Distinct Role of Long 3′UTR BDNF mRNA in Spine Morphology and Synaptic Plasticity in Hippocampal Neurons (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 11.
SUMMARY
The brain produces two brain-derived neurotrophic factor (BDNF) transcripts, with either short or long 3′ untranslated regions (3′UTR). The physiological significance of the two forms of mRNAs encoding the same protein is unknown. Here we show that the short and long 3′UTR BDNF mRNAs are involved in different cellular functions. The short 3′UTR mRNAs are restricted to somata whereas the long 3′UTR mRNAs are also localized in dendrites. In a mouse mutant where the long 3′UTR is truncated, dendritic targeting of BDNF mRNAs is impaired. There is little BDNF in hippocampal dendrites despite normal levels of total BDNF protein. This mutant exhibits deficits in pruning and enlargement of dendritic spines, as well as selective impairment in long-term potentiation in dendrites, but not somata, of hippocampal neurons. These results provide insights into local and dendritic actions of BDNF and reveal a mechanism for differential regulation of subcellular functions of proteins.
INTRODUCTION
It has become increasingly clear that the generation of multiple transcripts from the same gene through alternative splicing is a general rule rather than an exception. It is relatively easy to appreciate the increase in functional diversity afforded by alternative splicing that produces mRNAs encoding different proteins. However, in many cases multiple transcripts encode exactly the same protein. Such is the case for brain-derived neurotrophic factor (BDNF), where the Bdnf gene is transcribed from at least 6 promoters, each of which drives transcription of a short 5′ exon alternatively spliced onto a common 3′ exon encoding the BDNF protein (Liu et al., 2006). A plausible explanation for having multiple promoters driving the expression of the same protein is that different transcripts are regulated by different signaling pathways (Lu, 2003). A more puzzling finding is that BDNF mRNAs are polyadenylated at either of two alternative sites, leading to distinct populations of mRNAs: those with a short 3′UTR and those with a long 3′UTR (Ghosh et al., 1994; Timmusk et al., 1993). It is not clear, however, why a neuron needs two species of BDNF mRNAs if they encode the same protein.
The 3′UTRs of some mRNAs, such as those for the α-subunit of Ca2+/calmodulin-dependent protein kinase II (CaMKIIα) and activity-regulated cytoskeleton-associated protein (Arc), have been shown to target transcripts to dendrites (Kobayashi et al., 2005; Rook et al., 2000), which can then serve as templates for local translation in response to synaptic activity (Bramham and Wells, 2007). BDNF mRNA is also localized in dendrites (Tongiorgi et al., 1997; Tongiorgi et al., 2004), although whether its 3′UTRs are involved in dendritic trafficking is unclear. Unlike CaMKIIα and Arc mRNAs that have a single dominant 3′UTR, the two BDNF mRNA species are found in comparable abundance in the cortex (Timmusk et al., 1993). We hypothesize that the two BDNF mRNA species may have different subcellular distributions in neurons, one in somata and the other in dendrites. In formulating this hypothesis we considered two unique features of the BDNF protein. First, BDNF elicits diverse cellular functions in the central nervous system (CNS), ranging from neuronal survival and morphological differentiation to synapse formation and plasticity (Reichardt, 2006). Second, the secretion of BDNF is primarily activity-dependent and its diffusion is relatively limited (Lu, 2003). Targeting a fraction of BDNF mRNAs to dendrites for local translation would facilitate differential regulation of BDNF functions in dendrites and somata.
In this work we present evidence for a role of the long 3′UTR, but not the short 3′UTR, in targeting BDNF mRNA to dendrites. By testing a mouse mutant that produces a truncated long BDNF 3′UTR, we have revealed unexpected roles for the long 3′UTR in controlling the abundance of dendritic BDNF protein and regulating pruning and enlargement of dendritic spines. Furthermore, we show a selective impairment in LTP at dendritic synapses, but not somatic synapses, in CA1 pyramidal neurons lacking dendritic BDNF mRNA. These results demonstrate the importance of the long 3′UTR in BDNF mRNA trafficking and dendritic functioning in CNS neurons.
RESULTS
Differential localization of short and long BDNF mRNAs in somata and dendrites
The short (0.35 kb) and long (2.85 kb) BDNF 3′UTRs arise from alternative polyadenylation (Fig. 1A and S1). Northern blot analyses of total RNA revealed that both long and short BDNF mRNAs were present in all examined brain regions (Fig. 1B). Interestingly, the ratio of long to short BDNF mRNAs (L/S ratio) varied greatly in different brain regions, with the highest ratio (1.3) in the cortex and the lowest ratio (0.5) in the brainstem. To examine whether transcription through specific promoters also affects selection between the two alternative polyadenylation sites, we performed Northern blot analyses of hippocampal RNA using exon 2, exon 4, or exon 6 of the Bdnf gene as probes (Fig. 1C and S1B). The L/S ratio for exon 2-containing mRNA was significantly higher than those for exon 4- and exon 6-containing mRNAs.
Figure 1. Localization of long 3′UTR BDNF mRNA to dendrites.
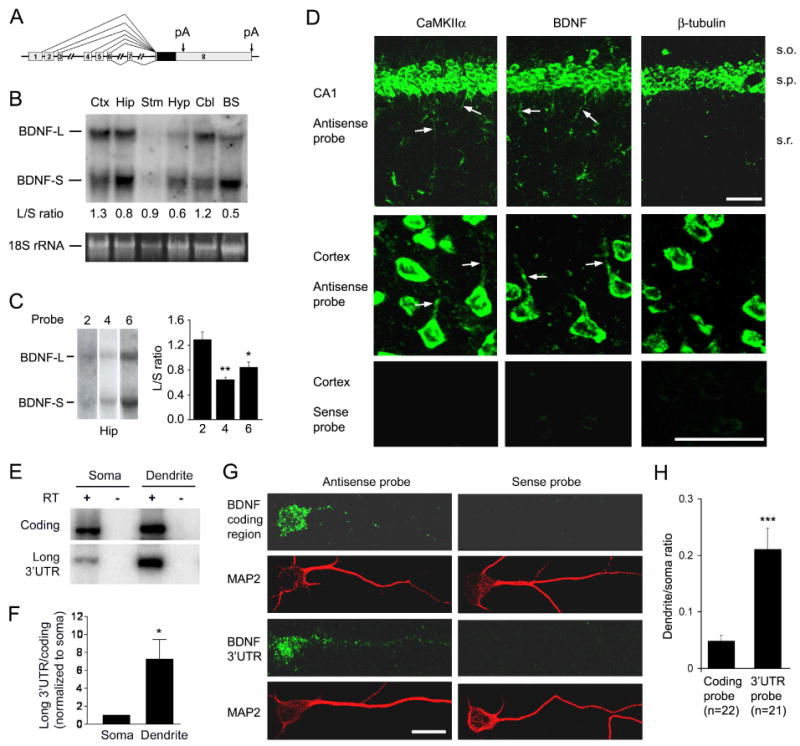
(A) Diagram of the mouse Bdnf gene depicting two alternative polyadenylation (pA) sites in exon 8 (arrows). Curved lines linking boxes (exons) indicate alternative splicing from the first seven exons to exon 8. The filled box within exon 8 represents the coding sequence.
(B) Long and short 3′UTR BDNF mRNAs in different brain regions. BDNF mRNAs with a short 3′UTR (BDNF-S) or a long 3′UTR (BDNF-L) were present in the cortex (Ctx), hippocampus (Hip), striatum (Stm), hypothalamus (Hyp), cerebellum (Cbl), and brainstem (BS). Lower panel: 18S rRNA as a loading control.
(C) Ratios of long and short BDNF mRNAs (L/S) derived from different Bdnf promoters. The same blot was used for all probes (n=4 mice).
(D) Localization of BDNF mRNAs in dendrites of hippocampal CA1 and cortical neurons. Riboprobes were derived from the coding regions for CaMKIIα, BDNF, and β-tubulin. CaMKIIα and BDNF mRNAs were detected in dendrites (arrows). Abbreviations in this and all other figures: s.o., stratum oriens; s.p., stratum pyramidale; s.r., stratum radiatum. Scale bars, 50 μm.
(E) Relative abundance of the long 3′UTR BDNF mRNA in the soma and dendrite fractions.
