohesin complex has been the subject of intense study over recent years. The architecture of the budding yeast cohesin complex resembles that of condensin. Cohesin’s Smc subunits, Smc1 and Smc3, dimerize at both the Smc hinge and the ATPase heads. The kleisin Scc1 associates with the Smc heads to stabilize their interaction and is required to recruit the remaining subunits, Scc3 and Pds5 (Haering et al. 2002; McIntyre et al. 2007). These biochemical features, combined with electron micrographs of human cohesin, have shown cohesin to form a large proteinaceous ring (Anderson et al. 2002; Haering et al. 2002). It has been suggested, and compelling evidence has been provided, that cohesin rings bind to chromosomes by topologically entrapping DNA (Haering et al. 2002; Ivanov and Nasmyth 2005). Cohesin loading onto chromosomes in vivo depends on a loading factor, the Scc2/4 complex (Ciosk et al. 2000). Scc2/4-binding sites along chromosomes are poorly characterized, but along budding yeast chromosome 6 correlate with strong transcriptional activity (Lengronne et al. 2004). Interaction of cohesin with the Scc2/4 complex may recruit cohesin to chromosomes, upon which ATP hydrolysis by cohesin’s Smc subunits is required for productive loading onto DNA (Arumugam et al. 2003; Weitzer et al. 2003). After loading, cohesin translocates away from the Scc2/4 complex to accumulate between convergent RNA polymerase II (pol II)-transcribed genes (Lengronne et al. 2004).
Unlike the cohesin rings, condensin shows a rod-like appearance on electron micrographs with the Smc2 and Smc4 coiled-coils juxtaposed (Anderson et al. 2002). Nevertheless, the subunit arrangement suggests that condensin also forms a ring that could bind DNA by topological embrace. Whether this is the case, and whether DNA binding of condensin in vivo also depends on a loading factor, is not known. A low-resolution, genome-wide localization analysis in budding yeast has found condensin at discrete sites, spaced on average every 10 kb (Wang et al. 2005). Chromosomal features that characterize the binding sites remained unknown. We performed high-resolution analysis of condensin chromatin immunoprecipitates on oligonucleotide tiling arrays. This confirmed a pattern of unique condensin-binding sites, concentrated at centromeres, that remained largely unchanged between interphase and mitosis. We find that condensin colocalizes with the cohesin loader Scc2/4 at tRNA genes, and additional sequences characterized by the RNA pol III transcription factor TFIIIC. TFIIIC and Scc2/4 contribute to productive condensin association at these sites, and an isolated B-box sequence element, recognized by TFIIIC, is sufficient to generate a condensin-binding site. This provides the first evidence for sequence-specific loading of condensin onto chromosomes and describes a new chromosomal feature with implications for both interphase and mitotic chromosome architecture.
Results
A largely unaltered chromosomal condensin pattern throughout the cell cycle
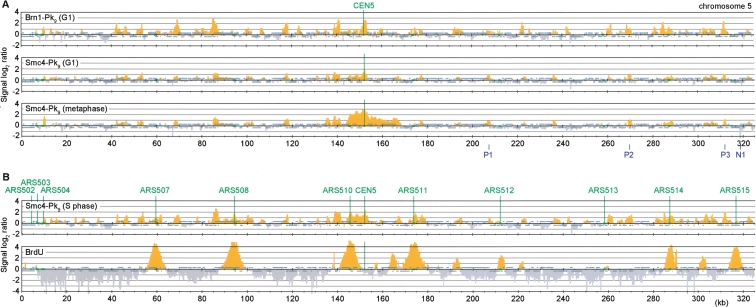
To investigate condensin binding to budding yeast chromosomes, we performed chromatin immunoprecipitation (ChIP) against two Pk epitope-tagged subunits of the condensin complex, Smc4 and Brn1. We first analyzed chromatin immunoprecipitates obtained from cells arrested in the G1 phase of the cell cycle by α-factor treatment. Consistent with an earlier report (Wang et al. 2005), peaks of association were enriched around the centromere. Additional peaks were found in frequent intervals along the chromosome arms (Fig. 1A shows a 325-kb segment of chromosome 5; a genome-wide condensin map can be found in Supplemental Fig. 1).
Figure 1.
A largely unaltered condensin pattern during the cell cycle. (A) Comparison of the condensin subunits Smc4-Pk9 and Brn1-Pk9 in G1 versus metaphase. Cultures of strain Y2315 (_MAT_a SMC4-Pk9) and Y2200 (_MAT_a BRN1-Pk9) were arrested in G1 by mating pheromone α-factor treatment, or in metaphase by addition of the spindle poison nocodazole. Uniform arrest was confirmed by flow cytometric analysis of DNA content. Enrichment of DNA fragments in condensin chromatin immunoprecipitates relative to a whole genome DNA sample is shown along a 325-kb region of budding yeast chromosome 5. Each bar represents the average of 16 oligonucleotide probes within adjacent 300-bp windows. Orange signals represent significant binding as described (Katou et al. 2003). Blue bars above and below the midline represent ORFs transcribed from left to right and opposite, respectively. The centromere and origins of replication are depicted in green. P1–P3 indicate three of the four positive, and N1 indicates one of the two negative, association sites that were confirmed by quantitative PCR (cf. Fig. 3). (B) Accumulation of Smc4 at stalled replication forks. Cells of strain Y2315 were synchronized in G1 by α-factor treatment and released into medium containing the replication inhibitor HU for 2 h. Cells of strain K5601 (_MAT_a GPD-TK) were treated in the same way, but were released from α-factor arrest in the presence of 400 μg/mL bromodeoxyuridine (BrdU). ChIP against Smc4-Pk9 or BrdU was performed.
