Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly (original) (raw)
. Author manuscript; available in PMC: 2008 Sep 3.
Summary
FliI is the peripheral membrane ATPase pivotal to the type III protein export mechanism underlying the assembly of the bacterial flagellum. Gel filtration and multiangle light scattering showed that purified soluble native FliI protein was in a monomeric state but, in the presence of ATP, FliI showed a propensity to oligomerize. Electron microscopy revealed that FliI assembles to a ring structure, the yield of which was increased by the presence of a non-hydrolysable ATP analogue. Single particle analysis of the resulting electron micrograph images, to which no symmetry was applied, showed that the FliI ring structure has sixfold symmetry and an external diameter of ≈ 10 nm. The oligomeric ring has a central cavity of 2.5–3.0 nm, which is comparable to the known diameter of the flagellar export channel into which export substrates feed. Enzymatic activity of the FliI ATPase showed positive co-operativity, establishing that oligomerization and enzyme activity are coupled. Escherichia coli phospholipids increased enzyme co-operativity, and in vitro cross-linking demonstrated that they promoted FliI multimerization. The data reveal central facets of the structure and action of the flagellar assembly ATPase and, by extension, the homologous ATPases of virulence-related type III export systems.
Introduction
Bacterial motility is commonly achieved by rotation of cell surface flagella. The well-characterized flagellum of Escherichia coli and Salmonella is a long helical filament, the propeller, attached to a flexible hook anchored to the cell envelope by the flagellar basal body (Macnab, 1996; Namba and Vonderviszt, 1997). The flagellum is assembled by polymerization of subunits that form the rod, hook, hook–filament junction, filament cap and filament at the distal end of the growing flagellar structure. These subunits diffuse through the channel at the centre of the flagellum, but they must first be exported from the cytosol by a dedicated type III export mechanism (Macnab, 1999; Aizawa, 2001).
The flagellar export apparatus comprises six integral membrane proteins, and it has been speculated that these could sit in the patch of membrane within the MS ring of the flagellar basal body (Fan and Macnab, 1996; Minamino and Macnab, 1999; Kihara et al., 2001). Delivery of structural subunits to the membrane apparatus is aided by several subunit-specific chaperones (Fraser et al., 1999; Auvray et al., 2002), but essential to the export mechanism is the flagellar ATPase FliI that provides energy to the export process (Vogler et al., 1991; Dreyfus et al., 1993), and is also assumed to be centrally involved in the series of protein–protein interactions underlying the translocation of substrates to the membrane apparatus. It is known that FliI interacts with another flagellar export component, FliH, in the cytosol and/or at the membrane and that this negatively regulates FliI activity (Minamino and Macnab, 2000a; Minamino et al., 2001), and that FliJ, which is proposed to be a general flagellar chaperone, interacts with FliH (Minamino et al., 2000; Minamino and Macnab, 2000b; Gonzalez-Pedrajo et al., 2002) and FliI (L. Claret, unpublished). Furthermore, interactions have been reported between both FliI and FliH and the cytosolic domains of the integral membrane proteins FlhA and FlhB (Minamino and Macnab, 2000b; Zhu et al., 2002). Despite the emerging view of these and other interactions, little is known about the behaviour of FliI, in particular how the flagellar ATPase activity is coupled to the export process. It has been reported that FliI is a cytosolic monomeric protein (Minamino and Macnab, 2000a), and it has been suggested that active FliI monomers ‘scavenge’ for substrates and target them for export at the flagellar basal body (Macnab, 1996) or, alternatively, FliI monomer could be held inactive by FliH in the cytosol until it docks as a complex with delivered substrate at the membrane machinery within the MS ring where ATP hydrolysis is triggered (Macnab, 1999; Minamino and Macnab, 2000a). We have subsequently shown that, although FliI lacks transmembrane domains, it behaves as a peripheral membrane protein when expressed physiologically in Salmonella (Auvray et al., 2002), and that purified FliI has an intrinsic affinity for E. coli phospholipids, i.e. even without the integral membrane machinery of the export pathway (Auvray et al., 2002). Our experiments also showed that these phospholipids stimulate FliI ATPase activity in vitro (Auvray et al., 2002). These experimental findings provided direct support for the view that FliI interacts with the membrane, while leaving open the many questions regarding which interactions with cytosolic and membrane proteins of the flagellar pathway precede or follow such interaction. By extension, the results would indicate a comparable membrane interaction by the homologous ATPases of virulence-related type III export systems (Venkatesan et al., 1992; Eichelberg et al., 1994; Woestyn et al., 1994).