(F) Ratio of the long 3′UTR signal to the coding region signal in the soma and dendrite fractions (n=3).
(G) Distribution of BDNF mRNA in cell bodies and dendrites of cultured rat cortical neurons. MAP2 immunostaining marks cell bodies and dendrites. Scale bar, 20 μm.
(H) Ratio of FISH signals in dendrites to those in cell bodies.
BDNF mRNA was detected in apical dendrites of hippocampal neurons after epileptogenic stimulation (Tongiorgi et al., 2004). To investigate whether BDNF mRNA is also present in dendrites in resting animals, we performed highly sensitive fluorescent in situ hybridization (FISH) on brain sections of untreated wild-type (WT) mice. In this experiment, we used dendritically targeted CaMKIIα mRNA as a positive control and soma-restricted β-tubulin mRNA as a negative control (Paradies and Steward, 1997). Hippocampal CA1 pyramidal neurons packed in the stratum pyramidale (s.p.) send their apical dendrites to the stratum radiatum (s.r.) and basal dendrites to the stratum oriens (s.o.). We detected CaMKIIα mRNA in the s.p. as well as in the s.r. and s.o. areas, but β-tubulin mRNA only in the s.p. (Fig. 1D). Similar to CaMKIIα mRNA, BDNF mRNA was also present in the s.r. and s.o. areas. Furthermore, BDNF mRNA and CaMKIIα mRNA were localized in dendrites of cortical neurons (Fig. 1D). In contrast, β-tubulin mRNA was restricted to the cell bodies of these neurons. Thus, BDNF mRNA is targeted to dendrites under physiological conditions.
To determine if either the long or short BDNF mRNA is preferentially targeted to dendrites, we separated cell bodies (soma fraction) from pre- and post-synaptic components (dendrite fraction) of cultured rat cortical neurons (Fig. S2). Two sets of primers were used simultaneously in semi-quantitative reverse transcription-polymerase chain reactions (RT-PCR) to measure the relative abundance of the BDNF coding sequence and the long 3′UTR in RNA samples isolated from the two fractions (Fig. 1E). The ratio of long 3′UTR to coding sequence was ∼7 fold higher in the dendrite fraction than in the soma fraction (Fig. 1F). This observation suggests that the long BDNF mRNA is preferentially targeted to dendrites. To corroborate this biochemical evidence, we carried out FISH on cultured rat cortical neurons using RNA probes derived from the BDNF coding region and a 1.9-kb cDNA fragment corresponding to the 3′ end of the long BDNF 3′UTR (Fig. 1G). The ratio of the FISH signal in the initial 50-μm segment of dendrites to the somatic FISH signal from the 3′UTR probe was ∼4 fold higher than that from the coding region probe (Fig. 1H), supporting the conclusion that the long BDNF mRNA is preferentially targeted to dendrites.
The long BDNF 3′UTR is sufficient for targeting mRNA to dendrites for local translation
We cloned the sequences encoding the two BDNF 3′UTRs from mouse genomic DNA and termed the sequence between the stop codon and the 1st polyadenylation site as “A” and the sequence between the 1st polyadenylation site and the 2nd polyadenylation site as “B”. Four constructs were generated by fusing the cDNA for green fluorescence protein (GFP) to a sequence encoding the 3′UTR for bovine growth hormone (BGH), sequence A (encoding the short 3′UTR), sequence AB (encoding the long 3′UTR), or sequence B (Fig. 2A). Fusion transcripts were expressed in cultured rat hippocampal neurons and examined using FISH with antisense probes corresponding to the GFP coding sequence. As expected, FISH signals were only detectable in proximal dendrites of neurons transfected with the GFP-BGH construct (Fig. 2B). Similarly, the short BDNF 3′UTR could not target GFP mRNA to distal dendrites (Fig. 2C). In contrast, GFP mRNA was present in distal dendrites in neurons transfected with either the GFP-AB or GFP-B construct (Fig. 2D, E).
Figure 2. Cis-acting sequence in the 3′UTR required for the dendritic localization.
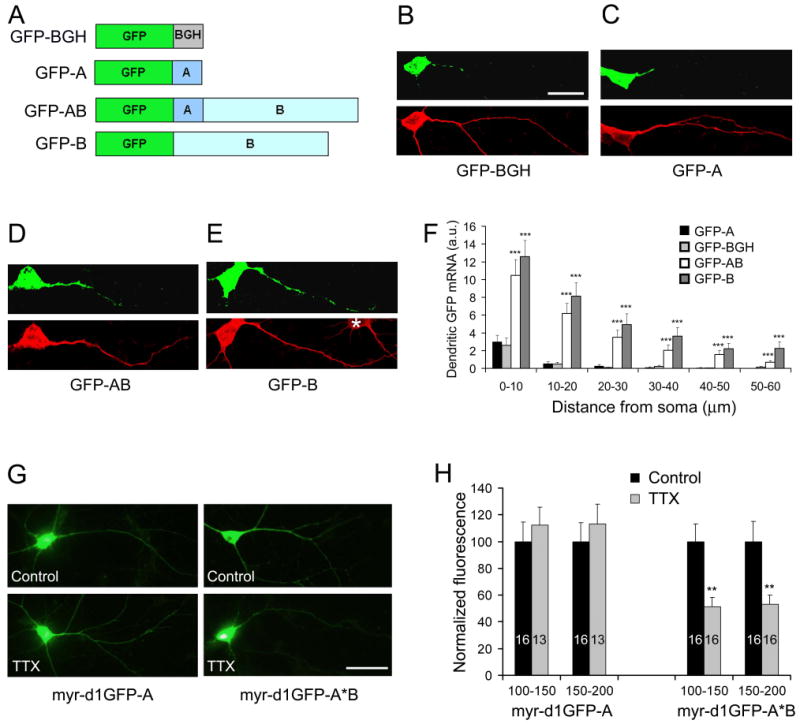
(A) Schematic of four GFP constructs with different 3′UTRs.
(B-E) Localization of GFP mRNA in neurons transfected with GFP-BGH (B), GFP-A (C), GFP-AB (D), or GFP-B (E). Top panels: FISH of cultured neurons with a GFP antisense riboprobe; Bottom panels: MAP2 immunocytochemistry; *, an untransfected neuron; Scale bar, 25 μm.
(F) Levels of GFP mRNAs along the main apical dendrites of transfected neurons. Data were obtained from 15-23 transfected neurons grown on 3 coverslips for each construct and presented in arbitrary unit (a.u.). GFP-A was compared with the other three constructs.
(G) Whole cell images of cultured hippocampal neurons expressing either myr-d1GFP-A or myr-d1GFP-A*B. The images were taken with a fluorescent microscope using a GFP filter. Scale bar, 50 μm.
(H) Quantification of myr-d1GFP fluorescence in dendrites. Fluorescence intensities on distal dendrites (100-150 μm and 150-200 μm away from somata) were measured and normalized to control levels. The number of neurons for each condition is indicated inside bars.
We then measured levels of GFP mRNA along one major dendrite of each transfected neuron. Levels of GFP mRNA at any 10-μm dendritic segment were significantly higher in neurons expressing GFP-AB or GFP-B than in neurons expressing GFP-A (Fig. 2F). GFP mRNA at dendrites 20 μm or more away from cell bodies was barely detectable in neurons transfected with either GFP-BGH or GFP-A, whereas its levels remained high in neurons expressing GFP-AB or GFP-B (Fig. 2F). There was no significant difference between neurons expressing GFP-BGH and GFP-A in levels of GFP mRNA along the main dendrite. These results indicate that neither the BGH 3′UTR nor the short BDNF 3′UTR is capable of targeting mRNA to distal dendrites. Although the difference was not statistically significant, neurons expressing GFP-B displayed a trend towards higher levels of GFP mRNA in dendrites than neurons expressing GFP-AB (Fig. 2F), which is likely due to the fact that GFP-AB generates two GFP mRNAs, one with the short BDNF 3′UTR and one with the long BDNF 3′UTR. Taken together, these results show that the BDNF 3′UTR sequence between the two polyadenylation sites is sufficient to target GFP mRNA to dendrites.