Chromosome condensation is at its lowest in G1 cells. We therefore compared this pattern to that in cells arrested in metaphase by treatment with the spindle poison nocodazole, when chromosomes are condensed (Guacci et al. 1994). The distribution of Smc4 along chromosome arms was indistinguishable from cells in G1, but condensin enrichment at the centromere was more pronounced (Fig. 1A). The total level of condensin on budding yeast chromosomes remains constant between G1 and metaphase (Wang et al. 2005), suggesting that mitotic chromosome condensation is not the consequence of condensin binding to specific sites, but of a reaction that condensin bound to these sites performs in a cell cycle regulated manner. Mitosis-specific centromeric enrichment opens the possibility that condensin contributes to structural integrity of mitotic budding yeast centromeres, as has been observed in metazoan cells (Ono et al. 2004; Gerlich et al. 2006a).
We also analyzed the condensin pattern in cells arrested in early S phase with the replication inhibitor hydroxyurea (HU). Again, much of the pattern was similar to cells in G1 (Fig. 1B). In addition, condensin accumulated at several sites of stalled replication forks in the HU arrest. These were visualized, after incorporation of the nucleotide analog 5-bromo-2′-deoxyuridine (BrdU), by ChIP with an α-BrdU antibody. We do not know whether condensin also associates with replication forks during an undisturbed S phase. The localization to HU-arrested forks is consistent with the observation that fission yeast condensin is required for the replication checkpoint response after HU treatment (Aono et al. 2002).
Intergenic enrichment of condensin, independent of a spacing rule
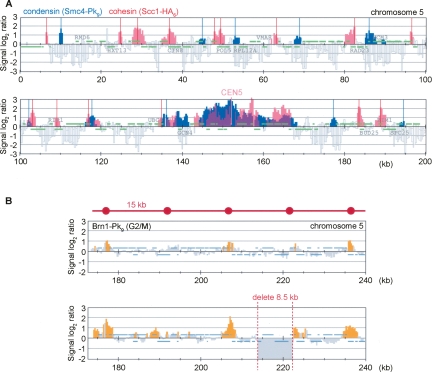
In our efforts to find a rule that underlies the condensin pattern, we first compared it with that of the related cohesin complex. Cytological analysis has suggested that, despite their similarity, the two complexes do not coincide in their binding to chromosomes (Ciosk et al. 2000). This was confirmed by our ChIP analysis. Although cohesin and condensin peaks partly overlapped at some sites, the patterns appeared largely distinct from each other (Fig. 2A). Both cohesin and condensin are enriched at centromeres, but even there the peak distribution was distinct. This suggests that the two related complexes differ in certain aspects of their chromosome binding.
Figure 2.
Condensin binding without a spacing rule. (A) Condensin binding sites are distinct from those of the related cohesin complex. The distribution of Smc4-Pk9 in nocodazole-arrested cells from the experiment in Figure 1 (significant peaks in blue) was overlaid with that of Scc1-HA6 (red) (Lengronne et al. 2004). The first 200 kb of chromosome 5 are shown. (B) Condensin binding does not follow a geometric spacing rule but adheres to sequence determinants. The pattern of Brn1-Pk9 is shown in strains Y2200 and Y2565 (_MAT_a BRN1-Pk9 ChrVΔ213,898-222,402) harboring an 8.5-kb deletion between two neighboring condensin-binding sites.
As a next step, we used a peak picking algorithm to assign 419 condensin-binding sites along the 16 budding yeast chromosomes, excluding a 20-kb region on either side of centromeres, where condensin enrichment might follow a mechanism different from that along chromosome arms (Supplemental Fig. 2). The number of condensin-binding sites on each chromosome was proportional to its length, consistent with earlier observations (Wang et al. 2005). We observed that condensin was preferentially found between ORFs. Twenty-five percent of peaks mapped within an annotated ORF, while almost 60% of the genome is covered by ORFs. This suggests that a mechanism exists that excludes condensin from ORFs (P < 10−44). There was no bias with respect to ORF orientation. When all condensin peaks were assigned to their closest inter-ORF region, we found that they equally likely mapped between convergent, divergent, and tandem ORFs (Supplemental Fig. 2). This is different from the association pattern of cohesin, which is preferentially found between convergent ORFs.
Along some, but not all, chromosome arms, we observed an apparently even spacing of peaks, ∼15 kb in distance from each other. This was interesting in light of mitotic chromosome models in which chromatin is folded into equally sized loops that are stabilized by condensin. We therefore tested whether a geometric spacing rule determines condensin association. We removed 8.5 kb, approximately half of the geometric unit, including four nonessential genes, between two condensin-binding sites on the right arm of chromosome 5. If a spacing rule controlled condensin association, we would expect peaks surrounding the deletion to change their position with respect to the underlying DNA sequence. Conversely, if chromosomal _cis_-acting features attracted condensin, we would expect the distance between the two neighboring condensin bindings sites to decrease. Figure 2B shows that the latter is the case. In response to the deletion, condensin maintained its association relative to the underlying sequence, and the distance between the two peaks decreased. This result does not exclude the possibility that condensin delineates units of folding within chromosomes, but suggests that such units can vary in size.
Condensin colocalization with the cohesin loader Scc2/4
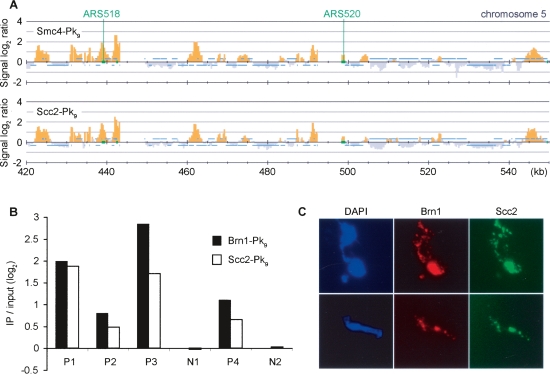
When comparing the chromosomal binding pattern of condensin with that of the cohesin loader Scc2/4, we noticed striking colocalization of the two. In cells arrested in early S phase with HU, the colocalization extended to both the constitutive chromosomal association sites as well as stalled replication forks (Fig. 3A). The coincidence of condensin and Scc2/4-binding sites was statistically significant (Supplemental Fig. 3). To confirm this observation, we validated four positive (P1–P4) and two negative (N1 and N2) (cf. Fig. 1A; Supplemental Fig. 1) binding sites by quantitative analysis of Brn1 and Scc2 chromatin immunoprecipitates using real-time PCR. This approach yielded results in good agreement with the peak intensities seen by our microarray analysis (Fig. 3B). As an independent approach, we used cytological staining of the condensin subunits Brn1 and Scc2 on spread metaphase chromosomes, which confirmed colocalization of the two proteins (Fig. 3C). Colocalization of condensin with Scc2/4 was observed on chromosome spreads from all stages of the cell cycle (data not shown). This opened the possibility that the Scc2/4 complex, in addition to its role as a cohesin loader, also participates in condensin’s function on chromosomes.