This view of membrane interaction appears to be strengthened by the similar peripheral membrane localization of ATPases energizing bacterial type IV transport across the cytoplasmic membrane despite lacking predicted transmembrane domains (Bhattacharjee et al., 2001). Provocatively, these ATPases have been shown to exist in an oligomeric state, specifically forming hexameric ring structures (Krause et al., 2000a,b; Yeo et al., 2000). These findings in another bacterial export pathway suggested to us the possibility that FliI activity might be coupled to the assembly of an active, multimeric form at the membrane. Therefore, we have assessed in this study whether the flagellar ATPase FliI can assemble to a multimeric state, if there might be a connection between such multimerization and enzymatic activation and whether this might be promoted by contact with phospholipid.
Results
FliI can form higher order oligomers
The full-length FliI protein was generated by thrombin cleavage of an affinity-purified glutathione _S_-transferase (GST) fusion. This FliI was soluble in buffer [20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM dithiothreitol (DTT)] and was subjected to gel filtration chromatography followed in line by a multiangle light scattering detector (Wyatt, 1993; Fleming et al., 1998). The elution profile (not shown) featured a single species of ≈45 kDa, approximating the deduced FliI monomer molecular mass of 49.3 kDa. These results confirm those of Minamino and Macnab (2000a). Repeated chromatography after the addition of ATP (to 1 mM) and MgCl2 (5 mM) to the buffer again revealed the prevalent form to be the ≈45 kDa monomer, constituting roughly 60% of the total protein (Fig. 1). However, other major peaks (containing 22.5% and 12% of the total protein respectively) indicated molecular masses of 120 kDa and 169 kDa. These data suggest that, although soluble FliI exists as a monomer, it is able to assemble to higher order oligomers in the presence of ATP.
Fig. 1.
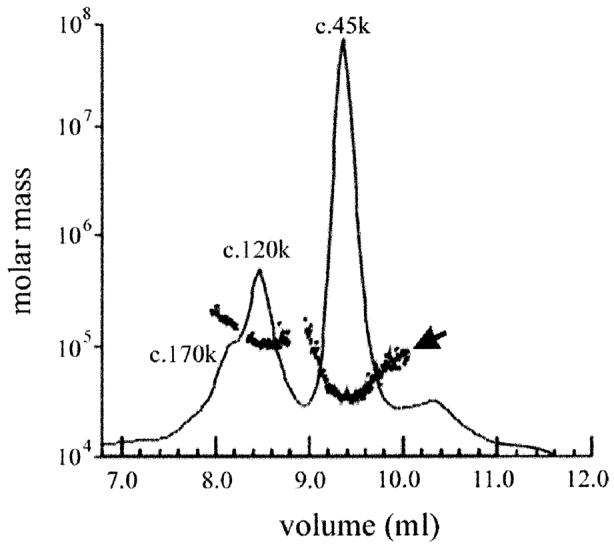
Multiangle light scattering of purified FliI in buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT, 1 mM ATP and 5 mM MgCl2). The continuous line represents refractive index on an arbitrary scale, with the height of peaks directly proportional to protein concentration. The dotted line (arrowed) indicates molecular mass, and corresponding mass determinations are shown above each peak.
FliI assembles to ring-shaped oligomers with sixfold symmetry and a central cavity
To pursue the architecture of FliI, the purified full-length protein was adsorbed to the hydrophobic surface of a carbon-coated transmission electron microscope grid and examined at 46 000× magnification. This revealed that FliI had a propensity to assemble to ring-shaped structures, but their incidence was low and matched by that of incomplete rings (not shown). In an attempt to increase the yield of the ring structures, FliI was preincubated with magnesium and ATP or the non-hydrolysable ATP analogue AMP-PNP, as had been reported for the TrbB ATPase underpinning type IV export and conjugative transfer of DNA (Krause et al., 2000a). In contrast to preincubation with ATP, which had only a marginal effect, AMP-PNP increased the yield of complete rings ≈20-fold, presumably by stabilization of assembled oligomers. Figure 2A and B shows the ring-shaped structures, which have a central depression into which negative stain penetrates.
Fig. 2.
