Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line (original) (raw)
Abstract
Epigenetic reprogramming is commonly observed in cancer, and is hypothesized to involve multiple mechanisms, including DNA methylation and Polycomb repressive complexes (PRCs). Here we devise a new experimental and analytical strategy using customized high-density tiling arrays to investigate coordinated patterns of gene expression, DNA methylation, and Polycomb marks which differentiate prostate cancer cells from their normal counterparts. Three major changes in the epigenomic landscape distinguish the two cell types. Developmentally significant genes containing CpG islands which are silenced by PRCs in the normal cells acquire DNA methylation silencing and lose their PRC marks (epigenetic switching). Because these genes are normally silent this switch does not cause de novo repression but might significantly reduce epigenetic plasticity. Two other groups of genes are silenced by either de novo DNA methylation without PRC occupancy (5mC reprogramming) or by de novo PRC occupancy without DNA methylation (PRC reprogramming). Our data suggest that the two silencing mechanisms act in parallel to reprogram the cancer epigenome and that DNA hypermethylation may replace Polycomb-based repression near key regulatory genes, possibly reducing their regulatory plasticity.
Keywords: spatial clustering, DNA methylation, MeDIP normalization, Polycomb
Biochemical processes including DNA cytosine methylation and histone modifications interact with each other to ensure the stability of epigenetic states. These processes are clearly altered in cancer and contribute to the establishment and maintenance of the malignant phenotype (1). Recent discoveries and technological developments have greatly expanded our understanding of the cell's epigenetic makeup, revealing a rich repertoire of histone modifications and protein complexes that regulate them (2, 3). Chromatin regulating complexes, and most notably the family of Polycomb repressive complexes (PRCs), which mediate trimethylation at H3K27, are highly active in cancer cells (4). Studies of Polycomb activity in pluripotent embryonic stem cells (ESC) (5–7) revealed broad patterns of Polycomb-based repression near key developmental regulators, many of which are known DNA hypermethylation targets in cancer (8). It is therefore pertinent to take an integrated look at both DNA methylation and histone modifications simultaneously in a coherent cell set in which the appropriate presumed normal cell counterpart is compared to its malignant state.
The evidence on interaction between PRCs and the DNA methylation machinery is partial and sometimes conflicting. The correlations between ESC PRC targets and cancer hypermethylation (8), or the reported enrichment of H3K27me3 marks at specific hypermethylated CpG islands (9), have led to multiple mechanistic hypotheses on the underlying process. The lack of DNA hypermethylation at PRC occupied regions in embryonic carcinoma cells (10) and of DNA hypomethylation after knockdown of a PRC2 component (EZH2) in cancer cells (11) further demonstrate that the interaction between PRC and DNA methylation is probably not as direct as previously suggested (12). Indeed recent analyses of embryonic stem cells have suggested that DNA methylation and PRCs target different gene sets for repression (13, 14). Similarly, systematic analysis of prostate cancer cells shows that promoters with high H3K27me3 levels in cancer cells are usually not hypermethylated (15). Evolutionary analyses revealed a tight association between PRC occupancy and CpG islands (16) but suggested PRC activity in ES cells is antagonistic to methylation in the germ line, rather than permissive to it. Taken together, the current evidence leaves many questions open as to the nature of the interaction between DNA methylation and the Polycomb systems.
Results
Integrated Array-Based Epigenomic Profiling.
We developed a systematic approach to characterize the combined epigenetic states of a carefully chosen set of 1,800 promoters in primary prostate epithelial cells (PrEC) and the PC3 prostate cancer cell line [supporting information (SI) Fig. S1]. We selected balanced groups of promoters having high and low CpG content and four PC3/PrEC expression profiles (PC3 repressed, constitutive inactive, constitutive active, and PC3 induced, see Methods). Regions surrounding these promoters [2.5 kb upstream and 1 kb downstream of their annotated transcription start site (TSS)] were tiled on a high-density array at a 20-bp resolution. Additional half megabase contiguous regions around the HOXA and ApoB clusters were included for additional control.
We developed an enhanced technique for DNA methylation profiling by combining 5-methylcytosine immunoprecipitation [MeDIP (17–19)] of target DNA and DNA that was treated with M.SssI to full methylation. The M.SssI data provided us with calibration for the experimental readout at regions with different G + C contents and for the saturation of enrichment at CpG dense loci. Combined with a new computational normalization scheme, this allowed accurate and carefully controlled quantification of DNA methylation levels at high resolution (Methods, Figs. S2 and S3).