To test whether the long 3′UTR mRNA targeted to dendrites is translated locally, we generated two protein synthesis reporter constructs by attaching the myr-d1GFP coding sequence to sequence A (myr-d1GFP-A) or sequence A*B (myr-d1GFP-A*B, where the first polyadenylation signal of sequence AB was mutated). The myristoylation peptide (myr) and the short half life of the destabilized d1GFP protein impede GFP synthesized in somata from diffusing to distal dendrites, so that GFP in distal dendrites reflects local protein synthesis (Aakalu et al., 2001). Blockade of action potentials with tetrodotoxin (TTX) has been shown to decrease dendritic local protein synthesis in hippocampal neurons (Sutton et al., 2004). We found that TTX significantly decreased dendritic expression of myr-d1GFP-A*B but not myr-d1GFP-A at distal dendrites 100-200 μm away from somata (Fig. 2G and H). Furthermore, TTX increased the amount of GFP in cell bodies transfected by myr-d1GFP-A*B (100 ± 15% for control vs. 155 ± 17% for TTX, _p_=0.024), but not by myr-d1GFP-A (100 ± 9% for control vs. 96 ± 9% for TTX, _p_=0.759). These results support the notion that the long BDNF 3′UTR is able to direct dendritic local protein synthesis and that spontaneous action potentials enhance local BDNF synthesis at least in part by stimulating dendritic targeting of the long 3′UTR BDNF mRNA.
Requirement of the long 3′UTR for dendritic targeting of BDNF mRNA in vivo
To examine dendritic targeting of BDNF mRNA in vivo, we took advantage of a previously generated Bdnf knockin strain. In this strain, a loxP site was inserted into the small 5′UTR within exon 8 and another loxP site into a BssHII site 936 bp downstream of the stop codon (Gorski et al., 2003). In addition, three tandem SV40 polyadenylation signal sequences were inserted immediately upstream of the second loxP site, which is followed by a lacZ gene (Fig. 3A). In this Bdnf allele, termed Bdnfklox here, insertion of the three tandem SV40 polyadenylation signal sequences results in truncation of the long BDNF 3′UTR. Indeed, in Bdnfklox/klox mice, the short BDNF mRNA was not affected, whereas the long BDNF mRNA was absent (Fig. 3B). One new species of BDNF mRNAs appeared in Bdnfklox/klox mice. It was longer than the short BDNF mRNA, presumably derived from polyadenylation at the three SV40 polyadenylation sites. However, the total amount of hippocampal BDNF mRNA in Bdnfklox/klox mice was equivalent to that in WT mice (100 ± 15% for WT mice vs. 105 ± 17% for Bdnfklox/klox mice, n=3 pairs of mice, _p_=0.833). These results show that Bdnfklox/klox mice have a negligible amount of the long 3′UTR BDNF mRNA, although their total amount of BDNF mRNAs remains similar.
Figure 3. Impairment of dendritic targeting of BDNF mRNA in 3′UTR mutant mice.
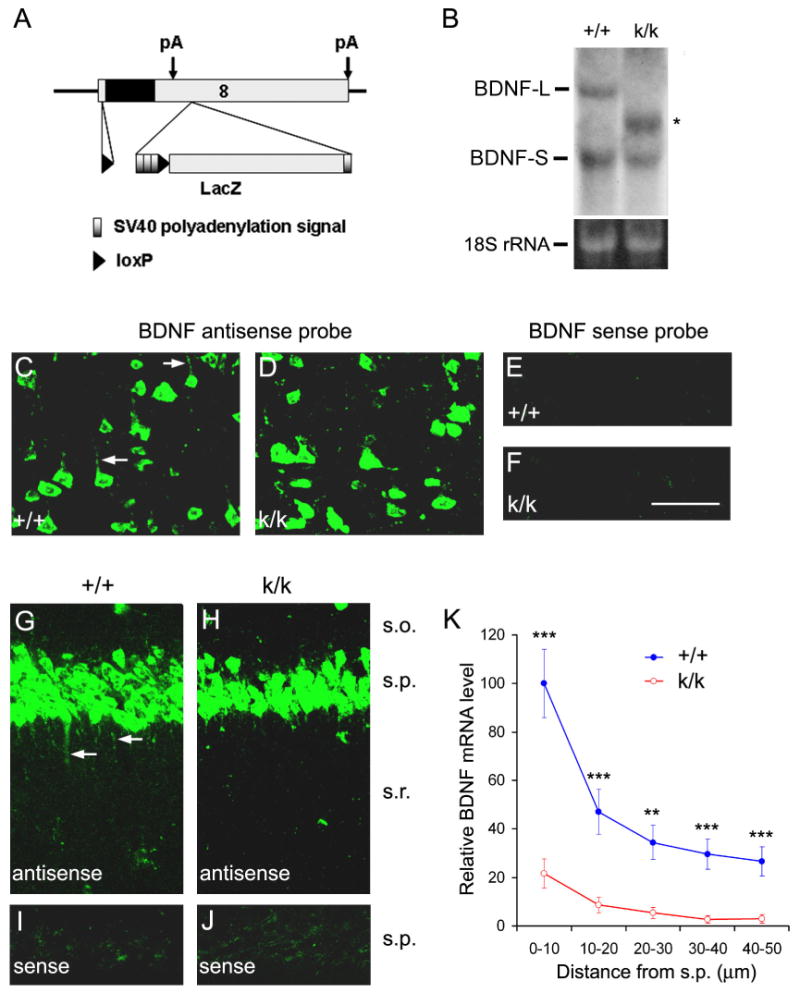
(A) The Bdnfklox allele generated by insertion of a lox P sequence into the 5′UTR within exon 8 and into a site 3′ to the first polyadenylation site of a sequence containing three tandem SV40 polyadenylation signals and a lacZ gene.
(B) Northern blot analysis of BDNF mRNA from the cortex of WT (+/+) and Bdnfklox/klox (k/k) mice. Total RNA (10 μg) was loaded onto each lane. The asterisk denotes a new BDNF mRNA species. Lower panel: 18S rRNA as a loading control.
(C-J) In situ hybridization revealing defects in dendritic targeting of BDNF mRNA in k/k mice. In layer 5 of the cortex (C, D) and the hippocampal CA1 region (G, H), there is a marked reduction in levels of dendritic BDNF mRNA in k/k mice as compared to +/+ mice. Arrows denote representative dendrites containing BDNF mRNA. Also note many small puncta containing BDNF mRNA in G. The sense probe did not produce significant signals in the cortex (E, F) or hippocampal CA1 region (I, J). Scale bar, 50 μm.
(K) Quantification of BDNF mRNA in situ hybridization signals in the s.r. of +/+ and k/k mice (n=4 mice each; 3 sections per mouse).
We next performed in situ hybridization to determine if the truncation of the long 3′UTR impairs dendritic localization of BDNF mRNA. Strikingly, virtually all cortical neurons in Bdnfklox/klox mice lacked BDNF mRNA in apical dendrites (Fig. 3D, F). This was in marked contrast to what was observed in WT cortical neurons (Fig. 3C, E). Similarly, BDNF mRNAs were rarely detected in the dendrites of hippocampal CA1 neurons in Bdnfklox/klox mice (Fig. 3G-J). Quantitative analysis indicates that levels of BDNF mRNA in the s.r. at various distances away from the s.p. were greatly reduced in Bdnfklox/klox mice as compared to WT mice (Fig. 3K). In contrast, Bdnfklox/klox mice appeared to have elevated levels of BDNF mRNA in somata of both cortical and CA1 pyramidal neurons (Fig. 3C-J). These results demonstrate the importance of the long 3′UTR in dendritic targeting of BDNF mRNA in vivo.
Deficits in dendritic BDNF protein localization in neurons lacking long 3′UTR mRNA
To identify BDNF proteins on immunoblots, we compared cortical protein extracts prepared from WT mice and Bdnf transgenic (BTg) mice that overexpress BDNF in the dorsal forebrain (Huang et al., 1999). Two proteins of 15 kDa and 32 kDa were detected in the WT extract and their abundance was increased in the BTg extract (Fig. 4A), indicating that the two bands correspond to the precursor (pro-BDNF) and protease-cleaved mature BDNF (Lee et al., 2001). Using the same antibody, we detected no alterations in levels of pro-BDNF and total BDNF (pro-BDNF plus mature BDNF) in the cortex, hippocampus, and striatum of Bdnfklox/klox mice (Fig. 4B, C). These results indicate that Bdnfklox/klox mice synthesize the same amount of BDNF in the brain.