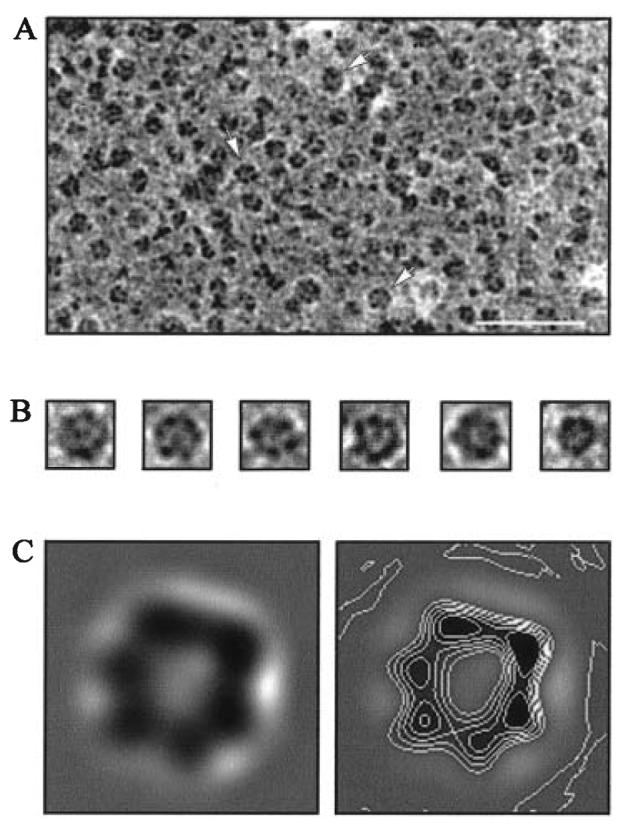
Electron microscopy of FliI. Purified FliI, preincubated with AMP-PNP, was stained with 2% uranyl acetate and visualized by the transmission electron microscope.
A. Typical field of view at 46 000× magnification. The white scale bar indicates 50 nm. Arrows highlight representative particles.
B. Selected images of individual ring-like particles.
C. Averaged images lowpass band filtered to their final resolution. The size of each frame is 25 × 25 nm.
A total of 7705 FliI images generated after AMP-PNP treatment were extracted from micrographs and were aligned, classified and averaged using imagic-v software (van Heel et al., 1996). The particles were centred by translational alignment to a rotationally averaged sum of images. Centred images were classified into distinct groups, and the images within each group were averaged together to generate ‘class averages’. Five distinct sets of class averages were generated, producing four, eight, 12, 24 or 48 classes respectively. In each case, the class averages appeared to be similar in size and shape, suggesting that a single view or closely related views had been obtained. Therefore, the FliI oligomers predominantly adopted a preferred orientation on the surface of the grid. For this reason, four representative reference class averages were selected from the set of 12, and the particles were translationally and rotationally aligned to these references and then averaged together into a single class. All images were realigned to this sum, and the summation was repeated. This cycle was repeated four times until no change was observed in the resultant average. At no stage was any symmetry applied to the particles, but the images belonged to a single class of ring structures with sixfold symmetry (Fig. 2C). The external diameter of the ring is 10 nm (±10%), and the average image comprises six regions of electron density surrounding a central low-density region 2.5–3 nm in diameter. The results confirm the propensity of FliI to oligomerize, and indicate that it possibly assembles to a hexameric state. The central depression would be compatible with the existence of a central channel.
Coupling of FliI multimerization and ATPase activity
To investigate the significance of FliI multimerization, in particular in influencing the enzyme activity, we measured FliI specific activity at different concentrations of protein. Figure 3 showing the relationship between concentration and activity suggests positive co-operativity. This characteristic was examined further in experiments in which initial velocities of ATP hydrolysis by FliI were measured as a function of the ATP substrate concentration. A constant high concentration of FliI (0.5 μM) was used, which is greater than the concentration required to achieve V max (Fig. 3A). The resulting K 0.5 for ATP was found to be 0.65 ± 0.10 μM and the V max was 0.28 ± 0.10 μmol ATP hydrolysed min−1 mg−1 FliI protein. The Hill's co-operativity coefficient calculated from the Hill's plot (Fig. 3B, inset) was 1.92 ± 0.05. As the FliI monomer contains a single ATP-binding motif (Dreyfus et al., 1993), this value of the Hill coefficient illustrates positive co-operativity in FliI activity as a result of interaction between FliI monomers (a value of 1.0 would indicate independent behaviour of catalytic sites). The results suggest that oligomerization stimulates the activity of the enzyme.