The quantitative DNA methylation profiles were combined with high-resolution data on occupancy of a PRC component (SUZ12) and the PRC histone mark H3K27me3 in the two cell types, and the integrated profile was dissected by a new algorithm. The use of a single experimental platform to probe the distributions of several epigenetic marks was crucial to the interpretation of the data. Using our experimental design, the improved DNA methylation profiling method and the integrated experimental and analytical strategy, we were able to comprehensively characterize the epigenetic reprogramming that differentiates normal prostate epithelial cells from the PC3 cancer line.
Extensive PC3 DNA Hypermethylation in Silenced CpG Islands.
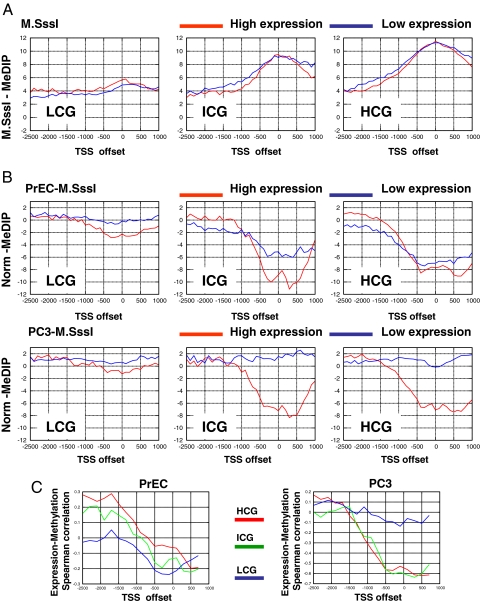
We first studied the correlation between gene expression and spatial patterns of DNA methylation. We compared methylation in promoters with different CpG contents (19), using the M.SssI-treated controls as a baseline that reflects full potential methylation (Fig. 1A). We observed activity-dependent depletion of methylation (Fig. 1B and Fig. S4) around the TSS for promoters with low CpG content (LCG) in both cell types. Following a similar trend, active promoters with intermediate (ICG) and high (HCG) CpG content were unmethylated in both PC3 and PrEC. However, while inactive HCG promoters in normal cells were also generally unmethylated, PC3 cells exhibited almost complete methylation at these promoters (79% of the probes at CpG islands of low-expression genes have PC3 normalized MeDIP values > −5, Fig. S5). Thus, these cells have almost fully methylated all of their inactive promoters (both those that were inactive already in PrEC cells and those that were de novo repressed, see below). Such hypermethylation is observed regardless of the genes' CpG content and in contrast to normal cells in which high CpG content promoters are unmethylated.
Fig. 1.
Methylation and gene expression. (A) M.SssI controls. Shown are medians of normalized methylation levels of immunoprecipitated M.SssI-treated DNA in spatial bins relative to the TSSs. We plot separately promoters with three CpG content ranges [high CpG content (HCG), intermediate CpG content (ICG), and low CpG content (LCG)] (19). In each group, we compare genes with high expression (log2 expression >10) to genes with low expression (log2 expression < 7). As expected, the controls show similar methylation capacity for active and inactive genes in each of the three sets. We used the M.SssI data to compensate for the variable CpG content when normalizing MeDIP data. (B) Spatial methylation patterns. Shown are medians of PrEC (Upper) and PC3 (Lower) normalized methylation values in spatial bins relative to the TSSs. The normal cells show activity-dependent lack of methylation near the start site in LCGs and to a lesser extent in ICGs. Cancer cells show methylation associated with inactivity in all CpG content ranges. (C) Expression–methylation correlation. Shown are Spearman correlation coefficients computed between methylation in bins of 200 bp relative to the TSS and gene expression. The graphs confirm the negative correlation between methylation and expression at low CpG-content TSSs in both cell types. The data also demonstrate the strong methylation–expression negative correlation at high CpG-content promoters established in the cancer cells.