Figure 4. Altered subcellular distribution of BDNF protein in Bdnfklox/klox neurons.
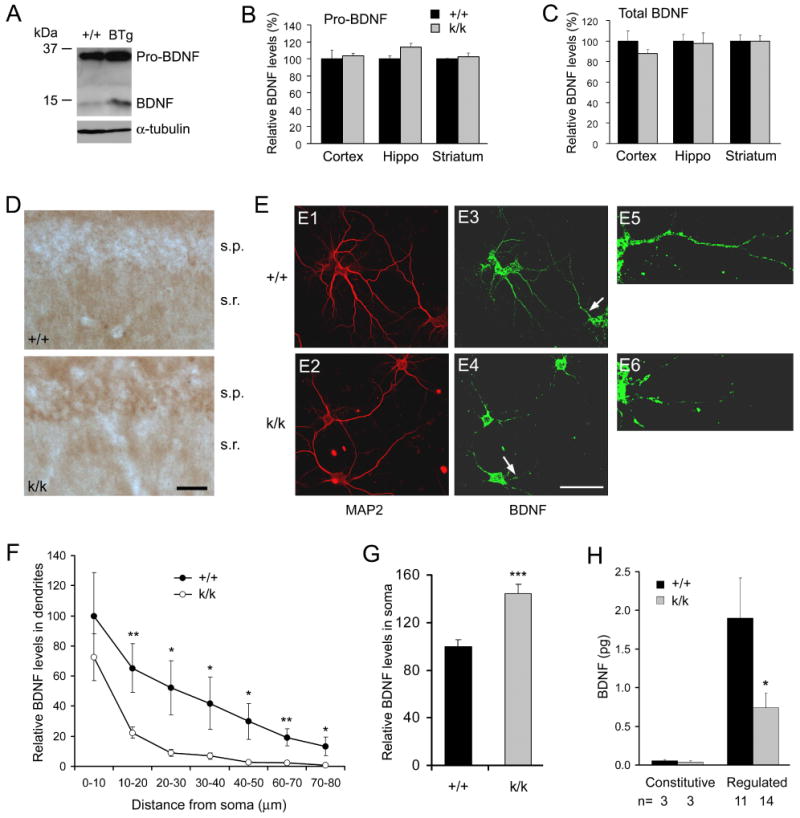
(A) Western blot analysis of cortical extracts from WT and BTg mice. The blot was probed with antibodies to BDNF and α-tubulin.
(B) Relative levels of pro-BDNF in the cortex, hippocampus and striatum as determined by immunoblotting (n=3 mice each).
(C) Relative levels of total BDNF (mature BDNF + pro-BDNF) as determined by immunoblotting (n=3 mice each).
(D) BDNF immunohistochemistry showing a decrease in the s.r. but an increase in the s.p. in levels of BDNF protein in hippocampal sections from k/k mice as compared with +/+ mice. Scale bar, 50 μm.
(E) BDNF immunocytochemistry showing a decrease in dendritic BDNF protein in cultured hippocampal neurons derived from k/k mice. Dendrites are marked by MAP2 immunofluorescence (E1 and E2). Panels E5 and E6 are high-magnification images of the dendrites denoted by arrows in panels E3 and E4 to show BDNF immunofluorescence along dendritic shafts. Scale bar, 50 μm.
(F) Relative levels of BDNF protein along apical dendrites of cultured hippocampal neurons. The amount of BDNF immunofluorescence along apical dendrites (10 μm intervals) was measured on +/+ neurons (n=9) and k/k neurons (n=12) and normalized to the level at the first 10-μm dendrites of WT neurons.
(G) Quantification of BDNF protein in cell bodies of cultured hippocampal neurons. Levels of BDNF protein were obtained by measuring total BDNF immunofluorescence in cell bodies of 71 +/+ and 71 k/k neurons.
(H) Impairment of regulated BDNF secretion from k/k hippocampal neurons. Amounts of secreted BDNF were normalized to 15 min.
A deficiency in dendritic targeting of BDNF mRNA may or may not lead to alterations in dendritic localization of BDNF protein, since BDNF synthesized in somata may be transported to dendrites. It is therefore important to examine the localization of BDNF protein in Bdnfklox/klox dendrites. Immunostaining of many hippocampal sections showed that BDNF levels were higher in the s.p. and lower in the s.r. in the Bdnfklox/klox hippocampus, as compared to those in the WT hippocampus (Fig. 4D). Quantification of immunostaining in brain sections is difficult. Moreover, the difference in dendritic and somatic BDNF contents between WT and Bdnfklox/klox mice may be reduced by BDNF-containing afferents projecting to the CA1 region. To directly examine BDNF levels in dendrites and somata, we used cultured hippocampal neurons isolated from WT and Bdnfklox/klox pups at postnatal day 1 or 2. After 10 days in vitro (DIV), neurons were stained with antibodies to MAP2 and BDNF. In WT neurons, BDNF protein could be detected in distal dendrites. In Bdnfklox/klox neurons, however, BDNF protein was only detectable in proximal dendrites and its level in dendrites was lower in comparison with WT neurons (Fig. 4E). Although the amount of BDNF protein in the first 10 μm of dendrites was similar for WT neurons and Bdnfklox/klox neurons, immunofluorescence declined from proximal to distal dendrites in a much steeper fashion in Bdnfklox/klox neurons as compared to WT neurons (Fig. 4F). In addition, BDNF immunofluorescence in somata was 44% higher in Bdnfklox/klox neurons than in WT neurons (Fig. 4G). Thus, impairment of dendritic targeting of BDNF mRNA seen in Bdnfklox/klox mice is associated with more BDNF protein in cell bodies and less BDNF protein in dendrites, without changes in the total amount of BDNF protein in the brain. It is believed that activity-dependent BDNF secretion occurs primarily in dendritic spines (Lu, 2003). In support of this view, we found that regulated BDNF secretion was impaired in hippocampal neurons isolated from Bdnfklox/klox mice (Fig. 4H).
Dysmorphogenesis of dendritic spines in neurons lacking the long 3′ UTR mRNA
To test whether the lack of dendritic BDNF mRNA in Bdnfklox/klox neurons affects dendritic growth, dendrites of Golgi-stained CA1 pyramidal neurons from 2-month old mice were traced using the Neurolucida software. Dendritic arbors of reconstructed WT CA1 pyramidal neurons did not appear different from those of Bdnfklox/klox neurons (Fig. 5A). Numbers of primary and secondary basal dendrites were also similar between genotypes (Fig. 5B). In agreement with these observations, Bdnfklox/klox mice had normal cytoarchitecture in the cortex and the hippocampus (Fig. S3).
Figure 5. Dysmorphogenesis of dendritic spines in Bdnfklox/klox hippocampal neurons at 2 months of age.
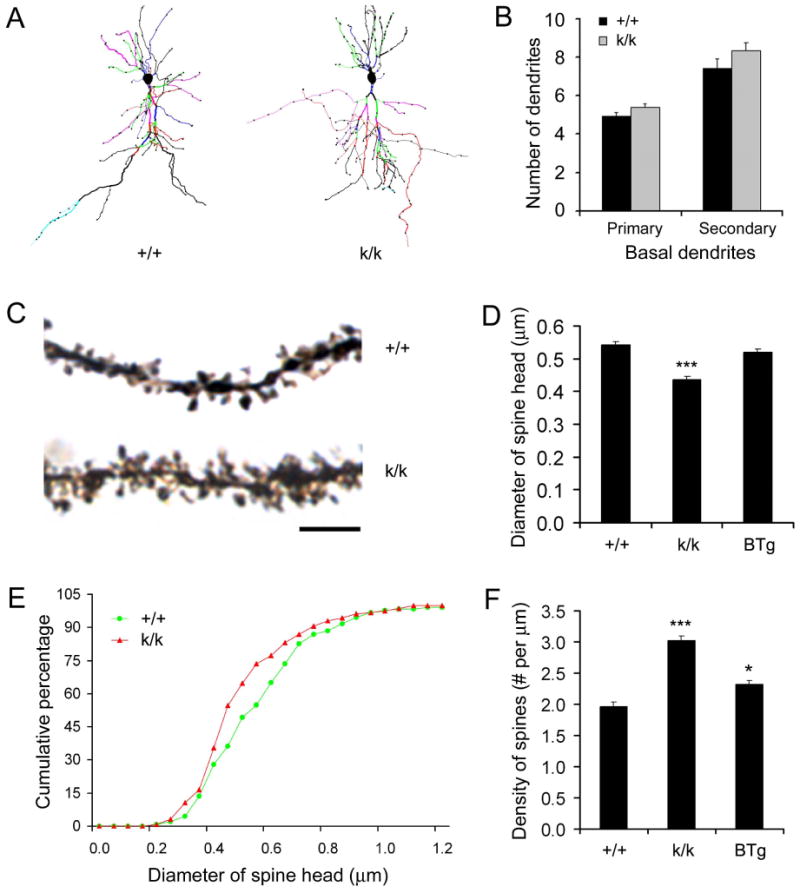
(A) Representative dendritic arbors of CA1 pyramidal neurons, reconstructed with the Neurolucida software from Golgi-impregnated brain sections of WT (+/+) and Bdnfklox/klox (k/k) mice.