Fig. 3.
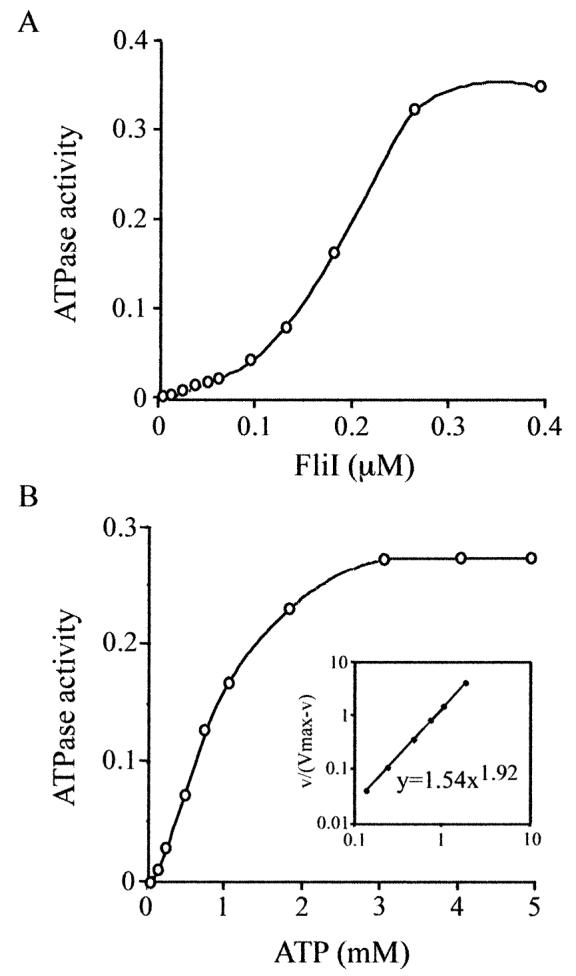
Dependence of FliI ATPase activity on protein and substrate concentration. Specific activity of FliI was measured at (A) different enzyme concentrations in the presence of 5 mM ATP, and (B) different ATP concentrations with 0.5 μM FliI. The plot of the logarithmic form of the Hill equation is shown inset and the Hill coefficient and K 0.5 for ATP are derived from the formula. Activity is expressed as μmol of Pi released min−1 mg−1 FliI.
Phospholipid-promoted multimerization and activation of FliI
We have shown that FliI is a peripheral membrane protein that has intrinsic affinity for E. coli phospholipids and, furthermore, that these increase FliI activity in vitro (Auvray et al., 2002). We therefore assessed whether FliI multimerization is influenced in vitro by the presence of phospholipid, and whether it is this that promotes enzyme activity.
In vitro cross-linking was applied, using disuccinimidyl glutarate (DSG), which has a short side-arm of 7.7 Å and has been used previously to demonstrate the multimerization of eukaryotic and bacterial ATPases (Pan et al., 1998; Xu et al., 1999). When cross-linking was applied to purified FliI protein in solution, i.e. in the absence of phospholipid, no multimers could be discerned on acrylamide gels containing 0.1% SDS, even when preincubated with ATP or AMP-PNP. All FliI was seen as the monomeric form (≈ 49 kDa), as it was in the absence of cross-linker (Fig. 4). In contrast, when the protein was preincubated in the same way but with E. coli phospholipid, several higher bands were seen, suggesting the formation of a dimer, trimer (the strongest band), tetramer and a higher order multimer above 230 kDa (Fig. 4). This experiment demonstrates that multimerization of FliI is promoted by the presence of bacterial phospholipids.
Fig. 4.