As shown in Fig. 1C, significant negative correlation between methylation and expression is observed in both cell types for low CpG-content promoters [PrEC, P < 10−8; PC3, P < 3 × 10−4 (Spearman)]. This result is at odds with a recent study (19) performed on fibroblasts that suggested that methylation of low CpG-content TSS regions was not correlated with RNA PolII activity. We show that renormalization of the data in the previous study according to our enhanced protocol settles this contradiction (SI Text and Fig. S4). We also provide experimental validations (bisulfite sequencing and RNA PolII ChIP) confirming this correlation for specific examples of low CpG-content promoters in PC3 cells and in fibroblasts (Fig. S4). We conclude that low-density DNA methylation around the TSS infrequently cooccurs with PolII occupancy and/or active transcription. Additional observations on hyper- and hypomethylation at the HOX clusters suggest that epigenetic reprogramming can span regions beyond a single gene (SI Text).
PRC2 and DNA Methylation Are Switched in PC3 Cells.
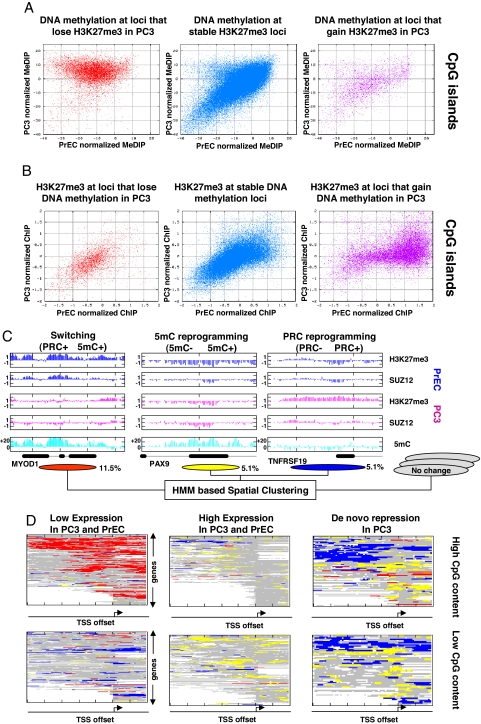
We next studied the levels of three Polycomb-related proteins (H3K27me3 and the PRC2 components SUZ12 and EZH2) in a line of reference germ cell tumor (NCCIT) and the prostate cells. We found that the overall abundances of both PRC2 components and the H3K27me3 mark are substantially higher in NCCIT and PC3 cells relative to the normal cells (Fig. 2A). We then used our array to study the distributions of H3K27me3 and SUZ12 occupancy and compared them to our DNA methylation profiles. Somewhat unexpectedly, we detected both cases of decreased and increased PRC occupancy on our array, even though the Polycomb system is globally more active in PC3 (Fig. 2B). Remarkably, our data showed sweeping DNA hypermethylation at almost all loci that lose H3K27me3 in PC3 cells relative to PrECs (Fig. 3A Left for CpG islands, Fig. S6 for non-CpG islands). Eighty-eight percent of the loci with at least a twofold decrease in H3K27me3 marks are methylated in PC3 (methylation > −5 units), compared to only 26% of the loci without observed change in H3K27me3 levels (Fig. 3A Middle, statistics for probes with PrEC methylation< −10 units, P < 10−150, KS test, significance is independent of the thresholds). Sharp changes in DNA methylation are not observed in loci that gain H3K27me3 marks (Fig. 3A Right). Conversely, loci that gain DNA hypermethylation are frequently, but not always, depleted from H3K27me3 marks in PC3 (Fig. 3B Right). In loci with unchanged or decreased PC3 DNA methylation (Fig. 3B Center and Left) we generally observe unchanged H3K27me3 marks.
Fig. 2.
Gain and loss of Polycomb marks in PC3 cells. (A) PRC components and marks in NCCIT, PC3, and PrEC cells. Shown are Western blot results, depicting the levels of SUZ12, EZH2, and H3K27me3 in PrEC, PC3, and reference NCCIT (germ cell tumor) cells. Levels of these enzymes and the H3K27me3 mark are high in PC3, comparable to the levels in the pluripotent, undifferentiated NCCIT cells. Polycomb components and marks in PrEC cells are considerably lower, although still expressed. (B) Gain and loss of H3K27me3/SUZ12 in PC3 cells. Two genomic regions are shown. The FAM58A region demonstrates gain of PRC marks in PC3 together with minimal change in DNA methylation. The PAX7 region is depleted of PRC marks in PC3, and gains extensive DNA hypermethylation.