Figure 3.
Condensin colocalization with the cohesin loader Scc2/4. (A) Colocalization seen by ChIP. The Smc4-Pk9 pattern along a 130-kb fragment of chromosome 5 in HU-arrested cells is compared with the distribution of Scc2-Pk9 obtained from strain Y2422 (_MAT_a SCC2-Pk9) under the same conditions. The locations of the early firing origins ARS518 and ARS520 are indicated. (B) Confirmation of colocalization by quantitative PCR analysis. Four positive (P1–P4) and two negative (N1 and N2) association sites identified by microarray analysis (cf. Fig. 1; Supplemental Fig. 1) were examined. Enrichment of Brn1-Pk9 (strain Y2200) and Scc2-Pk9 chromatin immunoprecipitates over a whole genome DNA sample is depicted, normalized to the average of the two negative association sites. 1.58% and 1.45% of input DNA at P1 was recovered in the Brn1 and Scc2 immunoprecipitates, respectively. (C) Colocalization on chromosome spreads. Strain Y2717 (_MAT_a BRN1-myc9 SCC2-HA6) was arrested in metaphase by nocodazole treatment. Chromosome spreads were stained to detect the epitope-tagged condensin subunit Brn1-myc9 and Scc2-HA6.
Scc2/4 contributes to condensin association with chromosomes
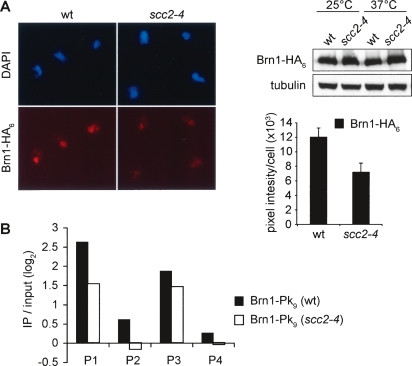
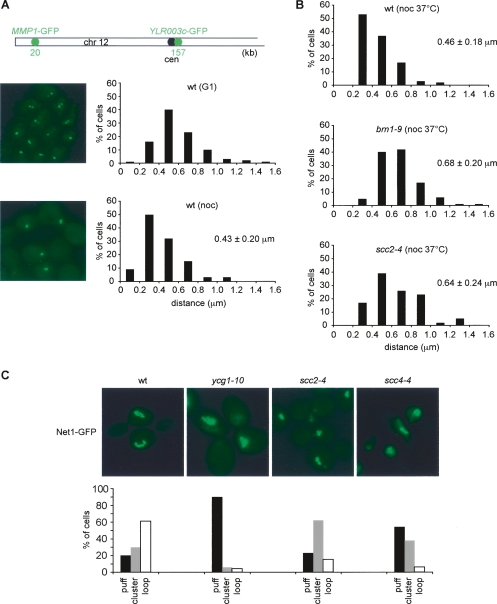
To test whether Scc2/4 was required for condensin binding to chromosomes, we compared wild-type and scc2-4 mutant cells arrested in metaphase by nocodazole treatment. Both cultures were shifted for 1 h to 37°C in the arrest, a restrictive temperature for the scc2-4 allele. To analyze condensin association with chromosomes, we immunostained spread metaphase chromosomes with an antibody against the HA epitope tag fused to Brn1. After inactivation of the scc2-4 allele, the Brn1 signal was reduced by approximately half compared with the signal before temperature shift, or with wild-type cells (Fig. 4A). The total cellular Brn1 levels remained unaffected by Scc2 inactivation, as visualized by Western blotting of cell extracts before and after the temperature shift. In a complementary approach, we analyzed condensin chromatin immunoprecipitates from metaphase arrested wild-type or scc2-4 mutant cells after shift to restrictive temperature by real-time PCR. This revealed that condensin binding to the four positive binding sites described in Figure 3B was similarly reduced, but not abolished, after Scc2 inactivation (Fig. 4B). This suggests that while Scc2 is not essential for condensin recruitment to chromosomes, it promotes its full level of association.
Figure 4.
A contribution of Scc2/4 to condensin association with chromosomes. (A) Chromosome spreads of nocodazole-arrested cells of strains Y2521 (_MAT_a BRN1-HA6 SCC1-Pk9) and Y2423 (_MAT_a scc2-4 BRN1-HA6 SCC1-Pk9) shifted for 1 h to 37°C were stained for the condensin subunit Brn1-HA6. The mean staining intensity and standard error for at least 50 cells in each sample are shown. The decrease of Brn1 on chromosomes was not due to degradation of the subunit, as confirmed by Western blotting of whole cell extracts. (B) Quantification of condensin association under the same conditions by ChIP followed by real-time PCR analysis of the four binding sites (P1–P4) characterized in Figure 3B. The two negative sites (N1 and N2) were used for normalization.
A protein interaction between Scc2/4 and cohesin has been reported that might be involved in the recruitment of cohesin to chromosomes (Arumugam et al. 2003). We therefore tested whether we could observe a similar interaction between Scc2/4 and condensin. By coimmunoprecipitation, we could confirm the interaction of Scc2/4 with cohesin, but not condensin (Supplemental Fig. 4). This suggests that while Scc2/4 is required for full levels of condensin binding to chromosomes, the two complexes interact only indirectly, or less stably than Scc2/4 and cohesin.