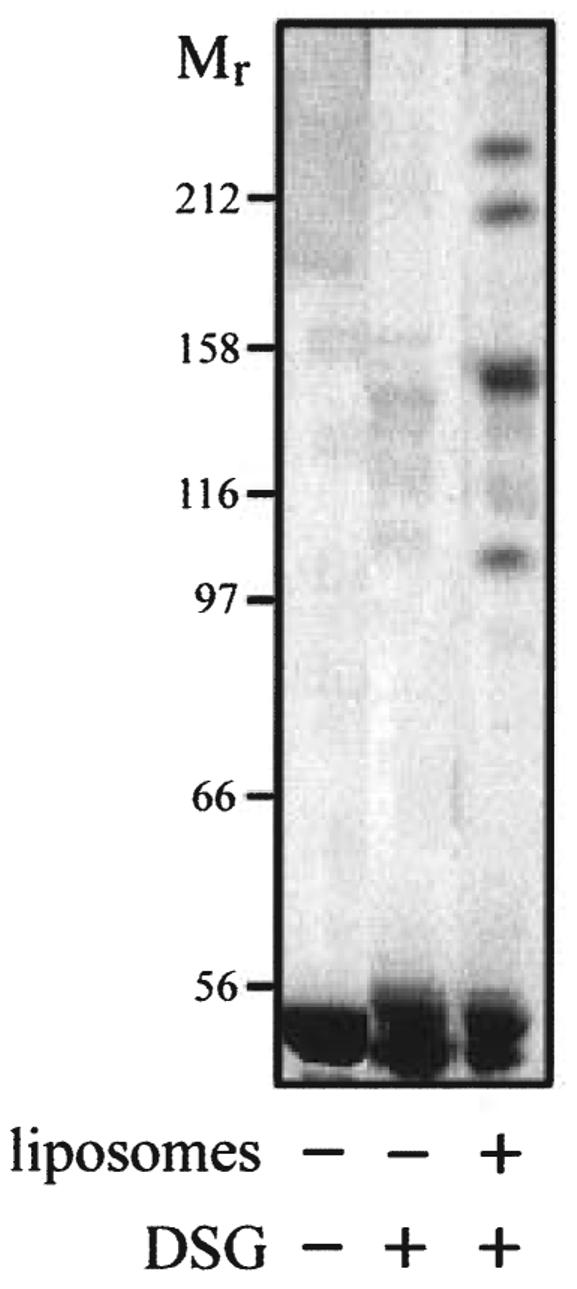
In vitro cross-linking of FliI. Purified FliI protein was incubated in cross-linking buffer either alone or with E. coli liposomes, before DSG was added to 0.1 mM. Aliquots were precipitated and subjected to electrophoresis through 4–10% acrylamide gradient gels containing 0.1% SDS.
To assess further the influence of phospholipids on FliI ATPase activity, the specific activity of the enzyme was measured in the presence of E. coli phospholipids at different concentrations of ATP (Fig. 5), as was shown in Fig. 3B (using the same concentration of FliI, 0.5 μM). The K 0.5 for ATP was 0.86 ± 0.10 mM, and the V max was 2.30 ± 0.10 μmol ATP hydrolysed min−1 mg−1 FliI protein compared, respectively, with 0.65 ± 0.10 mM and 0.28 ± 0.10 in the absence of phospholipid. These results confirm that E. coli phospholipids stimulate FliI activity in vitro, and the Hill's co-operativity coefficient (inset) was 2.83 ± 0.05, significantly higher than the 1.92 ± 0.05 measured without phospholipid, which indicated that positive co-operativity had increased.
Fig. 5.

Substrate dependence of FliI ATPase activity in the presence of E. coli phospholipids. ATP hydrolysis was measured as a function of ATP concentration in the presence of 0.5 μM FliI plus 300 μg ml−1 phospholipids. The plot of the logarithmic form of the Hill equation is shown inset and the Hill coefficient and K 0.5 for ATP are derived from the formula.
Discussion
The flagellum-specific type III export pathway targets structural subunits to the flagellum assembling on the bacterial cell surface. Central to the process is the dedicated, essential ATPase FliI (Vogler et al., 1991). We have established previously that this export ATPase associates physiologically with the inner membrane in Salmonella typhimurium, and that in vitro FliI has intrinsic affinity for E. coli phospholipids, which increase its ATPase activity 10-fold (Auvray et al., 2002). These findings support the view that FliI is activated at the cytosolic membrane. To resolve further the poorly established mechanics of this and other type III protein export systems, it is necessary to define in greater detail the structure and activity of the ATPase, and the lipid and protein interactions that locate and regulate it.