Fig. 3.
Epigenetic switching and reprogramming. (A) Differential DNA methylation as a function of H3K27me3 changes. Shown are normalized PrEC (_x_-axis) and PC3 (_y_-axis) MeDIP values for probes that reflect a twofold decrease (Left), no change (Center), or a twofold increase (Right) of their H3K27me3 occupancy in PC3 relative to PrECs. H3K27me3 depletion implies hypermethylation in 90% of the cases (Left). Data are shown for probes in CpG islands only; see Fig. S6 for the trends in low CpG-content regions. (B) Differential H3K27me3 as a function of DNA methylation changes. Shown are PrEC (_x_-axis) and PC3 (_y_-axis) H3K27me3 values for CpG island probes that reflect DNA hypomethylation (>10 normalized MeDIP units, Left), unchanged DNA methylation (Center), and DNA hypermethylation (>15 normalized MeDIP units, Right). Depletion of H3K27me3 marks is observed for a significant fraction, but not all of the hypermethylated loci. See Fig. S6 for data on non-CpG island probes. (C) Trends of epigenetic switching and reprogramming. To generalize probabilistically the observations shown in the above scatter plots, and to integrate information from adjacent probes, we developed a new HMM-based algorithm that assigned each probe on the array to an epigenetic mode (Methods and Figs. S7 and S8). Shown are genomic examples for three modes of epigenetic change that were identified in the scatter plots and confirmed by the algorithm. Regions that lose PRC marks and gain DNA methylation in PC3 are defined as switching loci. Regions that gain either PRC marks or 5mC marks independently in PC3 are defined as PRC- or 5mC-reprogramming loci, respectively. The percentage of probes on the array that were annotated as part of each of the three modes is shown below the genomic examples; note that these do not necessarily represent the genomic percentages because CpG islands are overrepresented on the array. (D) Functional analysis of spatial epigenetic modes. Shown are genes' promoters (rows) color coded according to the epigenetic modes associated with them (red, switching; blue, PRC reprogramming; yellow, 5mC reprogramming). We plot data for distinct groups of promoters with high (Upper) and low (Lower) CpG contents and different expression properties (Methods). White boxes represent missing data (at repetitive sequences or promoters that were only partially covered on our array). The data show that epigenetic switching is a dominant effect at high CpG-content promoters that have constitutively low expression in PrEC and PC3 cells. Loci that are subject to PRC and 5mC reprogramming are enriched at genes that are de novo repressed in PC3.
Systematic Characterization of Switching and Reprogramming in PC3 Cells.
The remarkable correlation between reprogramming of DNA methylation and H3K27me3 in PC3 cells can be summarized into three patterns or modes (see examples in Fig. 3C). Loci that lose H3K27me3 marks gain DNA methylation (epigenetic switching). Additional DNA hypermethylation is observed without change in H3K27me3 (5mC reprogramming). Finally, de novo H3K27me3 is observed without change in DNA methylation (PRC reprogramming). To further characterize these patterns and to ensure they generalize all global modes of epigenetic changes in our data we developed a new unsupervised analytic methodology to dissect an integrated epigenetic profile. Our new algorithm (“spatial clustering”) models the data using several modes, each of them representing particular levels of Polycomb marks and DNA methylation in the two lines. The algorithm further assumes that adjacent loci are likely to be represented by the same mode and learns a set of modes and hidden Markov model (HMM)-based connections between them such that the data are optimally explained (Methods). Importantly, the optimal model learned by our algorithm fits the three trends outlined above (in addition to several static epigenetic patterns) (Figs. S7 and S8). We conclude that on a large scale, the epigenetic differences between PrECs and PC3 are accounted for by three regimes of correlated and independent changes in DNA hypermethylation and H3K27me3 levels. Additional trends (such as DNA hypomethylation) can also be observed, but in our data reflect more localized and specific cases.
Switching Occurs at Constitutively Repressed Genes, de Novo PRC2 and DNA Methylation Correlate With de Novo Gene Repression.