Scc2/4 promotes chromosome condensation
The relatively small effect of Scc2 inactivation on chromosomal condensin levels could mean that condensin functions largely independently of Scc2/4. Alternatively, the reduced condensin levels could reflect a difference in the manner by which condensin binds to chromosomes in the absence of Scc2/4. We therefore tested whether chromosome-bound condensin in the scc2-4 mutant was proficient in mitotic chromosome condensation. To this end, we established a chromosome condensation assay based on two lac operator arrays spaced 137 kb from each other on the left arm of chromosome 12, visualized by expression of a lacI-GFP fusion protein. In G1 cells, the average distance between the two GFP-marked loci, measured in three dimensions, was 0.60 ± 0.24 μm. This distance decreased in metaphase arrested cells to 0.43 ± 0.20 μm (Fig. 5A). These values agree well with fluorescent in situ hybridization analysis of similarly spaced loci on spread chromosomes (Guacci et al. 1994), confirming 1.5-fold lengthwise mitotic condensation of budding yeast chromosome arms. We next arrested wild-type, condensin brn1-9, and scc2-4 mutant cells in metaphase by nocodazole treatment and then inactivated the mutant alleles by temperature shift. While condensation of wild-type chromosomes remained intact, the distance of the GFP-marked loci in both brn1-9 and scc2-4 mutant cells increased to values of uncondensed chromosome arms (Fig. 5B). This suggests that condensin as well as Scc2/4 are required to maintain mitotic chromosome condensation.
Figure 5.
Scc2/4 requirement for chromosome condensation. (A) A condensation assay for the chromosome 12 left arm. Strain Y2869 (_MAT_a lacOs∷YLR003c-1 lacOs∷MMP1 LacI-GFP) was arrested in G1 by α-factor, and in metaphase by nocodazole treatment. Chromosome condensation was assessed by measuring the distance between the two fluorescent loci in three dimensions. Examples of wild-type cells in G1 and metaphase are shown, as well as the distribution of measured distances. The mean distances, with their standard deviations, are given. (B) Scc2/4 is required to maintain chromosome arm condensation. Metaphase arrested cells of strains Y2869, Y3104 (as Y2869 but brn1-9) and Y2887 (as Y2869 but scc2-4) were shifted for 1 h to 35.5°C, and the condensation status of the chromosome 12 left arm was analyzed as in A. (C) Scc2/4 is required for condensation of the rDNA locus. Strains Y2727 (_MAT_a NET1-GFP), Y2729 (_MAT_α ycg1-10 NET1-GFP), Y2750 (_MAT_α scc2-4 NET1-GFP), and Y2728 (_MAT_α scc4Δ scc4-4 NET1-GFP) were arrested in S phase by HU treatment at 23°C, and released into nocodazole-containing medium at 35.5°C. rDNA condensation was analyzed by visualizing the rDNA-binding protein Net1 fused to GFP. Examples of the cells are shown, and the numbers of nuclei displaying puff-, cluster-, or loop-shaped rDNA loci (Lavoie et al. 2004) were counted in >100 cells in each sample.
To confirm these observations, we analyzed condensation of the rDNA locus by visualizing the rDNA-binding protein Net1 fused to GFP. In this experiment, we released wild-type, ycg1-10, scc2-4, and scc4-4 cells from HU-block in early S phase into nocodazole-imposed metaphase arrest at restrictive temperature. Sister chromatid cohesion, which in budding yeast is also required for mitotic chromosome condensation, was intact after cohesin loading in the HU block at permissive conditions. In wild-type metaphase cells, the rDNA condensed into characteristic loops (Guacci et al. 1994), while in the ycg1-10 mutant it maintained an uncondensed, puff-like appearance (Fig. 5C). The rDNA in scc2-4 and scc4-4 cells also did not form loops. Uncondensed rDNA puffs were prominent in the scc4-4 mutant, while scc2-4 cells contained puffs as well as compact rDNA structures similar to the clusters that have been described as an rDNA condensation intermediate (Lavoie et al. 2004). Together, these results indicate that Scc2/4 is required to establish and maintain chromosome condensation. The colocalization of Scc2/4 with condensin and the altered condensin levels on chromosomes in the scc2-4 mutant are consistent with the possibility that the Scc2/4 complex contributes to productive chromosome condensation by the condensin complex. We cannot exclude that the Scc2/4 complex plays a role in chromosome condensation in addition to, or independently of, condensin.
Condensin coincides with tRNA genes and other TFIIIC-binding sites
Our results on the colocalization of condensin and the Scc2/4 complex left open the question of how these binding sites are defined. During our analysis, we noticed a clear correlation of condensin peaks with tRNA genes (Fig. 6A). Of the 419 condensin peaks identified, 142 (34%) were assigned to within 1 kb of a tRNA gene, with an average distance of 66 bp from the start of the tRNA gene. This represents more than half of the 274 tRNA genes in Saccharomyces cerevisiae. On visual inspection, the remaining tRNA genes were also found associated with condensin, although the peaks were below the threshold for recognition by our peak picking algorithm (Supplemental Figs. 1, 2). tRNAs are transcribed by RNA pol III, and condensin peaks also decorated all other known pol III genes—SNR6, SCR1, RPR1, and _SNR52_—encoding small nuclear and cytoplasmic RNAs (Fig. 6A; Supplemental Fig. 1). This suggests that pol III-transcribed genes contain features that form condensin-binding sites. In addition to pol III-transcribed genes, we found condensin at all RNA pol II-transcribed genes encoding small nuclear and nucleolar RNAs (SNR genes), as well as protein components of the large and small ribosome subunits (RPL and RPS genes) (Fig. 6A; Supplemental Figs. 1, 2).
Figure 6.