A previous report has indicated that His-tagged FliI can exist in monomeric form (Minamino and Macnab, 2000a), and our gel filtration and multiangle light scattering analysis of the native soluble enzyme confirmed this earlier work; all FliI was recovered as the monomeric form. However, when we incubated the enzyme with its substrate ATP, it was evident that FliI has a tendency to oligomerize. Electron microscopy and two-dimensional image processing of FliI stabilized by the non-hydrolysable ATP analogue AMP-PNP revealed that the ATPase assembles to ring structures that exhibit a strong sixfold symmetry around a central cavity, which may represent a central channel. The data, achieved without the application of any symmetry, indicate that FliI is possibly a homohexamer. Such a structure would be comparable to that of the ATPases of the distinct type IV protein export pathway in Helicobacter pylori and E. coli, ascertained by comparable means (Krause et al., 2000a,b). Our data also revealed that ATPase activity of FliI displays positive co–operativity, indicating interactions between subunits, i.e. FliI oligomerization is coupled to activation of the enzyme. This coupling has been shown to be the case for the multimeric histidine permease and maltose transporter ATPases (Davidson et al., 1996; Liu and Ames, 1998). The finding that the FliI Hill's coefficient is significantly higher in the presence of phospholipids than in their absence indicates that phospholipids promote FliI co-operativity, i.e. increase self-interaction between FliI monomers. This was confirmed by in vitro cross-linking, which showed that phospholipids strongly encourage multimerization of FliI. We have no data on the kinetics of oligomerization, but our observation of smaller ‘intermediate’ multimers in our biochemical assays and partial rings by electron microscopy indicates some equilibrium between monomer and multimer.
Our data appear to advance substantially the view of FliI, and one can reasonably suggest other ATPases of the type III export pathway. They would be compatible with FliI monomers assembling to the active oligomer, most likely a hexamer, at the cytosolic membrane to provide the energy for assembly at the critical location. Whether this energy is used directly for substrate protein translocation and/or protein unfolding or interaction is not known. Crystal structures of the H. pylori and E. coli hexameric type IV export ATPases (Yeo et al., 2000; Gomis-Ruth et al., 2001) show that their N- and C-terminal domains form two rings, which together create a chamber open on one side and closed on the other, and it is proposed that these ATPases act as inner membrane gates for proteins, with rounds of ATP hydrolysis modulating a conformational shift to open each end of the channel in turn. It is possible that FliI hexamers might behave in a comparable way, but this remains to be seen when higher resolution structural data become available. It is possible that ATP hydrolysis is closely coupled to substrate export into the flagellar channel by oligomerization of the FliI ring in close proximity to the integral membrane components of the type III export machinery, which are proposed to sit in the patch of membrane within the MS ring structure of the flagellum basal body (Macnab, 1996; Fan et al., 1997). Provocatively, the single particle analysis data indicate that the FliI central cavity is 2.5–3.0 nm in diameter, a figure corresponding to the size of the central export channel running through the centre of the basal body and the growing flagellum (Namba et al., 1989). Nevertheless, this is conjecture, as little is known of the sequence of protein–protein interactions that determine the assembly of the type III export apparatus. Although in vitro phospholipid interaction and in vivo peripheral membrane location are not dependent upon the flagellar apparatus, it is not possible to say at what stage phospholipid contact(s) occur, and they may follow protein–protein interactions. The data do not negate early models in which FliI is assumed to engage with the integral membrane export machinery, components of which, e.g. FliP and FliR, are believed to be housed within the MS ring of the flagellum (Fan et al., 1997; Minamino and Macnab, 2000b; Zhu et al., 2002). They appear to lend them strength.
Understanding of how FliI and the many other proteins of the type III export pathway function to effect export is still at an early stage. FliI is envisaged to be involved in protein–protein interactions at the transition from cytosolic to membrane phases of the export pathway. The cytosolic stages are slowly emerging. FliI is bound with the protein FliH in a heterotrimeric complex H2-I (Minamino and Macnab, 2000a; Gonzalez-Pedrajo et al., 2002). FliH represses FliI activity and, like FliI, it behaves physiologically as a peripheral membrane protein and has an intrinsic affinity for phospholipids (Auvray et al., 2002). Our data prompt the consideration that FliH repression might include reduction of non-productive oligomerization, although we have as yet been unable to show that purified FliH influences oligomerization or co-operativity in vitro. It is possible that the FliHI complex is targeted by several components of the export pathway, including chaperoned flagellar subunits and cytosolic and membrane export components (Minamino and Macnab, 2000b; Zhu et al., 2002; L. Claret and S. R. Calder, unpublished), and such interactions might relieve FliH repression when substrate can be efficiently coupled to ATP hydrolysis. These and other possibilities await clarification by further experimental data.