How do epigenetic switching and reprogramming affect the regulation of nearby genes? To address this question we used our model for PC3/PrEC epigenetic reprogramming to study the distribution of epigenetic changes near genes with distinct regulatory dynamics. The results (Fig. 3D) reflect a tight association between epigenetic mode and gene regulation. Epigenetic switching shown in red was almost exclusively associated with constitutively repressed genes (P < 10−50 hypergeometric) and was rarely observed near genes that were de novo repressed in PC3. Interestingly, the genes associated with this epigenetic trend are PRC targets in human ESC (hESC) (see Fig. S7 and below), and include some well characterized markers of DNA hypermethylation in cancer (e.g., GATA4, MYOD1, and more, Table S1). The data therefore suggest that DNA hypermethylation of CpG islands that are targeted by PRC in hESCs is not associated with de novo gene repression in PC3 and cooccurs with depletion of PRC marks. On the other hand, PRC reprogramming shown in blue was observed near genes that were de novo repressed in PC3 cells (P < 10−50, hypergeometric test). Notably, this mode is characterized by unchanged levels of DNA methylation and its spatial distribution is biased to regions upstream of the TSS. 5mC reprogramming shown in yellow is also enriched at de novo repressed genes, but with a spatial distribution that is focused on the TSS itself. Close observation (Fig. 4C, Right) shows that the two reprogramming trends may sometimes occur over the same promoter, but are generally disjointed. [Note that hypermethylation that is mapped around constitutively high-expression genes is distributed away from the TSS (see Fig. S9)].
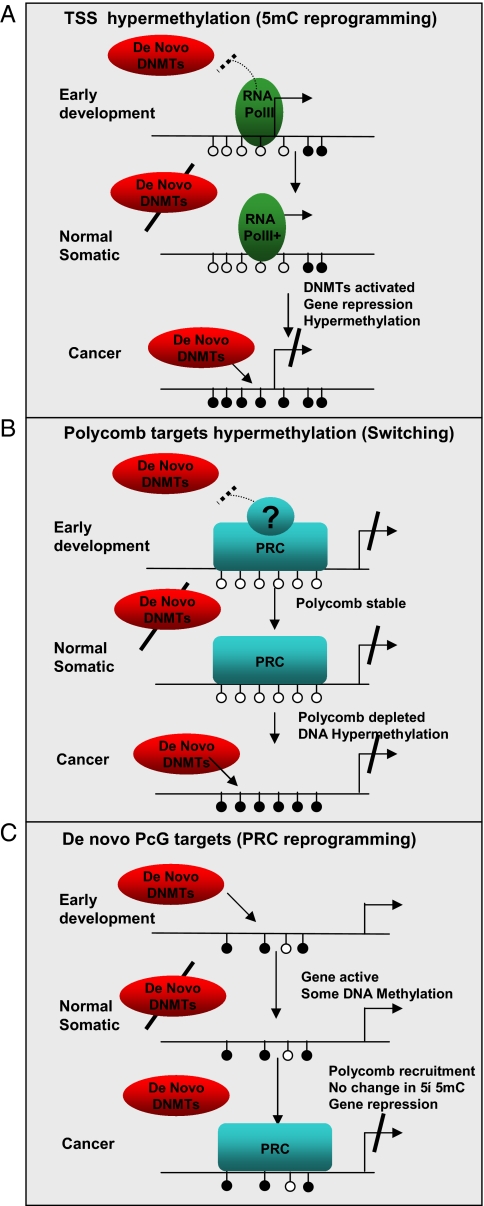
Fig. 4.
Model for DNA methylation and PRC recruitment. Shown are schematic promoters. Arrows represent transcription; filled (empty) circles represent methylated (unmethylated) CpG loci. (A and B) A schematic model for passive DNA methylation in PC3 cells. According to the model, the methylation system blindly methylates all CpGs that are not physically masked from it. Two possible masking mechanisms are shown, the first involving transcription initiation complexes (RNA PolII and related chromatin marks) around the TSS of active genes, and the second involves PRCs. Masking is effective only when the de novo DNA methylation machinery is active, either early in development [where masking decreases evolutionary CpG loss and contributes to the emergence of CpG islands (16)], or as part of an aberrant regulatory program in cancer. The experimental observations supporting this model include the sweeping hypermethylation at silenced TSSs in PC3 (Fig. 1) and the sweeping hypermethylation at loci that lose PRC marks in PC3 (Fig. 3A). An alternative active model for hypermethylation via Polycomb interaction should explain the reduction in PRC marks during or after the establishment of DNA hypermethylation. (C) Transcriptional silencing at de novo PRC targets. De novo PRC occupied regions are found upstream of the TSS of many of the repressed genes we studied, in regions that are not occupied by PRCs in hESCs and that are sometimes DNA methylated. Thus, according to our data, de novo repression of normally active genes in cancer PC3 cells occurs in conjunction with two systems that are spatially nonoverlapping [DNA methylation (A) and PRCs (C)]. Interactions between these two spatially separate mechanisms is possible (in some cases we do observe both over the same promoter), but the associated genes are not those that interact with PRCs in hESCs (panel B), and the regions that acquire PRC marks are physically separated from those that gain DNA methylation.