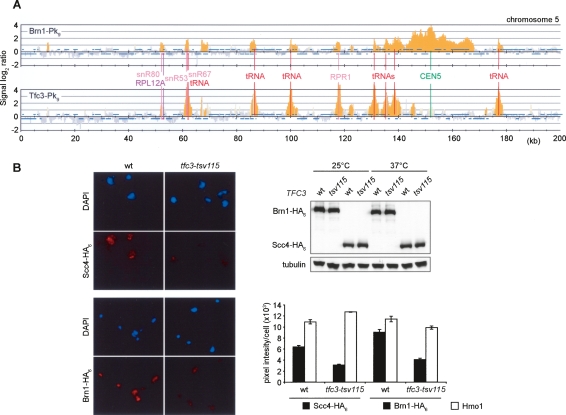
Condensin coincides with the RNA pol III transcription factor TFIIIC. (A) Colocalization of condensin with TFIIIC at tRNA genes, and genes encoding ribosomal protein components and small noncoding RNAs. Strains Y2200 and Y3096 (_MAT_α TFC3-Pk9 BRN1-HA6) were arrested in metaphase by nocodazole treatment, and ChIP against the condensin subunit Brn1-Pk9 and the TFIIIC subunit Tfc3-Pk9 was performed. A 200-kb region including the centromere of chromosome 5 is shown. (B) Reduced chromosome association of Scc2/4 and condensin in the temperature-sensitive TFIIIC mutant tfc3-tsv115. Cultures of strains Y3343 (_MAT_a SCC4-HA6), Y3344 (as Y3343 but tfc3-tsv115), Y3345 (_MAT_a BRN1-HA6 SCC1-Pk9), and Y3346 (as Y3345 but tfc3-tsv1) were shifted for 4 h to 37°C, and chromosome binding of Scc4-HA6 and Brn1-HA6 was quantified on stained spread chromosomes. Examples of the spreads and the measured mean staining intensity and standard error are shown.
tRNA and other pol III genes are strongly transcribed. However, inhibition of their transcription by the pol III inhibitor ML-60218 (Wu et al. 2003) did not alter condensin binding to these sites (data not shown). Instead, sequence elements present at these genes, or transcription factors that recognize them, might confer condensin binding. A core pol III promoter element is the B-box (GTTCxAxxC), recognized by the transcription factor TFIIIC. We therefore compared the chromosome binding patterns of TFIIIC and condensin. ChIP against the epitope-tagged TFIIIC subunit Tfc3 revealed that it colocalized with condensin at pol III genes (Fig. 6A; Supplemental Fig. 5), as well as several previously described “extra TFIIIC (ETC)” sites (Moqtaderi and Struhl 2004). We observed Tfc3 also at most other condensin-binding sites along chromosome arms that did not correspond to previously known TFIIIC-binding sites. Association at these sites was weaker when compared with pol III genes or ETC loci. We currently do not know whether Tfc3 at these sites weakly recognizes degenerate or partial B-box motifs that are found at frequent distances throughout the genome, or whether Tfc3 association is mediated indirectly; e.g., by interactions between neighboring condensin-binding sites in trans. The chromosome-wide colocalization of condensin with Tfc3 was statistically significant (Supplemental Fig. 3). Exceptions where condensin did not coincide with Tfc3 were centromeres where the enrichment of condensin was not reflected by Tfc3 and four of the ETC loci where strong Tfc3 binding was matched only by a weak condensin peak.
We next addressed whether TFIIIC contributes to the recruitment of Scc2/4 and condensin to its binding sites. To test this, we made use of a temperature-sensitive mutation in the TFIIIC subunit Tfc3, tfc3-tsv115 (Lefebvre et al. 1994). We shifted exponentially growing cultures of wild-type and tfc3-tsv115 mutant cells to a restrictive temperature of 37°C, and analyzed chromosome association of Scc2/4 and condensin by immunostaining of the epitope-tagged Scc4 and Brn1 subunits on spread chromosomes. Four hours after the shift, both mutant and wild-type cells still proliferated indistinguishably, and the cellular protein levels of Scc4 and Brn1 remained unchanged, suggesting that pol III transcripts were not yet limiting for cell growth. Only after longer times at restrictive temperature, after five to six cell divisions, proliferation of tfc3-tsv115 mutant cells ceased in a nonuniform growth arrest. After 4 h of inactivation, chromosome association of Scc4 and Brn1 was reduced twofold in the tfc3-tsv115 mutant compared with wild-type cells, while as a control the chromosomal levels of the high mobility group protein Hmo1 were not affected (Fig. 6B). This suggests that TFIIIC has a role in recruiting or stabilizing Scc2/4 and condensin on chromosomes in addition to its role as a pol III transcription factor.
A B-box forms a minimal condensin-binding site
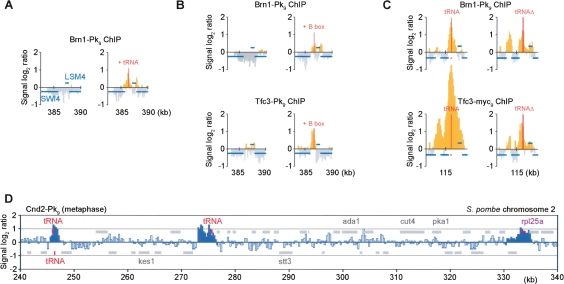
TFIIIC recognizes and directly binds to the B-box motif in pol III promoters (Huang and Maraia 2001). If TFIIIC indeed plays a role in recruiting condensin to chromosomes, ectopic tRNA genes containing a pol III promoter, or an isolated B-box element, should be sufficient to form a condensin-binding site. To test this, we transferred two tandem tRNA [tH(GUG)E1] genes, or only a consensus B-box element, to the intergenic region between the SWI4 and LSM4 genes on chromosome 5, which is normally devoid of condensin. ChIP experiments showed that transfer of these sequences was sufficient to create a binding site for both TFIIIC and condensin (Fig. 7A,B). TFIIIC and condensin binding to the ectopic B-box was less strong when compared with authentic tRNA genes (cf. Fig. 7C), indicating that features in addition to the B-box contribute to the formation of an efficient binding site. While TFIIIC binding was limited to sequences immediately surrounding the B-box insertion, condensin was seen both coinciding with TFIIIC, as well as downstream from the neighboring LSM4 gene. This opens the possibility that, after loading, condensin might be able to translocate along chromosomes in a transcription-dependent fashion similar to cohesin, albeit with reduced processivity.