(B) Similar dendritic numbers in CA1 pyramidal neurons from +/+ and k/k mice. Primary and secondary basal dendrites from 12 +/+ neurons and 13 k/k neurons were traced with the Neurolucida software and counted (n=4 mice for each genotype). _p_=0.12 for primary dendrites; _p_=0.19 for secondary dendrites.
(C) Representative dendritic spines on distal apical dendrites of +/+ and k/k mice. Scale bar, 5 μm.
(D) Reduction in spine size in k/k neurons. Average spine head diameter was calculated from measurements on 355 +/+ spines (3 mice), 355 k/k spines (3 mice), and 343 BTg spines (3 mice).
(E) Cumulative distribution curves for spine head diameter, using the data in D.
(F) Spine density on distal apical dendrites of CA1 pyramidal neurons (n=3 mice each).
In contrast to the relatively normal dendritic arbors, spines on distal apical dendrites of CA1 pyramidal neurons were noticeably altered in Bdnfklox/klox mice at 2 months of age. Dendritic spines on higher orders of Golgi-stained apical dendrites in Bdnfklox/klox mice appeared thinner and more numerous as compared to those in WT mice (Fig. 5C). The average spine head diameter in Bdnfklox/klox mice was 20% smaller than that in WT mice (Fig. 5D). There was a left-shift in cumulative distribution of spine head diameter, indicating that all classes of dendritic spines have smaller heads in Bdnfklox/klox neurons (Fig. 5E). Lack of the long 3′UTR BDNF mRNA also led to more spines in apical dendrites. Quantitative analysis revealed a 54% increase in spine density in Bdnfklox/klox neurons (Fig. 5F).
The spine phenotypes could result from either lack of dendritic BDNF synthesis or increased levels of BDNF in cell bodies in Bdnfklox/klox CA1 pyramidal neurons. To distinguish between these two possibilities, we used BTg mice that express 2-3 fold higher levels of BDNF mRNA in the hippocampus (Huang et al., 1999). Importantly, BDNF mRNA derived from this transgene contains an SV40 3′UTR and therefore should stay in the soma. Indeed, whereas the in situ hybridization signals for BDNF mRNA in the s.p. were higher in BTg neurons as compared to WT neurons (Fig. S4A), the distribution of BDNF mRNA along the apical dendrites was similar between WT and BTg mice (Fig. S4B). Unlike Bdnfklox/klox neurons, the average spine head diameter in BTg neurons was not reduced (Fig. 5D, _p_=0.143). The spine density in BTg mice was only slightly increased (18% over WT, _p_=0.022), and much lower than that in Bdnfklox/klox mice (Fig. 5F, _p_=0.0021). These results argue that it is the decrease in dendritic BDNF, rather than the increase in somatic BDNF, that caused spine dysmorphogenesis.
We further investigated whether the increase in spine density in Bdnfklox/klox neurons is due to spine overproduction or lack of spine pruning. Spine pruning occurs after the third postnatal week in mice (Zuo et al., 2005). Consistent with this, we found that the spine density was much higher in CA1 neurons in P21 juvenile WT mice than that in adult WT mice (3.43 ± 0.39 spines/μm at P21 vs. 1.96 ± 0.08 spines/μm at 2 months of age). If the lack of dendritic BDNF mRNA leads to spine overproduction, an increase in spine density is expected before spine pruning occurs. However, the spine density on distal apical dendrites of CA1 pyramidal neurons was similar between WT and Bdnfklox/klox mice at P21 (Fig. 6A, _p_=0.517). In addition, we found that dendritic spines in WT and Bdnfklox/klox mice at P21 had similar sizes (Fig. 6B) and distributions (Fig. 6C) of spine head diameter. These results suggest that BDNF mRNAs with the long 3′UTR are not essential for the formation and initial growth of dendritic spines but are required for their pruning and enlargement.
Figure 6. Normal dendritic spines in hippocampal neurons of juvenile Bdnfklox/klox mice.
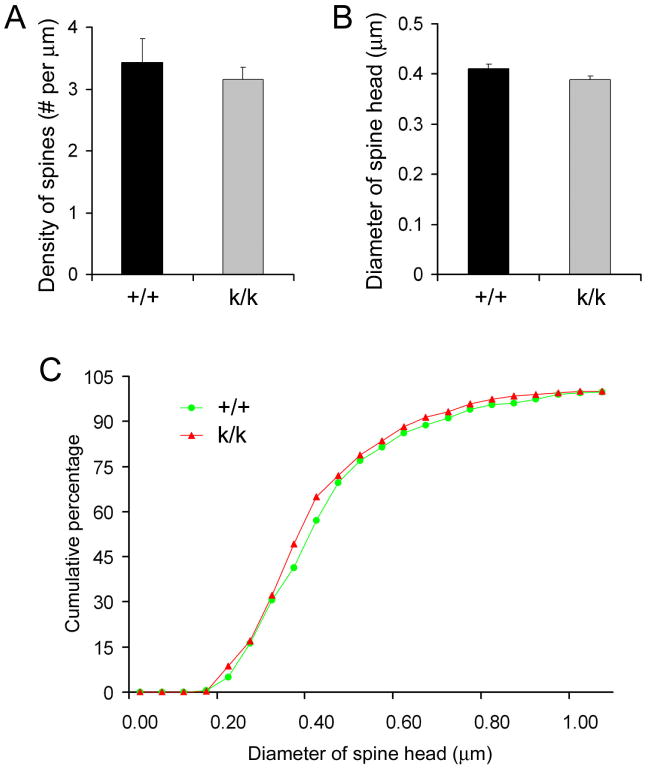
(A) Similar spine density on distal apical dendrites of CA1 pyramidal neurons in k/k and +/+ mice at 21 days of age (n=3 mice each).
(B) Spine head diameter. Average spine head diameter was calculated from measurements on 365 +/+ spines (3 mice) and 365 k/k spines (3 mice) at 21 days of age.
(C) Cumulative distribution curves for spine head diameter, using the data in B.
Selective impairment of hippocampal LTP at apical dendrites in Bdnfklox/klox mice
Deletion of the Bdnf gene results in marked impairment in LTP at hippocampal CA1 synapses (Korte et al., 1995; Patterson et al., 1996). It is not known, however, whether local BDNF synthesis in apical dendrites of CA1 pyramidal neurons is important for CA1 LTP. To address this issue, we performed electrophysiological recording using hippocampal slices derived from Bdnfklox/klox mice and WT littermates. Robust LTP was induced in the apical dendrite region of CA1 pyramidal neurons by applying tetanic stimulation (HFS, 2 × 100 Hz, 1 sec) to the Schaeffer collaterals in slices derived from WT animals (Fig. 7A). In contrast, slices derived from mutant mice did not show persistent LTP (Fig. 7A). Mean field EPSP (fEPSP) slopes at 60 min after HFS were 145 ± 4% and 113 ± 8% for WT and Bdnfklox/klox slices, respectively. In similar experiments, LTP induced by theta-burst stimulation (TBS) at CA1 synapses in the s.r. was also impaired in mutant mice as compared to WT mice (Fig. 7B). Thus, the long 3′UTR BDNF mRNA appears to be important for LTP expression in the dendritic region. As a control, input-output curves obtained by plotting the fEPSP slopes against fiber volley amplitudes (proportional to stimulation intensities) were indistinguishable between the genotypes, suggesting normal basal synaptic transmission (Fig. S5A). In addition, synaptic responses to a prolonged train of low-frequency repetitive stimulation (LFS), which results in a continuous decline of fEPSP slopes over time, also showed no differences between WT and Bdnfklox/klox slices (Fig. S5B).