Characterizing Switching and Reprogramming Genes in Cancer Cell Lines, Tumors, and Stem Cells.
How does the epigenetic reprogramming we characterized in PC3 and PrEC cells relate to normal and malignant prostate cells and to cancer in general? To gain initial insights on the universality of the switching process we tested the levels of H3K27me3 and DNA methylation in a panel of four switching genes (MYOD1, PAX7, EDNRB, and Onecut1) and 5 PRC reprogramming genes across several normal and cancer cell lines (Fig. S10_A_ and Table S2). Overall, we observed significantly lower levels of DNA methylation at loci with increased H3K27me3 levels than at loci with low H3K27me3 levels [P < 0.0002 (KS test)], suggesting DNA hypermethylation and PRC marks do not tend to cooccur. For example, the MYOD1 and PAX7 loci are both marked by H3K27me3 in the two normal cell lines we analyzed (PrEC and LD419), but lack this mark in all cancer lines (Table S2). Moreover, hypermethylation at these loci is observed in all of the cancer lines examined.
To further validate the regulatory dynamics observed in the PC3/PrEC model, we analyzed published gene expression data sets on tumors and normal prostate tissues and compared them to our models' expression data (Fig. S10_B_). We grouped genes according to the three defined epigenetic modes and tested their differential expression in PrEC and PC3 cells (Left) and in two tissue panels (20). We found the expression of the switching genes, which is unchanged between PC3 and PrECs, is also unchanged between normal and tumor tissues (Fig. S10_B_ Right). The expression of the two reprogramming groups, however, is significantly reduced in tumors [P < 10−6 (KS test)], similarly to the behavior in our model cells. As shown in Fig. S11, the switching genes are generally not expressed in prostate tissues, further supporting the view of epigenetic switching as occurring near constitutively silenced genes.
As shown in Fig. S10_C_ (see also Fig. S12), the epigenetic switching group includes genes that are targets of PRC in ESCs. Genes in the PRC and 5mC reprogramming groups are not occupied by Polycombs in ESCs. It can be concluded that the group of genes that is targeted by PRCs in ESCs is generally, beyond a few tissue-specific cases, maintained repressed and PRC occupied in normal prostate cells and is therefore not a primary target for de novo suppression in cancer. The epigenetic switching we observed at genes in this group may affect their regulatory flexibility more than their actual level of expression. This is because DNA methylation may constitute a more permanent and heritable epigenetic state. On the other hand, PRC reprogramming does not transform the epigenetic makeup of PC3 cells closer toward the ESC pattern, because it occurs at genes that are not normal targets of the Polycomb system in stem cells.
Discussion
Our results may be explained by several alternative mechanistic models. A key observation is the magnitude of hypermethylation in PC3 cells (Fig. 1), which occurs at almost all of the inactive CpG island genes (see enrichment of switching probes in Fig. 3D). A second key observation is that when PRC marks are depleted, we observe PC3 hypermethylation (Fig. 3A). One possible model that can explain these observations assumes that DNA methylation in PC3 cells sweeps all CpGs that are not physically protected (or “masked”) (Fig. 4). According to this “passive” methylation model, transcription initiation complexes at the TSS [RNA PolII and related chromatin marks (21)] can serve as one masking mechanism (Fig. 4A), and Polycomb repressive complexes can serve as another (Fig. 4B). This view is also supported by recent data on DNA methylation patterns of mouse ESCs (13). An alternative to the passive model suggests that hypermethylation is actively mediated by the presence of PRC marks. To rationalize this alternative, one would have to characterize a scenario involving first recruitment of DNA methyltransferases [as in Vire _et al._ (12)] and then clearing of H3K27me3 marks, so that the cooccurrence of PRC marks depletion and DNA hypermethylation in PC3 is accounted for. It is important to note that even if Polycomb complexes mask loci from de novo DNA methylation, it is possible that PRC activity (or recruitment) is not masked by DNA methylation (Fig. 4C). This may be the case in some of the de novo PRC targets we observe near genes that are repressed in PC3.