Figure 7.
A B-box as minimal condensin-binding site. (A) tRNA genes confer condensin binding. Brn1-Pk9 ChIP was performed in strains Y2200 and Y3528 [as Y2200, but two tandem tRNA tH(GUG)E1 genes inserted between SWI4 and _LSM4_] arrested in mitosis by nocodazole treatment. A 7.5-kb region around this locus is shown. All probes with a positive signal log2 ratio are depicted in yellow. (B) A B-box is sufficient to form a TFIIIC and condensin-binding site. As in A, but a B-box consensus sequence (GGTTCGAACCC) was inserted. Chromatin immunoprecipitates against Brn1-Pk9 (strains Y2200 and Y3485) or Tfc3-Pk9 (Y3096 and Y3475) were analyzed. (C) Additional determinants at an endogenous tRNA gene. Brn1-Pk9 and Tfc3-myc18 association around tRNA tE(UUC)J on chromosome 10 before (strains Y2200 and Y3096) and after deletion of the tRNA gene (Y3521 and Y3522) is shown. (D) Condensin association along fission yeast chromosome 2. Nda3-KM311 cells, Y253 (h- leu1 nda3-KM311 cnd2-Pk9), were arrested in metaphase by temperature shift for 6 h to 20°C, before ChIP against Cnd2-Pk9 was performed. The distribution over a 100-kb region of S. pombe chromosome 2 is shown. Peaks identified as detailed in Supplemental Figure 6 are indicated in dark blue.
If tRNA sequence elements, recognized by TFIIIC, determine condensin binding, deletion of this sequence should abolish binding from an existing site. We deleted the tRNA tE(UUC)J gene on chromosome 10 and analyzed the consequence on condensin distribution. In response to the deletion, condensin was still detected, although the peak height was reduced (Fig. 7C). In addition, small peaks of condensin association at neighboring inter-ORF regions increased in intensity. A similar change in the condensin pattern was observed at two additional tRNA genes that we deleted (data not shown). We also analyzed TFIIIC binding around the deleted tRNA tE(UUC)J locus and, as with condensin, found a reduction but not elimination of binding. This suggests that while a B-box element forms a minimal TFIIIC- and condensin-binding site, additional features that remain to be further characterized also contribute.
A conserved condensin-binding pattern in fission yeast
We addressed whether the pattern of condensin binding along budding yeast chromosomes is a feature specific to this organism or whether a similar pattern could be observed in the distantly related fission yeast Schizosaccharomyces pombe. We performed ChIP against the Pk epitope-tagged Cnd2 subunit of the fission yeast condensin complex, the ortholog of budding yeast Brn1 (Sutani et al. 1999). Hybridization of the chromatin immunoprecipitate to an oligonucleotide tiling array covering fission yeast chromosomes 2 and 3 revealed frequent condensin-binding sites along chromosome arms, with an average distance between peaks of 40 kb (Fig. 7C; Supplemental Fig. 6). As in budding yeast, tRNA genes and genes encoding ribosomal protein components were enriched for condensin. This suggests that features that define binding sites of the chromosomal condensin complex are conserved between budding and fission yeast, and probably other eukaryotes.
Discussion
Condensin is a chromosomal protein complex with functions in both mitotic chromosome structure and segregation, as well as gene regulation and replication checkpoint signaling in interphase. Here, we provide a detailed analysis of condensin’s binding pattern along budding yeast chromosomes. This uncovered an unexpected organizing principle, the binding to chromosomes together with the Scc2/4 complex at sites characterized by the presence of the pol III transcription factor TFIIIC. These findings have important implications for both interphase and mitotic chromosome architecture.
The Scc2/4 complex has first been characterized as a chromosomal loading factor for the cohesin complex, but it is not yet known how Scc2/4 loads cohesin onto DNA. Scc2/4 interacts with cohesin and may stimulate its low intrinsic ATPase activity, which is essential for cohesin loading onto DNA (Arumugam et al. 2003; Weitzer et al. 2003). Our results show that Scc2/4 is also required for chromosome condensation, and that in the scc2-4 mutant, condensin fails to reach its full level of association with chromosomes. Whether the involvement of Scc2/4 with condensin is in principle similar to its role in cohesin loading or contributes to chromosome condensation by a distinct mechanism will be an important question to be addressed. Cohesin association with chromosomes involves topological binding to DNA (Ivanov and Nasmyth 2005), and it appears likely that the Scc2/4-dependent loading step relates to achieving cohesin’s topological DNA embrace. Scc2/4 also contributes to chromosomal binding of the Smc5/6 complex (Betts Lindroos et al. 2006). Given the structural similarity of all three eukaryotic Smc complexes, one possibility, although a hypothetical one, is that Scc2/4 is involved in the topological loading of all Smc complexes onto DNA. In this case, the observed condensin binding to chromosomes in the scc2-4 mutant might reflect a pre-loading state. This could explain why condensation remains defective in scc2-4 cells despite the presence of condensin.
An evident difference, compared with cohesin, is that condensin does not translocate away from its loading site, or does so only with reduced processivity. While this difference accounts for the largely alternating pattern of cohesin and condensin along chromosomes, it may constitute a quantitative rather than qualitative difference. Condensin, like cohesin, appears to be excluded from ORFs, and induction of RNA pol I or II transcription within the rDNA locus has been shown to cause displacement of condensin from transcribed regions (Wang et al. 2006; Johzuka and Horiuchi 2007). We furthermore observed that ectopic condensin recruitment upstream of the actively transcribed LSM4 gene led to condensin accumulation both upstream and downstream of the gene. Thus, condensin appears able to translocate in response to transcription, but it may be dissociating from chromosomes at a faster rate than cohesin. Direct measurements by fluorescence recovery after photobleaching experiments revealed faster turnover of human condensin on chromosomes, as compared with cohesin (Gerlich et al. 2006a, b). Continuous cycles of loading and dissociation by budding yeast condensin would be consistent with the continued requirement for Scc2/4 to maintain chromosome condensation in mitosis.