Experimental procedures
FliI protein purification
GST–FliI fusion protein was overproduced from the pGEX4T3 expression vector (Pharmacia) into which the fliI sequence was inserted after polymerase chain reaction (PCR) amplification from S. typhimurium SJW1103 chromosomal DNA using Pfu DNA polymerase (Stratagene). E. coli C41 (Miroux and Walker, 1996) carrying pGEX4T3_fliI_ was grown to midexponential phase at 25°C in Luria–Bertani broth containing 2% glucose and 100 μg ml−1 ampicillin and induced with 0.2 mM IPTG. After incubation overnight at 25°C, cells were harvested, resuspended in buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT) and disrupted in a French press. Extract was clarified by high-speed centrifugation for 60 min. The soluble GST–FliI was purified by affinity chromatography on glutathione agarose (Pharmacia), and the GST protein was cleaved with thrombin protease. Contaminating thrombin protease was removed by chromatography as described by the manufacturer (Pharmacia). The FliI was maintained in the same buffer at 4°C.
ATPase activity measurements
Activity of purified FliI was determined essentially as described previously (Eichelberg et al., 1994).
Multiangle light scattering
Analytical gel filtration with multiangle light scattering was performed at a flow rate of 0.5 ml min−1 through a 300 × 7.8 mm TSK-Gel G3000SWXL column (Toso Haas) equilibrated with buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT, 1 mM ATP and 5 mM MgCl2). The column was followed in line by a Mini-DAWN light scattering detector at 690 nm and differential refractometry. Molecular masses were calculated using astra software (Wyatt Technologies) and the Debye fit method (Wyatt, 1993). Purified protein was diluted in equilibration buffer to a concentration of 2 mg ml−1.
Chemical cross-linking
Purified FliI (0.5 μM final concentration) was incubated in 100 μl of cross-linking buffer (20 mM Hepes, pH 8.0, 0.1 M NaCl, 0.1 mM EDTA, 1 mM DTT) containing 5 mM MgCl2 and 1 mM ATP, alone or in the presence of 100 μg ml−1 E. coli phospholipids (Avanti Polar Lipids). After 30 min at 37°C, disuccinimidyl glutarate (DSG) was added to a final concentration of 0.1 mM. The reaction was terminated by the addition of 0.2 M Tris-HCl and 10% TCA. After precipitation, protein samples were heated at 95°C for 5 min, separated by 0.1% SDS-4–10% PAGE and stained with Coomassie brilliant blue.
Electron microscopy and image processing
Purified protein (2 μg ml−1 final concentration) was incubated in buffer (100 mM NaCl, 20 mM Tris-HCl, pH 8.0, 5 mM AMP-PNP, 5 mM MgCl2, 1 mM DTT) for 30 min at 37°C and applied to a carbon-coated copper grid. After 1 min, excess solution was removed, the grid was washed twice with buffer and negatively stained with two washes of 2% (w/v) uranyl acetate. Grids were observed in a Phillips CM100 electron microscope under low-dose conditions at 46 000× magnification and 1–1.5 μm defocus. Micrographs were digitized on a UMAX MagicScan. A total of 7705 particle images were selected manually using ximdisp (Crowther et al., 1996), and image processing was carried out using imagic-v (van Heel et al., 1996) on a Silicon Graphics work station. Images were bandpass filtered to include information only within a resolution range of 30–140 Å. Particles were normalized and centred by translational alignment to a rotationally averaged sum of all images. Multivariance statistical analysis (MSA) sorted the centred images into classes of like views, which were summed together. Four of the resulting averages were used to align the entire data set translationally and rotationally. Four cycles of MSA led to stable classes of similar appearance, and further cycles did not lead to a decrease in variance. Finally, all particles were averaged into one class. At no stage in the image processing was any symmetry imposed on the particles.
Acknowledgements
We thank Emma McGhie for advice on multiangle light scattering, and Gillian Fraser and Jeyanthy Eswaran for critical reading of the manuscript. This work is supported by a Wellcome Trust Programme grant (C.H.) and Studentship (S.R.C.).