Figure 7. Selective impairments in LTP recorded at the Schaffer collateral-CA1 synapses of Bdnfklox/klox mice.
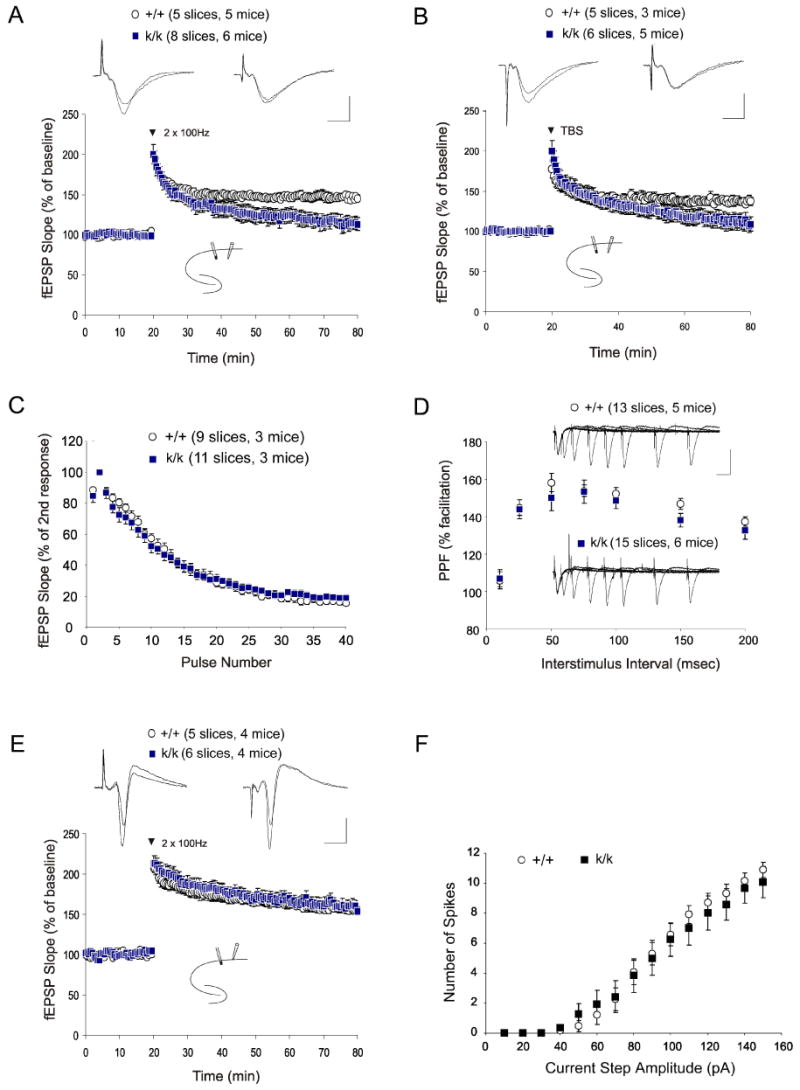
(A) Impairment of tetanus-induced LTP (2 × 100 Hz, 1 sec, spaced 20 sec apart) recorded in CA1 stratum radiatum. Test pulses and tetanic stimulation were administered to the Schaeffer collaterals.
(B) Impairment of TBS-induced LTP (8 bursts consisting of 4 pulses at 100Hz, spaced 200 msec apart) recorded in the s.r. of the CA1 region.
(C) Normal synaptic responses to brief high frequency stimulation (100 Hz, 40 stimuli) recorded in hippocampal CA1 in the presence of an NMDAR antagonist, APV.
(D) Normal paired pulse facilitation in k/k mice. No significant difference was observed when the PPF ratio (second fEPSP slope / first fEPSP * 100) was plotted against different inter-stimulus intervals.
(E) Normal tetanus-induced LTP was recorded in the s.p. area. Test pulses and tetanic stimulation were administered to the Schaeffer collaterals.
(F) Normal excitability of CA1 pyramidal neurons in k/k mice. Spike number in response to depolarizing current steps was not significantly different between +/+ and k/k mice at all current steps tested.
In addition to the LTP deficit, conventional Bdnf knockout mice exhibit impairments in paired pulse facilitation (PPF) and synaptic response to high frequency stimulation (HFS), both of which reflect changes in the properties of presynaptic terminals from CA3 neurons (Pozzo-Miller et al., 1999). We next assessed whether these forms of short-term presynaptic plasticity require BDNF derived from the apical dendrites of CA1 neurons. Surprisingly, synaptic response to a brief HFS (100 Hz, 1 sec) was completely normal in Bdnfklox/klox slices (Fig. 7C). Additionally, PPF ratios were also the same at all tested intervals in Bdnfklox/klox and WT slices (Fig. 7D). Taken together, these results suggest that BDNF derived from the long 3′UTR mRNA may not be important for the regulation of the readily releasable pool of synaptic vesicles.
Concomitant with the lack of dendritic BDNF mRNA, there is an increase in BDNF protein in the cell bodies of CA1 pyramidal neurons. To examine whether synaptic plasticity may be differentially regulated in the somata and apical dendrites of Bdnfklox/klox neurons, we recorded LTP in cell bodies. Evoked population spikes were monitored by placing a field recording electrode in the s.p. and stimulating the proximal area of the CA1 region. HFS induced robust LTP in WT animals (Fig. 7E). In contrast to the LTP deficit in the dendritic region, the magnitude and duration of LTP induced in the cell body region were completely normal in Bdnfklox/klox slices. The population spike amplitudes at 60 min after HFS were 155 ± 4% and 154 ± 5% for WT and Bdnfklox/klox slices, respectively. To further investigate whether an increase in BDNF protein in somata could alter neuronal excitability, we performed whole cell patch clamp recordings of CA1 neurons in hippocampal slices while all synaptic transmission was pharmacologically inhibited (Fig. S6A). The number of action potentials increased, paralleling the increasing depolarization steps, but no difference was found between WT and Bdnfklox/klox neurons (Fig. 7F). Additionally, the threshold, amplitude and half width of the first recorded action potentials were identical in both genotypes (Table S1). Finally, spike accommodation during a prolonged depolarization, defined by the ratio of 1st and 9th interspike interval (ISI), was similar between the two genotypes (Fig. S6B). Collectively, lack of the long BDNF mRNA altered neither synaptic function nor neuronal excitability in the soma region of CA1 pyramidal neurons.
DISCUSSION
Differential localization of mRNAs with short and long 3′UTRs
Our study provides an example where mRNAs containing the same coding sequence but distinct 3′UTRs can have distinct physiological functions due to their selective subcellular localization and translation. The control of translation through differential use of the two 3′UTRs may be important for key regulatory proteins such as BDNF, which regulates many aspects of neuronal structure and function. Translation of the long BDNF mRNA in dendrites may regulate synaptic structure and function locally, whereas that of the short BDNF mRNA in the soma may produce BDNF protein required for neuronal survival and maintenance. Furthermore, dendritic local BDNF synthesis may offer a mechanism by which a diffusible molecule such as BDNF achieves local and synapse-specific modulation.
Transcripts with different lengths of 3′UTRs encoding the same protein appear to be a general phenomenon. For example, the trkB gene encodes one full-length receptor (TrkB-F) and one truncated receptor (TrkB-T) that lacks the tyrosine kinase domain (Klein et al., 1990). TrkB mRNA has been detected in dendrites of cultured hippocampal neurons (Tongiorgi et al., 1997). While both TrkB-F and TrkB-T mRNAs have short and long 3′UTRs, it remains to be demonstrated whether they are targeted to different subcellular compartments. CaMKIIα mRNA also has a short 3′UTR and a long 3′UTR, although the long mRNA is much more abundant. The sequence required for dendritic targeting has been mapped downstream of the first polyadenylation site (Blichenberg et al., 2001), suggesting that the short CaMKIIα mRNA stays in somata while the long CaMKIIα mRNA is transported to dendrites. Thus, the two-3′UTR strategy may be commonly employed for genes that have important functions in both somata and dendrites.