Studies in organisms other than budding yeast have yielded mixed results as to a possible contribution of the Scc2/4 complex to chromosome condensation. A mutation in the fission yeast Scc2 ortholog, mis4-242, causes defects in sister chromatid cohesion, but not in chromosome condensation (Furuya et al. 1998). The Scc2 ortholog in the fungus Coprinus cinereus, encoded by its RAD9 gene, is required for meiotic chromosome condensation, but its relationship with the condensin complex has not been analyzed (Seitz et al. 1996). When Scc2/4 was depleted from Xenopus egg extracts (Gillespie and Hirano 2004), or its level reduced by RNAi in human cells (Watrin et al. 2006), no apparent chromosome condensation defect was observed. However, it should be noted that vertebrate chromosomes reach mitotic condensation even with largely reduced levels of condensin (Hudson et al. 2003; Ono et al. 2004; Gerlich et al. 2006a). It will therefore be interesting to analyze condensin levels, and the structural stability of vertebrate chromosomes, in the absence of Scc2/4. A mutation in the Drosophila Scc2 ortholog, Nipped-B, leads to structurally compromised mitotic chromosomes in larval neuroblasts (Rollins et al. 2004).
The Scc2/4 and condensin-binding sites along budding and fission yeast chromosomes follow an unexpected pattern, in that they are characterized by sequence elements and proteins involved in transcriptional regulation. Most notable is the association of Scc2/4 and condensin with all tRNA genes. Consistent with the function as cohesin and condensin loading sites, a tRNA gene has recently been identified as a _cis_-acting element for the establishment of sister chromatid cohesion at the budding yeast HMR mating-type locus (Dubey and Gartenberg 2007). In addition to tRNA genes, we found condensin also at genes encoding small noncoding RNAs and ribosomal proteins, as well as several additional genes that remain to be further characterized. While strong transcriptional activity is common to these genes, the process of transcription is probably not solely responsible for condensin recruitment. This is suggested by an unaltered condensin pattern after chemical inhibition of pol III transcription. Furthermore, an isolated B-box element, unlikely to promote transcription in the absence of additional promoter elements, was sufficient to create a minimal condensin-binding site. It could therefore be that transcription factors in common to these genes are mediating the recruitment of Scc2/4 and condensin. These include the pol III transcription factor TFIIIC that is recruited by the B-box element, and maybe additional factors that characterize ribosomal protein genes and other genes that act as condensin-binding sites.
Once bound to DNA, how does condensin exert its function on interphase and mitotic chromosomes? A hint as to condensin’s mode of action comes from the observation that tRNA genes and TFIIIC are found in clusters in interphase nuclei (Thompson et al. 2003; Noma et al. 2006). In an accompanying paper, Haeusler et al. (2008) show that condensin mediates the tRNA gene clustering. This exemplifies that condensin establishes interactions between its binding sites. This could occur by interactions between more than one condensin complex, each bound to distinct binding sites. Alternatively, individual condensin complexes are able to subsequently load at more than one site. While maintaining topological, or other, contact with one site, condensin might repeat a DNA-binding reaction at a second site. The establishment of contacts between condensin-binding sites provides a powerful means to influence chromosome architecture. tRNA genes are known to act as chromatin domain boundaries, and clustering of boundary elements has been linked with their function (Valenzuela and Kamakaka 2006). Domain boundary activity is consistent with condensin’s described role in gene regulation (Lupo et al. 2001; Bhalla et al. 2002). After replication fork stalling during S phase, recruitment of condensin might stabilize the fork within the nuclear context, thereby facilitating replication restart.
As cells enter mitosis, the in vitro DNA supercoiling activity of Xenopus condensin I is stimulated by Cdk phosphorylation of its XCAP-D2 subunit (Kimura et al. 1998), and the human condensin II complex slows down its turnover on chromosomes in vivo (Gerlich et al. 2006a). Both, increased DNA binding and slower turnover at binding sites, could now enhance condensin-mediated DNA interactions. In this model, chromosome condensation is mediated by increased interactions between neighboring condensin-binding sites. Whether, and how, condensin is able to discriminate between the many binding sites that may come within its reach on the same chromatid, its sister chromatid, or other chromosomes in the vicinity, will be interesting to address. The linkage of neighboring condensin binding sites in cis by the intervening DNA sequence may contribute to a preference of interactions within each sister chromatid. Our identification of specific binding sites for the chromosomal condensin complex opens new possibilities for probing eukaryotic chromosome architecture.
Materials and methods
Yeast strain construction
All strains were of W303 background. Epitope tagging of genes at their endogenous loci was performed by gene targeting using PCR products. Deletion of a 8.5-kb segment between two condensin-binding sites on the chromosome V right arm was achieved by replacing the region between coordinates 213,898 and 222,402 (as annotated in the Saccharomyces Genome Database) with a URA3 auxotrophic marker flanked by 142-bp direct repeats (Reid et al. 2002). This removed four nonessential genes: YPT31, FIR1, ZRG8, and YER034w. The URA3 insertion was then lost after counterselection on 5-fluoroorotic acid (5FOA), leaving one 142-bp repeat behind. Two copies of the tRNA gene tH(GUG)E1 plus 100 bp upstream and 50 bp downstream (207256–207477 on chromosome 5) were cloned in tandem into plasmid pWJ1007 (Reid et al. 2002). The tRNA genes and repeat-flanked URA3 were amplified by PCR, and integrated between SWI4 and LSM4 (at coordinate 386,500) on chromosome V. URA3 was lost as before after selection on 5FOA. Integration of a B-box consensus (GGTTCGAACCC) was achieved by PCR amplification of repeat-flanked URA3 using an oligonucleotide containing the consensus. The PCR product was integrated at the same locus on chromosome V and URA3 sequences again removed by 5FOA selection. The tRNA tE(UUC)J gene on chromosome X was replaced with a PCR product encompassing repeat-flanked URA3.