Regardless of the underlying mechanism, our data outline a fundamental change in the epigenetic programming of PC3 cells, in which many genes that are originally silenced by PRCs acquire DNA methylation as an alternative silencing mechanism, and other genes are de novo repressed by Polycomb recruitment, in agreement with another recent study (15). Many of the genes affected by this shift are central developmental regulators that are associated with PRC complexes in hESCs. One can hypothesize that the epigenetic switch to DNA methylation-mediated repression reduces the plasticity of the regulatory program, locking silencing of key regulators and contributing to the cell's abnormal growth potential. The epigenome of the PC3 cancer line is therefore fundamentally different from that of its normal somatic cell counterpart from hESCs (for example, the de novo Polycomb domains we observe are typically not associated with PRC in hESCs, Figs. S9 and S10_C_). If the PC3 regulatory program has stem cell features, these must be achieved through a variation on the normal epigenetic mechanisms for immortality and pluripotency, a variation that may be critically less flexible.
The applicability of our observations to prostate tumors (and cancer cells in general) should be further investigated. Even though there are limitations to the use of short-term cultures of epithelial cells for these studies, they at least represent a relatively homogeneous population of the normal cell counterpart of PC3 cells. While PC3 cells have certainly accumulated many additional genetic and epigenetic changes in culture, previous studies in our lab have shown that these often represent enhancement of existing defects in the tumors rather than de novo acquisition of such changes (22). This is supported by analysis of gene expression in uncultured tumors and by the data we obtained for a panel of cell lines. Our study stresses the importance of an integrative approach for the characterization of the cancer epigenome and demonstrates the benefits of using a coherent experimental system to map multiple epigenetic factors. It will be intriguing to apply the methodology developed here to additional cell systems and epigenetic marks and to use a more extensive genomic coverage to uncover additional processes that contribute to the epigenetic reprogramming of cancer cells.
Methods
MeDIP.
MeDIP was performed as previously described (16) with the following alterations: 10 μg of sonicated genomic DNA (100–400 bp in length) was denatured, incubated O/N at 4°C with 20 μg of anti-methyl cytosine antibody (Diagenode, Belgium), and subsequently with 60 μl of protein A/G beads (Upstate Biotechnologies) for 2 h at 4°C. The beads were washed and incubated with digestion buffer and proteinase K for 3 h and the DNA extracted by phenol-chloroform and EtOH precipitation. For array experiments the output from 3 MeDIP reactions was combined (total of 30 μg starting DNA) to constitute one replicate to eliminate the need for whole genome amplification. The sonicated DNA served as input. MeDIP arrays were done in triplicates.
For the M.SssI controls, human genomic DNA was treated with M.SssI (4 units per 1 μg DNA, New England Biolabs) for 4 h before sonication and immunoprecipitation as described above. The output of 2 MeDIP reactions performed on M.SssI-treated DNA (total of 20 μg starting DNA) was combined to constitute one replicate. Sonicated M.SssI-treated DNA served as the input. Labeling and hybridization to our custom array was performed by Nimblegen (Madison, WI). Validation of M.SssI and the MeDIP experiments are shown in Figs. S13 and S14.
Complete data are available at accession number GSEX. Additional details are available in SI Text and Fig. S15and Table S3–S5.
Supplementary Material
Supporting Information
Acknowledgments.
This work was supported by National Institutes of Health Grant RO1CA 82422 (to P.J.) and the ISF Converging Technologies Program (A.T.). A.T. is an Alon Fellow.
Footnotes
The authors declare no conflict of interest.
Data deposition: Array data is available on NCBI/GEO (accession no. GSE12334).
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 5.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squazzo SL, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 9.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 10.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 12.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 13.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo Y, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 16.Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci USA. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 18.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 19.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 20.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 21.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markl ID, et al. Global and gene-specific epigenetic patterns in human bladder cancer genomes are relatively stable in vivo and in vitro over time. Cancer Res. 2001;61:5875–5884. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information