A unique system to study dendritic BDNF mRNA in vivo
A growing number of mRNAs have been localized to neuronal dendrites (Steward and Schuman, 2003). It is generally believed that dendritic local protein synthesis is important in long-lasting synaptic plasticity (Huber et al., 2000; Kang and Schuman, 1996; Miller et al., 2002). However, physiological functions of dendritically localized mRNAs remain undetermined due to the technical challenge of selectively abolishing dendritic local protein synthesis without affecting the total protein level in a neuron. For example, a mouse mutant where the native 3′UTR of CaMKIIα mRNA is replaced with the BGH 3′UTR exhibits impairment in dendritic localization of CaMKIIα mRNA as well as a significant reduction in overall expression of CaMKIIα (Miller et al., 2002). Thus, it is difficult to attribute behavioral abnormalities of the mutant to the lack of local CaMKIIα synthesis.
In Bdnfklox/klox mice, the long 3′UTR mRNA is truncated, and thus all BDNF transcripts are converted to short 3′UTR mRNAs, leading to decreased dendritic BDNF mRNA. Importantly, levels of total BDNF mRNA and protein in the brain are unchanged, making it an ideal system to examine the physiological function of dendritically localized mRNAs. Using this unique model, we provide evidence that dendritically localized BDNF mRNAs are required for maintenance of a physiological level of BDNF protein in distal dendrites, maturation of dendritic spines, and plasticity of synapses located on dendrites.
Local protein synthesis and pruning of dendritic spines
Spine pruning is as important as spine growth in controlling the number and location of functional synapses. The pruning process is dependent on sensory experience (Zuo et al., 2005) and has been implicated in activity-dependent refinement of synaptic connections. We show here that Bdnfklox/klox mice have the same spine density on distal dendrites of hippocampal neurons as WT mice at P21 but do not show a reduction in spine density with maturation. These results suggest that Bdnfklox/klox mice have deficits in spine pruning and implicate BDNF in the control of spine pruning. Whether BDNF is truly involved in spine pruning requires validation by future experiments using in vivo time-lapse imaging.
The deficit in spine pruning in distal dendrites of Bdnfklox/klox neurons appears to be due to decreased BDNF in dendrites, rather than an increase in the soma. This phenotype is very different than the small increase seen in the number of spines for BTg neurons. In the latter case, there is a global increase in BDNF levels in the entire dendritic field due to an increase in BDNF transported to dendrites from cell bodies, which leads to a small increase in spine formation. This is consistent with the published observation that application of BDNF to cultured neurons stimulates spine formation (Ji et al., 2005). In addition to the different timings for spine formation and pruning, our findings underscore the differential effects of global increases in BDNF signaling and local changes in BDNF concentration in dendrites.
How does locally synthesized BDNF regulate spine morphology and synaptic plasticity?
Pro-BDNF is secreted and has unique biological activities in neurons (Lee et al., 2001; Woo et al., 2005), although the amount of secreted pro-BDNF remains controversial (Lee et al., 2001; Matsumoto et al., 2008). Since the majority of dendrites lack Golgi-like organelles to process secreted proteins (Horton et al., 2005), a significant portion of dendritically translated BDNF is likely secreted as pro-BDNF, which can be converted to mature BDNF extracellularly by proteases such as tPA (Pang et al., 2004). Since tPA is only secreted from stimulated spines (Lochner et al., 2006), the action of mature BDNF is limited to the stimulated spines by activating TrkB in an autocrine manner. We speculate that TrkB signaling induces local synthesis of proteins associated with spine growth and plasticity within and/or underneath the stimulated spines. These locally synthesized proteins promote actin dynamics and AMPA receptor trafficking in spines, which leads to growth of the spine head and formation of stable LTP. In parallel, pro-BDNF diffused nearby should promote pruning of unstimulated spines, which lack tPA, by binding to the sortilin/p75NTR receptor complex with high affinity (Lee et al., 2001). This model is based on several previous observations in addition to our findings described here. First, the activation of TrkB-mediated signaling cascades has been shown to induce local synthesis of several proteins, including Arc, CaMKIIα, Homer2, and LIMK1 (Kang and Schuman, 1996; Schratt et al., 2004; Yin et al., 2002). While CaMKIIα is a key regulator of AMPA receptor trafficking at synapses (Derkach et al., 2007), the other three proteins promote actin polymerization required for LTP consolidation (Bramham and Wells, 2007) and the maturation and enlargement of spine heads (Meng et al., 2002; Sala et al., 2001). Second, the gradual phase of spine enlargement induced by synaptic stimulation is dependent on protein synthesis and BDNF action (Tanaka et al., 2008). Finally, the p75NTR receptor is localized in postsynaptic sites in the CA1 area (Woo et al., 2005) and deletion of the gene for the receptor results in increased spine density on apical dendrites of hippocampal neurons (Zagrebelsky et al., 2005). Our model predicts that selective inhibition of dendritic BDNF synthesis would reduce the size of spine heads but increase spine number, the phenotypes that we observed in Bdnfklox/klox mice.
While application of exogenous BDNF has been shown to increase the size of EPSPs (Kang and Schuman, 1995), deletion of the Bdnf gene or inhibition of TrkB signaling generally impairs only LTP without affecting basal synaptic transmission (Figurov et al., 1996; Patterson et al., 1996; Xu et al., 2000). Here we show that inhibition of dendritic local BDNF synthesis also affects early-phase LTP but not basal transmission in apical dendrites. The selective impairment in dendritic LTP is consistent with the size reduction in spine heads observed on apical dendrites of CA1 neurons in Bdnfklox/klox mice. The increase in spine density in the mutant mice may not alter synaptic transmission and plasticity, as these thin spines may not form functional connections with presynaptic terminals.
Given that early-phase LTP does not require newly synthesized proteins during its induction and expression (Kelleher et al., 2004), it is unlikely that the LTP impairment in the Bdnfklox/klox synapses is due to a failure in stimulation-induced BDNF synthesis in dendrites. Rather, a reduced basal level of BDNF in dendrites due to deficits in local BDNF synthesis may lead to accumulated deficiencies in LTP-generating signaling cascades at CA1 synapses prior to LTP recording, which impair LTP expression in Bdnfklox/klox hippocampal slices. Moreover, the LTP deficits are less severe as compared to those in Bdnf knockout mice. Several factors could contribute to the difference between the two types of Bdnf mutant mice. First, despite lack of locally synthesized BDNF, there is a small amount of soma-derived BDNF in Bdnfklox/klox dendrites, which could alleviate some of the LTP deficit. Second, the LTP phenotype in Bdnf knockout mice may results from both pre- and post-synaptic deficits (Gartner et al., 2006; Kovalchuk et al., 2002; Xu et al., 2000). However, Bdnfklox/klox mice exhibit normal PPF and response to HFS. Taken together, our data demonstrate the distinct roles of BDNF synthesized in somata vs. dendrites in synaptic plasticity.
EXPERIMENTAL PROCEDURES
Animals
Mice were maintained on standard mouse chow (Purina diet 5001) at 22 °C on a 12 h light/12 h dark cycle. Both Bdnfklox/klox and BTg mouse strains were in the C57BL6/J genetic background. All animal procedures were approved by the Georgetown University Animal Care and Use Committee.
Culture and transfection of primary neurons
Hippocampal and cortical neurons were isolated and cultured according to a previously described procedure (Sala et al., 2000) from E18.5 rat embryos or mouse pups at P0-P2. We used lipofectamine 2000 or lipofectamine LTX to transfect cultured neurons.
In situ hybridization
In situ hybridization of brain sections and cultured neurons was performed using DIG-labeled riboprobes and the TSA Plus Fluorescein System (PerkinElmer, Waltham, MA) according to previously described procedures (Marz et al., 1998; Muddashetty et al., 2007) with modifications. Brain sections at 40 μm were obtained with a sliding microtome from brains fixed in 4% paraformaldehyde-20% sucrose for 3 days. Sections were digested with 10-30 μg/ml proteinase K in PBS plus 0.1% Tween 20 for 20 min at room temperature to expose mRNAs before hybridization. After hybridization, sections or cells were treated with RNase A at 37 °C to remove un-annealed probes.
GFP constructs and local protein synthesis assay
The EGFP coding sequence from the pEGFPN1 vector (BD Bioscience, San Jose, CA) was inserted into pcDNA3.1(+) (Invitrogen, Carlsbad, CA) to generate the GFP-BGH construct. The mouse genomic sequences encoding 3′UTRs A, B and AB were obtained by PCR and used to replace the BGH 3′UTR for generation of GFP-A, GFP-B, and GFP-AB constructs.