ChIP
ChIP was performed as described (Lengronne et al. 2004). Immunoprecipitates were hybridized to Affymetrix S. cerevisiae chromosomes 3–6 (SC3456a520015F; P/N 520,015), or whole genome forward oligonucleotide tiling arrays (P/N 520,286), or to S. pombe chromosomes 2–3 oligonucleotide tiling arrays (S_pombea520106F, P/N 520,106). BrdU incorporation to mark stalled replication forks in HU-treated early S-phase cells was as described (Katou et al. 2003). Quantitative analysis of the chromatin immunoprecipitates was performed using an MJ Research Chromo 4 real-time PCR system (MJ). All primer sequences are available upon request. Duplicate samples in two dilutions were analyzed. Relative enrichment over the input sample was compared between the four positive and two negative association sites. The microarray data contained in this manuscript have been deposited with the Gene Expression Omnibus (accession no. GSE12149).
Chromosome condensation assay
Lac operator repeats were inserted at the MMP1 and YLR003c-1 loci on chromosome XII, at a distance of ∼137 kb from each other, and visualized by expression of a lacI-GFP fusion protein as described (Sullivan et al. 2004). Cells were fixed in 100% ethanol and mounted on a 2% agarose patch, and images were acquired as z-stacks of 10 frames at 0.15 μm distance with an Axioplan 2 imaging microscope (Zeiss) equipped with a 100× (NA = 1.45) Plan-Neofluar objective. Distance measurements were then performed in three dimensions using Volocity (Improvision) software in at least 100 cells, and the distance distribution analyzed using JMP 5.1 software (SAS Institute). rDNA condensation was analyzed in ethanol-fixed cells expressing a Net1-GFP fusion protein.
Other techniques
The analysis of Scc2, Scc4, condensin, and Hmo1 on chromosome spreads was performed as described (Ciosk et al. 2000). Signals from >50 chromosome spreads, imaged under identical exposure conditions, were quantified using the analysis tools provided by the Volocity software (Improvision), and background staining in adjacent regions of the same size was subtracted. The antibodies used for ChIP, staining of chromosome spreads, and Western blotting were α-Pk clone SV5-Pk1 (Serotec), α-HA clone 3F10 (Roche), α-myc clone 9E10, α-BrdU clone 2B1D5F5H4E2 (MBL), α-tubulin clone YOL1/34 (Serotec), and α-Hmo1 antiserum (a kind gift from S. Brill).
Acknowledgments
We thank A. Lengronne for her help at the outset of this study; Neil Brooks for biostatistics support; S. Brill, B. Lavoie, and O. Lefebvre for reagents; J. Cooper, T. Hirano, T. Hirota, and members of our laboratory for discussion and critical comments on the manuscript; and R. Haeusler and D. Engelke for communicating unpublished results. This work was supported by the Human Frontier Science Program.
Footnotes
References
- Anderson D.E., Losada A., Erickson H.P., Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Kimura K., Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell. 2002;9:1183–1190. doi: 10.1016/s1097-2765(02)00546-4. [DOI] [PubMed] [Google Scholar]
- Betts Lindroos H., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A.W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M.A., Philp A.V., Glover D.M., Bellen H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:1–20. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Dubey R.N., Gartenberg M.R. A tDNA establishes cohesion of a neighbouring silent chromatin domain. Genes & Dev. 2007;21:2150–2160. doi: 10.1101/gad.1583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Takahashi K., Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes & Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.-M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006a;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Gerlich D., Koch B., Dupeux F., Peters J.-M., Ellenberg J. Live-cell imaging reveals a stable cohesin–chromatin interaction after but not before DNA replication. Curr. Biol. 2006b;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- Gillespie P.J., Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H., Löwe J., Hochwagen A., Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haeusler R.A., Pratt-Hyatt M., Good P.D., Gipson T.A., Engelke D.R.2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes Genes & Dev. (this issue) 10.1101/gad.1675908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hirano T., Mitchison T.J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.-M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Huang Y., Maraia R.J. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D.F., Vagnarelli P., Gassmann R., Earnshaw W.C. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- Ivanov D., Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells. 2007;12:759–771. doi: 10.1111/j.1365-2443.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bandoh M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Hogan E., Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes & Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O., Rüth J., Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. J. Biol. Chem. 1994;269:23374–23381. [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo R., Breiling A., Bianchi M.E., Orlando V. Drosophila chromosome condensation proteins topoisomerase II and barren colocalize with polycomb and maintain Fab-7 PRE silencing. Mol. Cell. 2001;7:127–136. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- MacCallum D.E., Losada A., Kobayashi R., Hirano T. ISWI remodeling complexes in Xenopus egg extracts: Identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre J., Muller E.G.D., Weitzer S., Snydsman B.E., Davis T.N., Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Noma K., Cam H.P., Maraia R.J., Grewal S.I.S. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Onn I., Aono N., Hirano M., Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. Spatial and temporal regulation of condensin I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R.J.D., Sunjevaric I., Kedacche M., Rothstein R. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast. 2002;19:319–328. doi: 10.1002/yea.817. [DOI] [PubMed] [Google Scholar]
- Rollins R.A., Korom M., Aulner N., Martens A., Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Sutani T., Yamashita Y., Saitoh S., Takeuchi M., Nakaseko Y., Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz L.C., Tang K., Cummings W.J., Zolan M.E. The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics. 1996;142:1105–1117. doi: 10.1093/genetics/142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A., Hogan E., Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Higuchi T., Katis V.L., Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sutani T., Yuasa T., Tomonaga T., Dohmae N., Takio K., Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R.T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Wang B.-D., Eyre D., Basrai M., Lichten M., Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.-D., Butylin P., Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E., Schleiffer A., Tanaka K., Eisenhaber F., Nasmyth K., Peters J.-M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Weitzer S., Lehane C., Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Wu L., Pan J., Thoroddsen V., Wysong D.R., Blackman R.K., Bulawa C.E., Gould A.E., Ocain T.D., Dick L.R., Errada P., et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot. Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]