References
- Aizawa SI. Bacterial flagella and type III secretion systems. FEMS Microbiol Lett. 2001;202:157–164. doi: 10.1111/j.1574-6968.2001.tb10797.x. [DOI] [PubMed] [Google Scholar]
- Auvray F, Ozin AJ, Claret L, Hughes C. Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J Mol Biol. 2002;318:941–950. doi: 10.1016/S0022-2836(02)00172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Davidson AL, Laghaeian SS, Mannering DE. The maltose transport system of Escherichia coli displays positive co-operativity in ATP hydrolysis. J Biol Chem. 1996;271:4858–4863. [PubMed] [Google Scholar]
- Dreyfus G, Williams AW, Kawagishi I, Macnab RM. Genetic and biochemical analysis of Salmonella typhimurium FliI, a flagellar protein related to the catalytic subunit of the F0F1 ATPase and to virulence proteins of mammalian and plant pathogens. J Bacteriol. 1993;175:3131–3138. doi: 10.1128/jb.175.10.3131-3138.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg K, Ginocchio CC, Galan JE. Molecular and functional characterisation of Salmonella typhimurium genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Macnab RM. Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- Fan F, Ohnishi K, Francis NR, Macnab RM. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Hohl TM, Yu RC, Muller SA, Wolpensinger B, Engel A, et al. A revised model for the oligomeric state of the N-ethylmaleimide-sensitive fusion protein, NSF. J Biol Chem. 1998;273:15675–15681. doi: 10.1074/jbc.273.25.15675. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Bennett JC, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;33:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pedrajo B, Fraser GM, Minamino T, Macnab RM. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol Microbiol. 2002;45:967–982. doi: 10.1046/j.1365-2958.2002.03047.x. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Kihara M, Minamino T, Yamaguchi S, Macnab RM. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J Bacteriol. 2001;183:1655–1662. doi: 10.1128/JB.183.5.1655-1662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Barcena M, Pansegrau W, Lurz R, Carazo JM, Lanka E. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. Proc Natl Acad Sci USA. 2000a;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000b;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P-Q, Ames GF-L. In vitro disassembly and reassembly of an ABC transporter, the histidine permease. Proc Natl Acad Sci USA. 1998;95:3495–3500. doi: 10.1073/pnas.95.7.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edn. Washington, DC: American Society for Microbiology Press; 1996. pp. 123–145. [Google Scholar]
- Macnab RM. Reversible rotary propeller and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol Microbiol. 2000a;37:1494–1503. doi: 10.1046/j.1365-2958.2000.02106.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000b;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Chu R, Yamaguchi S, Macnab RM. Role of FliJ in flagellar protein export in Salmonella. J Bacteriol. 2000;182:4207–4215. doi: 10.1128/jb.182.15.4207-4215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Tame JR, Namba K, Macnab RM. Proteolytic analysis of the FliH/FliI complex, the ATPase component of the type III flagellar export apparatus of Salmonella. J Mol Biol. 2001;312:1027–1036. doi: 10.1006/jmbi.2001.5000. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Namba K, Vonderviszt F. Molecular architecture of the bacterial flagellum. Quart Rev Biophys. 1997;30:1–65. doi: 10.1017/s0033583596003319. [DOI] [PubMed] [Google Scholar]
- Namba K, Yamashita I, Vonderviszt F. Structure of the core and central channel of bacterial flagella. Nature. 1989;342:648–654. doi: 10.1038/342648a0. [DOI] [PubMed] [Google Scholar]
- Pan W, Ko YH, Pedersen PL. Delta subunit of rat liver mitochondrial ATP synthase: molecular description and novel insights into the nature of its association with the F1-moiety. Biochemistry. 1998;19:6911–6923. doi: 10.1021/bi9800698. [DOI] [PubMed] [Google Scholar]
- Venkatesan MM, Buysse JM, Oaks EV. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AP, Homma M, Irikura VM, Macnab RM. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woestyn S, Allaoui A, Wattiau P, Cornelis GR. YscN, the putative energiser of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt PJ. Submicrometer particle sizing by multiangle light scattering following fractionation. J Colloid Interface Sci. 1993;197:9–20. doi: 10.1006/jcis.1997.5215. [DOI] [PubMed] [Google Scholar]
- Xu T, Vasilyeva E, Forgac M. Subunit interactions in the clathrin-coated vesicle vacuolar H(+)-ATPase complex. J Biol Chem. 1999;274:28909–28915. doi: 10.1074/jbc.274.41.28909. [DOI] [PubMed] [Google Scholar]
- Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1462–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- Zhu K, Gonzalez-Pedrajo B, Macnab RM. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry. 2002;41:9516–9524. doi: 10.1021/bi0203280. [DOI] [PubMed] [Google Scholar]