A sequence encoding the Src myristoylation peptide was added to the 5′-end of a PCR-amplified d1EGFP insert from plasmid pd1EGFP-N1, which was inserted into a plasmid downstream the human synapsin promoter, generating phSYN-myr-d1GFP. The mouse sequences A and A*B (where the first polyadenylation signal AATAAA was changed to TTTTTT) were cloned into phSYN-myr-d1GFP, generating phSYN-myr-d1GFP-A (myr-d1GFP-A) and phSYN-myr-d1GFP-A*B (myr-d1GFP-A*B). These two constructs were transfected into cultured rat hippocampal neurons at 13 DIV. One day after transfection, the cultured neurons were treated with 1 μM TTX or vehicle for 6 hours and then fixed. The longest dendrites of transfected neurons were analyzed by quantifying the fluorescent intensity of a line drawn through the center of the dendrite, and mean intensity values for each condition were calculated using 50 μm bins.
Golgi impregnation
Golgi staining was performed using the FD Rapid GolgiStain Kit (FD NeuroTechnologies, Ellicott City, MD). We used the Neurolucida software (MicroBrightField, Williston, VT) to trace dendrites. To measure spine density, we took images of distal dendrites with similar diameters in the stratum radiatum, enlarged the images with Photoshop software (Adobe Systems, San Jose, CA), and counted spines on printouts. All small protrusions on dendrites were considered as spines. Spine density for each animal was obtained from many distal dendrites with a total length of 1000-2000 μm. Dendritic images from each mouse were randomly used for measurement of spine size. The densely stained area at the tip of a spine was considered as a spine head.
Immunohistochemistry, Northern blots, and Immunoblotting
Immunohistochemistry of brain sections and cultured neurons was carried out as previously described (Gharami et al., 2008). The antibody to BDNF (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:2,000 for brain sections, at 1:500 for cultured neurons, and at 1:1,000 for immunoblotting. The mouse monoclonal antibody to MAP2 (1:500 dilution) was purchased from Chemicon International (Temecula, CA). The probes for Northern blots were labeled using the Rediprime II DNA labeling system (Amersham Bioscience, Piscataway, NJ).
BDNF secretion
Hippocampal neurons were seeded at a density of 5 × 105 cells per well in a 12-well plate. For measurement of constitutive BDNF secretion, cells at 10 DIV were incubated with Neurobasal medium supplemented with 0.1 μg/ml bovine serum albumin, 15 μM TTX, 20 μM D-2-amino-5-phosphonovaleric acid, and 20 μM 6-cyano-7-nitroquinoxaline-2,3-dione for 16 hours. For measurement of regulated BDNF secretion, cells at 10 DIV were treated with Neurobasal medium supplemented with 0.1 μg/ml bovine serum albumin and 50 mM KCl for 15 min. The amount of BDNF in each medium sample was determined using the BDNF Emax ImmunoAssay System (Promega, Madison, WI).
Electrophysiological recording
Transverse hippocampal slices (400 μm) were prepared from Bdnfklox/klox and age-matched WT mice (8-10 weeks old). Extracellular and whole cell recordings were performed as previously described (Xu et al., 2000).
Statistical analysis
All data are expressed as mean ± SEM. Data were analyzed using an unpaired Student's _t_-test; *, p<0.05; **, p<0.01; ***, p<0.001.
SUPPLEMENTAL DATA
01
Supplemental Data include Experimental Procedures, six figures, and one table and can found with this article online.
Acknowledgments
We thank S. Tonegawa for BTg mice, G. Bassell for sharing FISH protocols, Y. Chang for the pd1EGFP-N1 plasmid, D. Pak for sharing rat neurons, and L. Reichardt for critical reading of the manuscript. This work was supported by the grants from the NIH (NS050596), the American Heart Association, the Whitehall Foundation, and the American Diabetes Association to BX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- Figurov A, Pozzo-Miller LD, Olafsson P, Wang T, Lu B. Regulation of synaptic responses to high-frequency stimulation and LTP by neurotrophins in the hippocampus. Nature. 1996;381:706–709. doi: 10.1038/381706a0. [DOI] [PubMed] [Google Scholar]
- Gartner A, Polnau DG, Staiger V, Sciarretta C, Minichiello L, Thoenen H, Bonhoeffer T, Korte M. Hippocampal long-term potentiation is supported by presynaptic and postsynaptic tyrosine receptor kinase B-mediated phospholipase Cgamma signaling. J Neurosci. 2006;26:3496–3504. doi: 10.1523/JNEUROSCI.3792-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington's disease phenotypes in mice. J Neurochem. 2008;105:369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Ji Y, Pang PT, Feng L, Lu B. Cyclic AMP controls BDNF-induced TrkB phosphorylation and dendritic spine formation in mature hippocampal neurons. Nat Neurosci. 2005;8:164–172. doi: 10.1038/nn1381. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Klein R, Conway D, Parada LF, Barbacid M. The trkB tyrosine protein kinase gene codes for a second neurogenic receptor that lacks the catalytic kinase domain. Cell. 1990;61:647–656. doi: 10.1016/0092-8674(90)90476-u. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Yamamoto S, Maruo T, Murakami F. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. Eur J Neurosci. 2005;22:2977–2984. doi: 10.1111/j.1460-9568.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk Y, Hanse E, Kafitz KW, Konnerth A. Postsynaptic induction of BDNF-mediated long-term potentiation. Science. 2002;295:1729–1734. doi: 10.1126/science.1067766. [DOI] [PubMed] [Google Scholar]
- Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lochner JE, Honigman LS, Grant WF, Gessford SK, Hansen AB, Silverman MA, Scalettar BA. Activity-dependent release of tissue plasminogen activator from the dendritic spines of hippocampal neurons revealed by live-cell imaging. J Neurobiol. 2006;66:564–577. doi: 10.1002/neu.20250. [DOI] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz P, Cheng JG, Gadient RA, Patterson PH, Stoyan T, Otten U, Rose-John S. Sympathetic neurons can produce and respond to interleukin 6. Proc Natl Acad Sci U S A. 1998;95:3251–3256. doi: 10.1073/pnas.95.6.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Rauskolb S, Polack M, Klose J, Kolbeck R, Korte M, Barde YA. Biosynthesis and processing of endogenous BDNF: CNS neurons store and secrete BDNF, not pro-BDNF. Nat Neurosci. 2008;11:131–133. doi: 10.1038/nn2038. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, Lu B. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Paradies MA, Steward O. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J Neurobiol. 1997;33:473–493. doi: 10.1002/(sici)1097-4695(199710)33:4<473::aid-neu10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Pozzo-Miller LD, Gottschalk W, Zhang L, McDermott K, Du J, Gopalakrishnan R, Oho C, Sheng ZH, Lu B. Impairments in high-frequency transmission, synaptic vesicle docking, and synaptic protein distribution in the hippocampus of BDNF knockout mice. J Neurosci. 1999;19:4972–4983. doi: 10.1523/JNEUROSCI.19-12-04972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Sala C, Rudolph-Correia S, Sheng M. Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J Neurosci. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, Paradiso B, Steward O, Cattaneo A, Simonato M. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J Neurosci. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo NH, Teng HK, Siao CJ, Chiaruttini C, Pang PT, Milner TA, Hempstead BL, Lu B. Activation of p75NTR by proBDNF facilitates hippocampal long-term depression. Nat Neurosci. 2005;8:1069–1077. doi: 10.1038/nn1510. [DOI] [PubMed] [Google Scholar]
- Xu B, Gottschalk W, Chow A, Wilson RI, Schnell E, Zang K, Wang D, Nicoll RA, Lu B, Reichardt LF. The role of brain-derived neurotrophic factor receptors in the mature hippocampus: modulation of long-term potentiation through a presynaptic mechanism involving TrkB. J Neurosci. 2000;20:6888–6897. doi: 10.1523/JNEUROSCI.20-18-06888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW. The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc Natl Acad Sci U S A. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Holz A, Dechant G, Barde YA, Bonhoeffer T, Korte M. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25:9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005;436:261–265. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Supplemental Data include Experimental Procedures, six figures, and one table and can found with this article online.